Abstract
Mycobacterium tuberculosis can exist in the actively growing state of the overt disease or in a latent quiescent state that can be induced, among other things, by anaerobiosis. Eradication of the latent state is particularly difficult with the available drugs and requires prolonged treatment. DevS is a member of the DevS-DevR two-component regulatory system that is thought to mediate the cellular response to anaerobiosis. Here we report cloning, expression, and initial characterization of a truncated version of DevS (DevS642) containing only the N-terminal GAF sensor domain (GAF-A) and of the full-length protein DevS. The DevS truncated construct quantitatively binds heme in a 1:1 stoichiometry, and the complex of the protein with ferrous heme reversibly binds O2, NO, and CO. UV-visible and resonance Raman spectroscopy of the wild-type protein and the H149A mutant confirm that His149 is the proximal ligand to the heme iron atom. While the heme-CO complex is present as two conformers in the GAF-A domain, a single set of [Fe-C-O] vibrations is observed with the full-length protein, suggesting that interactions between domains within DevS influence the distal pocket environment of the heme in the GAF-A domain.
Mycobacterium tuberculosis remains one of the most dreaded pathogens, accounting for 9 million new cases and about 2 million deaths worldwide in 2004 (1). The currently available therapeutic schemes require at least six months for completion (2), a situation that accelerates the development of resistant strains (3). There are 450,000 new cases of multi-drug resistant tuberculosis each year while the total number of such infections is estimated at one million. HIV co-infection poses additional difficulties in the control of tuberculosis (2, 4, 5), as it accelerates the emergence of resistant strains and the combination of HIV and tuberculosis therapeutic regimens is oftentimes problematic (5, 6).
Mycobacterium tuberculosis, under the influence of environmental stimuli such as hypoxia or NO, has been shown to undergo a metabolic transformation to a different state previously labeled as non-replicating persistence (7–9). Hypoxia shifts Mycobacterium tuberculosis to this non-replicating persistent state in which its sensitivity to most drugs is diminished (7). One third of the world’s population is estimated to carry Mycobacterium tuberculosis in this dormant form (9, 10). The scale of the latent tuberculosis infection and the fact that it responds poorly to the available medications renders eradication difficult.
Clearly, tuberculosis represents a therapeutic challenge. The development of new, more efficient anti-infective agents against Mycobacterium tuberculosis depends on the identification of new targets within this bacterium. In this context, there is a need to unravel the mechanisms that allow Mycobacterium tuberculosis to enter the dormant state. DevS-DevR, one of the 11 two-component systems of Mycobacterium tuberculosis (11), is hypothesized to mediate the entry of this pathogen into dormancy and its adjustment to unfavorable environmental conditions such as hypoxia and NO.
DevR has been identified in human monocytes infected with Mycobacterium tuberculosis but it is absent in uninfected cells (12). DevR is required for expression of the α-crystallin gene that is promptly induced upon exposure to either low oxygen or NO (13). α-Crystallin, a heat shock protein, is likely to be involved in stabilizing essential proteins in Mycobacterium tuberculosis, thus reducing the need for the biosynthesis of new molecules for its survival (7). Other changes occur as the bacteria enter the nonreplicating persistent stage, including thickening of the cell wall and a modified metabolic activity (8, 14). Replication of the bacilli is halted. However, Mycobacterium tuberculosis from anaerobic media can be cultured in the presence of oxygen and the bacteria resume their growth under these favorable circumstances (7).
Hypoxic conditions are present in many tuberculosis lesions in patients (9). Another means of limiting tuberculosis infections is the production of NO by the host. Exposure to NO causes expression of a 16 kDa heat shock protein, sHsp16, a homolog of α-crystallin (15). The same two component system may be involved in detection of both hypoxia and NO in Mycobacterium tuberculosis, since the coordinated expression of 48 genes is similarly affected by these two environmental stimuli (16). Nitric oxide synthase-2 (NOS-2 or iNOS), the inducible form, has been identified in the alveolar macrophages of tuberculosis patients and the administration of iNOS inhibitors to infected mice exacerbates their condition (17–19). These facts suggest that NO assists the host in controlling infection by Mycobacterium tuberculosis and that the ability of the bacteria to adapt to its presence greatly enhances their chances of survival.
How does Mycobacterium tuberculosis successfully detect environmental cues such as hypoxia and NO to undergo metabolic changes allowing its survival in these unfavorable conditions? The two sensors that function in conjunction with DevR are DevS and DosT. DosT and DevS were described as membrane anchored kinases (12, 20), although most sequence topology tools do not predict transmembrane domains in either of these proteins (22). Using the database search engine SMART (http://wsmart.embl-heidelberg.de/), both DosT and DevS appear as modular proteins with two N-terminal GAF (cGMP, adenylyl cyclase, FhlA) domains, one HisKA (histidine kinase phosphor-acceptor) domain, and one C-terminal His ATPase (histidine kinase-like ATPase) domain (20, 21). Recently, the first GAF domain of DevS (GAF-A), was reported to bind heme (22). Moreover, site-specific mutagenesis of conserved histidines within GAF-A domains of DevS orthologs showed that His149 is important for heme binding, leading Sardiwal et al. (22) to propose that His149 acts as the proximal iron ligand in DevS. Yet, other histidine mutants such as H89A studied by Sardiwal et al. also display severely affected heme binding properties. Because Sardiwal et al. solely present UV-vis spectra of “as isolated” GAF-A domains, most likely corresponding to heme Fe(II)-O2 complexes, the spin and coordination states of the heme in DevS in the absence of an exogenous ligand remain unknown. Moreover, the fidelity of the heme-binding pocket in the truncated GAF-A to that of the full-length protein remains to be demonstrated. In the present study we examine the N-terminal GAF-A domain and full-length DevS using UV-vis and resonance Raman (RR) spectroscopies.
We characterize the wild-type (wt) GAF-A domain and the H149A mutant in terms of heme binding to the protein and coordination of diatomic gases to the heme iron. We provide resonance Raman evidence that residue His149 is, indeed, the proximal heme ligand in the wt protein, and show that the H149A mutation causes a notable decrease in heme-binding affinity. The proximal ligand His149 is essential for the formation of a stable Fe(II)-O2 complex, and whereas a 6-coordinate heme-NO complex is formed in the wt protein, exposure of the H149A mutant to NO produces a 5-coordinate Fe-NO species. RR experiments also probed the hydrogen-bond interactions of the diatomic gases within the distal pocket that provide a potential framework for the signaling pathway. Finally, the spectroscopic analysis carried out with the GAF-A domain was reproduced with the full length protein. The near identity of the UV-vis and high-frequency RR spectra in the Fe(III), Fe(II), Fe(II)-CO, and Fe(II)-O2 forms of these two proteins confirms that the environment of the heme in DevS is conserved in the truncated GAF-A protein. However, the presence of a single Fe(II)-CO conformer in the full length DevS contrasts with the detection of two Fe(II)-CO conformers in the truncated protein, which suggests that inter-domain interactions have an impact on non-covalent interactions in the distal pocket.
MATERIALS AND METHODS
Materials
Mycobacterium tuberculosis H37Rv genomic DNA was obtained from Dr. Jeffery Cox of the University of California, San Francisco. Ampicillin was purchased from Fisher Biotech while chloramphenicol was obtained from Roche, hemin from Fluka Biochemica, dithionite from JT Baker, and IPTG from Promega. Lysozyme and phenylmethylsulfonyl fluoride were from Sigma. Protease inhibitors (antipain, leupeptin and pepstatin), chloramphenicol, Fast Start High Fidelity PCR system, and Amplitaq polymerase were obtained from Roche. A 1 kbplus DNA ladder was purchased from Life technologies. Argon and CO were from Matheson Tri-Gas.
Simply Blue Safe Stain, In Vision Histag In-Gel Stain, See Blue Plus 2 protein markers, S.O.C. medium, along with the gel apparatus (Novex MiniCell), NuPage 12% Bis-Tris gels and NuPage MOPS SDS running buffer were purchased from Invitrogen. The E. coli XL-10 gold, BL21DE3, and XL-1 Blue cells were from Stratagene.
The TOPO TA cloning kit was obtained from Invitrogen and the QuikChange Mutagenesis kit from Stratagene. DNA purification was carried out using QIAgen kits. The restriction enzymes, T4 DNA ligase, and alkaline phosphatase were from New England Biolabs. HisTrap TM HP columns were obtained from GE Healthcare, while dialysis tubing was purchased from Spectrum laboratories, Inc.
The pET23a+ plasmid was purchased from Novagen. The pTGroE plasmid was kindly provided by Dr. Shunsuke Ishii from the Laboratory of Molecular Genetics, Riken Tsukuba Life Science Center, Japan.
The primers were designed using GCG software package version 10.3 for UNIX from the University of Wisconsin. Custom primers were synthesized by Invitrogen. Amino acid analyses were performed at the University of California, Davis. DNA samples were sequenced at the University of California, Berkeley.
Cloning the Full-length DevS Gene
The DevS gene was amplified from Mycobacterium tuberculosis H37Rv genomic DNA using a PCR PTC-200 Peltier Thermal Cycler from MJ Research and the Fast Start High Fidelity PCR system from Roche. A 10 min. initial denaturation at 94 °C was followed by 35 cycles: 94 °C 1 min., 45 °C 1.5 min, 72 °C 3 min and a 10 min final extension at 72 °C. The forward primer (GGATAGGGCATATGACAACAGGGGGCCTC) included an NdeI cleavage site while the reverse primer (CCAAACGGGGATCCGAAGAGCTACTGCGACAAC) contained a BamH1 site. The reaction mixture contained 4% DMSO. The product obtained in this reaction was reamplified using the same conditions in order to obtain sufficient amounts of PCR product.
TOPO TA Cloning
3’ Poly-adenine overhangs were added to the purified PCR product using Amplitaq DNA polymerase. The TOPO TA cloning reaction was performed for 30 min at room temperature, according to the manufacturer’s instructions using the pCR2.1-TOPO vector. Plasmid DNA was purified from several colonies of TOP10 cells transformed with the TOPO TA cloning reaction product. Insertion of the DevS gene was verified using restriction digests (either NcoI or NotI) and the correct sequence of the DevS gene was confirmed by DNA sequencing.
Subcloning of DevS642
The first 642 bp which code for the N-terminal GAF domain were amplified using the same PCR conditions and the following primers: CCGCCGCCATATGCATCATCATCATCATCACGAGAACTTATATTTTCAAGGAATGACAACAGGGGGCCTCGTCGAC (forward primer containing a 6-His tag and an NdeI cleavage site) and CGGACAAGCTTCTATTACGACTGACGCGCCTTAGCCTGCTG (the reverse primer that includes a HindIII site). The purified PCR product was digested with NdeI and HindIII and ligated into pET23a+ cleaved with the same restriction enzymes as well as with alkaline phosphatase. XL-10 gold ultracompetent cells were transformed with the ligation product. After restriction analysis, the plasmid DNA was also submitted for sequencing in order to confirm the correct sequence.
Mutagenesis of His149 of DevS642
The QuikChange Mutagenesis kit from Stratagene was used to mutate His149 to an alanine according to the manufacturer’s instructions. The mutagenic primers used were: CGATTGGTTTTCCGCCGTATGCCCCGCCGATGCGTACCTTCCT (forward primer) AGGAAGGTACGCATCGGCGGGGCATACGGCGGAAAACCAATCG (reverse primer). The constructs were submitted for DNA sequencing in order to confirm the correct sequence.
Expression of DevS642 and Full-length DevS
BL21gold DE3 cells were co-transformed with pET23a+DevS642 and pT-GroE. The cells were grown on LB plates containing both ampicillin (50 μg/mL) and chloramphenicol (34 μg/mL). Five starter cultures were then inoculated with one colony each. The five cultures were then pooled and used to inoculate flasks containing 1.5 L LB medium as well as the antibiotics ampicillin (100 μg/mL) and chloramphenicol (34 μg/mL). The cells were grown to an optical density of OD600 0.8 at 37 °C and 230 rpm. Hemin was added before induction (45 mg/4.5 mL NaOH 0.1 N for each 1.5 L culture). Protein expression was induced with IPTG at a final concentration of 1 mM and the flasks were kept at 18 °C for 20 h. The cells were harvested by centrifugation at 5000 rpm for 25 min. Besides E. coli BL21DE3, two other cell lines were tested: Rosetta 2 and DH5α. DH5α cells did not express the desired protein either when they were transformed only with the plasmid coding for DevS642 or when they were co-transformed in order to provide the GroEL/ES complex as well. Rosetta 2 cells were also tested, because several codons of lower usage in E. coli were identified inside the gene. It was found, however, that the expression level was not significantly enhanced. The full-length DevS was cloned into pET23a+ using the same strategy as in the case of the wt DevS642 protein. The full-length DevS was co-expressed with the GroEL/ES complex and then purified using the same conditions.
Expression of ApoDevS642
The same procedure was followed as for the heme bound protein except i) no hemin was added, and ii) extra steps were taken to remove iron and cobalt from the culture medium. A published procedure was used and modified to exclude cobalt salts from the mineral mix (23).
Expression of the H149A Mutant of DevS642
The mutant could be expressed both as the heme bound protein and as the apoprotein following the method employed in the case of wt DevS642. In order to obtain the apoprotein, the lack of hemin addition was sufficient and no further steps were required in order to decrease contamination with the heme bound recombinant protein. The apoprotein of the H149A mutant was obtained using LB medium.
Protein Purification
The cells were lysed in phosphate buffer pH 7.6 (50 mM NaH2PO4, 10% glycerol, 200 mM NaCl, 1% Triton X-100, 0.5 mg/mL lysozyme, protease inhibitors: antipain 1 μg/mL, leupeptin 1 μM, pepstatin 1 μM, PMSF 0.1 mM). Then the mixture was incubated with shaking at 37 °C for 10 min. The cell membranes were disrupted by repeated sonication cycles at 50% using a Branson sonifier 450 from VWR Scientific, while cooling on ice. The insoluble fraction was isolated by centrifugation at 35000 rpm for 1 h at 4 °C. The cell lysate was applied to a 5 mL His trap column at a rate of 1 mL/min. The column was then washed with 20 and 50 mM imidazole in phosphate buffer (50 mM NaH2PO4, 10% glycerol, 500 mM NaCl, 50 mL at a rate of 2 mL/min). The recombinant protein eluted with 200 mM imidazole in phosphate buffer (50 mL, 1 mL/min). The sample was then dialyzed and the fractions that were not pure were repurified on a His trap column to provide the pure protein. This procedure was successful for the heme containing wt DevS642 and H149A, and also for the apoprotein of the H149A mutant. The buffer was replaced with 20 mM Hepes buffer (150 mM NaCl pH 8) for the isolation of apoDevS642.
Protein Quantification
The protein content for the wt DevS642 was determined by amino acid analysis at the University of California, Davis. The yield of soluble protein was 16 mg/L culture in the case of co-expression with the heat shock proteins GroEL/ES. The yield of soluble protein when it was expressed without chaperones was 2.3 mg/L culture.
Heme Content Determination of Wt DevS642
Sodium hydroxide 5 N (5 μL) and pyridine (18 μL) were added to the protein sample (80 μL of the ferric complex). The oxidized spectrum was recorded and then sodium dithionite (a few crystals) was added to obtain the reduced spectrum. The absorbance at 539 and 556 nm was used to determine the heme content according to the published protocol (24). The heme content was determined to be 97%.
Size Exclusion Chromatography of Wt DevS642
The oligomeric state of the recombinant protein DevS642 was established using a Superdex 75 (FPLC) column in a phosphate buffer system (50 mM phosphate, 200 mM NaCl pH 7.6). The absorbance of the eluate was monitored at two wavelengths, 280 and 406 nm. Dextran (1 mg/ml) was used to determine the void volume. The calibration curve was obtained with the following protein standards: albumin (MW 66000, 10 mg/mL), alcohol dehydrogenase (MW 150000, 5 mg/mL), carbonic anhydrase (MW 29000, 3 mg/mL), cytochrome c (MW 12400, 2 mg/mL). DevS642 is 96% monomeric (MW 30269 as estimated from the gel filtration assay) and 4% dimeric (MW 61518).
Hemin Titrations of Wt ApoDevS642
A 500 μL protein sample (typically 5 μM concentration in Hepes 20 mM, NaCl 150 mM, 10% glycerol buffer) was titrated with a hemin solution. Aliquots of 0.5 μL of a 0.106 mg/mL hemin solution were added and the difference spectrum was recorded 10 min after each addition (the reference cuvette contained buffer and the same amounts of hemin.) To obtain the dissociation constant, ΔAbsorbance 407-350 nm was plotted as a function of hemin concentration and the data points in the titration were fit to the following equation: ΔAbsorbance = Amax/1 + KD/[hemin] (25).
Cyanide and Azide Titrations of Wt DevS642
The concentration of DevS642 was determined based on the intensity of the Soret peak at 407 nm. A 500 μL sample (usual concentration 5 μM) was used in these assays. The initial spectrum of the protein in phosphate buffer (50 mM phosphate and 200 mM sodium chloride) was recorded and then difference spectra were acquired 5 min. after each addition (typically 0.5 μL) of the ligand to both the reference cuvette and the cuvette containing the protein solution. Sodium azide and potassium cyanide were prepared as aqueous solutions in the same buffer. The increase in absorbance at 424 nm (cyanide) or 422 (azide) was plotted as a function of ligand concentration and the data were fit to a rectangular hyperbola (the azide titration) or the quadratic tight binding equation in the case of cyanide (26).
Electronic Absorption and Resonance Raman Spectroscopy
Typical enzyme concentrations used were ~100–300 μM. Biomax-10 ultrafiltration devices (Millipore) were used for buffer exchange and for concentrating the protein. Reduction to the ferrous state was achieved by adding microliter aliquots of 25–35 mM sodium dithionite solution to an argon-purged sample in the Raman capillary cell and was monitored by UV-visible spectroscopy directly in the capillary using a Cary 50 spectrometer. 12CO (Airgas) and 13CO (99% 13C, ICON Stable Isotopes) adducts were obtained by injecting CO through a septum-sealed capillary containing argon-purged, reduced protein (~20 μL). O2 (Airgas), 18O2 (99% 18O, ICON Stable Isotopes), NO (Aldrich), and 15N18O (98% 15N and 95%18O, Aldrich) adducts were generated using the same procedure after excess dithionite was removed from the reduced sample with desalting spin columns (Zebra™ 0.5 mL, Pierce). These procedures were performed in a glovebox with a controlled atmosphere of less than 1-ppm of O2 (Omni-Lab System, Vacuum Atmospheres Company). RR spectra were obtained using a custom McPherson 2061/207 spectrograph (0.67 m with variable gratings) equipped with a Princeton Instruments liquid N2-cooled CCD detector (LN-1100PB). Kaiser Optical supernotch filters were used to attenuate Raleigh scattering. A Krypton laser (Innova 302, Coherent) and a He/Cd laser (Liconix 4240NB) were used for the 413- and 442-nm excitations, respectively. Spectra were collected in a 90° scattering geometry on samples at room temperature. Frequencies were calibrated relative to indene and CCl4 and are accurate at ±1 cm−1. CCl4 was also used to check the polarization conditions. The integrity of the RR samples, before and after laser illumination, was confirmed by direct monitoring of their UV-visible spectra in the Raman capillaries.
RESULTS
Protein Expression and Purification
The truncated protein DevS642 comprises the first 214 amino acids of DevS, which includes the putative N-terminal GAF domain from residue 63 to 210. The DevS gene was cloned from genomic Mycobacterium tuberculosis H37Rv DNA and the first 642 base pairs of the gene along with an N-terminal 6 histidine tag were subcloned into a pET23a+ expression vector. The construct was expected to provide high levels of protein expression, but the strong IPTG inducible T7 promoter in pET vectors is also known to provide the recombinant proteins as insoluble inclusion bodies (21). To increase the yield of soluble protein, DevS642 was co-expressed with the GroEL/ES chaperones and the expression was carried out at a relatively low temperature (18 °C) for an extended period of time (20 h). Experiments varying the IPTG concentration from 1 to 0.4 mM were also performed, but the lower IPTG concentration resulted in lower levels of recombinant protein in BL21DE3 cells (Fig. S1 and S2). Comparable amounts of recombinant protein were produced in cells that expressed only DevS642 and cells where DevS642 and GroEL/ES were co-expressed, but the co-expression of GroEL/ES resulted in a 7-fold yield increase in the purification of soluble DevS642. Purification of the wt protein was achieved using a metal affinity column (Fig. 1). Size exclusion chromatography indicated that the recombinant protein exists predominantly as a monomer in the buffer system employed. A pyridine hemochrome assay revealed that DevS642 binds heme in stoichiometric amounts with one heme molecule bound per GAF domain.
FIGURE 1.
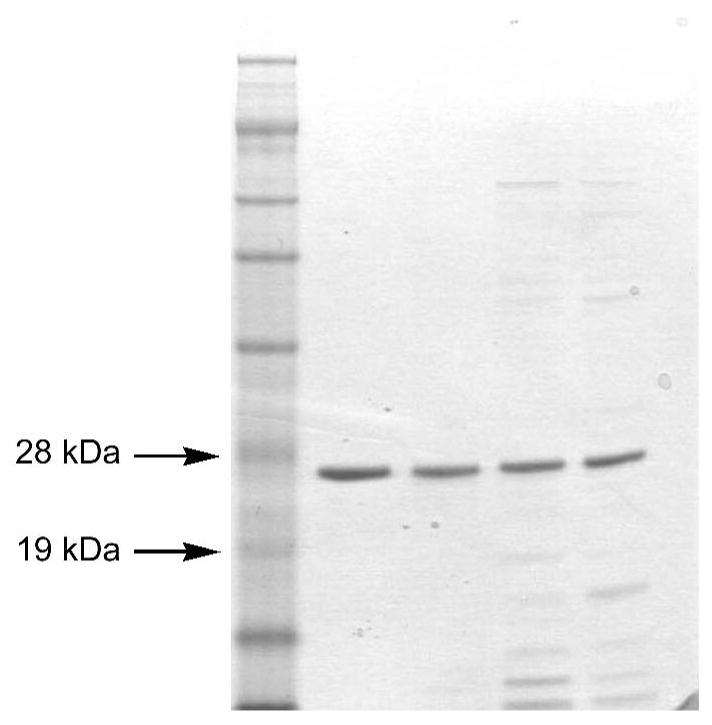
SDS-PAGE of wt DevS642 (molecular weight 25330 Da) and H149A mutant after purification by affinity chromatography. In each lane about 0.5 μg of protein was loaded on a 12% BT Novex NuPage gel. Lane 1, SeeBlue Plus 2 prestained protein standard (7 μL); lane 2, DevS642 expressed in the presence of hemin, lane 3, H149A mutant expressed in the presence of hemin; lane 4, DevS 642 apoprotein; lane 5, H149A mutant apoprotein.
The full-length DevS and the H149A DevS642 variant were expressed and purified following the same procedure used for the wt DevS642 protein. Despite the mutation, the variant protein is purified as a heme-containing protein when the growth medium is complemented with hemin before induction. The variant could also be purified as the apoprotein when the expression occurs in the usual Luria Bertani culture medium (which includes both iron and cobalt) but in the absence of added hemin. The affinity for hemin in the mutant could not be precisely determined due to spectral overlap difficulties but is clearly lower than that of wt DevS642 (Kd 3.1±2.0 μM).
Spectroscopic Characterization of Wt and H149A DevS642
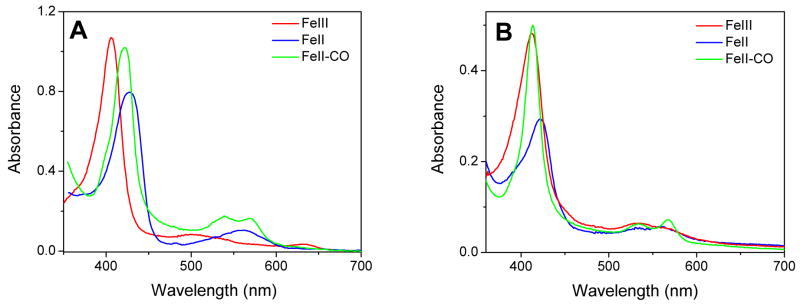
UV-vis absorption spectra of wt DevS642 (see Fig. 2A and Table 1; also see Fig. S3A) clearly establish that its heme forms stable complexes with O2, CO, and NO. The formation of a stable oxy-complex is consistent with its potential role as a gas sensor in vivo. Additionally, DevS642 in the ferric state binds cyanide with high affinity (Kd = 0.37 ± 0.2 μM). DevS 642 also binds azide (Kd = 8.81 mM ± 0.76), but fails to interact with the typical P450 azole inhibitors clotrimazole, econazole, fluconazole, miconazole, sulconazole, or ketoconazole. NO binds to both oxidation states of the iron, while CO and O2 only bind to the reduced protein. The histidine to alanine substitution in the H149A variant has serious consequences on the heme absorption (see Fig 2B and Table 1; also see Fig. S3B). Most importantly, H149A DevS642 does not form a stable oxy complex, as exposure of the reduced form to O2 leads to autooxidation, producing the oxidized protein. The H149A variant also responds differently to NO than the wt protein. Specifically, the ferrous heme in H149A DevS642 binds NO to form a 5-coordinate Fe(II)-NO complex with a characteristic Soret absorption near 400 nm (Fig. S3B and Table 1). When the ferric heme is exposed to NO, the Soret absorption is slightly broader with a maximum at 402 nm which suggests the presence of a transient species with a red-shifted Soret, either an Fe(III)-NO or a 6-coordinate Fe(II)-NO, but the sample quickly evolves to produce the 5-coordinate Fe(II)-NO complex (Fig. S3B). High-frequency RR spectra of the nitrosyl adducts in the H149A variant confirm the UV-vis analyses (data not shown).
FIGURE 2.
UV-visible spectra of the ferric, ferrous, and Fe(II)-CO complexes of wt DevS642 (A) and the H149A DevS642 mutant (B).
Table 1.
UV-vis spectroscopic data for wt and H149A GAF-A domains of DevS and their complexes
| Heme state | wt DevS | H149A DevS | ||
|---|---|---|---|---|
| Soret band (nm) | Visible bands (nm) | Soret band (nm) | Visible bands (nm) | |
| Fe(III) | 406 | 500, 630 | 412 | 535 |
| Fe(II) | 428 | 562 | 422 | 559 |
| Fe(II)-CO | 422 | 540, 570 | 413 | 533, 567 |
| Fe(II)-NO | 419 | 545, 574 | 400 | 541, 568 |
| Fe(III)-NO | 420 | 539, 569 | unstable | unstable |
The coordination number, spin state, and oxidation state of the heme iron of the wt and H149A proteins are revealed by the frequency of the ν4, ν3, ν2, and ν10 modes in the high-frequency region of the RR spectra obtained with Soret excitation (Table 2) (27). At room temperature and neutral pH, ferric wt DevS642 exhibits ν4, ν3, ν2, and ν10 at 1370, 1479, 1557, and 1609 cm−1, respectively. The frequencies of these bands are characteristic of a 6-coordinate high-spin (6cHS) heme (Fig. 3A). A minor 6-coordinate low-spin (6cLS) species is evidenced by a shoulder at 1507 cm−1 in the range of ν3 modes. The intense high-spin charge-transfer marker band at 630 nm in the UV-vis spectrum (Fig. 2A) and the dominance of high-spin marker bands in the RR spectra (Fig. 3A) support the assignment of the high-spin state as the major conformer in the wt protein. The low-spin population remains nearly constant when the pH is raised to 9.5 or when the temperature is lowered to 100 K (data not shown). Upon reduction with dithionite, the ferrous wt DevS642 adopts a pure 5-coordinate high-spin (5cHS) configuration with ν4, ν3, ν2, and ν10 at 1354, 1470, 1557, and 1603 cm-1, respectively (Fig. 3B).
Table 2.
Heme iron coordination and Raman frequencies of porphyrin skeletal modes for wt and H149A DevS642
| Protein | Heme state | ν4 (cm−1) | ν3 (cm−1) | ν2 (cm−1) | ν10 (cm−1) |
|---|---|---|---|---|---|
| Fe(III) wt DevS642 | 6cHS/6cLS | 1370 | 1479, 1507 | 1557, 1579 | 1609, 1639 |
| Fe(III) H149A DevS642 | 5cHS/6cLS | 1374 | 1506, 1491 | 1581, 1568 | 1640,1624 |
| Fe(II) wt DevS642 | 5cHS | 1354 | 1470 | 1557 | 1603 |
| Fe(II) H149A DevS642 | 5cHS/4cIS | 1359 | 1471, 1501 | 1557, 1583 | 1603, 1638 |
FIGURE 3.
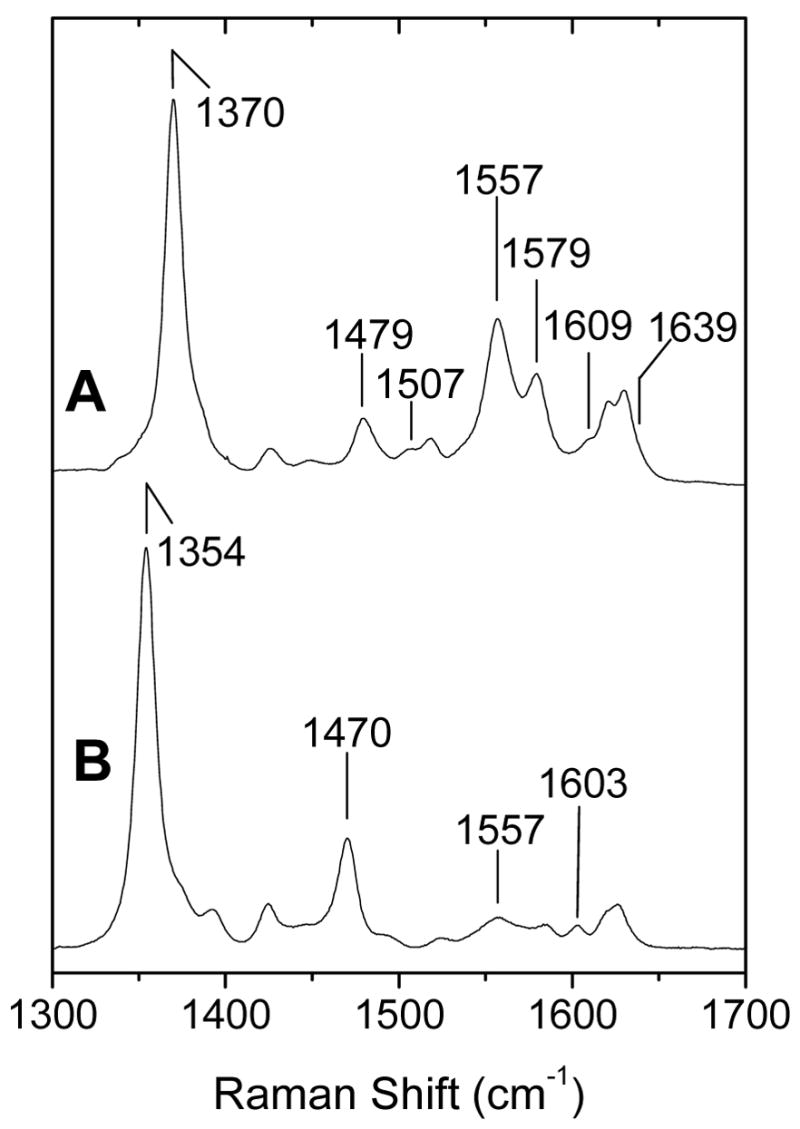
High-frequency RR spectra of ferric (A) and ferrous (B) wt DevS642 at room temperature (λexc = 413 nm; 5 mW).
In contrast to the wt protein, the ferric heme in H149A DevS642 exists as a mixture of 5cHS and 6cLS states with ν3 at 1491 and 1506 cm-1, and ν10 at 1624 and 1640 cm−1, respectively (Fig. 4A). The ferrous heme-H149A DevS exists as a mixture of spin and coordination states where a 5cHS species is indicated by ν4, ν3, ν2, and ν10 at 1359, 1471, 1557, and 1603 cm-1, respectively (Fig. 4B), and contributions from a 4-coordinate intermediate-spin (4cIS) species are also apparent with ν3 and ν10 at 1501 and 1638 cm-1, respectively (28–30). Determining spin-state populations from relative intensities in the RR spectra could be misleading since the ferric and ferrous variant proteins exhibit broad Soret absorptions, raising the possibility of significant differences in resonance enhancement for individual spin states. As with the wt protein, varying the pH has little effect on the RR spectra of the H149A variant protein (data not shown).
FIGURE 4.
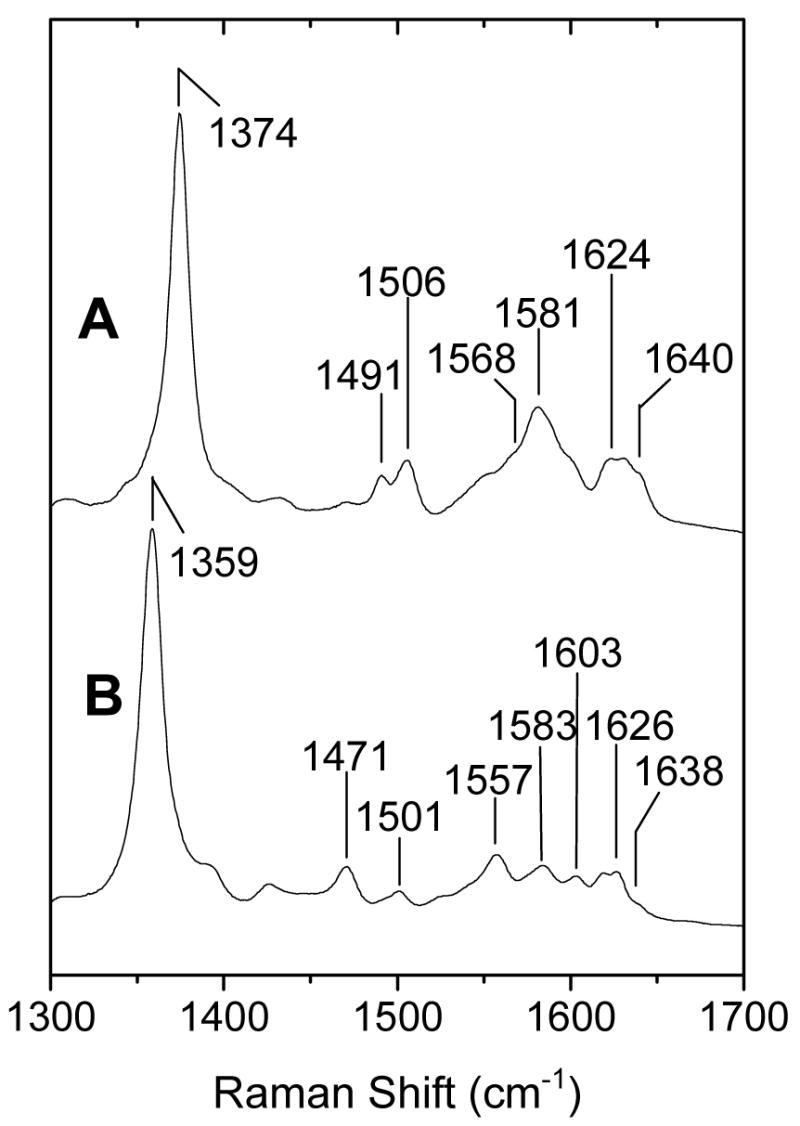
High-frequency RR spectra of ferric (A) and ferrous (B) H149A-DevS642 at room temperature (λexc = 413 nm; 5 mW).
When 442-nm excitation is used, the low-frequency region of the RR spectrum of the reduced wt protein displays a strong RR signal at 214 cm−1 (Fig. 5A). The vibrational frequency and the intensity of this band are characteristic of a ν(Fe-NHis) mode from a neutral proximal histidine that is free of any strong hydrogen bonds (31). The intensity of the ν(Fe-NHis) is large compared to the out-of-plane porphyrin vibrations and ν(C-C-C) from porphyrin peripheral groups observed within this 250–450 cm−1 frequency window (Fig. 5A), and more alike to what is seen in ferrous heme oxygenases than in deoxymyoglobin (30–33).
FIGURE 5.
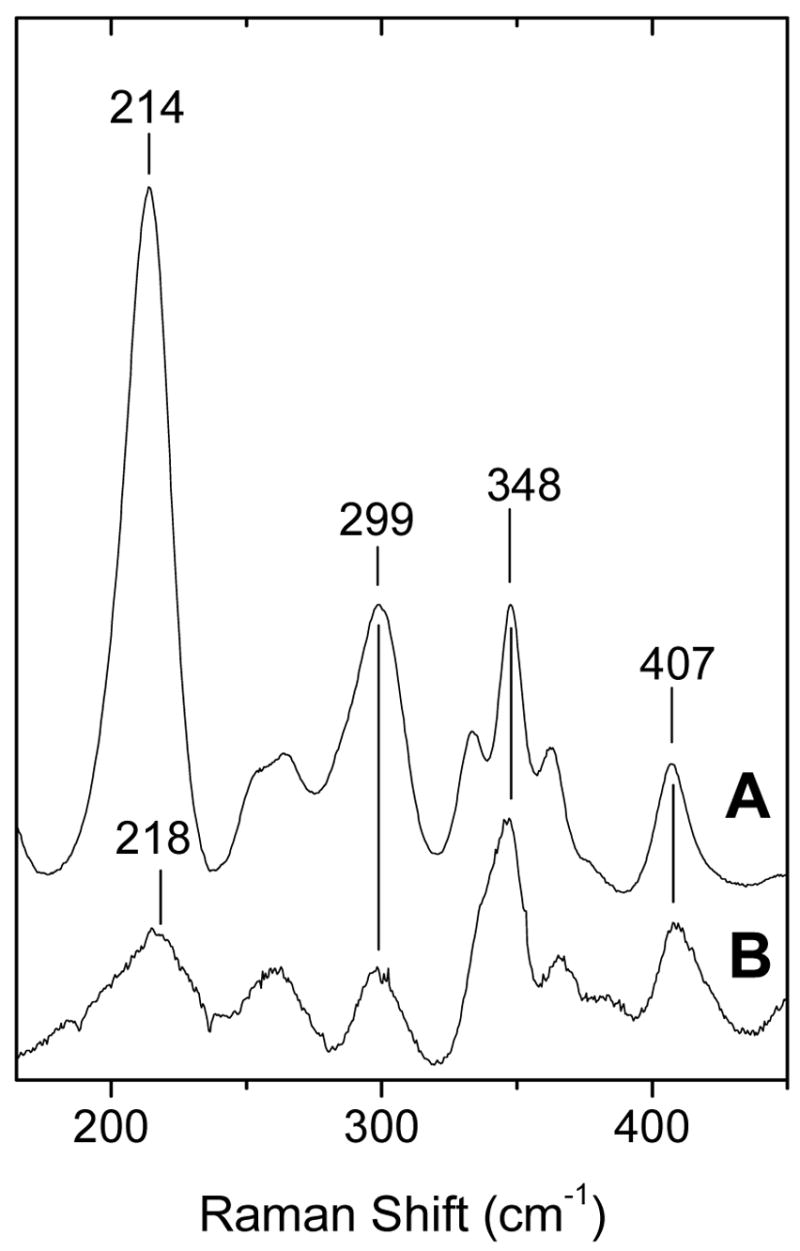
Low-frequency RR of ferrous wt DevS642 (A) and H149A-DevS642 (B) at room temperature (λexc = 442 nm; 15 mW).
While differences in the ν(Fe-NHis) intensities may reflect variations in displacement of the iron from the heme plane and/or changes in tilt angle of the Fe-NHis bond from the heme normal (34, 35), such analysis is beyond the scope of this article. While other low frequency modes are only marginally perturbed in the ferrous H149A variant, the ν(Fe-NHis) band is not observed (Fig. 5B). The origin of the weak signal at 218 cm−1 observed in the RR spectrum of the variant protein is unknown. This signal may correspond to an Fe(II)-N stretching mode or an out-of-plane porphyrin deformation mode.
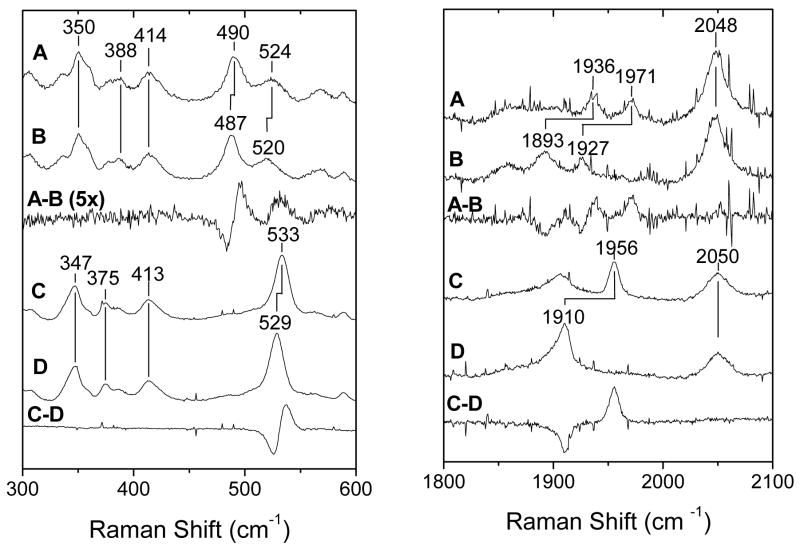
The identification of His149 as the proximal ligand to the iron in the wt protein was further confirmed by the RR characterization of the carbonyl complexes. Vibrations from the [Fe-CO] unit were unambiguously identified by 12C/13C isotopic substitution (Fig. 6). In wt DevS642, RR bands at 524 and 490 cm−1 that downshift by 4 and 3 cm−1 upon 13CO substitution, respectively, are assigned to two distinct ν(Fe-CO) bands. Corresponding ν(C-O) bands appear at 1936 and 1971 cm−1, with 12C/13C shifts of -43 and -44 cm−1, respectively. These observed frequencies are characteristic of heme-carbonyl complexes with a neutral histidine coordinating trans to the CO group (36). Effects of the trans ligand on the Fe-C-O vibrations are well understood in terms of competition from σ-bond donation to the iron and are easily visualized in graphs correlating ν(Fe-CO) to ν(C-O) frequencies (Fig. 7). The two sets of Fe-C-O vibrations observed in the wt protein fall on the correlation line for complexes with trans histidine ligands, and thus, suggest two species with distinct distal pocket conformations. These two conformers appear to be present at equivalent concentration as their respective ν(C-O) vibrations show comparable intensities. The position of a correlation point along the line for an imidazole trans ligand is governed by the degree of backbonding from the filled Fe d orbital to the empty π* orbital of the bound CO. The extent of backbonding is a good indicator of the electrostatic environment in the distal pocket where positive polarity and/or hydrogen bonding interactions at the CO group promote backbonding while negative polarity inhibits backbonding (36). The low correlation points originating from the ν(Fe-CO) and ν(C-O) at 490 and 1971 cm−1, respectively, are very similar to those measured with the H64L variant of human myoglobin (i.e., 490 and 1965 cm−1, respectively) (37, 38), suggesting that in this conformer, the CO group lacks any hydrogen-bond interaction (Fig. 7). In contrast, the high correlation point with ν(Fe-CO) and ν(C-O) at 524 and 1936 cm−1, respectively, falls nearer to values obtained with the V68N-myoglobin variant (i.e., 526 and 1922 cm−1, respectively) (37, 39), where an additional hydrogen bond donor is substituted for a hydrophobic residue in the distal pocket (Fig. 7). These observations suggest that this CO conformer in wt DevS642 engages strong hydrogen bond interaction(s) with distal pocket residue(s).
FIGURE 6.
Low-frequency (right) and high frequency (left) RR spectra of wt DevS642-12CO (A), wt DevS642-13CO (B), H149A-DevS64212CO (C), and H149A-DevS64213CO (D) at room temperature (λexc = 413 nm, 0.5 mW).
FIGURE 7.
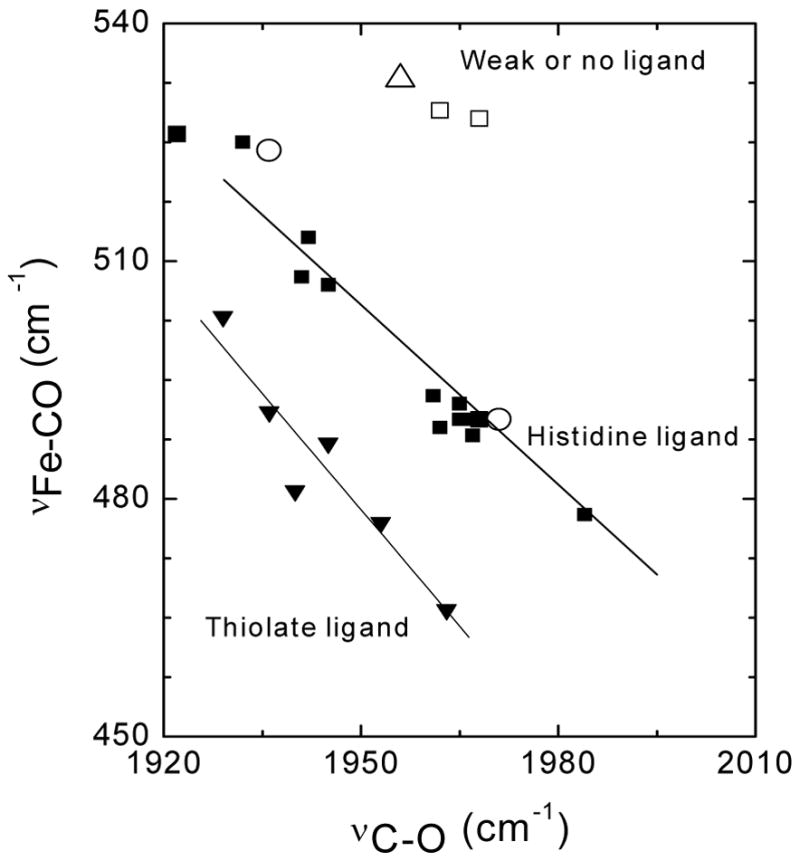
ν(C-O) versus ν(Fe-CO) plots of hemoprotein-CO complexes. Data points for wt DevS642 (○) and H149A (△) are compared with values for wt and distal mutants of myoglobin from (64) (◆), and from cytochrome P450 and NO synthase (←) (36, 68). Also shown are proximal histidine mutants of heme oxygenase (□) (29, 30).
The RR spectra of the carbonyl complex in the H149A variant reveal a single conformer, with a ν(Fe-CO) at 533 cm−1 (Δ13C = −4 cm−1) and a ν(C-O) at 1956 cm−1 (Δ13C = −46 cm−1) (Fig. 6). These Fe-C-O stretching frequencies are characteristic of a complex with a very weak or no proximal ligand (Fig. 7), supporting the identification of His149 as the proximal histidine ligand in the wt protein.
Spectroscopic Characterization of the Full-length Wt DevS
To confirm the relevance of the spectroscopic data obtained with the heme-containing GAF-A domain of DevS, some of the RR experiments were reproduced with the full-length protein wt DevS. The UV-vis spectra of the full length protein (Fig. S4) are virtually the same as those of wt DevS642 (Fig. 2A). A comparison of the high-frequency RR spectra of the ferric and ferrous proteins revealed no significant differences between the GAF-A and the full-length protein (Fig. S5). Moreover, the reduced full-length protein displays a strong ν(Fe-NHis) which is identical within experimental error (±1 cm−1) to that observed in the truncated protein (Fig. S6). Thus, it appears that the proximal pocket in the full-length protein is not affected by interactions of the GAF-A domain with other domains of DevS.
In contrast with the identity of the proximal pocket in truncated and full-length DevS, the characterization of the CO complex in the full-length protein reveals differences in the distal pocket of the isolated GAF-A domain and the full-length protein. Specifically, the CO conformation lacking the hydrogen bond interaction is the only conformer stabilized in the full-length protein (Fig. 8). Thus, it appears that inter-domain interactions in the full-length protein eliminate the structural flexibility that leads to the presence of two CO conformers. Interactions between the heme sensing domain and the kinase domains are anticipated, since conformational changes accompanying ligand binding to the heme domain must be transduced to the kinase domain in order to modulate its activity.
FIGURE 8.
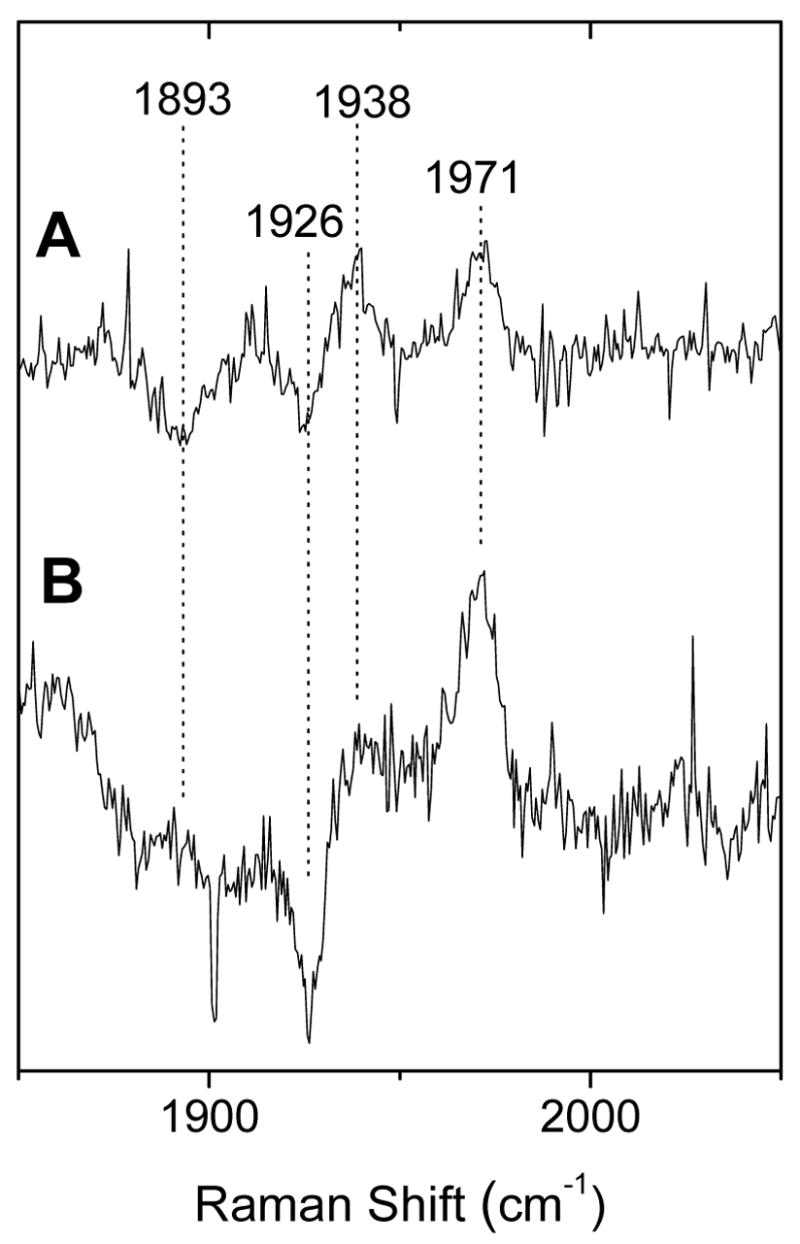
High-frequency RR difference spectra of the reduced-12CO minus reduced-13CO complexes of DevS642 (A) and full-length DevS (B) (λexc = 413 nm; 0.5 mW)
DISCUSSION
Analysis of the DevS amino acid sequence using different algorithms leads to conflicting results regarding the presence of transmembrane segments. Based mainly on the SwissProt database of proteins and their transmembrane helices, TMPred (European Molecular Biology Network, Swiss node) (40) identifies three hypothetical transmembrane helices in DevS, one within the GAF A domain (TM1 187–206), and two within the GAF B domain (TM2 276–295 and TM3 327–351). In contrast, TMHMM (Center for Biological Sequence Analysis, Technical University of Denmark) predicts no transmembrane domain within DevS (41). As a result, different views exist in the literature concerning transmembrane regions within DevS (12, 20–22). With the aid of chaperones, we were able to express the first GAF domain of DevS and the full-length protein as soluble, well-behaved proteins. These proteins showed no unusual propensity to aggregation, an observation that argues against the presence of transmembrane segments in DevS.
The bulk of the full-length DevS and truncated versions were expressed in the earlier study as insoluble proteins (22). We counteracted the anticipated difficulties of inclusion bodies by co-overexpressing the N-terminal GAF domain and full-length DevS with the GroES/EL complex. The accumulation of recombinant protein in inclusion bodies in E. coli as a consequence of protein expression using a strong promoter such as T7 is well documented. Previous attempts to obtain other proteins in their biologically active soluble form included chaperone co-expression methods (42–47). Expression of heat shock proteins is often induced shortly before or at the same time as the recombinant protein of interest (48–53). Moreover, in vitro studies indicated that an excess of GroES/EL favors refolding of recombinant proteins (54). Consistent with this observation, over-expression of these chaperones proved to be beneficial in our situation as well, as the GroES/EL chaperones apparently assisted proper folding of the DevS proteins.
Recently, Sardiwal et al. reported that the GAF-A domain of DevS binds heme (22). However, the UV-vis spectra suggested that DevS possesses a low affinity for heme, since a relatively low fraction of the protein appears to be heme-bound. The GAF-A domain is thought to bind heme in a pocket similar to that which binds cGMP in the GAF-B domain of phosphodiesterase 2A, for which a crystal structure is available (22, 55). With our expression system, the protein-to-heme ratio deduced from amino acid quantitation and pyridine hemochrome assay indicates that heme binds in a stoichiometric amount to the GAF-A domain. The improvement in heme incorporation in DevS642 is probably due to our addition of hemin before induction of the recombinant-protein expression. While the early addition of hemin could facilitate proper folding of the protein, we assign our increased yield of soluble protein to the presence of GroEL/ES, since in its absence our yield of soluble protein is comparable to that of Sardiwal et al. (22).
We characterized the binding of DevS642 to several nonvolatile ligands. The wt DevS642 has a high affinity for cyanide (Kd = 0.37 ± 0.2 μM), as expected for a heme protein with a water accessible active site. It also binds azide albeit with much lower affinity (Kd = 8.81 ± 0.76 mM). Azole antifungals, most notably econazole, have been found to be active against Mycobacterium tuberculosis both in vitro and in vivo (56–58). M. tuberculosis CYP121 and CYP51 are two possible hypothetical targets, as these enzymes have been shown to bind azole antifungals (59–61). DevS624 does not interact with the azole P450 inhibitors clotrimazole, econazole, ketoconazole, miconazole, sulconazole, or fluconazole, ruling out a direct role for this protein in inhibiting the growth of Mycobacterium tuberculosis when exposed to these agents.
The previous report on the H149A mutant of DevS suggested that this protein did not bind heme at all, a finding that was used as evidence for a role of His149 as a heme axial ligand (22). In our system, the H149A variant incorporates heme and displays an intense Soret band suggesting a well-defined environment governed by heme-protein interactions other than direct coordination of the His149 imidazole ring to the iron atom. While these heme-protein interactions in the H149A variant are sufficient to generate a homogenous complex, they remain weak, and allow easy acquisition of apo-H149A-DevS642 under conditions where the wt DevS642 remains fully loaded with heme. Indeed, the omission of added hemin suffices to produce the apo-form of H149A, whereas wt DevS642 retains 50%-heme occupancy under similar conditions even when steps are taken to exclude iron from the culture medium and the iron chelator 2, 2′-dipyridyl must be used to obtain the apo-wt protein.
Our RR analysis provides direct evidence for proximal coordination of the heme iron to His149. The observed ν(Fe-NHis) vibrations in wt DevS642 and in full-length DevS are in the expected range for a proximal histidine with a neutral imidazole ring free of strong hydrogen bonding interactions (31). A neutral proximal histidine is also present in the heme domain of B. japonicum FixL, another oxygen sensing protein (62). Signal transduction in FixL is believed to be dominated by an expansion of the heme pocket and salt bridge interactions engaged by the heme propionate groups (62, 63). This mechanism of gas sensing in FixL contrasts with the allosteric control that takes place in hemoglobin, where O2-binding results in a subtle shift of the proximal histidine and F-helix. Proximal rearrangements are also important in soluble guanylate cyclase (sGC), where binding of NO results in cleavage of the heme iron-histidine bond and activation of the catalytic activity located on a different subunit (64–66). While proximal histidine movement cannot be ruled out as a source of signal transduction in wt DevS, the UV-vis and RR characterization (data not shown) of its nitrosyl complex excludes a labile histidine as a possible mechanism.
The ferric wt DevS exists predominantly as a 6cHS species with minor contributions from a 6cLS species. The independence of the RR spectra to changes in pH and temperature argue against a water molecule as the sixth iron ligand. These observations suggest that a protein residue may ligate the heme iron on the distal side of the heme, and the existence of two spin-states may be explained as an equilibrium between weakly and strongly bound forms of this ligand. Alternatively, the equilibrium could exist between two endogenous ligands of differing field strength competing for the bond to the heme iron. When His149 is replaced by an alanine, the sixth ligand may still interact with the ferric heme iron to produce a 5cHS species as well as a 6cLS species, which suggests that a second ligand can substitute for His149 at the proximal site. However, this set of axial ligands is unstable since the ferrous protein appears as a mixture of 5cHS and 4cIS. Most importantly, upon exposure to CO, H149A appears as a homogeneous population that lacks a strong ligand trans to the carbonyl group. The presence of an endogenous but labile sixth ligand in the wt protein supports this interpretation.
In wt DevS642, two distinct CO conformers are detected. These two conformers are present at comparable concentrations and reflect geometries which differ in terms of the hydrogen bond interactions engaged by the CO group in the distal pocket. In contrast, in the full-length protein only the CO conformation lacking the hydrogen bond is observed. A similar behavior has been reported for the nitrosyl adducts of FixL, as the isolated heme domain displays two distinct N-O stretches but only one is present in the functional heme kinase protein (67). Thus in DevS, like in FixL, the heme binding domain presents a better-defined distal pocket in the full-length protein, and it is reasonable to propose that this decrease in flexibility originates from intramolecular interactions between the heme domain and other domains present only in the full-length enzyme.
In conclusion, the truncated version of DevS including the N terminal GAF domain was characterized using UV-visible and RR spectroscopy and compared to the H149A mutant as well as to the full-length DevS. Although the mutation of His149 allows the resulting protein to bind heme and display an intense Soret peak, the heme environment is clearly altered and the interaction with the prosthetic group is weakened. The absence of the ν(Fe-NHis) band at 214 cm−1 from the RR spectrum of the reduced H149A mutant following 442 nm excitation (a band that is intense in the case of the wt) clearly demonstrates that His149 coordinates to the heme iron. The first GAF domain of DevS binds heme in stoichiometric amounts and interacts, in the ferrous form, with diatomic gases such as O2, CO, and NO. This truncated version of the wt protein also binds NO as the ferric complex. Two conformations have been detected by RR for the CO complex of the ferrous DevS GAF-A domain, yet in the full-length protein only one of these two conformations is present. The first GAF domain has limited flexibility in the full-length protein, probably due to inter-domain interactions within DevS. Overall, our results provide support for the role of DevS as a gas sensor responsible for mediating the adaptation of Mycobacterium tuberculosis to stress factors.
Supplementary Material
Supplementary SDS PAGE data on different expression systems, UV-visible spectra of Fe-NO complexes, and low-frequency and high-frequency RR spectra of ferrous DevS642 and full-length DevS are provided. This material is available free of charge via the Internet at http://pubs.acs.org.
Footnotes
This work was supported by grant PO1 GM56531 (PROM) and GM74785 (PML) from the National Institutes of Health.
Abbreviations: IPTG, isopropyl-β-D-thiogalactopyranoside; resonance Raman, RR; wt, wild-type; GAF domain, protein domain conserved in cyclic GMP-specific and stimulated phosphodiesterases, adenylate cyclases, and E. coli formate hydrogenlyase transcriptional activator (Pfam accession number PF01590)
References
- 1.World Health Organization. Global tuberculosis control - surveillance, planning, financing. 2006. [Google Scholar]
- 2.Sbarbaro JA. Tuberculosis in the 1990s. Epidemiology and therapeutic challenge. Chest. 1995;108:58S–62S. doi: 10.1378/chest.108.2_supplement.58s. [DOI] [PubMed] [Google Scholar]
- 3.Bloom BR, Murray CJ. Tuberculosis: commentary on a reemergent killer. Science. 1992;257:1055–1064. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- 4.Coker RJ. Review: multidrug-resistant tuberculosis: public health challenges. Trop Med Int Health. 2004;9:25–40. doi: 10.1046/j.1365-3156.2003.01156.x. [DOI] [PubMed] [Google Scholar]
- 5.de Jong BC, Israelski DM, Corbett EL, Small PM. Clinical management of tuberculosis in the context of HIV infection. Annu Rev Med. 2004;55:283–301. doi: 10.1146/annurev.med.55.091902.103753. [DOI] [PubMed] [Google Scholar]
- 6.Corbett EL, Watt CJ, Walker N, Maher D, Williams BG, Raviglione MC, Dye C. The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch Intern Med. 2003;163:1009–1021. doi: 10.1001/archinte.163.9.1009. [DOI] [PubMed] [Google Scholar]
- 7.Wayne LG, Sohaskey CD. Nonreplicating persistence of Mycobacterium tuberculosis. Annu Rev Microbiol. 2001;55:139–163. doi: 10.1146/annurev.micro.55.1.139. [DOI] [PubMed] [Google Scholar]
- 8.Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64:2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parrish NM, Dick JD, Bishai WR. Mechanisms of latency in Mycobacterium tuberculosis. Trends Microbiol. 1998;6:107–112. doi: 10.1016/s0966-842x(98)01216-5. [DOI] [PubMed] [Google Scholar]
- 10.Bloom BR, Small PM. The evolving relation between humans and Mycobacterium tuberculosis. N Engl J Med. 1998;338:677–678. doi: 10.1056/NEJM199803053381008. [DOI] [PubMed] [Google Scholar]
- 11.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 12.Dasgupta N, Kapur V, Singh KK, Das TK, Sachdeva S, Jyothisri K, Tyagi JS. Characterization of a two-component system, devR-devS, of Mycobacterium tuberculosis. Tuber Lung Dis. 2000;80:141–159. doi: 10.1054/tuld.2000.0240. [DOI] [PubMed] [Google Scholar]
- 13.Sherman DR, Voskuil M, Schnappinger D, Liao R, Harrell MI, Schoolnik GK. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha -crystallin. Proc Natl Acad Sci U S A. 2001;98:7534–7539. doi: 10.1073/pnas.121172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cunningham AF, Spreadbury CL. Mycobacterial stationary phase induced by low oxygen tension: cell wall thickening and localization of the 16-kilodalton alpha-crystallin homolog. J Bacteriol. 1998;180:801–808. doi: 10.1128/jb.180.4.801-808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garbe TR, Hibler NS, Deretic V. Response to reactive nitrogen intermediates in Mycobacterium tuberculosis: induction of the 16-kilodalton alpha-crystallin homolog by exposure to nitric oxide donors. Infect Immun. 1999;67:460–465. doi: 10.1128/iai.67.1.460-465.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voskuil MI, Schnappinger D, Visconti KC, Harrell MI, Dolganov GM, Sherman DR, Schoolnik GK. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med. 2003;198:705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicholson S, Bonecini-Almeida Mda G, Lapa e Silva JR, Nathan C, Xie QW, Mumford R, Weidner JR, Calaycay J, Geng J, Boechat N, Linhares C, Rom W, Ho JL. Inducible nitric oxide synthase in pulmonary alveolar macrophages from patients with tuberculosis. J Exp Med. 1996;183:2293–2302. doi: 10.1084/jem.183.5.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan J, Tanaka K, Carroll D, Flynn J, Bloom BR. Effects of nitric oxide synthase inhibitors on murine infection with Mycobacterium tuberculosis. Infect Immun. 1995;63:736–740. doi: 10.1128/iai.63.2.736-740.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacMicking JD, North RJ, LaCourse R, Mudgett JS, Shah SK, Nathan CF. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc Natl Acad Sci U SA. 1997;94:5243–5248. doi: 10.1073/pnas.94.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saini DK, Tyagi JS. High-throughput microplate phosphorylation assays based on DevR-DevS/Rv2027c 2-component signal transduction pathway to screen for novel antitubercular compounds. J Biomol Screen. 2005;10:215–224. doi: 10.1177/1087057104272090. [DOI] [PubMed] [Google Scholar]
- 21.Saini DK, Pant N, Das TK, Tyagi JS. Cloning, overexpression, purification, and matrix-assisted refolding of DevS (Rv 3132c) histidine protein kinase of Mycobacterium tuberculosis. Protein Expr Purif. 2002;25:203–208. doi: 10.1006/prep.2002.1628. [DOI] [PubMed] [Google Scholar]
- 22.Sardiwal S, Kendall SL, Movahedzadeh F, Rison SC, Stoker NG, Djordjevic S. A GAF domain in the hypoxia/NO-inducible Mycobacterium tuberculosis DosS protein binds haem. J Mol Biol. 2005;353:929–936. doi: 10.1016/j.jmb.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 23.Ghiladi RA, Knudsen GM, Medzihradszky KF, Ortiz de Montellano PR. The Met-Tyr-Trp cross-link in Mycobacterium tuberculosis catalase-peroxidase (KatG): autocatalytic formation and effect on enzyme catalysis and spectroscopic properties. J Biol Chem. 2005;280:22651–22663. doi: 10.1074/jbc.M502486200. [DOI] [PubMed] [Google Scholar]
- 24.Frey AD, Farres J, Bollinger CJ, Kallio PT. Bacterial hemoglobins and flavohemoglobins for alleviation of nitrosative stress in Escherichia coli. Appl Environ Microbiol. 2002;68:4835–4840. doi: 10.1128/AEM.68.10.4835-4840.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilks A, Schmitt MP. Expression and characterization of a heme oxygenase (Hmu O) from Corynebacterium diphtheriae. Iron acquisition requires oxidative cleavage of the heme macrocycle. J Biol Chem. 1998;273:837–841. doi: 10.1074/jbc.273.2.837. [DOI] [PubMed] [Google Scholar]
- 26.Monzani E, Bonafe B, Fallarini A, Redaelli C, Casella L, Minchiotti L, Galliano M. Enzymatic properties of human hemalbumin. Biochim Biophys Acta. 2001;1547:302–312. doi: 10.1016/s0167-4838(01)00192-3. [DOI] [PubMed] [Google Scholar]
- 27.Spiro TG, Li XY. Resonance Raman spectroscopy of metalloporphyrins. In: Spiro TG, editor. Biological Applications of Raman Spectroscopy. Vol. 3. Resonance Raman spectra of hemes and metalloproteins. Vol. 3. John Wiley and Sons; New York: 1988. pp. 1–37. [Google Scholar]
- 28.Andersson LA, Mylrajan M, Sullivan EP, Jr, Strauss SH. Modeling low-pH hemoproteins. J Biol Chem. 1989;264:19099–19102. [PubMed] [Google Scholar]
- 29.Sun J, Loehr TM, Wilks A, Ortiz de Montellano PR. Identification of histidine 25 as the heme ligand in human liver heme oxygenase. Biochemistry. 1994;33:13734–13740. doi: 10.1021/bi00250a026. [DOI] [PubMed] [Google Scholar]
- 30.Wilks A, Moenne-Loccoz P. Identification of the proximal ligand His-20 in heme oxygenase (Hmu O) from Corynebacterium diphtheriae. Oxidative cleavage of the heme macrocycle does not require the proximal histidine. J Biol Chem. 2000;275:11686–11692. doi: 10.1074/jbc.275.16.11686. [DOI] [PubMed] [Google Scholar]
- 31.Kitagawa T. Heme protein structure and the iron-histidine stretching mode. In: Spiro TG, editor. Biological Applications of Raman Spectroscopy. Vol. 3. Resonance Raman spectra of hemes and metalloproteins. Vol. 3. John Wiley and Sons; New York: 1988. pp. 97–131. [Google Scholar]
- 32.Sun J, Wilks A, Ortiz de Montellano PR, Loehr TM. Resonance Raman and EPR spectroscopic studies on heme-heme oxygenase complexes. Biochemistry. 1993;32:14151–14157. doi: 10.1021/bi00214a012. [DOI] [PubMed] [Google Scholar]
- 33.Takahashi S, Wang J, Rousseau DL, Ishikawa K, Yoshida T, Takeuchi N, Ikeda-Saito M. Heme-heme oxygenase complex: structure and properties of the catalytic site from resonance Raman scattering. Biochemistry. 1994;33:5531–5538. doi: 10.1021/bi00184a023. [DOI] [PubMed] [Google Scholar]
- 34.Bangcharoenpaurpong O, Schomacker KT, Champion PM. A resonance Raman investigation of myoglobin and hemoglobin. J Am Chem Soc. 1984;106:5688–5698. [Google Scholar]
- 35.Stavrov SS. The effect of iron displacement out of the porphyrin plane on the resonance Raman spectra of heme proteins and iron porphyrins. Biophys J. 1993;65:1942–1950. doi: 10.1016/S0006-3495(93)81265-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ray GB, Li XY, Ibers JA, Sessler JL, Spiro TG. How far can proteins bend the FeCO unit? Distal polar and steric effects in heme proteins and models. J Am Chem Soc. 1994;116:162–176. [Google Scholar]
- 37.Li T, Quillin ML, Phillips GN, Jr, Olson JS. Structural determinants of the stretching frequency of CO bound to myoglobin. Biochemistry. 1994;33:1433–1446. doi: 10.1021/bi00172a021. [DOI] [PubMed] [Google Scholar]
- 38.Ling J, Li T, Olson JS, Bocian DF. Identification of the iron-carbonyl stretch in distal histidine mutants of carbonmonoxymyoglobin. Biochim Biophys Acta. 1994;1188:417–421. doi: 10.1016/0005-2728(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 39.Anderton CL, Hester RE, Moore JN. A chemometric analysis of the resonance Raman spectra of mutant carbonmonoxy-myoglobins reveals the effects of polarity. Biochim Biophys Acta. 1997;1338:107–120. doi: 10.1016/s0167-4838(96)00194-x. [DOI] [PubMed] [Google Scholar]
- 40.Hoffmann K, Stoffel W. TMBase - A database of membrane spanning protein segments. Biol Chem Hoppe-Seyler. 1993;347:166. [Google Scholar]
- 41.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 42.Elleby B, Svensson S, Wu X, Stefansson K, Nilsson J, Hallen D, Oppermann U, Abrahmsen L. High-level production and optimization of monodispersity of 11beta-hydroxysteroid dehydrogenase type 1. Biochim Biophys Acta. 2004;1700:199–207. doi: 10.1016/j.bbapap.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 43.Lee KH, Kim HS, Jeong HS, Lee YS. Chaperonin GroESL mediates the protein folding of human liver mitochondrial aldehyde dehydrogenase in Escherichia coli. Biochem Biophys Res Commun. 2002;298:216–224. doi: 10.1016/s0006-291x(02)02423-3. [DOI] [PubMed] [Google Scholar]
- 44.Lamark T, Ingebrigtsen M, Bjornstad C, Melkko T, Mollnes TE, Nielsen EW. Expression of active human C1 inhibitor serpin domain in Escherichia coli. Protein Expr Purif. 2001;22:349–358. doi: 10.1006/prep.2001.1445. [DOI] [PubMed] [Google Scholar]
- 45.Yanase H, Moriya K, Mukai N, Kawata Y, Okamoto K, Kato N. Effects of GroESL coexpression on the folding of nicotinoprotein formaldehyde dismutase from Pseudomonas putida F61. Biosci Biotechnol Biochem. 2002;66:85–91. doi: 10.1271/bbb.66.85. [DOI] [PubMed] [Google Scholar]
- 46.Mitsuda M, Iwasaki M. Improvement in the expression of CYP2B6 by co-expression with molecular chaperones GroES/EL in Escherichia coli. Protein Expr Purif. 2006;46:401–405. doi: 10.1016/j.pep.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 47.Kim SG, Kweon DH, Lee DH, Park YC, Seo JH. Coexpression of folding accessory proteins for production of active cyclodextrin glycosyltransferase of Bacillus macerans in recombinant Escherichia coli. Protein Expr Purif. 2005;41:426–432. doi: 10.1016/j.pep.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 48.Yasukawa T, Kanei-Ishii C, Maekawa T, Fujimoto J, Yamamoto T, Ishii S. Increase of solubility of foreign proteins in Escherichia coli by coproduction of the bacterial thioredoxin. J Biol Chem. 1995;270:25328–25331. doi: 10.1074/jbc.270.43.25328. [DOI] [PubMed] [Google Scholar]
- 49.Herrmann S, Ma Q, Johnson MS, Repik AV, Taylor BL. PAS domain of the Aer redox sensor requires C-terminal residues for native-fold formation and flavin adenine dinucleotide binding. J Bacteriol. 2004;186:6782–6791. doi: 10.1128/JB.186.20.6782-6791.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kojouharova MS, Panchev ID, Tchorbadjieva MI, Reid KB, Hoppe HJ. Differential binding of IgG and of a HIV gp41 peptide by the B chain and A chain globular head sequences of C1q, respectively. J Immunol. 1998;161:4325–4331. [PubMed] [Google Scholar]
- 51.Marana SR, Terra WR, Ferreira C. The role of amino-acid residues Q39 and E451 in the determination of substrate specificity of the Spodoptera frugiperda beta-glycosidase. Eur J Biochem. 2002;269:3705–3714. doi: 10.1046/j.1432-1033.2002.03061.x. [DOI] [PubMed] [Google Scholar]
- 52.Marana SR, Mendonca LM, Andrade EH, Terra WR, Ferreira C. The role of residues R97 and Y331 in modulating the pH optimum of an insect beta-glycosidase of family 1. Eur J Biochem. 2003;270:4866–4875. doi: 10.1046/j.1432-1033.2003.03887.x. [DOI] [PubMed] [Google Scholar]
- 53.Knop M, Miller KJ, Mazza M, Feng D, Weber M, Keranen S, Jantti J. Molecular interactions position Mso1p, a novel PTB domain homologue, in the interface of the exocyst complex and the exocytic SNARE machinery in yeast. Mol Biol Cell. 2005;16:4543–4556. doi: 10.1091/mbc.E05-03-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mizobata T, Akiyama Y, Ito K, Yumoto N, Kawata Y. Effects of the chaperonin GroE on the refolding of tryptophanase from Escherichia coli. Refolding is enhanced in the presence of ADP. J Biol Chem. 1992;267:17773–17779. [PubMed] [Google Scholar]
- 55.Martinez SE, Wu AY, Glavas NA, Tang XB, Turley S, Hol WG, Beavo JA. The two GAF domains in phosphodiesterase 2A have distinct roles in dimerization and in cGMP binding. Proc Natl Acad Sci USA. 2002;99:13260–13265. doi: 10.1073/pnas.192374899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahmad Z, Sharma S, Khuller GK. In vitro and ex vivo antimycobacterial potential of azole drugs against Mycobacterium tuberculosis H37Rv. FEMS Microbiol Lett. 2005;251:19–22. doi: 10.1016/j.femsle.2005.07.022. [DOI] [PubMed] [Google Scholar]
- 57.Ahmad Z, Sharma S, Khuller GK. The potential of azole antifungals against latent/persistent tuberculosis. FEMS Microbiol Lett. 2006;258:200–203. doi: 10.1111/j.1574-6968.2006.00224.x. [DOI] [PubMed] [Google Scholar]
- 58.Ahmad Z, Sharma S, Khuller GK. Azole antifungals as novel chemotherapeutic agents against murine tuberculosis. FEMS Microbiol Lett. 2006;261:181–186. doi: 10.1111/j.1574-6968.2006.00350.x. [DOI] [PubMed] [Google Scholar]
- 59.McLean KJ, Cheesman MR, Rivers SL, Richmond A, Leys D, Chapman SK, Reid GA, Price NC, Kelly SM, Clarkson J, Smith WE, Munro AW. Expression, purification and spectroscopic characterization of the cytochrome P450 CYP121 from Mycobacterium tuberculosis. J Inorg Biochem. 2002;91:527–541. doi: 10.1016/s0162-0134(02)00479-8. [DOI] [PubMed] [Google Scholar]
- 60.McLean KJ, Marshall KR, Richmond A, Hunter IS, Fowler K, Kieser T, Gurcha SS, Besra GS, Munro AW. Azole antifungals are potent inhibitors of cytochrome P450 mono-oxygenases and bacterial growth in mycobacteria and streptomycetes. Microbiology. 2002;148:2937–2949. doi: 10.1099/00221287-148-10-2937. [DOI] [PubMed] [Google Scholar]
- 61.Guardiola-Diaz HM, Foster LA, Mushrush D, Vaz AD. Azole-antifungal binding to a novel cytochrome P450 from Mycobacterium tuberculosis: implications for treatment of tuberculosis. Biochem Pharmacol. 2001;61:1463–1470. doi: 10.1016/s0006-2952(01)00571-8. [DOI] [PubMed] [Google Scholar]
- 62.Gong W, Hao B, Chan MK. New mechanistic insights from structural studies of the oxygen-sensing domain of Bradyrhizobium japonicum FixL. Biochemistry. 2000;39:3955–3962. doi: 10.1021/bi992346w. [DOI] [PubMed] [Google Scholar]
- 63.Gong W, Hao B, Mansy SS, Gonzalez G, Gilles-Gonzalez MA, Chan MK. Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction. Proc Natl Acad Sci U S A. 1998;95:15177–15182. doi: 10.1073/pnas.95.26.15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stone JR, Marletta MA. Soluble guanylate cyclase from bovine lung: activation with nitric oxide and carbon monoxide and spectral characterization of the ferrous and ferric states. Biochemistry. 1994;33:5636–5640. doi: 10.1021/bi00184a036. [DOI] [PubMed] [Google Scholar]
- 65.Deinum G, Stone JR, Babcock GT, Marletta MA. Binding of nitric oxide and carbon monoxide to soluble guanylate cyclase as observed with resonance Raman spectroscopy. Biochemistry. 1996;35:1540–1547. doi: 10.1021/bi952440m. [DOI] [PubMed] [Google Scholar]
- 66.Stone JR, Marletta MA. Spectral and kinetic studies on the activation of soluble guanylate cyclase by nitric oxide. Biochemistry. 1996;35:1093–1099. doi: 10.1021/bi9519718. [DOI] [PubMed] [Google Scholar]
- 67.Lukat-Rodgers GS, Rodgers KR. Characterization of ferrous FixL-nitric oxide adducts by resonance Raman spectroscopy. Biochemistry. 1997;36:4178–87. doi: 10.1021/bi9628230. [DOI] [PubMed] [Google Scholar]
- 68.Fan B, Wang J, Stuehr DJ, Rousseau DL. NO synthase isozymes have distinct substrate binding sites. Biochemistry. 1997;36:12660–12665. doi: 10.1021/bi9715369. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary SDS PAGE data on different expression systems, UV-visible spectra of Fe-NO complexes, and low-frequency and high-frequency RR spectra of ferrous DevS642 and full-length DevS are provided. This material is available free of charge via the Internet at http://pubs.acs.org.