Abstract
The human estrogen receptor α (ER α) has been tagged at its amino terminus with the S65T variant of the green fluorescent protein (GFP), allowing subcellular trafficking and localization to be observed in living cells by fluorescence microscopy. The tagged receptor, GFP-ER, is functional as a ligand-dependent transcription factor, responds to both agonist and antagonist ligands, and can associate with the nuclear matrix. Its cellular localization was analyzed in four human breast cancer epithelial cell lines, two ER+ (MCF7 and T47D) and two ER− (MDA-MB-231 and MDA-MB-435A), under a variety of ligand conditions. In all cell lines, GFP-ER is observed only in the nucleus in the absence of ligand. Upon the addition of agonist or antagonist ligand, a dramatic redistribution of GFP-ER from a reticular to punctate pattern occurs within the nucleus. In addition, the full antagonist ICI 182780 alters the nucleocytoplasmic compartmentalization of the receptor and causes partial accumulation in the cytoplasm in a process requiring continued protein synthesis. GFP-ER localization varies between cells, despite being cultured and treated in a similar manner. Analysis of the nuclear fluorescence intensity for variation in its frequency distribution helped establish localization patterns characteristic of cell line and ligand. During the course of this study, localization of GFP-ER to the nucleolar region is observed for ER− but not ER+ human breast cancer epithelial cell lines. Finally, our work provides a visual description of the “unoccupied” and ligand-bound receptor and is discussed in the context of the role of ligand in modulating receptor activity.
INTRODUCTION
Steroid hormones elicit diverse biological responses, important during growth, differentiation, inflammation, pregnancy, and homeostasis among many other processes. The genomic actions of steroid hormones are mediated by steroid receptors, members of the nuclear receptor superfamily of ligand-dependent transcription factors. In the absence of hormone, steroid receptors exist in a complex with chaperone proteins capable of high-affinity binding to steroid hormones. Hormone binding leads to a conformational change in the receptor that results in its dissociation from chaperone proteins and ultimately in the binding of the receptor as a homodimer to cognate sites in steroid-responsive genes (reviewed in Tsai and O’Malley, 1994; Mangelsdorf et al., 1995; Beato et al., 1996).
Immunohistochemistry and biochemical fractionation show the unoccupied steroid receptors to reside predominantly in the cytoplasm, the nucleus, or both compartments, depending on the receptor, in a complex with chaperone proteins (Jensen, 1991; DeFranco et al., 1995; Beato et al., 1996; Pratt and Toft, 1997). For the predominantly nuclear receptors, such as the estrogen receptor (ER), the unoccupied receptor exists in the nucleus either bound or not bound to its cognate site in target genes. Hormone binding leads to activation of the receptor and transcriptional regulation of the responsive genes (Press et al., 1989; Picard et al., 1990; Parker, 1992; Tsai and O’Malley, 1994).
We and others have previously shown that the subcellular localization and trafficking of the glucocorticoid receptor (GR) can be followed with a green fluorescent protein (GFP) fusion (Ogawa et al., 1995; Carey et al., 1996; Htun et al., 1996; Rizzuto et al., 1996). The chimeric receptor GFP-GR can be fully functional as a ligand-dependent transcription factor and shows the ligand specificity of GR. The ligand-dependent translocation of GFP-GR from the cytoplasm to nucleus can be observed in real time in a single cell. Most interestingly, we observed an intranuclear pattern and distribution of GFP-GR that reflects the type of ligand, either agonist or antagonist, used to activate the receptor (Htun et al., 1996).
In this report, we have chosen a similar strategy with the human ER α, referred to throughout the paper as ER, to see whether ligand affects the nuclear distribution of this receptor. Previous biochemical studies described the existence of two biochemically distinct forms of ER (Gorski et al., 1968; Jensen et al., 1968). In the absence of ligand, the “unoccupied” ER with a sedimentation coefficent of 9S is “loosely” associated with the nucleus; ligand causes a biochemical transformation to a complex with a sedimentation of 5S that associates more “tightly” with the nucleus (Greene and Press, 1986; Press et al., 1989; Jensen, 1991). To determine whether the biochemical difference is reflected by a change in the intranuclear distribution of ER, we have directly visualized ER in living cells by tagging the receptor with the S65T variant of the naturally fluorescent protein GFP.
MATERIALS AND METHODS
Cell Lines and Plasmids
Human breast cancer epithelial cell lines MCF-7, T47D, and MDA-MB-231 were obtained from American Type Culture Collection (Rockville, MD). MDA-MB-435A is a derivative of MDA-MB-435 (Yee et al., 1996). Unless otherwise noted, cells were maintained in a 225-cm2 cell culture flask (Costar, Cambridge, MA) in 35 ml of Dulbecco’s modified Eagle’s medium (DMEM) with phenol red as pH indicator (Life Technologies, Grand Island, NY; catalog number 11960-044), supplemented with 10% FBS (Life Technologies; catalog number 10437-028), 2 mM l-glutamine, 4.8 μg/ml insulin, and 100 U of penicillin G/100 μg of streptomycin sulfate/ml at 37°C in a 5% CO2-water jacketed incubator. Medium was changed every 2 d. At confluence, cells were harvested by first washing with Dulbecco’s PBS (D-PBS) without calcium or magnesium and then treating with 0.05% trypsin-0.02% EDTA without phenol red. Cells were reseeded in a fresh flask at about a one-to-four dilution. Four days before transfection, cells were placed in DMEM lacking phenol red (Life Technologies; catalog number 31053-028) and supplemented as described above, except dextran/charcoal–treated FBS (Hyclone, Logan, UT; catalog number SH30068.03) was used in place of FBS. The reporter gene, pERE-tk-CAT, contains two copies of a perfect estrogen response element (ERE) in tandem, upstream of the thymidine kinase (tk) minimal promoter hooked up to the chloramphenicol acetyltransferase (CAT) reporter gene (Seiler-Tuyns et al., 1986). The human ER expression vector pSG5-HEGO (Tora et al., 1989), contains a wild-type human ER under the control of the SV40 early promoter. The GFP-ER expression plasmid pCI-nGL1-HEGO (GenBank database; accession number AF061181), was prepared by first replacing the S65T GFP coding region in the plasmid pCI-nGFP-C656G (Htun et al., 1996) with an S65T GFP coding region optimized for expression in mammalian cells from pGreen Lantern-1 (Life Technologies) and then replacing the rat GR with the human ER coding region in pSG5-HEGO, previously mutated with the Chameleon site-directed mutagenesis kit (Stratagene, La Jolla, CA) to introduce an MluI site in the first three amino acids of the ER coding region with the oligonucleotide 5′-TGGTGTGGAGGGTCAACGCGTTGGTCCGTGGCCGCG-3′. To enrich for cells transfected with the expression plasmids, the plasmid pCMV-IL2R, which expresses the human interleukin 2 receptor (IL2R), was used in the transfection experiments, as previously described (Htun et al., 1996).
Transfections
Plasmid DNA was transiently introduced into cells either by calcium phosphate coprecipitation, electroporation, or liposome-mediated gene transfer. For ERE reporter assays, MDA-MB-435 cells were plated in 100-mm dishes in DMEM lacking phenol red with 5% twice charcoal-stripped FBS 2 d before transfection and were given fresh media 24 h before transfection. Cells were maintained at 37°C in a 5% CO2 incubator before transfection. Cells were transfected by the CaPO4/BES precipitation method (Kingston et al., 1995). One ml of precipitate contained 4 μg of pCH110 (β-galactosidase reporter plasmid) as an internal control, 5 μg of ERE-tk-CAT reporter plasmid, pCI-nGL1-HEGO in the amount indicated, and pCEP4 as carrier DNA to a total of 15 μg of DNA. Cells were in contact with precipitate for 14 h and then washed twice with D-PBS and treated with fresh phenol red–free media plus 5% twice charcoal-stripped FBS containing 17β-estradiol, ICI 182780, or 4-hydroxytamoxifen as indicated. Cells were harvested 30 h after the removal of the calcium phosphate precipitates. All transfections were done in triplicate. CAT assays (Kingston et al., 1995) and β-galactosidase assays (Sambrook et al., 1989) were done using the method essentially as described. CAT activity was normalized to β-galactosidase activity, and the mean and SEM for three sets of data were plotted. For electroporation, cells were electroporated with the indicated amount of pCI-nGL1-HEGO DNA with or without 5 μg of pCMV-IL2R DNA for 2 × 107 cells in 0.2 ml of cold phenol red–free DMEM at 250 V and 1100 μF in a 0.4-cm electrode gap electroporation chamber supplied with the Cell-Porator electroporation system (Life Technologies; catalog number 71600-019), left to recover on ice for 5 min, and then diluted in phenol red–free DMEM supplemented with 10% dextran/charcoal–treated FBS before plating. Cells were then grown for 12–16 h in 37°C, 5% CO2 incubators before imaging. Liposome-mediated gene transfer was used as directed by the manufacturer of DOSPER liposomal transfection reagent (Boehringer Mannheim, Indianapolis, IN; catalog number 1 811 169) for nuclear matrix isolation experiments.
Enrichment of Transfected Cells and Analysis of Total Cell Lysates with Human ER Monoclonal Antibody H226
Approximately 18 h after electroporation with 5 μg of pCMV-IL2R and various amounts of pCI-nGL1-HEGO, cells were washed twice with PBS and then sorted with magnetic beads coated with antibody to the IL2R, as previously described (Htun et al., 1996). IL2R-positive cells were washed several times and then divided into two equal aliquots. One aliquot was lysed with an NP-40 lysis solution (0.5% NP-40, 50 mM Tris-HCl, 1 mM EDTA, 200 mM NaCl, pH 7.5, and a mixture of protease inhibitors) after incubating on ice for 5 min and then centrifuged for 5 min in a microfuge to remove the insoluble debris. The protein concentration in the soluble extract was determined by the Bradford method (Bradford, 1976) using a Bio-Rad (Hercules, CA) protein assay reagent. The other half of the cells was used to prepare total cellular lysate with 1× SDS gel-loading buffer (50 mM Tris-Cl, pH 6.8, 100 mM dithiothreitol, 2% SDS, 0.1% bromophenol blue, and 10% glycerol; Sambrook et al., 1989) in the same volume as the NP-40 lysis solution. Based on the protein concentration, a volume equivalent to 30–40 μg was removed from the total cellular lysate, heated to 90°C for 5 min, subjected to denaturing polyacrylamide gel electroporesis in 8% polyacrylamide (30 acrylamide:1 bisacrylamide) containing 0.1% SDS, and analyzed by the Western blotting method, essentially as described previously (Sambrook et al., 1989), using a rat monoclonal antibody, H226, raised against the human ER α (a gift from Geoffrey Greene, The Ben May Institute for Cancer Research, University of Chicago, Chicago, IL) as a primary antibody and donkey anti-rat antibody conjugated to horseradish peroxidase as a secondary antibody with the Pierce (Rockford, IL) Super Signal chemiluminescent substrate.
Extraction of Nuclear Matrix
A modified procedure was used for the extraction of nuclear matrix (Jackson and Cook, 1988; Belgrader et al., 1991; Berezney, 1991). Nuclei from frozen MCF-7 cell pellets, which had been transiently transfected with 15 μg of pCI-nGL1-HEGO HEGO in 90 μl of DOSPER liposomal transfection reagent for 1 × 106 cells and grown with DMEM in CSFBS without phenol red, were resuspended in TNM buffer (10 mM Tris, pH 8.0, 300 mM sucrose, 100 mM NaCl, 2 mM MgCl2, 1%, vol/vol, thiodiglycol, and 1 mM PMSF) and homogenized three times in a Dounce homogenizer with a Teflon pestle. Triton X-100 was added to a final concentration of 0.5%, vol/vol, and nuclei were collected by centrifugation at 500 × g for 10 min at 4°C. Nuclei were then resuspended in TNM buffer, homogenized, and pelleted as described above. The isolated nuclei were then resuspended to a concentration of 20 A260/ml in DIG buffer (10 mM Tris, pH 8.0, 300 mM sucrose, 50 mM NaCl, 3 mM MgCl2, 1%, vol/vol, thiodiglycol, and 1 mM PMSF) and digested with 168 U/ml DNase I (D5025; Sigma, St. Louis, MO) for 20 min at room temperature. Ammonium sulfate was added to a final concentration of 0.25 M, and nuclear matrix was collected by centrifugation. The pellet NM-IF1 was then resuspended in DIG buffer, followed by the addition of an equal volume of 4 M NaCl. The nuclear matrix was then collected by centrifugation and re-extracted with 2 M NaCl as above. The final pellet NM-IF2 was resuspended in 1× SDS gel-loading buffer, as previously described. Supernatants from the 0.25 M ammonium sulfate and 2 M NaCl extractions were dialyzed overnight against double-distilled H2O and 1 mM PMSF and then lyophilized. All four samples were subjected to denaturing polyacrylamide gel and Western blot analyses, essentially as described above, using an antibody (BabCo, Berkeley, CA) against the hemagglutinin (HA) tag (YPYDVPDYA) at the amino terminus of GFP-ER.
Microscopy
For differential interference contrast, the cells electroporated with only pCI-nGL1-HEGO were grown on glass coverslips overnight. The coverslips were rinsed with D-PBS and placed inverted on a microscope slide. Cells were illuminated by white light from a tungsten light source and viewed under a 63×, 1.4 numerical aperture Plan-Apochromat oil immersion objective in a Zeiss Axiophot microscope (Carl Zeiss, Thornwood, NY) under Nomarski optics. For green fluorescence, the same cells were examined through a 480- to 490-nm excitation, a 510-nm dichroic mirror, and a 515- to 565-nm emission filter using a 100-W mercury bulb light source. Images were recorded on Kodak (Rochester, NY) Elite 200 color slide film. In the case of confocal laser scanning microscopy, cells were imaged through a 100×, 1.4 numerical aperture Plan-Apochromat oil immersion objective by excitation with the 488-nm line from a krypton–argon laser, and the emission was viewed through a 506- to 538-nm band pass filter. Images were collected on a Zeiss Axiovert 135 platform attached to a Bio-Rad MRC 1024 confocal imaging system using Bio-Rad LaserSharp software.
Image Analysis
Image analyses and representation in Figure 2 were performed on an Apple (Cupertino, CA) Power Macintosh 8600/200 computer using the public domain NIH Image program version 1.61 (developed at the National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image). To generate Table 1, additional image analyses were performed by determining mean pixel value and SE for each area containing a nucleus but excluding the nucleoli using IPLab Spectrum software (Signal Analytics, Fairfax, VA) operating on a Power Macintosh 8600/200 computer. To obtain the coefficient of variation for fluorescence intensity, the SD of the pixel values for each nucleus was divided by the mean pixel value. The mean of the coefficient of variation for the population and the SD were determined for each set of cells exposed to the same ligand and are provided in Table 1. Statistical significance was determined using the Z test (Chase and Bown, 1992).
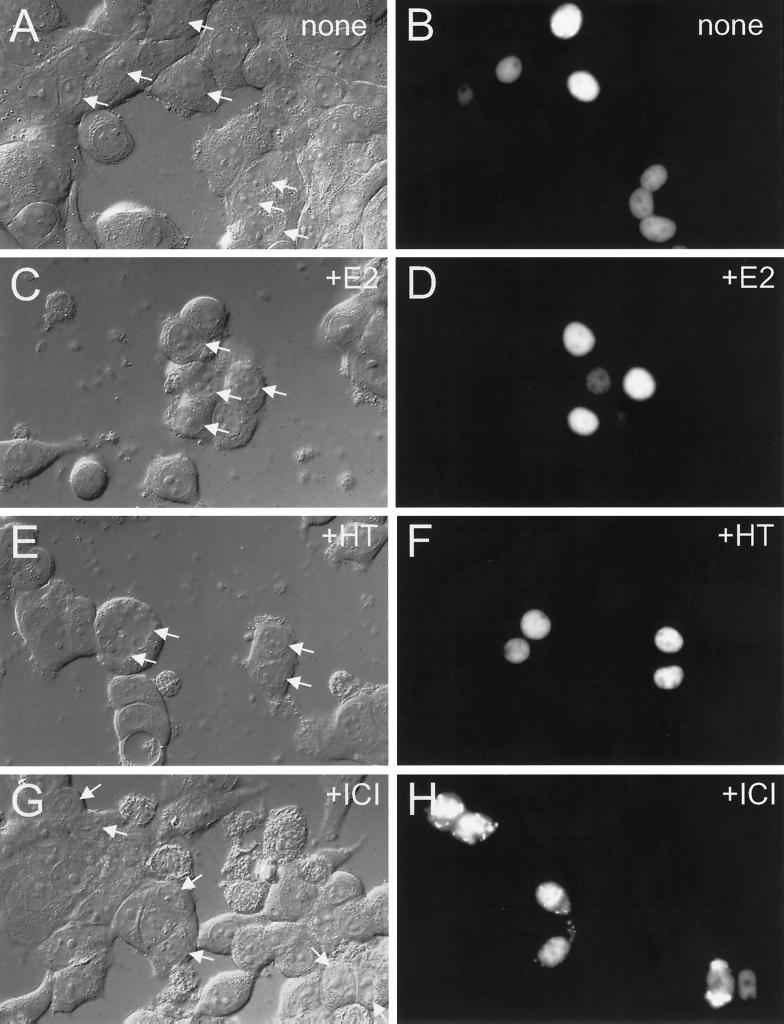
Figure 2.
Effect of ligand treatment on GFP-ER localization in MCF-7 cells. Cells from a human breast cancer epithelial cell line, MCF-7, were electroporated with 0.2 μg of GFP-ER expression plasmid, pCI-nGL1-HEGO, and cultured on coverslips for 12–16 h before visualization by differential interference contrast (A, C, E, and G) or epifluorescence with a standard set of FITC filters (B, D, F, and H). Cells were treated at the time of culturing with nothing (A and B), 10 nM 17β-estradiol (C and D), 10 nM 4-hydroxytamoxifen (E and F), or 10 nM ICI 182780 (G and H). The arrow in the left panel indicates the nucleus of a cell exhibiting green fluorescence, as seen in the right panel.
Table 1.
Mean coefficient of variation and its standard deviation obtained from the frequency distribution of nuclear fluorescence intensity for a population of cells treated in a similar manner
| None | 17β-Estradiol | 4-Hydroxytamoxifen | ICI 182780 | |
|---|---|---|---|---|
| MCF7 | 0.225 ± 0.042 (164) | 0.365 ± 0.068 (82) | 0.329 ± 0.059 (108) | 0.326 ± 0.057 (109) |
| T47D | 0.245 ± 0.050 (76) | 0.365 ± 0.060 (60) | 0.370 ± 0.084 (56) | 0.373 ± 0.058 (34) |
| MDA-MB-231 | 0.245 ± 0.040 (118) | 0.353 ± 0.057 (78) | 0.315 ± 0.078 (54) | 0.342 ± 0.074 (49) |
| MDA-MB-435A | 0.257 ± 0.040 (84) | 0.301 ± 0.040 (53) | 0.287 ± 0.042 (44) | 0.294 ± 0.041 (35) |
Coefficient of variation is determined from the frequency distribution of the pixel intensity within each optical section of a nucleus but ignoring the nucleolus, using IPLab Spectrum software (Signal Analytics). From the coefficient of variation, the mean and its SD were determined for the number of nuclei indicated within parenthesis and are separated by the ± sign, respectively.
RESULTS
Expression and Functional Analysis of the GFP-ER Fusion Protein
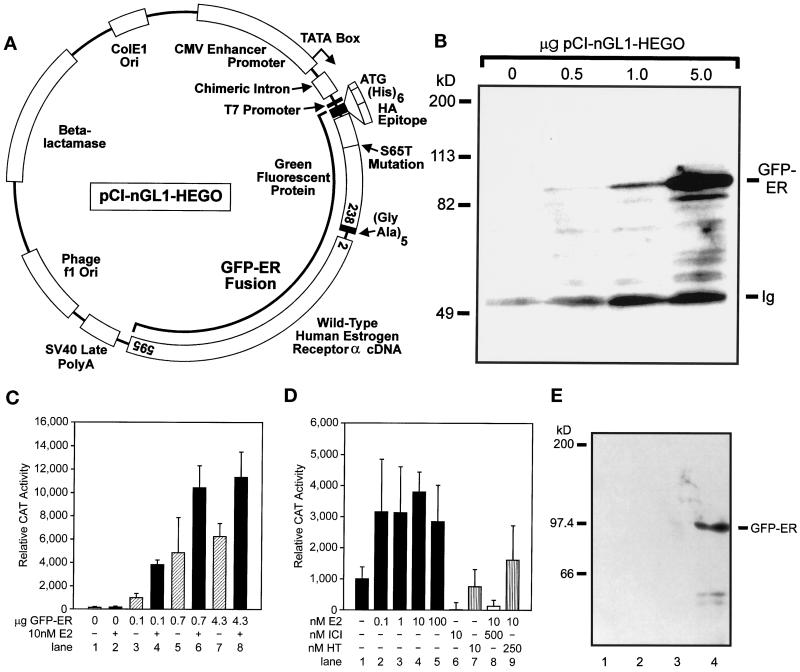
To follow the subcellular localization and trafficking of the ER in living cells, we tagged the amino terminus of the human ER α (ER) with the S65T variant of GFP. Figure 1A shows the cytomegalovirus (CMV) enhancer/promoter–driven GFP-ER expression vector. Functionality of GFP-ER as a ligand-dependent transcription factor was assayed on an estrogen response element–containing reporter gene in transient transfection experiments in a human breast cancer epithelial cell line, MDA-MB-435A, which lacks endogenous ER (Yee et al., 1996). GFP-ER activates the reporter gene in a dose-dependent manner and shows additional activation in the presence of agonist ligand 17β-estradiol (Figure 1C). Significant ligand-independent activation has previously been reported for the ER (Danielian et al., 1992). Similar results were obtained with the unsubstituted ER (our unpublished results). Maximal activation of GFP-ER by 17β-estradiol was observed at a 10 nM concentration of ligand (Figure 1D; lanes 1–5), consistent with the previously reported subnanomolar Kd for ER (Kuiper et al., 1997). ICI 182780, a pure antagonist for ER (Wakeling et al., 1991), completely inhibited GFP-ER activation of the reporter gene (Figure 1D, compare lanes 1 and 6). When ER antagonists were present in 25- to 50-fold molar excess, the action of 17β-estradiol was inhibited, albeit the pure antagonist ICI 182780 compound was more effective than the partial antagonist 4-hydroxytamoxifen (Figure 1D, compare lanes 4 and 8 and lanes 4 and 9). Thus, GFP-ER functions as a transcriptional activator, and its activity is fully regulated by ER ligands.
Figure 1.
Construction and characterization of GFP-ER. (A) Map of GFP-ER expression plasmid pCI-nGL1-HEGO. The human wild-type ER is fused to the carboxyl terminus of an S65T variant of GFP whose codons have been optimized for translation in mammalian cells. A small linker region containing five glycine-alanine repeats separates the two coding regions. The S65T GFP is tagged with a (his)6 and HA epitope at the amino terminus. Amino acids present in the fusion protein are indicated in the open boxes for GFP and ER. (B) Expression of GFP-ER in MCF-7 cells. MCF-7 cells were electroporated with the indicated amount of GFP-ER expression plasmid pCI-nGFL1-HEGO and 5 μg of IL2R expression plasmid pCMV-IL2R and allowed to express for 16 h. The population of transiently transfected cells was identified by the presence of the IL2R cell surface marker and isolated by magnetic beads coated with the anti-IL2R monoclonal antibody, as previously described (Htun et al., 1996). The total cellular lysate prepared from these cells was analyzed for the presence of GFP-ER by Western blot analysis using a rat monoclonal antibody, H226, against human ER as primary antibody, donkey anti-rat antibody coupled to horseradish peroxidase as second antibody, and Pierce Super Signal chemiluminescent substrate. Location of GFP-ER is indicated along with that of the Ig, which is present in the extract because of the cell-sorting procedure and is also detected by the secondary antibody. Molecular weight markers are indicated on the left. (C) GFP-ER–dependent and 17β-estradiol–dependent transcriptional activation of the ERE-containing reporter gene pERE-tk-CAT in MDA-MB-435A cells. Human breast cancer epithelial cells, MDA-MB-435A, were transfected with various amounts of the GFP-ER expression plasmid as indicated along with 5 μg of pERE-tk-CAT reporter gene and 4 μg of β-galactosidase expression plasmid pCH110 by the calcium phosphate coprecipitation method. After replacement with fresh medium the next day, cells were treated (gray bars) or not treated (stippled bars) with 10 nM 17β-estradiol for 30 h. Extracts were prepared subsequently and assayed for CAT and β-galactosidase activity. The relative CAT activity was calculated using the β-galactosidase activity to normalize for transfection efficiency. (D) Effect of 17β-estradiol concentration and type of ligand on GFP-ER activation of the pERE-tk-CAT reporter gene in MDA-MB-435A cells. Cells were transfected as described in C using 0.1 μg of pCI-nGL1-HEGO expression plasmid, 5 μg of pERE-tk-CAT reporter gene, and 4 μg of β-galactosidase expression plasmid pCH110. After transfection, cells were treated with no additional ligand (lane 1), 0.1 nM 17β-estradiol (lane 2), 1 nM 17β-estradiol (lane 3), 10 nM 17β-estradiol (lane 4), 100 nM 17β-estradiol (lane 5), 10 nM ICI 182780 (lane 6), 10 nM 4-hydroxytamoxifen (lane 7), 500 nM ICI 182780 and 10 nM 17β-estradiol (lane 8), and 250 nM 4-hydroxytamoxifen and 10 nM 17β-estradiol (lane 9) for 30 h before analysis. (E) Association of GFP-ER with the nuclear matrix in MCF-7 cells. MCF-7 cells were grown in charcoal-stripped FBS. Lane 1, supernatant after extraction of isolated nuclei with 0.25 M ammonium sulfate; lane 2, supernatant after the first extraction with 2M NaCl; lane 3, supernatant after the second extraction with 2 M NaCl; lane 4, nuclear matrix and associated intermediate filaments. Immunodetection of GFP-ER was done using the HA antibody to the HA tag located on the N terminus of the GFP-ER fusion protein.
Presence of the GFP tag results in a receptor that is ∼27 kDa larger than the untagged ER. An immunoblot using a rat monoclonal ER antibody, H226, of total cellular lysates prepared from MCF-7 cells transfected with GFP-ER expression vector pCI-nGL1-HEGO shows the presence of a protein with a molecular mass of ∼94 kDa, the expected molecular mass of the fusion protein (Figure 1B). Furthermore, from the width and intensity of the band, inclusion of a greater amount of GFP-ER expression plasmid in a transfection results in more fusion protein being expressed in the transfected cells.
Previous investigations have shown that unliganded ER associates loosely with the nucleus but that the liganded ER associates more tightly with the nucleus (Greene and Press, 1986; Press et al., 1989; Jensen, 1991). Several studies have also suggested that steroid receptors are associated with the nuclear matrix (Barrack and Coffey, 1980; Alexander et al., 1987; Samuel et al., 1998). In addition, transcribing chromatin has been reported to be selectively associated with the nuclear matrix (Davie, 1995; Davie et al., 1997). These considerations led us to examine the potential association of GFP-ER with the matrix. Nuclear matrix was prepared from MCF-7 cells transfected with the GFP-ER expression plasmid. Figure 1E shows the result of two successive 2 M NaCl extractions of a nuclear matrix preparation from MCF-7 cells whose nuclei have been digested with DNase I and extracted with 0.25 M ammonium sulfate. From the Western blot analysis using an anti-HA antibody to detect the HA epitope at the amino terminus of GFP-ER, the GFP-ER that remains associated with the initial nuclear matrix preparation is tightly bound to the matrix and resistant to 2 M NaCl extraction (Figure 1E, compare lanes 1–3 with lane 4). The initial cytosolic fraction contains soluble nuclear protein and significant amounts of GFP-ER (our unpublished results). Comparable results were obtained when cells were grown in the presence of estrogens (our unpublished results). These findings are consistent with the earlier description of ER as cytosolic, based on biochemical fractionation experiments (Gorski et al., 1968; Jensen et al., 1968; Greene and Press, 1986; Press et al., 1989; Jensen, 1991), as well as a nuclear matrix-binding protein (Barrack and Coffey, 1980; Alexander et al., 1987).
Cellular Localization of GFP-ER
To determine the localization of the tagged ER, we examined various human breast cancer epithelial cell lines transfected with the GFP-ER expression vector. In particular, we were interested in the effect of ligand as well as cellular structure and milieu on GFP-ER subcellular localization in the ER-positive (ER+) and ER-negative (ER−) cell lines. Four representative human breast cancer epithelial cell lines were examined, two ER+ (MCF-7 and T47D) and two ER− (MDA-MB-231 and MDA-MB-435A). Green fluorescence can be detected by conventional fluorescence microscopy in MCF-7 cells after electroporation with the GFP-ER expression plasmid, indicating that the GFP chromophore in the fusion protein is functional (Figure 2B). Comparison with the differential interference contrast image shows green fluorescence to be restricted to the nucleus of a few cells successfully transfected with the GFP-ER expression plasmid (Figure 2A, white arrows, also compare A and B). In the cells with a lower level of green fluorescence, nucleolar outlines are observed, which is consistent with GFP-ER being excluded from the nucleolus (Figure 2B, two leftmost nuclei and three lower nuclei). Thus, GFP-ER localization is consistent with that previously reported for the endogenous ER in MCF-7 cells (King and Greene, 1984; Welshons et al., 1988).
Inclusion of 10 nM ligand, either agonist 17β-estradiol (Figure 2, C and D) or partial antagonist 4-hydroxytamoxifen (Figure 2, E and F), during the time of transfection and culturing of the cells for 20 h had no apparent effect on the nucleocytoplasmic compartmentalization of GFP-ER. In contrast, when GFP-ER–expressing cells were treated with 10 nM ICI 182780, a pure ER antagonist, green fluorescence was observed not only in the nucleus but also in the cytoplasm (Figure 2, G and H). This effect of ICI 182780 on the nucleocytoplasmic compartmentalization of GFP-ER is similar to that previously reported for the untagged ER (Dauvois et al., 1993).
Although a similar effect of ligand on GFP-ER nucleocytoplasmic compartmentalization was observed in T47D, MDA-MB-231, and MDA-MB-435A cells (our unpublished results), we observed variability in the proportion of cells showing cytoplasmic green fluorescence after overnight treatment with 10 nM ICI 182780. In T47D and MCF-7 cells, cytoplasmic green fluorescence was seen in a minority of the cells within 30 min of 10 nM ICI 182780 treatment and peaked at 6–8 h with ∼90% of the cells showing some degree of cytoplasmic fluorescence (e.g., Figure 3B). In the case of the ER− cell lines, cytoplasmic accumulation was observed in ∼10% of the population despite prolonged treatment (20 h) with 10 nM ICI 182780 (our unpublished results). However, in no case did total cytoplasmic green fluorescence exceed nuclear fluorescence (e.g., Figures 2H and 3B).
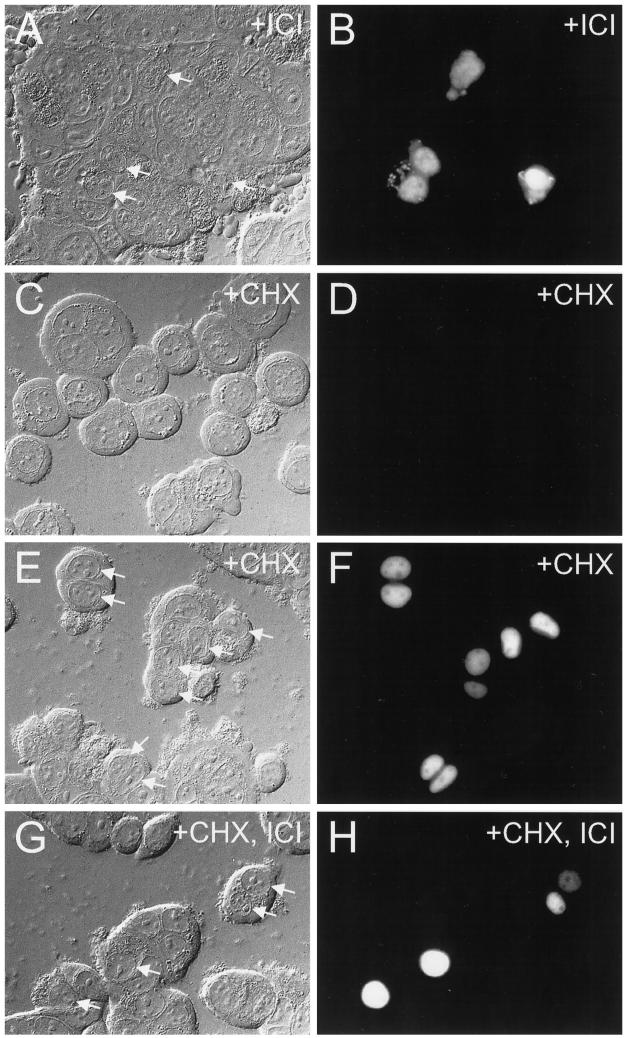
Figure 3.
Effect of cycloheximide treatment on ICI 182780-induced accumulation of GFP-ER in the cytoplasm. MCF-7 cells were electroporated with 0.5 μg of pCI-nGL1-HEGO DNAs and left to express for 12 h before 8 h of treatment with 10 nM ICI 182780 (A and B), 200 μg/ml cycloheximide (E and F), or 200 μg/ml cycloheximide and 10 nM ICI 182780 (G and H). Alternatively, the cells were treated with 200 μg/ml cycloheximide immediately after electroporation for 20 h (C and D). Cells were visualized by differential interference contrast (A, C, E, and G) or epifluorescence using a standard FITC filter set (B, D, F, and H). Arrows in the left panels point to nuclear fluorescence observed in the right panels.
Effect of Cycloheximide on ICI 182780-induced Cytoplasmic Accumulation of GFP-ER
The protein synthesis inhibitor cycloheximide was used to block protein synthesis, and its effect on cytoplasmic accumulation of GFP-ER was examined after 10 nM ICI 182780 treatment. In MCF-7 cells treated with 200 μg/ml cycloheximide for 20 h immediately after electroporation, no green fluorescence was seen (compare Figure 3, C and D, with no cycloheximide treated cells in Figure 2, A and B), indicating that no GFP-ER was synthesized in the cycloheximide-treated cells. In contrast, allowing protein synthesis to continue for the first 12 h after transfection and then halting it for 8 h through the addition of cycloheximide to the culture medium showed green fluorescence in the nucleoplasm but not in the nucleolus (Figure 3, E and F). Simultaneous inclusion of both cycloheximide and 10 nM ICI 182780 12 h after transfection but 8 h before microscopy failed to reveal any cell with cytoplasmic green fluorescence (Figure 3, G and H). Because absence of the protein synthesis inhibitor results in cytoplasmic accumulation of GFP-ER in the presence of ICI 182780 (Figure 3, A and B), continued protein synthesis appears to be required for ICI 182780-induced cytoplasmic accumulation of GFP-ER.
Confocal Laser Scanning Microscopic Examination of GFP-ER Nuclear Distribution
The ability of ICI 182780 to affect the cytoplasm–nucleus partitioning of GFP-ER suggests that other ER ligands might affect the localization of the receptor. In particular, although no change in the nuclear versus cytoplasmic compartmentalization of GFP-ER was observed for cells treated with either 17β-estradiol or 4-hydroxytamoxifen (Figure 2, compare B with D and F), the localization of GFP-ER might have been altered within the nucleus. To determine whether changes occurred in the nuclear localization of GFP-ER upon the addition of ligand, the GFP-ER-expressing MCF-7 cells were examined by high-resolution fluorescent microscopy using a confocal laser scanning microscope.
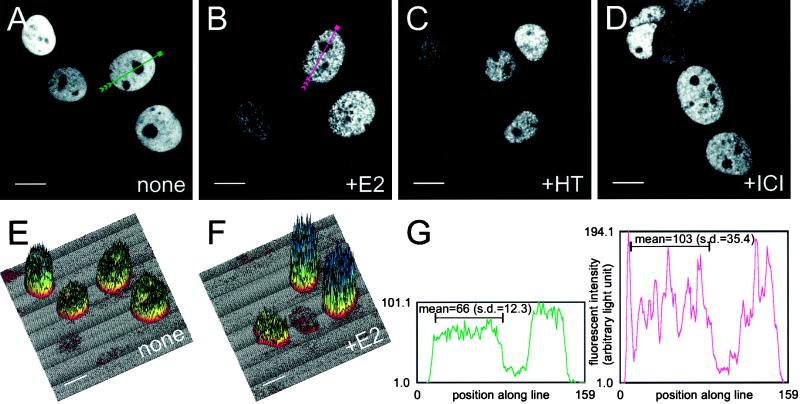
Figure 4A shows an optical section of four live MCF7 cells grown in the absence of ligand, obtained through a confocal laser scanning microscope. Note the increased resolution of the images (also other images in Figures 4 and 5) over the conventional epifluorescent images in Figures 2 and 3. Despite differences in the overall level of brightness attributable to cell-to-cell variation in the level of GFP-ER expression (e.g., Figure 4A, top left corner compared with the adjacent nucleus or the two rightmost pair of nuclei), the tagged receptor is found to be present in a reticular pattern evenly distributed throughout the nucleus. It is, however, excluded from the nucleoli. In contrast, treatment with 10 nM 17β-estradiol, 4-hydroxytamoxifen, or ICI 182780 for 1 h leads to the redistribution of the receptor, resulting in the nucleus appearing punctate and highly structured (Figure 4, B–D). Note that the ICI 182780-treated cells (1 h) failed to show cytoplasmic GFP-ER, because at this time point, few cells accumulate GFP-ER in the cytoplasm (Figures 4D and 5, D, H, and L; our unpublished results). This result is in contrast to the cytoplasmic accumulation seen after an overnight treatment (Figure 2H). Thus, intranuclear localization of the ER is altered by treatment with ER ligands (Figure 4, B–D).
Figure 4.
Effect of ligand treatment on the intranuclear distribution of GFP-ER in MCF-7 cells. MCF-7 cells were electroporated with 0.2 μg of GFP-ER expression plasmid pCI-nGL1-HEGO and cultured on coverslips overnight. The next day, media were changed, and the cells were treated with nothing (A), 10 nM 17β-estradiol (B), 10 nM 4-hydroxytamoxifen (C), or 10 nM ICI 182780 (D) for 1 h before visualization by confocal laser scanning microscopy on a Bio-Rad MRC 1024 system. Representation of fluorescence intensity in A and B is as three-dimensional plots in E and F, respectively, with the greater the fluorescence, the higher the peaks, and the cooler the colors. Bar, 10 μm. (G) Graph of fluorescence intensity along the green line for the nucleus in A (left graph with green curve) and red line for the nucleus in B (right graph with red curve). Arrowheads in A and B point to the direction of the plot from left to right in H. The black bar within each graph in H marks the segment of the line for which the mean fluorescence intensity and SDs are calculated.
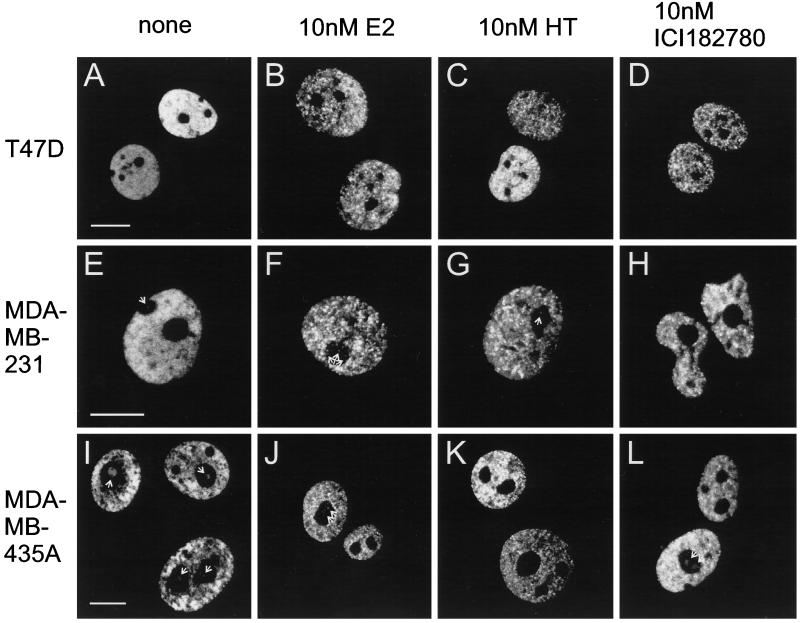
Figure 5.
Effect of ligand treatment on the intranuclear distribution of GFP-ER in three human breast cancer epithelial cell lines. Three human breast cancer epithelial cell lines, T47D, MDA-MB-231, and MDA-MB-435A, were electroporated with 0.2 μg of pCI-nGL1-HEGO and treated and visualized as in Figure 3. Bar in each panel for the first column (A, E, and I), same scale bar for the other panels in the row, 10 μm. Note the presence of fluorescence in the nucleolar regions (indicated by white arrows) for the some of the MDA-MDA-231 cells (e.g., E–G), which can be quite prominent in MDA-MB-435A cells (e.g., I, J, and L).
Representation of Confocal Laser Scanning Microscopic Images
Because the confocal laser scanning microscope records images in a digital format, these images can be analyzed by a number of procedures. Figure 4, E and F, shows three-dimensional color surfaces in which height and color are used to represent the light intensity in the panel corresponding to the untreated or 17β-estradiol–treated cells (Figure 4, A and B). For example, the brightest nucleus in the untreated sample and the two brightest in the 17β-estradiol–treated sample are represented by the tallest nuclei whose surface color either approaches blue or is blue in Figure 4, E and F, respectively.
The fluctuation in light intensity is readily apparent when a perpendicular slice through the surface plot of the optical section for the cell is made and examined. Figure 4G, left diagram, shows a slice from an untreated nucleus (Figure 4A, green line), whereas the right diagram shows a slice from a 17β-estradiol–treated nucleus (Figure 4B, red line). In tracing along the green line from left to right in Figure 4A and examining the left diagram in the same manner for Figure 4G, it is clear that the fluorescence intensity is zero outside of the nucleus. As the line enters the nucleus, the fluorescence intensity value suddenly rises and remains fairly constant in the nucleus, except where it drops dramatically at the nucleolus, which is a little over halfway into the nucleus. The fluorescence intensity again recovers outside the nucleolus and subsequently drops to the baseline upon exiting the nucleus. A similar trend is seen for the slice of the 17β-estradiol–treated nucleus indicated in Figure 4B (right diagram); however, in this case, dramatic changes in the fluorescence intensity values are seen for the nuclear region excluding the nucleolus. Such changes mirror the punctation observed for the 17β-estradiol–treated nuclei (Figure 4B); in contrast, minor fluctuation in fluorescence intensity values in the slice of the untreated nucleus (Figure 4B, left diagram) reflects the more even, reticular distribution of GFP-ER observed in Figure 4A.
Quantitative Analysis of the Effect of Ligand on GFP-ER Nuclear Distribution
To quantitatively compare the magnitude of fluctuation in fluorescence intensity, the coefficient of variation was determined from the mean and SD of the fluorescence intensity for a segment of each line traversing the nucleus (Figure 4G, portion under the two black bars). In the case of the untreated sample, the mean fluorescence intensity was 66 with an SD of 12.3, whereas the 17β-estradiol sample had a mean of 103 with an SD of 35.4. Normalization of the SD by the mean yielded the coefficient of variation, which allows direct comparison of the magnitude in the variation of the fluorescence intensity for samples with different mean values. Indeed, the coefficient of variation was nearly twofold higher for the 17β-estradiol–treated sample than for the untreated sample (0.344 vs. 0.186), indicating a substantial fluctuation in fluorescence intensity for the 17β-estradiol–treated cells than untreated cells.
To determine whether a statistically significant difference exists in the coefficient of variation after ligand treatment, the mean coefficient of variation was determined from a large population of nuclei treated in a similar manner. In the current instance, the coefficient of variation was computed in a similar manner as described in the earlier example, but now encompassing the entire nucleus in the optical section minus the nucleolus. From the coefficient of variation, a mean coefficient of variation along with its SD was calculated for each population treated in a similar manner. As seen in Table 1, the mean coefficient of variation for 164 MCF-7 cells not treated with ligand is 0.225, whereas that for 82 MCF-7 cells treated with 17β-estradiol is 0.365. Statistical Z test gives a value >26. This value indicates a chance of <1 in 10 million that these two means would show such a difference on the basis of chance alone and thereby establishes a high degree of statistical significance to these results (Chase and Bown, 1992). Similarly, treatment with 4-hydroxytamoxifen or ICI 182780 resulted in a 1.4-fold increase in the mean coefficient of variation, which is also a high statistical difference from the untreated sample.
Nuclear distribution of GFP-ER was also analyzed in another ER+ cell line, T47D, and two ER− cell lines, MDA-MB-231 and MDA-MB-435A, to determine whether the 1-h ligand treatment also caused a statistically significant redistribution of GFP-ER. In each case, ligand treatment results in an increase in the mean coefficient of variation (Table 1, compare leftmost number with others in each row) to a value that would occur by chance in <1 of 10 million cases. Thus, for each cell line, a significant redistribution of GFP-ER occurs upon treatment with ligand (Figure 5, compare first panel with other panels in each row).
Ligand– and Cell Line–specific Distribution of GFP-ER in the Nucleus
Although it is apparent that no two GFP-ER–expressing cells look identical (Figures 4 and 5, compare the nuclear appearance of the group of cells in each panel), each cell line has a characteristic distribution of GFP-ER and responds in a characteristic manner to ligand. For example, in the absence of ligand, GFP-ER is distributed evenly in a reticular pattern in MCF-7 cells (Figure 3A). However, deviation from this pattern occurs in T47D, MDA-MB-231, and MDA-MB-435A cells, resulting in a small but statistically significant increase in the mean coefficient of variation (Table 1, compare the value for MCF-7 with others in the first column). In the case of T47D cells, GFP-ER can be seen to concentrate at a few nuclear sites over a reticular background (more evident for the brighter nucleus in Figure 5A). The two other cell lines show an uneven distribution of GFP-ER. In the case of the MDA-MB-231 cell line, the unevenness results in the left and lower left half of the nucleus being brighter than the other half (Figure 5E). For MDA-MB-435A cells, the unevenness is very apparent from the “patchy” GFP-ER patterns within the nucleus (Figure 5I).
Despite the fact that GFP-ER undergoes a dramatic redistribution after ligand treatment, the magnitude of the change differs between different cell lines as each cell line responds in a characteristic manner to ligand. 17β-Estradiol treatment leads to a high degree of punctation in the distribution of GFP-ER for MCF-7 and T47D cells, less so in MDA-MB-231 cells, and least in MDA-MB-435A cells (Table 1). The extent of punctation observed for MCF-7 cells is greater for 17β-estradiol than for the antagonists, but for T47D cells, it is not significantly different. Although the GFP-ER patterns for the two antagonists are significantly different for MDA-MB-231 cells (p < than 1 in 100 that the mean coefficients of variation are those in Table 1), these patterns appear not to be significantly different for MDA-MB-435A cells.
GFP-ER Presence in the Nucleolar Region
In the two ER− cell lines, expression of GFP-ER results not only in its accumulation within the nucleus but also in its presence within the nucleolar region (Figure 5, E–G, I, J, and L). This is most evident for the MDA-MB-435A cells. The extent of “nucleolar” accumulation differs between different cells in the population, independent of ligand treatment. Thus, unlike the ER+ cell lines, GFP-ER can accumulate in the nucleolar region of ER− cells.
DISCUSSION
Early attempts at localization of the ER by biochemical fractionation led to the two-step model of steroid hormone action. Binding of steroid by cytosolic steroid hormone receptor leads to its transformation and subsequent translocation to the nucleus where it regulates gene expression (Gorski et al., 1968; Jensen et al., 1968). This view was revised when the human ER was shown to be in the nucleus independent of ligand by immunocytology (King and Greene, 1984) and hormone-binding assays of cytoplast and nucleoplast fractions from cytochalasin B-induced enucleation of intact cells (Welshons et al., 1984). Steroid hormone transformed a “loosely” bound nuclear ER to a more “tightly” bound nuclear form, which regulated gene expression (Greene and Press, 1986; Press et al., 1989; Jensen, 1991). However, attempts at defining these two biochemically distinct states of ER by immunocytology failed to reveal any notable difference in the intranuclear localization of the loosely and more tightly bound forms of ER (Press et al., 1985; Vazquez-Nin et al., 1991; Yamashita, 1995).
In our current work, we have revisited this issue of the role of ligand in ER localization, using a direct visualization approach in living cells based on GFP tagging. We have previously shown this approach to be extremely useful for observing the subcellular localization of the GR. Using the GFP-tagging approach, we saw for the first time differences in the intranuclear distribution of the receptor that reflected the type of ligand, agonist or antagonist, bound to the receptor. That work represents the first report of the importance of ligand or signal in affecting the distribution of a steroid hormone and nuclear receptor within the nucleus (Htun et al., 1996).
When ER is tagged at its amino terminus with GFP, the tagged receptor is functional by a number of criteria. First, the receptor is capable of transcriptional activation of the ERE-containing reporter gene. Second, GFP-ER responds to ligands, ER agonist or antagonist, similar to ER. Third, GFP-ER is nuclear in the absence of any added ligand, as has been reported for ER. Fourth, the pure antagonist ICI 182780 causes partial cytoplasmic accumulation of GFP-ER, as has been reported for ER (Dauvois et al., 1993). Finally, GFP-ER can associate with the nuclear matrix, similar to that reported for the wild-type ER (Barrack and Coffey, 1980; Alexander et al., 1987; Samuel et al., 1998). Although we have not quantitatively investigated this issue, we do not see a large effect of hormone stimulation on the interaction with matrix. This could suggest that the receptor is present on different matrix sites in the presence and absence of ligand.
Using this GFP-ER, we find the receptor to be distributed in a reticular pattern in the absence of ligand. This pattern suggests that the majority of the unliganded GFP-ER is not freely diffusing in the nucleus but is rather associated with some nuclear meshwork present throughout the nucleus. Ligand causes a dramatic redistribution of the receptors to numerous nuclear sites, giving a punctate nuclear pattern. These two distinct nuclear localization patterns provide the first visual evidence of the changes in receptor activity by hormone and may reflect the loose and tighter binding nuclear forms of ER (Greene and Press, 1986; Press et al., 1989; Jensen, 1991). These findings also complement our earlier work with the GR, which demonstrated the importance of ligand on the nucleocytoplasmic compartmentalization and intranuclear distribution of the receptor (Htun et al., 1996). Despite the fact that the current work was performed on human breast cancer epithelial cells, the nuclear redistribution of ER is expected to be a general feature of all ER-containing cells and is likely a consequence of hormone-dependent transformation of ER from a loose to tighter nuclear form.
Closer examination of the various human breast cancer epithelial cell lines shows a cell line–specific intranuclear distribution of GFP-ER. Small but significant differences in GFP-ER localization can be observed in the absence of ligand. In MCF-7 cells, GFP-ER can be seen to localize in a reticular pattern evenly distributed throughout the nucleus, excluding the nucleolus (Figure 4A). This is not so for T47D cells where a composite pattern emerged as a result of the accumulation of the receptor in a reticular pattern and to a low level of concentration at numerous nuclear sites present throughout the nucleus (Figure 5A). Despite this alteration in the nuclear pattern, comparison of different portions of the nucleus in these ER+ cells shows no remarkable difference in the fluorescent intensity and localization patterns, indicative of an even, intranuclear distribution of GFP-ER. In contrast, ER− cell lines lack this even distribution. In MDA-MB-231 cells, the receptor is slightly more abundant on one half over the other half of the nucleus (Figure 5E), whereas in MDA-MB-435A cells, the unevenness in GFP-ER distribution results in a patchy nuclear appearance (Figure 5I). Thus, each cell line shows a characteristic distribution of GFP-ER in the absence of ligand.
In comparing the confocal laser scanning microscopic images of GFP-ER-expressing cells, it is clear that no two nuclei show an identical distribution of GFP-ER despite the cells being treated and handled in a similar manner (Figures 4 and 5, compare nuclei within each panel). Differences in the phase of the cell cycle, physical characteristics, local external environment, and stochastic nature of some biological regulatory processes may contribute to a unique nuclear appearance. To describe the distribution of GFP-ER in a quantitative manner, the coefficient of variation was determined from the frequency distribution of fluorescence intensity. Although the coefficient of variation analysis ignores the precise spatial organization of GFP-ER, changes in the GFP-ER patterns are often accompanied by changes in the size and number of GFP-ER clusters and hence the frequency distribution of fluorescence intensity. Under most circumstances, the coefficient of variation will be different, and the significance of this difference can be addressed statistically. However, in cases in which the values of the coefficient of variation are similar, direct visual assessment is required to address the issue of similarity or difference in the GFP-ER patterns. Thus, the coefficient of variation serves as an indirect measure of the spatial distribution of GFP-ER through its effect on the frequency distribution of fluorescence intensity.
When cells not exposed to ER ligand are examined, a reticular pattern can be observed throughout the nuclear volume excluding the nucleolus. However, subtle differences exist (Figures 4A and 5, A, E, and I) that can be evaluated quantitatively and analyzed statistically. When the coefficient of variation was determined from the nuclear fluorescence but excluding the nucleolus, the mean coefficient of variation was smallest for MCF-7 cells, which showed an even and reticular distribution of GFP-ER (Table 1; see Figure 4A). The mean coefficient was largest for the cell line MDA-MB-435A, which deviated furthest from this distribution, as evident from the patchy appearance of the nuclei (Table 1; see Figure 5I). Intermediate values were obtained for the untreated T47D and MDA-MB-231 cells, which had a low level of deviation from the MCF-7 nuclear pattern (Table 1; see Figure 5, A and E). Heterogeneity in the observed distribution of GFP-ER is partly reflected by the SD, as indicated in Table 1. From the mean and SD, the statistical Z test established the statistical significance of the difference in the mean coefficient of variation among the different cell lines, except between T47D and MDA-MB-231 cells, and hence the existence of a unique and characteristic GFP-ER distribution pattern, at least on the average. For T47D and MDA-MB-231 cells, the mean coefficient of variation is similar; however, examination of the confocal sections has shown differences in the nuclear patterns between T47D and MDA-MB-231 cells, as discussed earlier. Thus, to a first approximation, the mean coefficient of variation for the most part adequately summarizes the different GFP-ER localization patterns and helps define a characteristic nuclear distribution of GFP-ER for each cell line.
In the presence of ligand, GFP-ER redistributes within the nucleus. Analysis of the number of cells, indicated within parentheses in Table 1, shows that for all ligands, the greatest redistribution occurs in T47D cells, and the least occurs in MDA-MB-435A cells. The response in MCF-7 and MDA-MB-231 cells is intermediate and ligand dependent. In MCF-7 cells, 17β-estradiol caused a greater redistribution than 4-hydroxytamoxifen, which elicited a similar response as ICI 182780. However, in MDA-MB-231 cells, both 17β-estradiol and ICI 182780 caused a similar but greater change than 4-hydroxytamoxifen. Thus, each cell line not only has a characteristic GFP-ER distribution pattern but responds in a characteristic manner to ER ligands.
Responsiveness of human breast cancer to hormonal therapy correlates well with ER status in which up to 60% of ER+ tumors respond to anti-estrogen therapy, in contrast to 10% of ER− tumors (Allegra et al., 1980; Samaan et al., 1981; Williams et al., 1987). Interestingly, treatment of ER− cells, made to express exogenous ER, with 17β-estradiol inhibited cell growth and proliferation, contrary to what is normally observed in ER+ cells (Garcia et al., 1992; Jiang and Jordan, 1992; Zajchowski et al., 1993; Levenson and Jordan, 1994). This differential response to the activation of ER by 17β-estradiol suggests important differences in cellular and/or nuclear content and/or structure that affects ER function. Comparing GFP-ER localization between the ER+ and ER− cell lines revealed three characteristic differences. First, GFP-ER is distributed more evenly throughout the nuclear volume minus the nucleolus in ER+ than ER− cells. Second, ER− cells had GFP-ER in the nucleolar region unlike ER+ cells, more so in MDA-MB-435A than MDA-MB-231 cells. Third, after 6–8 h of treatment with ICI 182780, GFP-ER is seen in the cytoplasm of ∼90% of the ER+ cells but only 10% of ER− cells. Thus, the cell line variations in the subcellular localization of GFP-ER demonstrate that ER function is affected by cellular, nuclear, or structural differences. The importance of this alteration on the receptor’s role in inhibiting the growth and proliferation of ER− cells, transfected with ER expression vector, remains to be elucidated.
Dauvois et al. (1993) proposed ER to constantly shuttle between the nucleus and the cytoplasm despite its predominantly nuclear location. They further reported that nuclear uptake was energy dependent and that ICI 182780 disrupted this process, resulting in the accumulation of ER in the cytoplasm. We observed that the ability of ICI 182780 to cause cytoplasmic localization of GFP-ER was prevented when breast cancer cells were incubated with both ICI 182780 and cycloheximide, a protein synthesis inhibitor. A possible explanation for this observation is that ICI 182780 prevents the nuclear uptake of newly synthesized GFP-ER. Alternatively, a labile protein factor could be required for ER to be exported from the nucleus to the cytoplasm. Variation in the abundance of this labile factor could account for the observed differences between the rate of cytoplasmic accumulation of GFP-ER in ER+ breast cancer epithelial cells and mouse ER in COS-1 cells (Dauvois et al., 1993), and absence of this labile factor in 90% of the ER− cells might explain why these cells failed to show cytoplasmic green fluorescence after ICI 182780 treatment.
From our current work, it is clear that the effectiveness of ICI 182780 as an antagonist is independent of its ability to induce cytoplasmic accumulation of GFP-ER. In the ER+ human breast cancer epithelial cells, most of the GFP-ER remained in the nucleus. In an ER− human breast cancer epithelial cell, MDA-MB-435A, ICI 182780 treatment effectively suppressed GFP-ER transcriptional activation of a reporter gene; however, only 10% of these cells showed any GFP-ER accumulation in the cytoplasm. Thus, the mechanism of ICI 182780 antagonism does not depend on the nuclear–cytoplasmic recompartmentalization of the receptor but, rather, must occur within the nuclear compartment at a step required for activation of transcription by ER.
Previous studies have shown that in the absence of hormone, steroid receptors are thought not to be bound to hormone-responsive elements in target genes (Kumar and Chambon, 1988; Wijnholds et al., 1988; Pham et al., 1991b; McDonnell et al., 1992; also reviewed in Tsai and O’Malley, 1994; Shibata et al., 1997). Hormone binding causes a conformational change in the receptor that allows it to bind to its cognate sites and to regulate gene transcription. Agonist ligands induce a receptor conformation that can interact with the general transcription factors or transcriptional coactivators to establish a productive transcriptional preinitiation complex. Antagonist ligands, on the other hand, induce a different receptor conformation that interferes with the ability of the receptor to bind to DNA or alternatively to prevent formation of a productive transcriptional preinitiation complex by abrogating its interaction with the general transcription factors and/or transcriptional coactivators (Martinez and Wahli, 1989; McDonnell et al., 1991; Pham et al., 1991a; Sabbah et al., 1991; McDonnell et al., 1994, 1995; Tsai and O’Malley, 1994; Mymryk and Archer, 1995; Vegeto et al., 1996; Brzozowski et al., 1997; Gallo and Kaufman, 1997; Shibata et al., 1997). Alternatively, antagonist ligands might promote receptor interaction with transcriptional corepressors to actively maintain a repressed transcriptional state (McDonnell et al., 1992, 1994; Smith et al., 1997; Lavinsky et al., 1998; Zhang et al., 1998). In the case of ER antagonists, antagonism appears to occur at a step subsequent to the binding of the receptor to the EREs of target genes than at the actual step of DNA binding (Martinez and Wahli, 1989; McDonnell et al., 1991, 1992, 1994, 1995; Pham et al., 1991a; Sabbah et al., 1991; Vegeto et al., 1996; Brzozowski et al., 1997; Gallo and Kaufman, 1997; Shibata et al., 1997). Our visual data favor a model of 4-hydroxytamoxifen and ICI 182780 antagonism in which ER binds to the hormone response elements of target genes, but its ability to activate transcription is partially or completely abolished, respectively. In support of this assertion, we find that, qualitatively, the punctate pattern observed after 4-hydroxytamoxifen or ICI 182780 treatment is no different from the pattern observed after 17β-estradiol treatment in the four human breast cancer epithelial cell lines. Quantitatively, in the case of T47D cells, no significant difference in the mean coefficient of variation can be established after 4-hydroxytamoxifen, ICI 182780, or 17β-estradiol treatment (Table 1), suggesting a similarity in the number of nuclear sites to which the ligand-bound receptor accumulates. Although we favor these sites of GFP-ER concentration as regions high in the concentration of EREs, we cannot rule out the possibility that these sites might be ER-processing sites, storage sites, or sites of interaction with the nuclear matrix.
Last, during mouse embryonic development, ER has been reported to preferentially accumulate within the nucleolar region of a specific subset of cells (Hou et al., 1996). Because this recompartmentalization is observed during the ontogeny of ER− from ER+ cells, it has been suggested that this recompartmentalization might be a consequence of the mechanism involved in ER down-regulation. In this regard, we have found ER− human breast cancer epithelial cells to show GFP-ER not only in the nucleus but also in the nucleolar region (Figures 4 and 5). The presence of GFP-ER in the nucleolar region of ER− human breast cancer epithelial cells further suggests the importance of ER localization for its function and points to parallel regulatory mechanisms governing ER localization during both early mouse embryonic development and human mammary gland development.
ACKNOWLEDGMENTS
We thank Pierre Chambon (Institut de Genetique et de Biologie Moleculaire et Cellulaire, France) for providing us with the pSG5-HEGO clone of the wild-type ER α, Geoffery L. Greene (The Ben May Institute for Cancer Research, University of Chicago, Chicago, IL) for providing us with ER antibodies H226 and H222, Leigh C. Murphy (University of Manitoba), William Whalen (Laboratory of Basic Research, National Cancer Institute), and Wendy J. Dixon (Center for Food Safety and Nutrition, Food and Drug Administration, Washington, DC) for useful discussion and advice, and Ravi Dhar (Laboratory of Basic Research, National Cancer Institute) and Mark Mortin (Laboratory of Biochemistry, National Cancer Institute) for the use of the Zeiss Axiophot and Bio-Rad MRC1024, respectively. This work was supported in part by grant PG-12809 from the Medical Research Council (MRC) of Canada to J.R.D. J.R.D is an MRC senior scientist. L.T.H. received support from U.S. Army Breast Cancer Postdoctoral Research Grant DAM17-96-1-6269.
REFERENCES
- Alexander RB, Greene GL, Barrack ER. Estrogen receptors in the nuclear matrix: direct demonstration using monoclonal antireceptor antibody. Endocrinology. 1987;120:1851–1857. doi: 10.1210/endo-120-5-1851. [DOI] [PubMed] [Google Scholar]
- Allegra JC, Barlock A, Huff KK, Lippman ME. Changes in multiple or sequential estrogen receptor determinations in breast cancer. Cancer. 1980;45:792–794. doi: 10.1002/1097-0142(19800215)45:4<792::aid-cncr2820450430>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Barrack ER, Coffey DS. The specific binding of estrogens and androgens to the nuclear matrix of sex hormone responsive tissues. J Biol Chem. 1980;255:7265–7275. [PubMed] [Google Scholar]
- Beato M, Chavez S, Truss M. Transcriptional regulation by steroid hormones. Steroids. 1996;61:240–251. doi: 10.1016/0039-128x(96)00030-x. [DOI] [PubMed] [Google Scholar]
- Belgrader P, Siegel AJ, Berezney R. A comprehensive study on the isolation and characterization of the HeLa S3 nuclear matrix. J Cell Sci. 1991;98:281–291. doi: 10.1242/jcs.98.3.281. [DOI] [PubMed] [Google Scholar]
- Berezney R. The nuclear matrix: a heuristic model for investigating genomic organization and function in the cell nucleus. J Cell Biochem. 1991;47:109–123. doi: 10.1002/jcb.240470204. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brzozowski AM, Pike AC, Dauter Z, Hubbard RE, Bonn T, Engstrom O, Ohman L, Greene GL, Gustafsson JA, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- Carey KL, Richards SA, Lounsbury KM, Macara IG. Evidence using a green fluorescent protein-glucocorticoid receptor chimera that the Ran/TC4 GTPase mediates an essential function independent of nuclear protein import. J Cell Biol. 1996;133:985–996. doi: 10.1083/jcb.133.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase W, Bown F. General Statistics. 2nd ed. New York: John Wiley & Sons; 1992. [Google Scholar]
- Danielian PS, White R, Lees JA, Parker MG. Identification of a conserved region required for hormone dependent transcriptional activation by steroid hormone receptors. EMBO J. 1992;11:1025–1033. doi: 10.1002/j.1460-2075.1992.tb05141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauvois S, White R, Parker MG. The antiestrogen ICI 182780 disrupts estrogen receptor nucleocytoplasmic shuttling. J Cell Sci. 1993;106:1377–1388. doi: 10.1242/jcs.106.4.1377. [DOI] [PubMed] [Google Scholar]
- Davie JR. The nuclear matrix and the regulation of chromatin organization and function. Int Rev Cytol. 1995;162A:191–250. doi: 10.1016/s0074-7696(08)61232-2. [DOI] [PubMed] [Google Scholar]
- Davie JR, Samuel S, Spencer V, Bajno L, Sun J-M, Chen HY, Holth LT. Nuclear matrix: application to diagnosis of cancer and role in transcription and modulation of chromatin structure. Gene Ther Mol Biol. 1997;1:509–528. [Google Scholar]
- DeFranco DB, Madan AP, Tang Y, Chandran UR, Xiao N, Yang J. Nucleocytoplasmic shuttling of steroid receptors. Vitam Horm. 1995;51:315–338. doi: 10.1016/s0083-6729(08)61043-2. [DOI] [PubMed] [Google Scholar]
- Gallo MA, Kaufman D. Antagonistic and agonistic effects of tamoxifen: significance in human cancer. Semin Oncol. 1997;24:S1-71–S1-80. [PubMed] [Google Scholar]
- Garcia M, Derocq D, Freiss G, Rochefort H. Activation of estrogen receptor transfected into a receptor-negative breast cancer cell line decreases the metastatic and invasive potential of the cells. Proc Natl Acad Sci USA. 1992;89:11538–11542. doi: 10.1073/pnas.89.23.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski J, Toft D, Shyamala G, Smith D, Notides A. Hormone receptors: studies on the interaction of estrogen with the uterus. Recent Prog Horm Res. 1968;24:45–80. doi: 10.1016/b978-1-4831-9827-9.50008-3. [DOI] [PubMed] [Google Scholar]
- Greene GL, Press MF. Structure and dynamics of the estrogen receptor. J Steroid Biochem. 1986;24:1–7. doi: 10.1016/0022-4731(86)90024-5. [DOI] [PubMed] [Google Scholar]
- Hou Q, Paria BC, Mui C, Dey SK, Gorski J. Immunolocalization of estrogen receptor protein in the mouse blastocyst during normal and delayed implantation. Proc Natl Acad Sci USA. 1996;93:2376–2381. doi: 10.1073/pnas.93.6.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Htun H, Barsony J, Renyi I, Gould DL, Hager GL. Visualization of glucocorticoid receptor translocation and intranuclear organization in living cells with a green fluorescent protein chimera. Proc Natl Acad Sci USA. 1996;93:4845–4850. doi: 10.1073/pnas.93.10.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Cook PR. Visualization of a filamentous nucleoskeleton with a 23 nm axial repeat. EMBO J. 1988;7:3667–3677. doi: 10.1002/j.1460-2075.1988.tb03248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen EV. Overview of the nuclear receptor family. In: Parker MG, editor. Nuclear Hormone Receptors: Molecular Mechanisms, Cellular Functions, Clinical Abnormalities. San Diego: Academic Press; 1991. pp. 1–13. [Google Scholar]
- Jensen EV, Suzuki T, Kawashima T, Stumpf WE, Jungblut PW, DeSombre ER. A two-step mechanism for the interaction of estradiol with rat uterus. Proc Natl Acad Sci USA. 1968;59:632–638. doi: 10.1073/pnas.59.2.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang SY, Jordan VC. Growth regulation of estrogen receptor-negative breast cancer cells transfected with complementary DNAs for estrogen receptor. J Natl Cancer Inst. 1992;84:580–591. doi: 10.1093/jnci/84.8.580. [DOI] [PubMed] [Google Scholar]
- King WJ, Greene GL. Monoclonal antibodies localize oestrogen receptor in the nuclei of target cells. Nature. 1984;307:745–747. doi: 10.1038/307745a0. [DOI] [PubMed] [Google Scholar]
- Kingston RE, Chen CA, Okayama H. Introduction of DNA into mammalian cells. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, editors. Current Protocols in Molecular Biology, chap. 9. K. Struhl, New York: John Wiley & Sons; 1995. [Google Scholar]
- Kuiper GG, Carlsson B, Grandien K, Enmark E, Haggblad J, Nilsson S, Gustafsson JA. Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors α and β. Endocrinology. 1997;138:863–870. doi: 10.1210/endo.138.3.4979. [DOI] [PubMed] [Google Scholar]
- Kumar V, Chambon P. The estrogen receptor binds tightly to its responsive element as a ligand-induced homodimer. Cell. 1988;55:145–156. doi: 10.1016/0092-8674(88)90017-7. [DOI] [PubMed] [Google Scholar]
- Lavinsky RM, et al. Diverse signaling pathways modulate nuclear receptor recruitment of N-CoR and SMRT complexes. Proc Natl Acad Sci USA. 1998;95:2920–2925. doi: 10.1073/pnas.95.6.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson AS, Jordan VC. Transfection of human estrogen receptor (ER) cDNA into ER-negative mammalian cell lines. J Steroid Biochem Mol Biol. 1994;51:229–239. doi: 10.1016/0960-0760(94)90035-3. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E, Wahli W. Cooperative binding of estrogen receptor to imperfect estrogen-responsive DNA elements correlates with their synergistic hormone-dependent enhancer activity. EMBO J. 1989;8:3781–3791. doi: 10.1002/j.1460-2075.1989.tb08555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell DP, Clemm DL, Hermann T, Goldman ME, Pike JW. Analysis of estrogen receptor function in vitro reveals three distinct classes of antiestrogens. Mol Endocrinol. 1995;9:659–669. doi: 10.1210/mend.9.6.8592512. [DOI] [PubMed] [Google Scholar]
- McDonnell DP, Clemm DL, Imhof MO. Definition of the cellular mechanisms which distinguish between hormone and antihormone activated steroid receptors. Semin Cancer Biol. 1994;5:327–336. [PubMed] [Google Scholar]
- McDonnell DP, Nawaz Z, O’Malley BW. In situ distinction between steroid receptor binding and transactivation at a target gene. Mol Cell Biol. 1991;11:4350–4355. doi: 10.1128/mcb.11.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell DP, Vegeto E, O’Malley BW. Identification of a negative regulatory function for steroid receptors. Proc Natl Acad Sci USA. 1992;89:10563–10567. doi: 10.1073/pnas.89.22.10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mymryk JS, Archer TK. Dissection of progesterone receptor-mediated chromatin remodeling and transcriptional activation in vivo. Genes & Dev. 1995;9:1366–1376. doi: 10.1101/gad.9.11.1366. [DOI] [PubMed] [Google Scholar]
- Ogawa H, Inouye S, Tsuji FI, Yasuda K, Umesono K. Localization, trafficking, and temperature-dependence of the Aequorea green fluorescent protein in cultured vertebrate cells. Proc Natl Acad Sci USA. 1995;92:11899–11911. doi: 10.1073/pnas.92.25.11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MG. Introduction. In: Parker MG, editor. Growth Regulation by Nuclear Hormone Receptors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1992. pp. 1–4. [Google Scholar]
- Pham TA, Elliston JF, Nawaz Z, McDonnell DP, Tsai MJ, O’Malley BW. Antiestrogen can establish nonproductive receptor complexes and alter chromatin structure at target enhancers. Proc Natl Acad Sci USA. 1991a;88:3125–3129. doi: 10.1073/pnas.88.8.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham TA, Hwung YP, McDonnell DP, O’Malley BW. Transactivation functions facilitate the disruption of chromatin structure by estrogen receptor derivatives in vivo. J Biol Chem. 1991b;266:18179–18187. [PubMed] [Google Scholar]
- Picard D, Kumar V, Chambon P, Yamamoto KR. Signal transduction by steroid hormones: nuclear localization is differentially regulated in estrogen and glucocorticoid receptors. Cell Regul. 1990;1:291–299. doi: 10.1091/mbc.1.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- Press MF, Nousek-Goebl NA, Greene GL. Immunoelectron microscopic localization of estrogen receptor with monoclonal estrophilin antibodies. J Histochem Cytochem. 1985;33:915–924. doi: 10.1177/33.9.4020102. [DOI] [PubMed] [Google Scholar]
- Press MF, Xu SH, Wang JD, Greene GL. Subcellular distribution of estrogen receptor and progesterone receptor with and without specific ligand. Am J Pathol. 1989;135:857–864. [PMC free article] [PubMed] [Google Scholar]
- Rizzuto R, Brini M, De Giorgi F, Rossi R, Heim R, Tsien RY, Pozzan T. Double labeling of subcellular structures with organelle-targeted GFP mutants in vivo. Curr Biol. 1996;6:183–188. doi: 10.1016/s0960-9822(02)00451-7. [DOI] [PubMed] [Google Scholar]
- Sabbah M, Gouilleux F, Sola B, Redeuilh G, Baulieu EE. Structural differences between the hormone and antihormone estrogen receptor complexes bound to the hormone response element. Proc Natl Acad Sci USA. 1991;88:390–394. doi: 10.1073/pnas.88.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaan NA, Buzdar AU, Aldinger KA, Schultz PN, Yang KP, Romsdahl MM, Martin R. Estrogen receptor: a prognostic factor in breast cancer. Cancer. 1981;47:554–560. doi: 10.1002/1097-0142(19810201)47:3<554::aid-cncr2820470322>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Samuel, S.K., Spencer, V.A., Bajno, L., Sun, J.-M., Holth, L.T., Oesterreich, S., and Davie, J.R. (1998). In situ cross-linking by cisplatin of nuclear matrix-bound transcription factors to nuclear DNA of human breast cancer cells. Cancer Res. (in press). [PubMed]
- Seiler-Tuyns A, Walker P, Martinez E, Merillat AM, Givel F, Wahli W. Identification of estrogen-responsive DNA sequences by transient expression experiments in a human breast cancer cell line. Nucleic Acids Res. 1986;14:8755–8770. doi: 10.1093/nar/14.22.8755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata H, Spencer TE, Onate SA, Jenster G, Tsai SY, Tsai MJ, O’Malley BW. Role of coactivators and corepressors in the mechanism of steroid/thyroid receptor action. Recent Prog Horm Res. 1997;52:141–165. [PubMed] [Google Scholar]
- Smith CL, Nawaz Z, O’Malley BW. Coactivator and corepressor regulation of the agonist/antagonist activity of the mixed antiestrogen, 4-hydroxytamoxifen. Mol Endocrinol. 1997;11:657–666. doi: 10.1210/mend.11.6.0009. [DOI] [PubMed] [Google Scholar]
- Tora L, Mullick A, Metzger D, Ponglikitmongkol M, Park I, Chambon P. The cloned human oestrogen receptor contains a mutation which alters its hormone binding properties. EMBO J. 1989;8:1981–1986. doi: 10.1002/j.1460-2075.1989.tb03604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MJ, O’Malley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- Vazquez-Nin GH, Echeverria OM, Fakan S, Traish AM, Wotiz HH, Martin TE. Immunoelectron microscopic localization of estrogen receptor on premRNA containing constituents of rat uterine cell nuclei. Exp Cell Res. 1991;192:396–404. doi: 10.1016/0014-4827(91)90057-2. [DOI] [PubMed] [Google Scholar]
- Vegeto E, Wagner BL, Imhof MO, McDonnell DP. The molecular pharmacology of ovarian steroid receptors. Vitam Horm. 1996;52:99–128. doi: 10.1016/s0083-6729(08)60408-2. [DOI] [PubMed] [Google Scholar]
- Wakeling AE, Dukes M, Bowler J. A potent specific pure antiestrogen with clinical potential. Cancer Res. 1991;51:3867–3873. [PubMed] [Google Scholar]
- Welshons WV, Cormier EM, Wolf MF, Williams PO, Jr, Jordan VC. Estrogen receptor distribution in enucleated breast cancer cell lines. Endocrinology. 1988;122:2379–2386. doi: 10.1210/endo-122-6-2379. [DOI] [PubMed] [Google Scholar]
- Welshons WV, Lieberman ME, Gorski J. Nuclear localization of unoccupied oestrogen receptors. Nature. 1984;307:747–749. doi: 10.1038/307747a0. [DOI] [PubMed] [Google Scholar]
- Wijnholds J, Philipsen JN, Ab G. Tissue-specific and steroid-dependent interaction of transcription factors with the oestrogen-inducible apoVLDL II promoter in vivo. EMBO J. 1988;7:2757–2763. doi: 10.1002/j.1460-2075.1988.tb03130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MR, Todd JH, Ellis IO, Dowle CS, Haybittle JL, Elston CW, Nicholson RI, Griffiths K, Blamey RW. Oestrogen receptors in primary and advanced breast cancer: an eight year review of 704 cases. Br J Cancer. 1987;55:67–73. doi: 10.1038/bjc.1987.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita S. Intranuclear localization of hormone-occupied and -unoccupied estrogen receptors in the mouse uterus: application of 1 nm immunogold-silver enhancement procedure to ultrathin frozen sections. J Electron Microsc. 1995;44:22–29. [PubMed] [Google Scholar]
- Yee D, McGuire SE, Brunner N, Kozelsky TW, Allred DC, Chen SH, Woo SL. Adenovirus-mediated gene transfer of herpes simplex virus thymidine kinase in an ascites model of human breast cancer. Hum Gene Ther. 1996;7:1251–1257. doi: 10.1089/hum.1996.7.10-1251. [DOI] [PubMed] [Google Scholar]
- Zajchowski DA, Sager R, Webster L. Estrogen inhibits the growth of estrogen receptor-negative, but not estrogen receptor-positive, human mammary epithelial cells expressing a recombinant estrogen receptor. Cancer Res. 1993;53:5004–5011. [PubMed] [Google Scholar]
- Zhang X, Jeyakumar M, Petukhov S, Bagchi MK. A nuclear receptor corepressor modulates transcriptional activity of antagonist-occupied steroid hormone receptor. Mol Endocrinol. 1998;12:513–524. doi: 10.1210/mend.12.4.0089. [DOI] [PubMed] [Google Scholar]