Abstract
Background
Variation in genomes among different closely-related organisms can be linked to phenotypic differences. A number of mechanisms, such as replication error, repeat expansion and contraction, recombination and transposition can contribute to genomic differences. These processes lead to generation of SNPs, different types of repeat-based and transposons or IS-element-based polymorphisms, inversions and duplications and changes in synteny. A database of all the variations in a group of organisms is not only useful for understanding genotype-phenotype relationship but also in clinical applications. There is no database available at present that provides information about detailed genomic variations among different strains and species of Mycobacterium tuberculosis complex, organisms responsible for human diseases.
Description
MGDD is a free web-based database that allows quick user friendly search to find different types of genomic variations among a group of fully sequenced organisms belonging to M. tuberculosis complex. The searches are based on data generated by pair wise comparison using a tool that has already been described. Different types of variations that can be searched are SNPs, indels, tandem repeats and divergent regions. The searches can be designed to find specific variations either in a given gene or any given location of the query genome with respect to any other genome currently available.
Conclusion
Web-based database MGDD can help to find all the possible differences that exists between two strains or species of M. tuberculosis complex. The search tool is very user-friendly and can be used by anyone not familiar with computational methods and will be useful to both clinicians and researchers working on tuberculosis and other Mycobacterial diseases.
Background
A large number of genomes of different strains and closely related species of pathogens have been sequenced and many others are in the process. A detailed analysis of these genomic sequences can help us to decipher and establish genotype to phenotype relationship. The organisms evolve through a series of molecular changes reflected in genomic sequences and some of these are evolutionarily selected based on survival in a specific ecological niche. Characterization of sequence alterations in closely related organisms can help us to understand genome evolution at the molecular level in short time span, for example emergence of new endemic strains in a few decades. Therefore it is important to catalog all the sequence differences between any two organisms so that these can be a basis for designing experiments linking phenotype to genotype. In pathogenic organisms such a database can be useful in identifying species and strain-specific markers that can be a basis for designing diagnostic reagents.
A number of molecular mechanisms have been described that are responsible for genomic changes [1]. These contribute to single nucleotide polymorphisms (SNP), variable number of tandem repeats, insertion/deletion with or without involving transposable elements and recombination. Many of these have been used as markers for identification of strains and diagnosis of pathogens [2-4]. M. tuberculosis is a major cause of morbidity and mortality throughout the world. Genomic variations in this organism have been used to type pathogenic strains in a limited scale [5,6]. A comprehensive database of all the genomic variations of M. tuberculosis is not currently available though some attempts have been made in this direction. For example, MTBreg (please see Availability & requirements for more information) covers variations that are detected using spoligotyping, MycoDB (please see Availability & requirements for more information) [7], MycoperonDB (please see Availability & requirements for more information) [8] and GenoMycDB (please see Availability & requirements for more information) [9] have some features that allow comparison between two genomes in a limited manner. In this report we describe a comprehensive database of genomic differences among strains and species of Mycobacteria belonging to the M. tuberculosis complex. The variations have been identified using ABWGC, a comparative genomic tool previously described by us [10]. We hope that this database will be useful to clinicians and basic scientists interested in understanding Mycobacterial diseases.
Construction and content
The database contains pre-computed data derived from full genome sequences of M. tuberculosis strains H37Rv, CDC1551, H37Ra, F11, Mycobacterium bovis AF2122/97 and M. bovis BCG str. Pasteur 1173P2 using ABWGC [10]. The variations are categorized as SNPs, insertions, divergent regions (based on lack of sequence identity) and tandem repeats. All computations have been carried out in a pair-wise fashion. In some cases, such as SNPs the results differ depending upon the genome that has been used as a query in a pair of genomes. The database contains two sets of data pertaining to using each genome as query sequence. Insertion in one genome can be considered as a deletion in another genome, so the database contains only the insertions. If the insertions are due to known insertion elements and phage sequences these have been pointed out so that the information can be used for devising methods for better diagnosis and strain identification. Tandem repeats were identified using ABWGC and verified by "Tandem Repeat Finder" [11].
MGDD is implemented by using three- tier architecture. The web based application is created by using Apache web server which is connected to the database using MYSQL through an application layer written in Perl-CGI.
The information from MGDD can be obtained by selecting a specific query using the "search option" given in the MGDD browser.
Utility and discussion
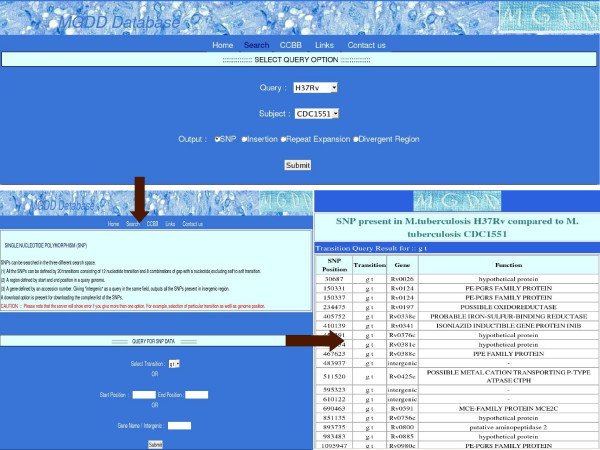
MGDD has a web interface for the retrieval of genomic diversity information. A search can be initiated by first selecting genomes from the "Query" and "Subject" scroll down menu-bar. Currently there is information about six organisms and these can be selected in a pair-wise manner (Fig. 1). These organisms are:
Figure 1.
A typical output of a query (SNP). The transition selected was 'gt' and M. tuberculosis H37Rv was compared with M. tuberculosis CDC1551.
M. tuberculosis CDC1551 (NC_002755.2)
M. tuberculosis F11 (NC_009565.1)
M. tuberculosis H37Ra (NC_009525.1)
M. tuberculosis strain H37Rv (NC_000962.2)
M. bovis AF2122/97 (NC_002945.3)
M. bovis BCG str Pasteur 1173P2 (NC_008769.1)
Each pair of organisms can be analyzed in two different ways by choosing each one as query and the other one as target genome. We recommend that a pair should be analyzed in both ways in order to get a comprehensive list of variations, particularly indels. After selection of organisms the type of variation needs to be selected from the search page. Currently there are four options available and one of these to be chosen among:
SNP, Insertion, Repeat expansion, Divergent regions
After submission of the selected information a detailed query page appears. For example, if SNP is selected the new page will ask for choosing one of the 20 different possible transitions in a user-defined menu-bar and the search can be made restrictive by specifying genomic coordinates or gene name (Fig. 1). The output would show all the indicated SNPs in the selected region along with annotation of genes that contain the SNPs (Fig. 1). For insertions, divergent regions and repeat expansion the query page has also the option of selecting output on the basis of size in nucleotides. There are four options at present and these are >10, 10–50, 50–100 and <100. Since one can select only one query at a time, an error message is displayed if more then one query is selected.
Table 1 gives the total data present in the database. However, the distribution of these changes among strains and species are different. For example, the number of SNPs between the two M. tuberculosis strains H37Ra and Rv are 588 and that between the two M. bovis strains are 1271 (Table 2). In general the number of variants, observed between the two M. tuberculosis strains were much less compared to that between the two M. bovis strains. This is consistent with the fact that M. tuberculosis strains H37Ra and Rv have been recently derived from H37 [12]. These differences are a result of evolutionary history of the organisms and can be useful to map all the potential mutation hotspots in these organisms.
Table 1.
Statistics and data composition of MGDD
| Type of Data | Total number of entries. |
| Divergence | 829 |
| Insertions | 7865 |
| Repeat expansion | 578 |
| SNP | 68768 |
Table 2.
Genomic variants in M. tuberculosis and M. bovis strains
| M. tuberculosis H37Rv compared to H37Ra | M. bovis compared to M. bovis strain BCG | |
| SNP | 582 | 1272 |
| Divergent Region | 1 | 4 |
| Repeat expansion | 3 | 12 |
| Insertions | 22 | 114 |
Conclusion
In this report we describe MGDD, a database of genomic variants computed from fully sequenced organisms belonging to the M. tuberculosis complex. It contains data pertaining to SNP, insertions, repeat expansion and regions that show sequence divergence. Since MGDD is modular information regarding new genomes can be incorporated as and when the sequences become available. The search tool is simple and user friendly and allows one to locate a specific variation in any part of the genome or a gene.
Availability and requirements
The web server can be accessed at http://mirna.jnu.ac.in/mgdd/.
MTBreg: http://www.doe-mbi.ucla.edu/Services/MTBreg/
MycoDB: http://xbase.bham.ac.uk/mycodb/about.pl
MycoperonDB: http://www.cdfd.org.in/mycoperondb/index.html
GenoMycDB: http://157.86.176.108/~catanho/genomycdb/
Authors' contributions
AV has implemented the ABWGC and identified the different divergent regions in two genomes and tested the database. AS has made the database and web based application. RR and AB helped in conceptualizing the database and writing the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
The authors thank Department of Biotechnology (COE and TB informatics project) for support.
Contributor Information
Anchal Vishnoi, Email: anchalv@gmail.com.
Alok Srivastava, Email: foralok@gmail.com.
Rahul Roy, Email: rahul@isid.ac.in.
Alok Bhattacharya, Email: alok0200@mail.jnu.ac.in.
References
- Whittam TS, Bumbaugh AC. Inferences from whole genome sequences of bacterial pathogens. Current Opinion in Genetics and Development. 2002;12:719–725. doi: 10.1016/S0959-437X(02)00361-1. [DOI] [PubMed] [Google Scholar]
- Van Soolingen D, de Haas PE, Hermans PW, Groenen PM, Dvan Embden J. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. Journal of Clinical Microbiology. 1993;31:1987–1995. doi: 10.1128/jcm.31.8.1987-1995.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RW, Athanassopoulos E, Tsiamis G, Mansfield JW, Sesma A, Arnold DL, Gibbon MJ, Murillo J, Taylor JD, Vivian A. Identification of a pathogenicity island, which contains genes for virulence and avirulence, on a large native plasmid in the bean pathogen Pseudomonas syringae pathovar phaseolicola. Proceeding of the National Academy of Sciences USA. 1999;96:10875–10880. doi: 10.1073/pnas.96.19.10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. Journal of Clinical Microbiology. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fomukong NG, Tang TH, Maamary al, Ibrahum WA, Ramayah S, Yates M, Zainuddin ZF, Dale JW. Insertion sequence typing of Mycobacterium tuberculosis: characterization of a widespread subtype with a single copy of IS6110. Tuber Lung Disease. 1994;75:435–40. doi: 10.1016/0962-8479(94)90117-1. [DOI] [PubMed] [Google Scholar]
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, III, Tekaia F, Badcock K, Basham D, Brown D, Chillingwoth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Sulston JE, Taylor K, Whitehead S, Barrel BG. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Chaudhuri RR, Pallen MJ. xBASE, a collection of online databases for bacterial comparative genomics. Nucleic Acid Research. 2006;34:D335–337. doi: 10.1093/nar/gkj140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan S, Gundu RK, Ranjan A. MycoperonDB: a database of computationally identified operons and transcriptional units in Mycobacteria. BMC Bioinformatics. 2006;7:S9. doi: 10.1186/1471-2105-7-S5-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanho M, Mascarenhas D, Degrave W, de Miranda AB. GenoMycDB: Database for Comparative Analysis of Mycobacterial Genes and Genomes. Genetics and Molecular Research. 2006;5:115–126. [PubMed] [Google Scholar]
- Vishnoi A, Roy R, Bhattacharya A. Comparative analysis of bacterial genomes: identification of divergent regions in mycobacterial genomes using an anchor based approach. Nucleic Acid Research. 2007;35:3654–3667. doi: 10.1093/nar/gkm209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Research. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenken W, Gardner LV. History of H37 strain of tubercle bacillus. Amer Rev Tuberc. 1946;54:52–66. doi: 10.1164/art.1946.54.1.62. [DOI] [PubMed] [Google Scholar]