Abstract
Background
The risk of hepatocellular carcinoma (HCC) increases with increasing level of hepatitis B virus (HBV) in serum (viral load). However, it is unclear whether genetic characteristics of HBV, including HBV genotype and specific genetic mutations, contribute to the risk of HCC. We examined the HCC risk associated with HBV genotypes and common variants in the precore and basal core promoter (BCP) regions.
Methods
From January 5, 1991, to December 21, 1992, baseline blood samples were collected from 2762 Taiwanese men and women who were seropositive for HBV surface antigen but had not been diagnosed with HCC; the samples were tested for HBV viral load by real-time polymerase chain reaction and genotyped by melting curve analysis. Participants who had a baseline serum HBV DNA level greater than 104 copies/mL (n = 1526) were tested for the precore G1896A and BCP A1762T/G1764A mutants by direct sequencing. Incident cases of HCC were ascertained through follow-up examinations and computerized linkage to the National Cancer Registry and death certification profiles. A Cox proportional hazards model was used to estimate the risk of HCC associated with HBV genotype and precore and BCP mutants after adjustment for other risk factors. All statistical tests were two-sided.
Results
A total of 153 HCC cases occurred during 33 847 person-years of follow-up. The HCC incidence rates per 100 000 person-years for participants infected with HBV genotype B or C were 305.6 (95% confidence interval [CI] = 236.9 to 388.1) and 785.8 (95% CI = 626.8 to 972.9), respectively. Among participants with a baseline HBV DNA level of at least 104 copies/mL, HCC incidence per 100 000 person-years was higher for those with the precore G1896 (wild-type) variant than for those with the G1896A variant (955.5 [95% CI = 749.0 to 1201.4] vs 269.4 [95% CI = 172.6 to 400.9]) and for those with the BCP A1762T/G1764A double mutant than for those with BCP A1762/G1764 (wild-type) variant (1149.2 [95% CI = 872.6 to 1485.6] vs 358.7 [95% CI = 255.1 to 490.4]). The multivariable-adjusted hazard ratio of developing HCC was 1.76 (95% CI = 1.19 to 2.61) for genotype C vs genotype B, 0.34 (95% CI = 0.21 to 0.57) for precore G1896A vs wild type, and 1.73 (95% CI = 1.13 to 2.67) for BCP A1762T/G1764A vs wild type. Risk was highest among participants infected with genotype C HBV and wild type for the precore 1896 variant and mutant for the BCP 1762/1764 variant (adjusted hazard ratio = 2.99, 95% CI = 1.57 to 5.70, P < .001).
Conclusions
HBV genotype C and specific alleles of BCP and precore were associated with risk of HCC. These associations were independent of serum HBV DNA level.
CONTEXT AND CAVEATS
Prior knowledge
The risk of hepatocellular carcinoma (HCC) increases with increasing level of hepatitis B virus (HBV) in serum. However, it is unclear whether HBV genotype and common variants in the precore and basal core promoter (BCP) regions contribute to the risk of HCC.
Study design
The incidence of HCC in association with HBV genotype and mutants was estimated for participants in a community-based cohort study with long-term follow-up.
Contribution
HBV genotype C and the BCP A1762T/G1764A mutant were independent risk factors for HCC, and the precore G1896A mutant was associated with a decreased risk of HCC.
Implications
HBV genotype and precore and BCP mutation status, in addition to other factors, including age, sex, and HBV viral load, may help identify those who are at an increased risk for liver disease progression.
Limitations
The analysis of HBV genotype and mutants was based on a single blood sample obtained at study entry. Uncommon mutations were detected but not analyzed because of small sample sizes.
From the Editors
Worldwide, more than 2 billion people have been infected with hepatitis B virus (HBV) and approximately 350 million remain chronically infected (1). Individuals with chronic HBV infection are at increased risk of developing end-stage liver disease (including cirrhosis, hepatic failure, and hepatocellular carcinoma [HCC]), with a cumulative lifetime incidence of 15%–40% (2,3). Other important risk factors for HCC include the presence of hepatitis B virus e antigen (HBeAg)—a surrogate marker of active viral replication—and the amount of HBV (ie, viral load) in serum (4,5). Several reports (6–9) have also suggested that the genetic characteristics of HBV, including HBV genotype and specific genetic mutations, are associated with the development of HCC.
Eight HBV genotypes (designated A through H) have been identified, and most display a distinct geographic distribution (10,11). Genotypes B and C are the predominant HBV genotypes in Eastern Asia, including Taiwan (7). Clinically important differences in outcomes are associated with the different HBV genotypes. HBV genotype C infection has been associated with later occurring and lower rates of spontaneous clearance of HBeAg in serum compared with HBV genotype B infection (12–15). In addition, genotype C is associated with higher levels of HBV DNA replication, more advanced liver disease, and a decreased rate of response to α-interferon therapy compared with genotype B (7,9,12,13,15–17).
HBV is a DNA virus that contains four overlapping open reading frames that encode the surface (viral envelope), core (nucleocapsid), X (a nonstructural protein that operates as a multifunctional regulator modulating gene transcription, cell responses to genotoxic stress, protein degradation, apoptosis, and several signaling pathways), and polymerase proteins. The basal core promoter (BCP), which is located in the overlapping X open reading frame, controls transcription of both the precore and core regions. The precore region encodes the precore protein, which is processed in the endoplasmic reticulum to produce secreted HBeAg. HBV can evolve distinct variants that tend to cluster in the precore and BCP regions (18,19). Mutations in the precore region, which were the first major common variants of HBV to be discovered (20), are first detectable around the time of HBeAg seroconversion and include mutations that prevent HBeAg synthesis without interfering with the production of infectious virions. The most common of these mutations is a G to A substitution at nucleotide 1896 (G1896A), which prevents the production of HBeAg by introducing a premature stop codon into the open reading frame of the precore region. This mutation (hereafter referred to as precore G1869A) is frequently detected in patients with HBeAg-negative chronic hepatitis B and in some patients with fulminant hepatitis B (21–23).
The other common class of HBV variants includes point mutations in the BCP, which overlaps with the open reading frame for gene X and regulates expression of both HBeAg and the core protein (24). The most frequent BCP mutation is a double mutation involving an A to T substitution at nucleotide 1762 and a G to A substitution at nucleotide 1764 (hereafter referred to as BCP A1762T/G1764A), which results in a substantial decrease in HBeAg expression but enhanced viral genome replication in vitro (25).
Although many cross-sectional and case–control studies (6,8,9,17,26–29) have suggested that HBV genotype and the precore and BCP mutants have a substantial impact on the progression of chronic hepatitis B, none has, to our knowledge, examined the prevalence of these variants in the general population of HBV hyperendemic areas. In addition, the incidence of HCC in association with HBV genotype and mutants has yet to be estimated in a community-based study with long-term follow-up. Our aim was to examine the prevalence of HBV genotype and precore and BCP mutants and their association with HCC risk after adjusting for well-known host and viral risk factors.
Participants and Methods
Cohort Enrollment and Subject Flow
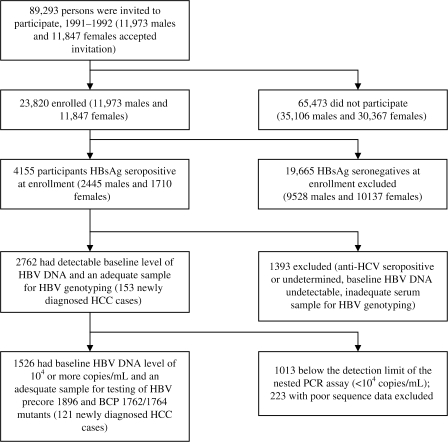
This study included participants in the established REVEAL-HBV cohort study, a community-based prospective cohort study for which the goal is to investigate the association between viral factors and the risk of liver diseases, including HCC. The enrollment of the study cohort has been described previously (4). Briefly, as shown in Figure 1, all 89 293 men and women between the ages of 30 and 65 years who were living in seven townships in Taiwan were invited by mail and telephone to participate in the study. A total of 23 820 residents (27%) agreed to participate and provided written informed consent for us to conduct interviews, health examinations, and blood collections at study entry and follow-up. We carried out laboratory tests, including tests for virological and serological biomarkers; medical record review; and computerized data linkage to national health profiles for each participant. All 23 820 participants were free of HCC at study entry as confirmed by baseline health examinations (abdominal ultrasonography and alpha-fetoprotein testing) and examination of records at the Taiwan National Cancer Registry. Hepatitis B surface antigen (HBsAg)–positive participants (n = 4155) were scheduled for follow-up every 6–12 months with abdominal ultrasonography and a medical examination. After excluding participants with antibodies against hepatitis C virus (anti-HCV, n = 191), unknown anti-HCV antibody status (n = 4), undetectable baseline serum HBV DNA levels (n = 882), or inadequate blood sample for HBV genotyping (n = 316), a total of 2762 participants (all with 100 copies or more of HBV DNA per milliliter) were included in this analysis. Of these, only those with 104 copies or more of HBV DNA per milliliter, the detection limit of the nested polymerase chain reaction (PCR) assay for mutant types (n = 1526), were tested for the precore 1896 and BCP 1762/1764 mutations. This study was approved by the Institutional Review Board of the College of Public Health, National Taiwan University in Taipei, Taiwan.
Figure 1.
Flow of participants in the study. HBsAg = hepatitis B surface antigen; HBV = hepatitis B virus; anti-HCV = antibodies against hepatitis C virus; BCP = basal core promoter; HCC = hepatocellular carcinoma; PCR = polymerase chain reaction.
Interview and Blood Collection
All of the participants were interviewed in person at enrollment by public health nurses who used a structured questionnaire to obtain information on sociodemographic characteristics, dietary habits, histories of cigarette smoking and alcohol consumption, personal medical and surgical history, family history of cancers and other major diseases, and gynecologic information (for female subjects). A 10-mL blood sample was collected from each participant at study entry and at each follow-up examination (every 6 to 12 months, maximum of 23 follow-ups). The samples were fractionated on the day of collection into several vials of plasma and buffy coat and the fractions were stored at −70°C.
Laboratory Tests
We used commercially available kits (all from Abbott Laboratories, North Chicago, IL) to test serum samples for HBsAg, HBeAg, and alpha-fetoprotein by radioimmunoassay and for anti-HCV by enzyme immunoassay. Serum alanine aminotransferase (ALT) levels—a marker of liver inflammation—were measured with the use of a serum chemistry autoanalyzer (model 736, Hitachi, Tokyo, Japan) and commercially available reagents (Biomérieux, Marcy L'Etoile, France).
HBV DNA was extracted from 200 mL of each plasma sample with the use of a High Pure Viral Nucleic Acid Kit (Roche Diagnostics Applied Science, Mannheim, Germany) as per the manufacturer's instructions. The viral titer and genotype of HBV were determined by a real-time PCR-based method that used fluorescent hybridization probes and a LightCycler PCR machine (Roche Diagnostics Applied Science). The method consisted of two steps that were carried out in a single tube: the first step used real-time PCR to quantify the viral DNA and the second step used melting curve analysis of the final PCR product to genotype the virus. Details of the design and experimental conditions of this assay have been published (30). The sequences for the primer set used for amplicon amplification were 5′-CCGATCCATACTGCGGAAC-3′ (forward) and 5′-GCAGAGGTGAAGCGAAGTGCA-3′ (reverse). The sequences for the probes used for generating the fluorescent signals were fluorescein–5′-TCTGTGCCTTCTCATCTGCCGGACC-3′-P (anchor probe, with its 3′ end phosphorylated to prevent probe elongation by Taq polymerase during PCR) and 5′-TCTTTACGCGGACTCCCC-LC-Red640-3′ (sensor probe) (TIB MOLBIOL, Berlin, Germany). This assay showed a broad linear distribution for HBV titers that ranged from 102 to 1011 copies/mL and a lower detection limit of 1–5 × 102 copies/mL (30). The results of this assay were strongly correlated (P < .001 based on linear regression analysis) with the results of three commercial assays for HBV titer: SuperQuant assay (National Genetics Institute, Culver City, CA; R2 = .9866), the Amplicor assay (Roche Diagnostics, Indianapolis, IN; R2 = .983), and the bDNA assay (Chiron Corporation, Emeryville, CA; R2 = .999). For genotype determination by melting curve analysis, different genotypes of HBV showed distinct melting temperature (Tm) values, that is, 59.1–62.7°C for genotype B and 53.0–56.6°C for genotype C (30). Our genotyping results yielded an inconsistency rate of less than 1% when compared with the gold standard method of direct sequencing and phylogenetic analysis (30). Samples that gave equivocal genotyping results by the melting curve analysis were subjected to DNA sequence analysis of the pre-S region of the HBV genome with the use of ABI PRISM BigDye sequencing kits and an ABI 3100 Genetic Analyzer (all from Applied Biosystems, Foster City, CA), according to the manufacturer's instructions as previously described (30), and the sequence was then processed for phylogenetic analysis for genotype determination.
Precore 1896 and BCP 1762/1764 mutations were assayed using direct DNA sequence analysis. PCR amplification was first conducted with the use of a T3 Thermocycler (Biometra, Whatman Corp., Gottingen, Germany) in a 15-mL reaction volume that contained either 4 mL of extracted serum DNA (for the first PCR) or 1 mL of a 20-fold dilution of the first PCR product (for the nested PCR) as the template DNA, 30 ng of each primer, and PCR buffer (20 mM Tris–HCl [pH 8.4], 50 mM KCl, 1.66 mM MgCl2, 200 mM of each dNTP, and 0.4 U Platinum Taq polymerase) (Invitrogen, Carlsbad, CA). The nested primer sets used for the PCR amplification were designed to flank the BCP and precore region of HBV genome: 5′-ACTCTTGGACTYTCAGCAATG-3′ (Y = C or T, forward primer) and 5′-GTCAGAAGGCAAAAAAGAGAG-3′ (reverse primer) for the first PCR reaction, and 5′-TCTCAGCAATGTCAACGACCG-3′ (forward primer) and 5′-AGAGAGTAACTCCACAGAWGCTC-3′ (W = A or T, reverse primer) for the nested PCR reaction. The thermal cycler conditions consisted of 5 minutes at 95°C, followed by 35 cycles of 95°C for 45 seconds, 60°C for 45 seconds, and 72°C for 30 seconds, followed by a final extension at 72°C for 10 minutes. The integrity of the final PCR products was checked by agarose gel electrophoresis, and the PCR products were then used as template for subsequent direct sequencing analysis as described above.
Follow-up and Ascertainment of HCC
Incident HCC cases were detected by follow-up health examinations, which included abdominal ultrasonography and serological tests for ALT and alpha-fetoprotein, and by computerized data linkage with profiles of the National Cancer Registry in Taiwan from January 1, 1991, through June 30, 2004. Data linkage with the profiles on the national death certification system was also performed to identify cases not registered in the cancer registry system. During 33 747 person-years of follow-up, 153 incident HCC cases were ascertained using the following criteria among the 2762 subjects who qualified for inclusion in this analysis set: a histopathologic examination (64 cases), a positive lesion detected by at least two different imaging techniques (abdominal ultrasonography, angiogram, and/or computed tomography, 81 cases), or a positive lesion by one imaging technique and a serum alpha-fetoprotein concentration of 400 ng/mL or higher (32 cases). Twenty-four cases were ascertained based on more than one criterion.
Statistical Analysis
Trends in the age-specific prevalence of HBV genotype C and precore G1896A and BCP A1762T/G1764A mutants were evaluated using the Cochran–Armitage trend test. Incidence rates of HCC by genotype and precore and BCP mutation status were calculated by dividing the number of incident HCC cases by the person-years of follow-up. The person-years of follow-up for each study subject were determined from the date of recruitment to the date of HCC detection, the date at death, or the last date of linked data available from the National Cancer Registry (up to June 30, 2004), whichever came first. Study subjects who were free of HCC at death or at the end of follow-up were censored.
A Cox proportional hazards model was used to estimate the risk of HCC associated with HBV genotype and precore and BCP mutants after adjustment for other risk factors for HCC, including sex, age at recruitment, cigarette smoking status, alcohol consumption status, serum levels of HBV DNA and ALT, and liver cirrhosis at enrollment. A 10-year interval and a base-10 logarithm were used for the cut point of age at recruitment and HBV DNA level, respectively, for ease of clinical application. The cut points for serum ALT level were chosen on the basis of our previous analysis using a trajectory model (31) that classified long-term ALT changes of participants into three groups. The trajectory model was used to identify subgroups in the population that have similar patterns of changes of serum ALT levels over time (32). Multivariable-adjusted hazard ratios (HRadjs) and 95% confidence intervals (CIs) were derived for each risk factor. The assumption of proportionality for the Cox analysis was tested by examining the interaction between the year of follow-up with HBV genotype and mutants, and no violation of this assumption was observed. The hazard ratio of HCC by HBV DNA and ALT levels was further investigated in subgroup analyses by using a Cox proportional hazards model with stratification according to HBV genotypes. The combined effects of HBV genotype and precore 1896 and BCP 1762/1764 mutants on the risk of HCC were further investigated using a Cox proportional hazards regression model. Statistical significance level was defined as P < .05 by two-tailed tests. SAS software (version 8.01; SAS Institute Inc., Cary, NC) was used for all analyses.
Results
Characteristics of the Study Cohort
As shown in Table 1, the majority of the study participants were male; 68.8% of study participants did not smoke cigarettes and 88.6% did not consume alcohol. Most participants were HBeAg seronegative, had very low serum ALT levels (<15 U/L), had elevated serum HBV DNA (≥104 copies/mL), and had no evidence of liver cirrhosis by ultrasonography.
Table 1.
Baseline characteristics of the study cohort (N = 2762)*
| Variable | No. (%) |
| Sex | |
| Female | 1187 (43.0) |
| Male | 1575 (57.0) |
| Age, y | |
| 30–39 | 927 (33.6) |
| 40–49 | 785 (28.4) |
| 50–59 | 798 (28.9) |
| 60–65 | 252 (9.1) |
| Cigarette smoker† | |
| No | 1898 (68.8) |
| Yes | 862 (31.2) |
| Alcohol drinker‡ | |
| No | 2442 (88.6) |
| Yes | 314 (11.4) |
| Serum ALT level, U/L | |
| <15 | 1659 (60.1) |
| 15–44 | 933 (33.8) |
| ≥45 | 170 (6.2) |
| HBeAg serostatus | |
| Negative | 2235 (80.9) |
| Positive | 527 (19.1) |
| HBV DNA level, copies/mL | |
| 100–9999 | 1013 (36.7) |
| 10 000–99 999 | 746 (27.0) |
| 100 000–999 999 | 380 (13.8) |
| ≥106 | 623 (22.6) |
| Liver cirrhosis at entry | |
| No | 2692 (97.5) |
| Yes | 70 (2.5) |
Some percentages do not total 100 because of rounding. ALT = alanine aminotransferase; HBeAg = hepatitis B virus e antigen; HBV = hepatitis B virus.
Data not available for two subjects.
Data not available for six subjects.
Prevalence of HBV Genotype and Mutants
Overall, 1773 subjects (64.2%) were infected with HBV genotype B alone, 889 (32.2%) were infected with genotype C alone, and 100 (3.6%) were infected with both genotypes (Table 2). The precore and BCP mutations were determined only in subjects whose baseline HBV DNA level was 104 copies/mL or higher (n = 1526). As shown in Table 2, the most common precore variant was the 1896A mutation (47.0%), and the most common BCP 1762/1764 variant was the double wild type (ie, A1762/G1764) (57.4%). Several less common BCP 1762/1764 variants were also observed, each of which had a prevalence of less than 6%. During follow-up, HCC developed in 6 (22.2%) of 27 subjects with the A1762/A1764 variant, 8 (9.8%) of 82 subjects with the H1762/H1764 variant, and 6 (7.7%) of 78 subjects with the D1762/D1764 variant.
Table 2.
Prevalence of hepatitis B virus genotype and precore 1896 and BCP 1762/1764 mutants in the study cohort*
| HBV characteristic | No. (%) |
| Genotype | |
| B | 1773 (64.2) |
| B and C | 100 (3.6) |
| C | 889 (32.2) |
| Precore 1896† | |
| G (wild type) | 641 (42.0) |
| A (predominant mutant) | 717 (47.0) |
| G and A (mixed type) | 168 (11.0) |
| BCP (1762/1764)†,‡ | |
| A/G (wild type/wild type) | 876 (57.4) |
| T/A (predominant mutant/predominant mutant) | 436 (28.6) |
| H/A | 6 (0.4) |
| H/H | 82 (5.4) |
| A/A | 27 (1.8) |
| A/H | 11 (0.7) |
| A/T | 1 (0.1) |
| H/G | 3 (0.2) |
| G/A | 2 (0.2) |
| T/G | 2 (0.1) |
| H/C | 1 (0.1) |
| G/H | 1 (0.1) |
| D/D | 78 (5.1) |
Some percentages do not total 100 because of rounding. HBV = hepatitis B virus; BCP = basal core promoter.
Among subjects with HBV DNA levels of at least 104 copies/mL (n = 1526).
H = both wild type and predominant mutants were detectable; D = deletion. Categories other than wild type and predominant mutant were classed as mixed type.
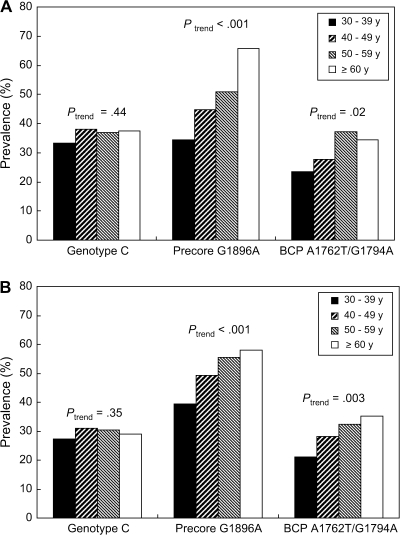
As shown in Figure 2, the prevalence of HBV genotype C remained constant across all four age groups in both men (Ptrend = .44) and women (Ptrend = .35). A higher proportion of women than men were infected with HBV genotype C (36.1% vs 29.3%; P < .001). The prevalence of the precore G1869A mutant increased with age in both males and females (Ptrend < .001 for both). The prevalence of the BCP A1762T/G1764A double mutant also increased with age (Ptrend = .02 for females and Ptrend = .003 for males).
Figure 2.
Prevalence of hepatitis B virus genotype and precore G1896A and BCP A1762T/G1764A mutants by age at recruitment. A) Female participants. B) Male participants. BCP = basal core promoter. Ptrend (two-sided) values are from the Cochran–Armitage test.
The prevalence of the precore G1896A mutant was higher in participants infected with HBV genotype B than in those infected with HBV genotype C (57.5% vs 25.8%; P < .001). The prevalence of the BCP A1762T/G1764A double mutant was higher in participants infected with HBV genotype C than in those infected with HBV genotype B (43.0% vs 21.4%; P < .001). Participants with genotype C infection had a higher viral load than those with genotype B infection. The proportion of participants with a plasma HBV DNA level of at least 106 copies/mL was 17.6% for those with genotype B infection compared with 33.8% for those with genotype C infection (P < .001).
HCC Incidence Rates by HBV Genotype and Precore and BCP Mutations
Among participants with a detectable baseline level of HBV DNA, the HCC incidence rates per 100 000 person-years for genotypes B and C were 305.6 (95% CI = 236.9 to 388.1) and 785.8 (95% CI = 626.8 to 972.9), respectively (Table 3). Among participants with a baseline HBV DNA level of at least 104 copies/mL, those with the precore wild-type G1896 variant had a higher incidence of HCC than those with the G1896A mutant variant (955.5 [95% CI = 749.0 to 1201.4] vs 269.4 [95% CI = 172.6 to 400.9] per 100 000 person-years), and those with the BCP A1762T/G1764A double mutant had a higher incidence of HCC than those with BCP A1762/G1764 wild type (1149.2 [95% CI = 872.6 to 1485.6] vs 358.7 [95% CI = 255.1 to 490.4] per 100 000 person-years).
Table 3.
Incidence rate of HCC by hepatitis B virus genotype and precore and BCP mutants*
| Genotype or mutant | No. of cohort members | Person-years of follow-up | No. of HCC cases | Incidence rate per 100 000 person-years (95% CI) |
| HBV genotype | ||||
| B | 1773 | 21 921.9 | 67 | 305.6 (236.9 to 388.1) |
| B and C | 100 | 1235.0 | 2 | 161.9 (19.6 to 585.0) |
| C | 889 | 10 689.7 | 84 | 785.8 (626.8 to 972.9) |
| Precore 1896† | ||||
| Wild type | 641 | 7639.8 | 73 | 955.5 (749.0 to 1201.4) |
| G1896A | 717 | 8908.4 | 24 | 269.4 (172.6 to 400.9) |
| Mixed type | 168 | 1943.4 | 24 | 1234.9 (791.3 to 1837.5) |
| BCP 1762/1764† | ||||
| Wild type | 876 | 10872.3 | 39 | 358.7 (255.1 to 490.4) |
| A1762T/G1764A | 436 | 5047.0 | 58 | 1149.2 (872.6 to 1485.6) |
| Mixed type | 214 | 2572.2 | 24 | 933.1 (597.8 to 1388.3) |
HCC = hepatocellular carcinoma; CI = confidence interval; HBV = hepatitis B virus; BCP = basal core promoter.
Among subjects with HBV DNA levels of at least 104 copies/mL (n = 1526).
HCC Risk Adjusted for Other Covariates
Overall, infection with genotype C was associated with a higher risk of HCC than infection with genotype B, with an adjusted hazard ratio of 2.35 (95% CI = 1.68 to 3.30; P < .001) (Table 4). Among participants with a baseline HBV DNA level of 104 copies/mL or greater, genotype C infection was also associated with a higher risk of HCC, with an adjusted hazard ratio of 1.76 (95% CI = 1.19 to 2.61; P = .005). The adjusted hazard ratio of HCC for the BCP A1762T/G1764A mutation vs wild type was 1.73 (95% CI = 1.13 to 2.67; P = .013). By contrast, the precore G1896A mutation was associated with a lower risk of HCC than the precore wild type (HRadj = 0.34, 95% CI = 0.21 to 0.57; P < .001).
Table 4.
Cox proportional hazards regression analysis of HCC predictors*
| All participants (N = 2762) |
Participants with 104 copies or more HBV DNA per milliliter at entry (n = 1526) |
|||||||
| Variable | Crude HR (95% CI) | P† | Adjusted HR (95% CI)‡ | P† | Crude HR (95% CI) | P† | Adjusted HR (95% CI)§ | P† |
| Sex | ||||||||
| Female | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | ||||
| Male | 3.38 (2.26 to 5.07) | <.001 | 2.25 (1.41 to 3.60) | <.001 | 3.86 (2.43 to 6.13) | <.001 | 2.93 (1.72 to 5.01) | <.001 |
| Age, y | ||||||||
| 30–39 | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | ||||
| 40–49 | 3.93 (2.16 to 7.15) | <.001 | 4.23 (2.31 to 7.74) | <.001 | 4.10 (2.09 to 8.05) | <.001 | 4.73 (2.39 to 9.36) | <.001 |
| 50–59 | 5.81 (3.26 to 10.34) | <.001 | 6.46 (3.60 to 11.60) | <.001 | 6.07 (3.17 to 11.62) | <.001 | 6.74 (3.46 to 13.13) | <.001 |
| ≥60 | 8.89 (4.70 to 16.83) | <.001 | 9.45 (4.94 to 18.08) | <.001 | 8.53 (4.11 to 17.70) | <.001 | 10.81 (5.13 to 22.80) | <.001 |
| Cigarette smoker | ||||||||
| No | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | ||||
| Yes | 1.86 (1.35 to 2.56) | <.001 | 1.16 (0.80 to 1.68) | .44 | 1.97 (1.37 to 2.81) | <.001 | 1.05 (0.69 to 1.59) | .83 |
| Alcohol drinker | ||||||||
| No | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | ||||
| Yes | 2.77 (1.92 to 4.00) | <.001 | 1.51 (1.01 to 2.26) | .044 | 2.94 (1.95 to 4.43) | <.001 | 1.30 (0.83 to 2.05) | .25 |
| Serum ALT level, U/L | ||||||||
| <15 | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | ||||
| 15–44 | 3.30 (2.29 to 4.75) | <.001 | 1.95 (1.33 to 2.86) | <.001 | 2.95 (1.93 to 4.49) | <.001 | 1.82 (1.16 to 2.84) | .009 |
| ≥45 | 6.63 (4.11 to 10.68) | <.001 | 1.83 (1.07 to 3.11) | .026 | 5.23 (3.11 to 8.80) | <.001 | 1.96 (1.10 to 3.50) | .023 |
| Cirrhosis at study entry | ||||||||
| No | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | ||||
| Yes | 19.22 (13.15 to 28.11) | <.001 | 7.52 (4.97 to 11.39) | <.001 | 12.61 (8.05 to 19.77) | <.001 | 4.49 (2.69 to 7.49) | <.001 |
| HBV DNA level, copies/mL | ||||||||
| 100–9999 | 1.00 (referent) | 1.00 (referent) | – | – | ||||
| 10 000–99 999 | 1.54 (0.88 to 2.69) | .13 | 1.76 (1.00 to 3.12) | .05 | 1.00 (referent) | 1.00 (referent) | ||
| 100 000–999 999 | 4.04 (2.38 to 6.86) | <.001 | 3.44 (1.99 to 5.94) | <.001 | 2.40 (1.39 to 4.13) | .002 | 1.61 (0.91 to 2.84) | .10 |
| ≥106 | 5.28 (3.29 to 8.45) | <.001 | 4.42 (2.71 to 7.20) | <.001 | 3.00 (1.84 to 4.90) | <.001 | 1.50 (0.88 to 2.58) | .14 |
| HBV genotype | ||||||||
| B | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | ||||
| C | 2.59 (1.88 to 3.57) | <.001 | 2.35 (1.68 to 3.30) | <.001 | 2.63 (1.83 to 3.77) | <.001 | 1.76 (1.19 to 2.61) | .005 |
| B and C | 0.53 (0.13 to 2.16) | .38 | 0.65 (0.16 to 2.66) | .55 | 0.35 (0.05 to 2.55) | .30 | 0.33 (0.04 to 2.40) | .27 |
| Precore 1896 | ||||||||
| Wild type | 1.00 (referent) | 1.00 (referent) | ||||||
| G1896A | ‖ | ‖ | 0.28 (0.18 to 0.44) | <.001 | 0.34 (0.21 to 0.57) | <.001 | ||
| Mixed type | 1.30 (0.82 to 2.06) | .27 | 1.18 (0.71 to 1.96) | .52 | ||||
| BCP 1764/1764 | ||||||||
| Wild type | 1.00 (referent) | 1.00 (referent) | ||||||
| A1762T/G1764A | ‖ | ‖ | 3.22 (2.14 to 4.83) | <.001 | 1.73 (1.13 to 2.67) | .013 | ||
| Mixed type | 2.61 (1.57 to 4.34) | <.001 | 1.04 (0.59 to 1.84) | .88 | ||||
P values (two-sided) were from Cox proportional hazards models. HCC = hepatocellular carcinoma; HBV = hepatitis B virus; HR = hazard ratio; CI = confidence interval; ALT = alanine aminotransferase; – = not applicable; BCP = basal core promoter.
P values (two-sided) were from Cox proportional hazards models.
Multivariable-adjusted model controlled for the following variables: sex, age, cigarette smoking, alcohol drinking, serum ALT level, cirrhosis at study entry, HBV DNA level, and HBV genotype.
Multivariable-adjusted model for sex, age, cigarette smoking, alcohol drinking, serum ALT level, cirrhosis at study entry, HBV DNA level, HBV genotype, and precore and BCP mutants.
Not included due to lack of data on HBV mutants for subjects with entry HBV DNA levels of less than 104 copies/mL.
Risk of HCC According to HBV DNA and Serum ALT Levels and Stratified by HBV Genotype
Among participants infected with HBV genotype B, the hazard ratio of developing HCC was 1.48 (95% CI = 0.67 to 3.26) for those with DNA levels of 10 000–99 999 vs 100–9999 copies/mL, 2.81 (95% CI = 1.27 to 6.19) for those with 100 000–999 999 vs 100–9999 copies/mL, and 4.63 (95% CI = 2.17 to 9.87) for those with 106 copies/mL or more vs 100–9999 copies/mL. Among participants infected with HBV genotype C, the corresponding hazard ratios were 2.46 (95% CI = 1.06 to 5.72), 4.71 (95% CI = 2.12 to 10.46), and 5.19 (95% CI = 2.62 to 10.31), respectively. There was no statistically significant interaction between HBV genotype and baseline HBV DNA level (P = .51).
Among participants infected with HBV genotype B, the hazard ratios of HCC for those with baseline ALT levels of 15–44 U/L or 45 U/L or more vs those with ALT levels less than 15 U/L were 1.42 (95% CI = 0.83 to 2.45) and 1.00 (95% CI = 0.40 to 2.52), respectively. Among participants infected with HBV genotype C, the corresponding hazard ratios were 2.61 (95% CI = 1.46 to 4.66) and 3.04 (95% CI = 1.48 to 6.22), respectively. There was no statistically significant interaction between HBV genotype and baseline ALT level (P = .21).
Incidence and Risk of HCC for Combinations of HBV Genotype and Mutants
Finally, we examined the risk of HCC associated with combinations of HBV genotype, the precore G1896A mutant, and the BCP double mutants. Participants who were infected with HBV genotype C, had the wild-type precore 1896 variant, and had the BCP A1762T/G1764A double mutation had the highest incidence of HCC (2254.2 per 100 000 person-years), with an adjusted hazard ratio of 2.99 (95% CI = 1.57 to 5.70, P < .001) relative to participants who were infected with HBV genotype B and wild type for the precore 1896 and BCP 1762/1764 variants (Table 5). By contrast, participants who were infected with HBV genotype B, had the precore G1896A variant, and had the wild-type BCP 1762/1764 variant had the lowest incidence of HCC (173.9 per 100 000 person-years), with an adjusted hazard ratio of 0.20 (95% CI = 0.09 to 0.46, P < .001) relative to participants who were infected with HBV genotype B and wild type for the precore 1896 and BCP 1762/1764 variants.
Table 5.
Incidence rates and hazard ratios of HCC for the combination of HBV genotype and precore and BCP variants among the 1526 subjects with at least 104 copies/mL HBV DNA*
| HBV genotype | Precore 1896 variant | BCP 1762/1764 variant | No. of participants | Person-years of follow-up | No. of HCC cases | HCC incidence (per 100 000 person-years) | Adjusted HR† (95% CI) | P‡ |
| B | Wild type | Wild type | 198 | 2404.8 | 14 | 582.2 | 1.0 (referent) | |
| B | Wild type | A1762T/G1764A | 82 | 950.1 | 9 | 947.3 | 1.35 (0.58 to 3.12) | .48 |
| B | G1896A | Wild type | 413 | 5175.0 | 9 | 173.9 | 0.20 (0.09 to 0.46)† | <.001 |
| B | G1896A | A1762T/G1764A | 110 | 1319.9 | 7 | 530.3 | 0.62 (0.25 to 1.54) | .30 |
| C | Wild type | Wild type | 129 | 1633.2 | 6 | 367.4 | 0.73 (0.28 to 1.89) | .51 |
| C | Wild type | A1762T/G1764A | 115 | 1242.1 | 28 | 2254.2 | 2.99 (1.57 to 5.70)† | <.001 |
| C | G1896A | Wild type | 41 | 497.3 | 4 | 804.3 | 1.38 (0.45 to 4.20) | .57 |
| C | G1896A | A1762T/G1764A | 73 | 911.2 | 2 | 219.5 | 0.36 (0.08 to 1.59) | .18 |
HCC = hepatocellular carcinoma; HBV = hepatitis B virus; BCP = basal core promoter; HR = hazard ratio; CI = confidence interval.
Adjusted for sex and age at recruitment.
P values (two-sided) were from Cox proportional hazards models.
Discussion
The role of HBV genotype and mutants in disease progression continues to be of great interest. Unlike in hepatitis C virus (HCV) infection, the relationship between different HBV genotypes and risk of future disease is not well established. In this analysis of a subset of the REVEAL-HBV study cohort, we found that the majority of participants (64.2%) were infected with genotype B virus alone, consistent with other studies from Taiwan (7,9). Among participants with baseline HBV DNA levels of 104 copies or more per milliliter, the prevalences of precore and BCP typical mutants were 47.0% and 28.6%, respectively, at study entry and increased with age. As has been described by other studies (mostly from Asia), we also found that HBV genotype C infection was associated with a higher risk of HCC than HBV genotype B infection; in addition, we found that the BCP A1762T/G1764A mutant was associated with an increased risk of HCC compared with the double wild-type variant, whereas the precore G1896A mutation was associated with a decreased risk of HCC compared with the wild-type variant.
Our finding that the precore G1896A mutation was associated with a lower risk of HCC compared with wild-type virus appears to be at odds with several early case reports that found an association between the precore stop mutation and a marked increase in viral virulence and the development of fulminant hepatitis (21,22). However, another study (33)—in HBeAg-negative, HBV-infected persons—found that infection with HBV containing wild-type precore was associated with more hepatic inflammation and fibrosis than infection with precore mutant HBV. Also, severe clinical manifestations of chronic hepatitis in patients with abrupt elevation of serum ALT level (>200 U/L) are more frequently observed in those with the precore wild type than G1896A mutation (34). The negative association between the precore G1896A mutant and HCC that we observed in this study after adjustment for sex, age, cigarette smoking status, alcohol consumption status, serum ALT level, and cirrhosis at study entry, as well as HBV genotype, viral load, and BCP mutant, suggested that the precore G1896A mutation was associated with a low risk of HCC.
Several mechanisms of hepatocarcinogenesis relating to the BCP A1762T/G1764A mutant have been hypothesized. It has been proposed that the BCP A1762T/G1764A mutation may enhance HBV virulence by increasing host immune response (25,35,36), increasing viral replication (37–42), or altering the coding region for the X antigen (6,18,43). Circulating HBeAg may interfere with the clearance of infected hepatocytes and promote immune tolerance (25,36). By diminishing circulating HBeAg, mutant BCP may augment the host immune response to HBV-infected hepatocytes, hence increasing hepatocyte apoptosis and regeneration, which leads to liver injury (25,36). The BCP mutation appears to enhance the efficiency of viral replication either by modulating the relative levels of the precore and core RNAs or by creating a hepatocyte nuclear factor 1 transcription factor binding site (42). The X antigen of HBV has potential transactivation activity for both viral and host genes, and it has been shown to interact directly with p53 and the DNA repair enzyme XAP-1 (44). Mutations in the BCP region, which overlaps the coding sequence for the X antigen of HBV, may result in amino acid changes K130M and V131I in the X antigen. These amino acid changes may interfere with cell growth control and DNA repair and may cause HCC (44,45). In a multivariable Cox regression analysis, we found that the BCP A1762T/G1764A mutation was associated with an increased risk of HCC independent of viral load and other risk factors. Therefore, other mechanisms of hepatocarcinogenesis, such as the alteration of transactivation capability of X antigen induced by amino acid changes related to the BCP A1762T/G1764A mutation, should be considered in future studies.
The association between genotype C infection and a higher disease progression and HCC risk has been reported previously (9,16,46), but the reason for this association remains poorly understood. Individuals infected with genotype C HBV might have a higher circulating viral load, as indicated by our analysis (33.8% of genotype C–infected participants had 106 copies or more of HBV DNA per milliliter of plasma compared with 17.6% of genotype B–infected participants). However, in the Cox regression analysis shown in Table 4, the hazard ratio of HCC for genotype C compared with genotype B was similar with or without adjustment for the viral load and other risk factors, suggesting that the association between genotype C and the increased risk of HCC was not confounded by HBV viral load. Several studies (8,26) have also revealed that individuals infected with HBV genotype C are more likely to develop the BCP double mutant than those infected with genotype B. It has been suggested that the BCP double mutant, rather than HBV genotype C per se, is the primary risk factor for liver cancer (8,47). However, in this study we demonstrated that both genotype C and BCP A1762T/G1764A mutation status are independent risk factors for the development of HCC. Therefore, the association between HBV genotype C and HCC risk cannot be totally explained by the appearance of BCP mutants.
One strength of this study is that all virological factors were determined on samples that were collected before the development of HCC from subjects who were not treated with any anti-HBV therapies. Our study is also unique because of the large sample size, because both men and women were enrolled, and because it was a prospective population-based study. Previous studies of HBV genotype and/or mutants and disease progression have been limited because they were hospital based and subject to selection bias, had a small sample size, included only men, or had a short follow-up.
There are also some limitations that should be considered when interpreting these data. First, our analysis of HBV genotype and mutants was based on a single blood sample obtained at study entry; thus, we could not assess the effect of changes in HBV genotype or mutation status on the development of HCC. However, the cross-sectional age-specific prevalence of these viral factors (Figure 2) suggests that HBV genotype is stable across age groups and that both the HBV precore and the BCP double mutants accumulated with increasing age. Thus, to the extent that HBV mutants emerged after study entry, the associations between each HBV mutations and the risk of HCC would be underestimated. Second, although we detected several other mutations in addition to the common ones we analyzed, small sample sizes hindered a further analysis of them. These mutants, although uncommon, may provide clues for further studies on the role of BCP mutants during HBV-associated hepatocarcinogenesis.
In conclusion, HBV genotype C and the BCP A1762T/G1764A mutant were independent risk factors for HCC, and the precore G1896A mutant was associated with a decreased risk of HCC. Consistent with our previous report (4), we found that the association between HBV DNA level and HCC was independent of other factors, including HBV genotype and precore and BCP mutation status. These genetic features of HBV, in addition to age, sex, HBV viral load, and serum ALT levels, may help identify those who are at an increased risk for liver disease progression and would therefore potentially benefit from early interventions, such as regular screening to detect disease progression, and treatment.
Funding
Department of Health, Executive Yuan, Republic of China, Academa Sinica, and the National Taiwan University Hospital, Taipei, Taiwan. The research grants were awarded to Professor C-J Chen and Professor P-J Chen.
Footnotes
Drs H-I Yang, S-H Yeh, and P-J Chen contributed equally to this article. Dr U. H. Iloeje and J. Su own stock in Bristol-Myers Squibb; Dr C-J Chen received a research grant from Bristol-Myers Squibb; Dr H-I Yang received an honorarium from Bristol-Myers Squibb; and Dr Y-F Liaw is a paid consultant for Bristol-Myers Squibb, GSK, Novartis, Roche, Schering–Plough, and SciClone and received a grant or research support from Bristol-Myers Squibb, Idenix, Novartis, Roche, SciClone, and Gilead.
The following institutions and investigators, in addition to the authors, participated in the REVEAL-HBV Study Group as a part of Taiwan Community-Based Cancer Screening Project: National Taiwan University Hospital (C. Y. Hsieh, H. S. Lee, P. M. Yang, C. H. Chen, J. D. Chen, S. P. Huang, C. F. Jan); College of Public Health, National Taiwan University (T. H. H. Chen; Department of Public Health, National Defense Medical Center-CA Sun); Taipei City Psychiatric Center (M. H. Wu); Department of Public Health, Tzu Chi University (L. Y. Wang, S. Y. Chen); Shin Kong Wu Ho-Su Memorial Hospital (K. E. Chu); Huhsi Health Center, Penghu County (S. C. Ho, T. G. Lu); Provincial Penghu Hospital (W. P. Wu, TY Ou); Sanchi Health Center, Taipei County (C. G. Lin); Provincial Chutung Hospital (K. C. Shih); Provincial Potzu Hospital (W. S. Chung, C. Li); Kaohsu Health Center, Pingtung County (C. C. Chen); and Paihsa Health Center, Penghu County (W. C. How).
We thank Dr Ann Pluta for her comments and editing of the manuscript.
The funding sources of this study had no role in the study design, data analysis, data interpretation, or in the writing, reviewing, or approving of this report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
References
- 1.Lee WM. Hepatitis B virus infection. N Engl J Med. 1997;337(24):1733–45. doi: 10.1056/NEJM199712113372406. [DOI] [PubMed] [Google Scholar]
- 2.Beasley RP. Hepatitis B virus. The major etiology of hepatocellular carcinoma. Cancer. 1988;61(10):1942–56. doi: 10.1002/1097-0142(19880515)61:10<1942::aid-cncr2820611003>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 3.McMahon BJ. Hepatocellular carcinoma and viral hepatitis. In: Wilson RA, editor. Viral Hepatitis: Diagnosis, Treatment, Prevention. New York: Marcel Dekker; 1997. pp. 315–30. [Google Scholar]
- 4.Chen CJ, Yang HI, Su J, et al. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA. 2006;295(1):65–73. doi: 10.1001/jama.295.1.65. [DOI] [PubMed] [Google Scholar]
- 5.Yang HI, Lu SN, Liaw YF, et al. Hepatitis B e antigen and the risk of hepatocellular carcinoma. N Engl J Med. 2002;347(3):168–74. doi: 10.1056/NEJMoa013215. [DOI] [PubMed] [Google Scholar]
- 6.Baptista M, Kramvis A, Kew MC. High prevalence of 1762(T) 1764(A) mutations in the basic core promoter of hepatitis B virus isolated from black Africans with hepatocellular carcinoma compared with asymptomatic carriers. Hepatology. 1999;29(3):946–53. doi: 10.1002/hep.510290336. [DOI] [PubMed] [Google Scholar]
- 7.Kao JH, Chen PJ, Lai MY, Chen DS. Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology. 2000;118(3):554–9. doi: 10.1016/s0016-5085(00)70261-7. [DOI] [PubMed] [Google Scholar]
- 8.Kao JH, Chen PJ, Lai MY, Chen DS. Basal core promoter mutations of hepatitis B virus increase the risk of hepatocellular carcinoma in hepatitis B carriers. Gastroenterology. 2003;124(2):327–34. doi: 10.1053/gast.2003.50053. [DOI] [PubMed] [Google Scholar]
- 9.Yu MW, Yeh SH, Chen PJ, et al. Hepatitis B virus genotype and DNA level and hepatocellular carcinoma: a prospective study in men. J Natl Cancer Inst. 2005;97(4):265–72. doi: 10.1093/jnci/dji043. [DOI] [PubMed] [Google Scholar]
- 10.Norder H, Hammas B, Lofdahl S, Courouce AM, Magnius LO. Comparison of the amino acid sequences of nine different serotypes of hepatitis B surface antigen and genomic classification of the corresponding hepatitis B virus strains. J Gen Virol. 1992;73(pt 5):1201–8. doi: 10.1099/0022-1317-73-5-1201. [DOI] [PubMed] [Google Scholar]
- 11.Arauz-Ruiz P, Norder H, Robertson BH, Magnius LO. Genotype H: a new Amerindian genotype of hepatitis B virus revealed in Central America. J Gen Virol. 2002;83(pt 8):2059–73. doi: 10.1099/0022-1317-83-8-2059. [DOI] [PubMed] [Google Scholar]
- 12.Kao JH, Chen PJ, Lai MY, Chen DS. Clinical and virological aspects of blood donors infected with hepatitis B virus genotypes B. C. J Clin Microbiol. 2002;40(1):22–5. doi: 10.1128/JCM.40.1.22-25.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orito E, Ichida T, Sakugawa H, et al. Geographic distribution of hepatitis B virus (HBV) genotype in patients with chronic HBV infection in Japan. Hepatology. 2001;34(3):590–4. doi: 10.1053/jhep.2001.27221. [DOI] [PubMed] [Google Scholar]
- 14.Chu CJ, Hussain M, Lok AS. Hepatitis B virus genotype B is associated with earlier HBeAg seroconversion compared with hepatitis B virus genotype C. Gastroenterology. 2002;122(7):1756–62. doi: 10.1053/gast.2002.33588. [DOI] [PubMed] [Google Scholar]
- 15.Sumi H, Yokosuka O, Seki N, et al. Influence of hepatitis B virus genotypes on the progression of chronic type B liver disease. Hepatology. 2003;37(1):19–26. doi: 10.1053/jhep.2003.50036. [DOI] [PubMed] [Google Scholar]
- 16.Kao JH, Chen PJ, Lai MY, Chen DS. Genotypes and clinical phenotypes of hepatitis B virus in patients with chronic hepatitis B virus infection. J Clin Microbiol. 2002;40(4):1207–9. doi: 10.1128/JCM.40.4.1207-1209.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kao JH, Wu NH, Chen PJ, Lai MY, Chen DS. Hepatitis B genotypes and the response to interferon therapy. J Hepatol. 2000;33(6):998–1002. doi: 10.1016/s0168-8278(00)80135-x. [DOI] [PubMed] [Google Scholar]
- 18.Gunther S, Fischer L, Pult I, Sterneck M, Will H. Naturally occurring variants of hepatitis B virus. Adv Virus Res. 1999;52:25–137. doi: 10.1016/s0065-3527(08)60298-5. [DOI] [PubMed] [Google Scholar]
- 19.Bartholomeusz A, Locarnini S. Hepatitis B virus mutants and fulminant hepatitis B: fitness plus phenotype. Hepatology. 2001;34(2):432–5. doi: 10.1053/jhep.2001.26764. [DOI] [PubMed] [Google Scholar]
- 20.Carman WF, Jacyna MR, Hadziyannis S, et al. Mutation preventing formation of hepatitis B e antigen in patients with chronic hepatitis B infection. Lancet. 1989;2(8663):588–91. doi: 10.1016/s0140-6736(89)90713-7. [DOI] [PubMed] [Google Scholar]
- 21.Omata M, Ehata T, Yokosuka O, Hosoda K, Ohto M. Mutations in the precore region of hepatitis B virus DNA in patients with fulminant and severe hepatitis. N Engl J Med. 1991;324(24):1699–704. doi: 10.1056/NEJM199106133242404. [DOI] [PubMed] [Google Scholar]
- 22.Liang TJ, Hasegawa K, Rimon N, Wands JR, Ben Porath E. A hepatitis B virus mutant associated with an epidemic of fulminant hepatitis. N Engl J Med. 1991;324(24):1705–9. doi: 10.1056/NEJM199106133242405. [DOI] [PubMed] [Google Scholar]
- 23.Brunetto MR, Giarin MM, Oliveri F, et al. Wild-type and e antigen-minus hepatitis B viruses and course of chronic hepatitis. Proc Natl Acad Sci U S A. 1991;88(10):4186–90. doi: 10.1073/pnas.88.10.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okamoto H, Tsuda F, Akahane Y, et al. Hepatitis B virus with mutations in the core promoter for an e antigen-negative phenotype in carriers with antibody to e antigen. J Virol. 1994;68(12):8102–10. doi: 10.1128/jvi.68.12.8102-8110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunt CM, McGill JM, Allen MI, Condreay LD. Clinical relevance of hepatitis B viral mutations. Hepatology. 2000;31(5):1037–44. doi: 10.1053/he.2000.6709. [DOI] [PubMed] [Google Scholar]
- 26.Orito E, Mizokami M, Sakugawa H, et al. A case-control study for clinical and molecular biological differences between hepatitis B viruses of genotypes B and C. Japan HBV Genotype Research Group. Hepatology. 2001;33(1):218–23. doi: 10.1053/jhep.2001.20532. [DOI] [PubMed] [Google Scholar]
- 27.Lin CL, Liao LY, Wang CS, et al. Basal core-promoter mutant of hepatitis B virus and progression of liver disease in hepatitis B e antigen-negative chronic hepatitis B. Liver Int. 2005;25(3):564–70. doi: 10.1111/j.1478-3231.2005.01041.x. [DOI] [PubMed] [Google Scholar]
- 28.Liu CJ, Chen BF, Chen PJ, et al. Role of hepatitis B viral load and basal core promoter mutation in hepatocellular carcinoma in hepatitis B carriers. J Infect Dis. 2006;193(9):1258–65. doi: 10.1086/502978. [DOI] [PubMed] [Google Scholar]
- 29.Fang ZL, Yang J, Ge X, et al. Core promoter mutations (A(1762)T and G(1764)A) and viral genotype in chronic hepatitis B and hepatocellular carcinoma in Guangxi, China. J Med Virol. 2002;68(1):33–40. doi: 10.1002/jmv.10167. [DOI] [PubMed] [Google Scholar]
- 30.Yeh SH, Tsai CY, Kao JH, et al. Quantification and genotyping of hepatitis B virus in a single reaction by real-time PCR and melting curve analysis. J Hepatol. 2004;41(4):659–66. doi: 10.1016/j.jhep.2004.06.031. [DOI] [PubMed] [Google Scholar]
- 31.Chen CF, Lee MH, Lee WC, et al. Changes in serum alanine aminotransferase (ALT) level using a trajectory model and risk of hepatocellular carcinoma (HCC) in chronic hepatitis B (CHB): the REVEAL-HBV study. Gastroenterology. 2007;132(4 suppl 2):A762. [Google Scholar]
- 32.Muthen B. Latent variable analysis: growth mixture modeling and related techniques for longitudinal data. In: Kaplan D, editor. Handbook of Quantitative Methodology for the Social Sciences. Newbury Park, CA: Sage Publications; 2004. pp. 345–68. [Google Scholar]
- 33.Lindh M, Horal P, Dhillon AP, Furuta Y, Norkrans G. Hepatitis B virus carriers without precore mutations in hepatitis B e antigen-negative stage show more severe liver damage. Hepatology. 1996;24(3):494–501. doi: 10.1002/hep.510240305. [DOI] [PubMed] [Google Scholar]
- 34.Tsai WL, Lo GH, Hsu PI, et al. Role of genotype and precore/basal core promoter mutations of hepatitis B virus in patients with chronic hepatitis B with acute exacerbation. Scand J Gastroenterol. 2008;43(2):196–201. doi: 10.1080/00365520701745693. [DOI] [PubMed] [Google Scholar]
- 35.Miyakawa Y, Okamoto H, Mayumi M. The molecular basis of hepatitis B e antigen (HBeAg)-negative infections. J Viral Hepat. 1997;4(1):1–8. doi: 10.1046/j.1365-2893.1997.00101.x. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi K, Aoyama K, Ohno N, et al. The precore/core promoter mutant (T1762A1764) of hepatitis B virus: clinical significance and an easy method for detection. J Gen Virol. 1995;76(pt 12):3159–64. doi: 10.1099/0022-1317-76-12-3159. [DOI] [PubMed] [Google Scholar]
- 37.Buckwold VE, Xu Z, Chen M, Yen TS, Ou JH. Effects of a naturally occurring mutation in the hepatitis B virus basal core promoter on precore gene expression and viral replication. J Virol. 1996;70(9):5845–51. doi: 10.1128/jvi.70.9.5845-5851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scaglioni PP, Melegari M, Wands JR. Biologic properties of hepatitis B viral genomes with mutations in the precore promoter and precore open reading frame. Virology. 1997;233(2):374–81. doi: 10.1006/viro.1997.8594. [DOI] [PubMed] [Google Scholar]
- 39.Tang H, Raney AK, McLachlan A. Replication of the wild type and a natural hepatitis B virus nucleocapsid promoter variant is differentially regulated by nuclear hormone receptors in cell culture. J Virol. 2001;75(19):8937–48. doi: 10.1128/JVI.75.19.8937-8948.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu X, Mertz JE. Distinct modes of regulation of transcription of hepatitis B virus by the nuclear receptors HNF4alpha and COUP-TF1. J Virol. 2003;77(4):2489–99. doi: 10.1128/JVI.77.4.2489-2499.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baumert TF, Rogers SA, Hasegawa K, Liang TJ. Two core promotor mutations identified in a hepatitis B virus strain associated with fulminant hepatitis result in enhanced viral replication. J Clin Invest. 1996;98(10):2268–76. doi: 10.1172/JCI119037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J, Buckwold VE, Hon MW, Ou JH. Mechanism of suppression of hepatitis B virus precore RNA transcription by a frequent double mutation. J Virol. 1999;73(2):1239–44. doi: 10.1128/jvi.73.2.1239-1244.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kidd-Ljunggren K, Oberg M, Kidd AH. The hepatitis B virus X gene: analysis of functional domain variation and gene phylogeny using multiple sequences. J Gen Virol. 1995;76(pt 9):2119–30. doi: 10.1099/0022-1317-76-9-2119. [DOI] [PubMed] [Google Scholar]
- 44.Koike K. Hepatitis B virus HBx gene and hepatocarcinogenesis. Intervirology. 1995;38(3–4):134–42. doi: 10.1159/000150424. [DOI] [PubMed] [Google Scholar]
- 45.Sirma H, Giannini C, Poussin K, Paterlini P, Kremsdorf D, Brechot C. Hepatitis B virus X mutants, present in hepatocellular carcinoma tissue abrogate both the antiproliferative and transactivation effects of HBx. Oncogene. 1999;18(34):4848–59. doi: 10.1038/sj.onc.1202867. [DOI] [PubMed] [Google Scholar]
- 46.Kao JH, Chen PJ, Lai MY, Chen DS. Hepatitis B virus genotypes and spontaneous hepatitis B e antigen seroconversion in Taiwanese hepatitis B carriers. J Med Virol. 2004;72(3):363–9. doi: 10.1002/jmv.10534. [DOI] [PubMed] [Google Scholar]
- 47.Yuen MF, Tanaka Y, Mizokami M, et al. Role of hepatitis B virus genotypes Ba and C, core promoter and precore mutations on hepatocellular carcinoma: a case control study. Carcinogenesis. 2004;25(9):1593–8. doi: 10.1093/carcin/bgh172. [DOI] [PubMed] [Google Scholar]