Abstract
The c-myc proto-oncogene is rapidly activated by serum and regulates genes involved in metabolism and cell cycle progression. This gene is thereby uniquely poised to coordinate both the metabolic and cell cycle regulatory events required for cell cycle entry. However, this function of Myc has not been evaluated. Using a rat fibroblast model of isogenic cell lines, myc−/−, myc+/−, myc+/+ and myc−/− cells with an inducible c-myc transgene (mycER), we show that the Myc protein programs cells to utilize both oxidative phosphorylation and glycolysis to drive cell cycle progression. We demonstrate this coordinate regulation of metabolic networks is essential, as specific inhibitors of these pathways block Myc-induced proliferation. Metabolic events temporally correlated with cell cycle entry include increased oxygen consumption, mitochondrial function, pyruvate and lactate production, and ATP generation. Treatment of normal cells with inhibitors of oxidative phosphorylation recapitulates the myc −/−phenotype, resulting in impaired cell cycle entry and reduced metabolism. Combined with a kinetic expression profiling analysis of genes linked to mitochondrial function, our study indicates that Myc's ability to coordinately regulate the mitochondrial metabolic network transcriptome is required for rapid cell cycle entry. This function of Myc may underlie the pervasive presence of Myc in many human cancers.
Keywords: Myc, metabolism, mitochondria, cell cycle, time-course array
Introduction
Cell cycle entry requires high metabolic activity to fuel rapid accumulation of biomass. Increased bioenergetic requirements can be provided by increased mitochondrial function, and cycle entry is associated with increased mitochondrial mass, potential and respiration (Sweet and Singh 1999; Herzig et al. 2000). The importance of mitochondrial function has been demonstrated by selective inhibition or knockout of mitochondrial complexes, which results in cell cycle arrest, prompting a proposal for metabolic cell-cycle checkpoints (Mandal et al. 2005; Liao et al. 2006). However, while it is clear that proliferation requires regulation of both cell cycle progression and mitochondrial biogenesis, no single gene has ever been tested for the ability to coordinately regulate these events.
Mitochondrial biogenesis is a complex process, as the key respiratory complex can consist of over 41 different protein subunits derived from both mitochondrial and nuclear genomes (Poyton and McEwen 1996). Hence, the task of coordinately regulating the translation, import and assembly of these complexes requires signaling networks that respond to both extrinsic and intrinsic signals. Known regulators include the nuclear transcription factors NRF-1 and NRF-2, and the coactivators PGC-1 and PPRC-1 (Scarpulla 2006). However, none of these factors have been shown to independently activate cell cycle entry. In contrast, some cell cycle regulators, most notably the Myc transcription factor, can both initiate and maintain cell cycle progression (Eilers et al. 1991). Expression of Myc increases rapidly after serum stimulation and initiates a cascade of events, still only incompletely understood, leading to cell cycle progression and biomass accumulation. In vivo genetic models confirm the importance of Myc for development, proliferation and tumorogenesis (Arvanitis and Felsher 2006; Pirity et al. 2006). An in vitro model system developed in a rat fibroblast cell line using genetic knockouts of endogenous c-myc genes has provided unique insights into Myc's regulation of the cell cycle, for example, the temporal regulation of cyclin genes (Mateyak et al. 1999). However, the c-myc knockout phenotype can not be compensated by any one known Myc target gene, or by other powerful regulators such as E2F, E1A or v-Src (Berns et al. 2000; Prathapam et al. 2006). The only single Myc target genes known to provide partial rescue are serine hydroxymethyl transferase (SHMT2), a mitochondrial enzyme of folate metabolism (Nikiforov et al. 2002) and ATF3, a member of the cAMP responsive (CREB) family of transcription factors (Tamura et al. 2005). The SHMT2 gene is among many metabolic genes that have been discovered as Myc targets by expression profiling and chromatin immunoprecipitation analyses (O'Connell et al. 2003; Li et al. 2005; Zeller et al. 2006). However, the role of metabolic genes in the global regulation of Myc-induced proliferation is poorly understood.
We have shown that cytochrome oxidase 5b and cytochrome c, a key regulator of mitochondrial respiration, are Myc target genes (Morrish et al. 2003). Subsequently, other mitochondrial targets have come to light, and the role of Myc in mitochondrial biogenesis has been confirmed (Li et al. 2005). Several glycolytic genes are also targets of Myc, and Myc over-expression increases glucose metabolism (Osthus et al. 2000). These pathways are believed to act as metabolic rheostats for cell cycle entry as both mitochondrial function and access to nutrients, such as glucose, provide key signals that dictate arrest or cell cycle progression (Jones et al. 2005; Mandal et al. 2005; Liao et al. 2006).
In the rat fibroblast model, the absence of Myc results in profound G1 phase lengthening and a significant delay in progression through the restriction point (Schorl and Sedivy 2003). The lack of rescue by any known cell cycle regulators has lead to proposals that additional factors are involved in the promotion of cell cycle entry by Myc (Nikiforov et al. 2002; Morrish and Hockenbery 2003). Despite ample evidence that Myc profoundly affects metabolism, including both glycolysis and mitochondrial biogenesis (Osthus et al. 2000; Li et al. 2005; Zhang et al. 2007), there have been no studies to investigate how coordinate regulation of carbon metabolism may be linked to cell cycle entry. The goal of the current study was to elucidate the functional importance of metabolic gene regulation for Myc-induced cell cycle entry. We evaluated the response of cells containing zero, low, normal and high levels of Myc to small molecule inhibitors of metabolism during both exponential growth and serum-stimulated cell cycle entry. We performed time course analyses of multiple parameters to dissect responses to metabolic inhibitors in the presence and absence of Myc. Finally, we undertook a kinetic analysis of Myc induced gene expression changes. These studies address the link between genotype, metabolic flexibility and signaling for cell cycle entry in response to external stimuli and confirm that coupling of mitochondrial respiration and glucose metabolism are key components of rapid cell cycle entry induced by the Myc oncogene.
Results
The high rate of proliferation of Myc-expressing cells requires both oxidative phosphorylation and glycolysis
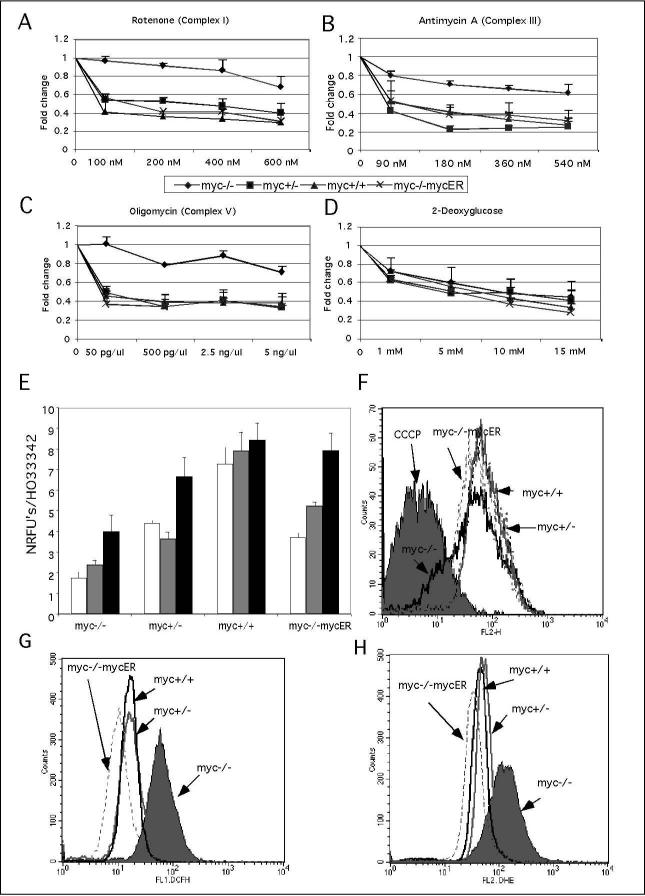
Myc regulates genes that function in both glycolysis and oxidative phosphorylation (Osthus et al. 2000; Li et al. 2005) and we hypothesized that the three-fold reduction in the doubling time of myc−/−cells (Mateyak et al. 1997) may be linked to reduced activity of these metabolic pathways. We determine the importance of these pathways by testing the effects of metabolic inhibitors. We evaluated the response of fibroblasts with the genotypes myc−/−, myc+/−, myc+/+ and myc−/−cells with reintroduced myc (myc-/mycER) to titrated doses of the OxPhos inhibitors rotenone, an inhibitor of respiratory complex II, antimycin A, an inhibitor of respiratory complex III and oligomycin, an inhibitor of respiratory complex V. The glycolysis inhibitor 2-deoxyglucose was also evaluated. We found that proliferation of all myc-positive cells (+/−, +/+, mycER) was highly sensitive to OxPhos inhibitors, while proliferation of myc−/−cells was unaffected at doses that induced a 50% decrease in growth for myc-positive cells, at 48 hours post addition of inhibitor (Figure 1A, B, C). This is not a simply a result of the decreased doubling time of myc−/−cells because there is a clear reduction of proliferation rates at higher doses of inhibitor. This interpretation is further substantiated by the contrasting effects on proliferation of the glycolysis inhibitor 2-deoxyglucose, which produced a similar dose response for all cells with IC50 of 5mM (Figure 1D). This pattern was consistent with other OxPhos and glycolysis inhibitors (data not shown) and suggests that the proliferation of myc−/−cells may rely predominantly on glycolysis rather than OxPhos, while myc-positive cells utilize both pathways for maximal proliferation. Confirmation that selective sensitivity to OxPhos inhibition is related to Myc expression is provided by the observation that myc−/−cells with re-introduced myc (myc−/−mycER) are susceptible to OxPhos inhibitors.
Figure 1. Myc sensitizes cells to OxPhos inhibitors and absence of Myc leads to mitochondrial dysfunction.
These experiments employed myc−/−, myc+/−, myc+/+ and myc−/mycER cell lines in asynchronous exponential growth culture. For the analysis of sensitization to inhibitors, proliferation was evaluated in the presence of (A) rotenone, (B) antimycin A, (C) oligomycin and (D) 2-deoxyglucose. Cell number was scored after 48 h of incubation by staining with the DNA dye HO33342 and values for controls relative to myc−/−cells were myc−/+ 1.6, myc+/+ 1.8, myc−/−MycER 1.4. Mitochondrial function analysis of (E) oxygen consumption was measured over periods of 3 h (white bars), 6 h (grey bars) and 8 h (black bars) using oxygen biosensor plates (Becton Dickinson), and is shown as fluorescence units normalized to cell number. (F) Mitochondrial membrane potential was measured in myc−/− (solid black line), myc+/(dashed black line), myc+/+ (solid grey line) and myc−/−mycER (dashed grey line) cells by staining with the dye JC-1 (5,5«,6,6«-tetrachloro-1,1«,3,3«-tetraethylbenzimidazolcarbocyanine iodide) followed by flow cytometric analysis. The overlap between myc−/cells and normal cells treated with the uncoupler CCCP (carbonyl cyanide m-chloro phenyl hydrazone) is indicative of mitochondrial de-polarization in the myc−/−cells. (G) Hydrogen peroxide levels were measured in myc−/− (grey fill), myc+/− (grey line), myc+/+ (black line) and myc−/−mycER ( dashed grey line) cells by staining with the dye DCF (2,7-dichlorofluorescein) followed by flow cytometric analysis. (H) Superoxide levels were measured in myc−/− (grey fill),, myc+/−(grey line), myc+/+ (black line) and myc−/mycER ( dashed grey line) cells by staining with the dye dihydroethidium and cells followed by flow cytometric analysis. Data are representative of two independent experiments performed in triplicates.
The selective inhibition of Myc-dependent cell proliferation by OxPhos inhibitors prompted us to compare mitochondrial function in myc−/−versus myc-positive cells. We evaluated oxygen consumption, mitochondrial membrane potential and the production of reactive oxygen species (ROS). In time-course studies, myc−/−cells had lower oxygen consumption rates when compared with myc-positive cells (Figure 1E). The low oxygen consumption of myc−/−cells likely resulted from reduced mitochondrial function due to de-polarized mitochondria (Figure 1F). This effect was alleviated on Myc induction as a clear increase in mitochondrial potential was observed in myc−/−MycER cells (Figure 1F). Further evidence for the presence of dysfunctional mitochondria in myc−/−cells is apparent from the increased production of hydrogen peroxide and superoxide (Figure 1G, H).
In summary, these results are consistent with reduced mitochondrial function in the absence of Myc. A consequent switch to glycolytic ATP production as an energy source for cell proliferation would explain the reduced impact of OxPhos inhibitors on myc−/−cells. Furthermore, our demonstration that re-introduction of myc into myc−/−cells (myc−/−mycER) restores mitochondrial membrane potential and oxygen consumption rates confirms the importance of Myc as a regulator of mitochondrial function.
Myc increases oxygen consumption and carbon metabolism during cell-cycle entry
The myc−/−cells show a prolonged delay in cell cycle entry that is correlated with reduced protein levels of Myc cell cycle target genes including cyclin D and cyclin E (Mateyak et al. 1999). However, over-expression of these genes in myc−/−cells fails to rescue this defect (Berns et al. 2000). Myc's demonstrated regulation of glucose consumption and glycolysis (Osthus et al. 2000), and the impaired mitochondrial function of myc-/fibroblasts described above, led us to hypothesize that metabolic coupling between glycolysis and OxPhos may be essential for the rapid cell cycle entry induced by Myc. To evaluate Myc's influence on metabolic responses, we examined oxygen consumption, carbon metabolism and ATP levels following serum addition in myc−/−, myc+/+ and myc−/−mycER cells. The 16 h time span during which myc-positive cells exit G0 and enter S phase was tested. Because myc−/−cells are still predominantly in G0/G1 at 16 h, we reasoned that differences in metabolism relevant to Myc-dependent cell cycle entry would be captured during this interval.
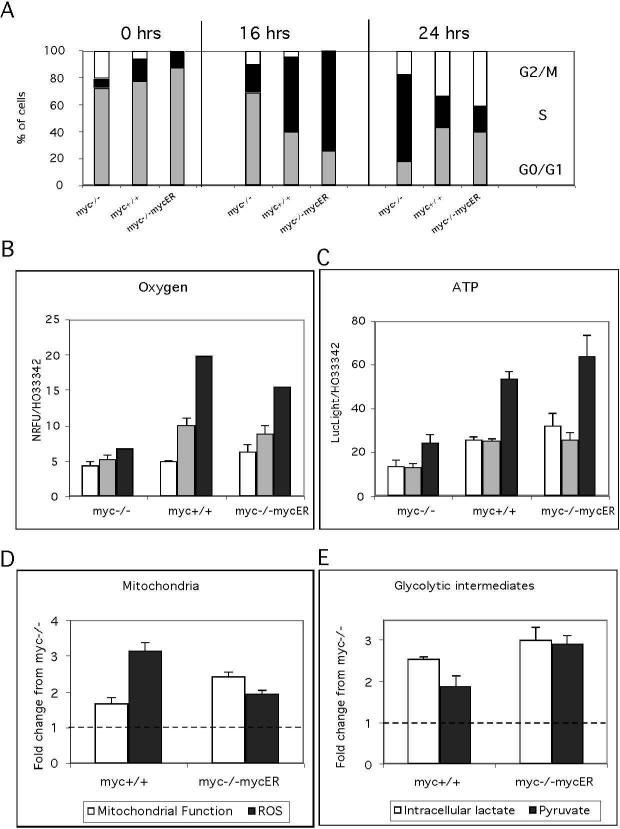
To demonstrate that we can recapitulate the effects of myc genotype on cell cycle entry previously reported we first evaluated S phase entry at 16 and 24 h (Figure 2A). These controls confirmed the delay in cell cycle entry reported for myc−/−cells. In our experiments, 55% of myc+/+ and 74% of myc−/−mycER cells were in S phase at 16 h after serum addition, compared to 20% for myc−/−cells. By 24 h, myc-positive cells had progressed into G2, while myc−/−cells were predominantly in S phase (Figure 2A).
Figure 2. Myc is required for rapid activation of metabolism following serum stimulation.
These experiments employed myc−/−, myc+/+and myc−/− mycER cultures and a serum deprivation-restimulation regimen to elicit synchronous cell cycle entry. (A) Cell cycle profiles at 0, 16 and 24 h after serum stimulation. Cells were stained with propidium iodide and analyzed by flow cytometry. The data are presented as the distribution (%) of cells in G0/G1 (grey bars), S (black bars) and G2/M (white bars) phases. The delay in cell cycle progression of myc−/−cells is evident by reduced S and G2/M phase content at the 16 h and 24 h time points, respectively. A representative of three independent experiments is shown. (B) Oxygen consumption was measured at 3 h (white bars), 6 h (grey bars) and 16 h (black bars) after serum stimulation using oxygen biosensor plates (Becton Dickinson), and is shown as fluorescence units normalized to cell number. Differences in oxygen consumption between myc-positive and myc-negative cells were highly significant (P<0.0001). (C) ATP levels were measured in cell extracts at 3 h (white bars), 6 h (grey bars) and 16 h (black bars) after serum stimulation using the CellTiterGlo assay (Promega). Data are normalized for cell number and expressed as the ratio of LucLight units (Promega) to HO33342 fluorescence units. Differences in ATP levels between myc-positive and myc-negative cells were highly significant (P<0.0001).
(D) Mitochondrial potential and mass were measured by staining cells with the dyes tetramethyl rhodamine methyl ester and non-acridine orange, respectively, followed by flow cytometric analysis. These data represent the change in mitochondrial function, as measured by the change in the ratio of potential over mass, between zero and sixteen hours post serum re-addition. ROS production was measured by staining with the dye DCF over the first 16 hours post serum readdition and is expressed as the fold change from myc-positive cells compared to myc−/−cells. (E) Intracellular lactate and pyruvate were measured in cell extracts using an Amplex red coupled assay [see methods]. The fold change over sixteen hours was calculated for both myc−/−and myc-positive cells and then the difference between myc-null and myc-positive was computed. These data represent the difference in the fold change in myc-positive cells compared to myc−/−cells. Data are representative of two to three independent experiments performed in triplicate.
In our metabolic measurements we found significant differences both in oxygen consumption rates and ATP production in myc-positive cells compared to myc−/−cells in the first 16 h post serum addition. While all cells had similar rates of oxygen consumption at 3 h, the myc+/+ and myc−/−mycER cells showed a 3−4 fold increase in oxygen consumption by 16 h compared with minimal changes for the myc−/−cells (Figure 2B). Similarly, these cells had a 2 fold greater level of ATP at 16 hr compared to the myc−/−cells (Figure 2C).
To determine if increased oxygen consumption rates in myc-positive cells were associated with other changes in mitochondrial function we evaluated mitochondrial membrane potential and ROS generation by FACS analysis. The data were expressed as the cumulative fold change over 16 h after addition of serum and graphed as the change in Myc positive cells compared to myc−/−cells. Mitochondrial function is expressed as the ratio of mitochondrial membrane potential to mitochondrial mass (Figure 2D). Both mycpositive cells had increased mitochondrial function compared to myc−/−cells, with a 1.5fold increase for myc+/+ and 2-fold increase for myc−/−mycER. A similar analysis of glycolysis showed increases increased intracellular lactate and pyruvate levels in mycpositive cells (Figure 3E).
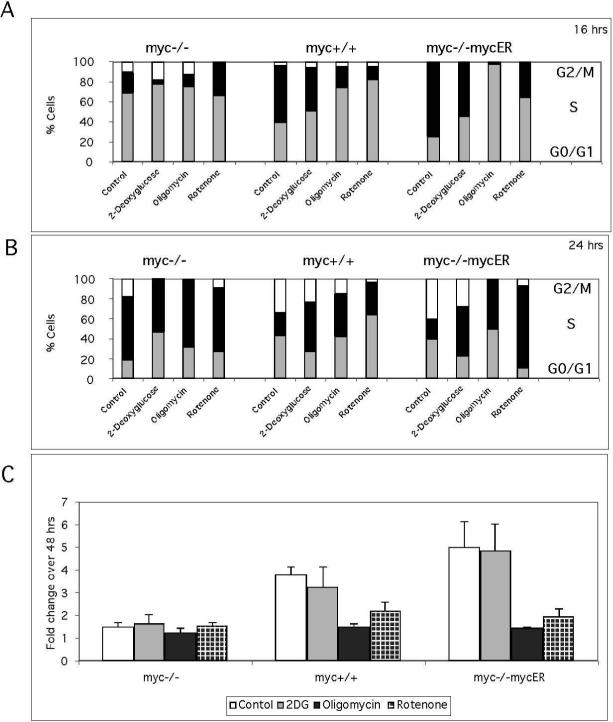
Figure 3. Metabolic inhibitors delay cell cycle entry and reduce proliferation.
These experiments employed myc−/−, myc+/+ and myc−/−mycER cultures and a serum deprivation and restimulation regimen to elicit synchronous cell cycle entry. Cell cycle distributions in the presence of the glycolysis inhibitor 2-deoxyglucose and the OxPhos inhibitors oligomycin (complex V) and rotenone (Complex I) were analyzed at 16 h (A) and 24 h (B) after serum stimulation, using flow cytometry as described in Fig. 3A. (C) Proliferative capacity in the presence of 2-deoxyglucose, oligomycin and rotenone was measured by staining with the DNA dye HO33342. Data are expressed as the fold change in fluorescence between 8 h and 48 h after serum stimulation. Differences in sensitivity to OxPhos inhibitors between myc-positive and myc−/−cells were highly significant (P<0.0001). Data are representative of two independent experiments performed in triplicate.
These studies of cell metabolism following serum stimulation demonstrate that, in comparison to myc−/−cells, bioenergetic metabolism in both myc+/+ and myc−/−mycER increased rapidly within the first 16 h. The significant, early increases in carbon metabolism, oxygen consumption, mitochondrial function and ATP production in Mycexpressing cells is consistent with Myc-induced activation of OxPhos and glycolysis. Myc expression can thereby rapidly induce the cellular bioenergetic activity and metabolite production required for cell proliferation. To confirm that upregulation of both OxPhos and glycolysis are essential to Myc-induced ATP production as well as cell cycle entry, we undertook studies with inhibitors specific for each metabolic pathway.
Cellular response to metabolic inhibitors during cell cycle entry
Cell cycle entry is a high energy-requiring process and metabolic perturbations delay cell cycle entry, activating metabolic checkpoints (Sweet and Singh 1995; Gardner et al. 2001; Jones et al. 2005). However, the response of all cells to metabolic perturbations is not equivalent and transcriptional reprogramming by oncogenes allows tumor cells to maintain cell proliferation under metabolic stress (Bi et al. 2005; Fantin et al. 2006; Kim et al. 2006). The following experiments assess the importance of glycolysis and respiration for Myc-induced cell cycle entry and analyze Myc's capacity to regulate metabolic flexibility in the presence of metabolic inhibitors. The inhibitors used were 2deoxyglucose (2-DG), oligomycin and rotenone. We confirmed that the inhibitors at the concentrations tested did not result in cell death (data not shown). Cell cycle was evaluated by flow cytometric analysis using propidium iodide staining of cells harvested at 16 and 24 h after serum addition. Cells were arrested by incubation for 48 h in 0.25% calf serum, and inhibitor-treated cells were compared to control untreated cells for all time points (Figure 3A, B). Because of the different timing of S phase entry of myc-/and myc-positive cells, the effects of inhibitors are manifested at different time points. Cell cycle progression at 16 h in the presence of inhibitors. At this time point, the presence of inhibitors reduced S phase entry or maintained G0/G1 arrest (Figure 3A). Due to the slow cell cycle entry of myc−/−cells, differences at this time point are subtle, with the most noticeable inhibitory effect being an increased G0/G1 arrest induced by 2DG. Cells with the highest levels of Myc were most sensitive to cell cycle arrest, as evidenced by the 97% G0/G1 arrest of myc−/−mycER cells by oligomycin. Comparing the inhibitors, there was a clear order in the response of myc+/+ cells, with Rot>OL>2DG in ability to elicit a G0/G1 arrest. Except for the oligomycin effect noted above, the trend was similar in the myc−/−mycER cells. Cell cycle progression at 24 h in the presence of inhibitors: By 24 h all inhibitors delayed cell cycle entry in myc−/−cells (Figure 3B). There were differences in the severity of the delay, as 2-DG produced a 30% increase in the number of cells in G0/G1 compared to 8−10% for oligomycin and rotenone. For myc-positive cells, control cells had entered G2/M and the differences between inhibitors observed at 16 h persisted, with lower reduction of G2/M cells with 2-DG (26%) in contrast to a 50 to 95% decrease for OxPhos inhibitors (Figure 3B). Note that oligomycin-treated myc−/−mycER cells overcame the G0/G1 block at 16 h and 50% of cells entered S phase by 24 h and rotenone treated cells also had an increased percentage of cells in S phase. These results demonstrate clear differences in cellular responses to metabolic inhibitors during cell cycle entry between myc-positive and myc-/-cells. The greater sensitivity of myc−/−cells to 2-DG suggests a dependence on glycolysis, which may be explained by diminished mitochondrial function in these cells (Figure 1). In contrast the higher sensitivity of mycpositive cells to OxPhos inhibitors suggests that respiration is essential to the rapid cell cycle entry induced in the presence of Myc. Measurements of cell growth were performed after 48 h of treatment with metabolic inhibitors (Figure 3C). OxPhos inhibitors selectively reduce growth in myc-positive cells with minimal effects on myc-/cells. The selective inhibition of myc-positive cell cycle entry and growth by OxPhos inhibitors suggests that an increase in cellular respiration is required for Myc-dependent cell cycle entry and growth, but not for myc−/−cells. To further determine how genotype influenced metabolic adaptation, we assessed oxygen and glucose consumption, the production of intermediary metabolites and ATP. We then tested for correlations between inhibitor-induced changes in metabolic parameters and the growth response of myc−/−and myc-positive cells.
Genotypic differences in metabolic adaptation in the presence of metabolic inhibitors
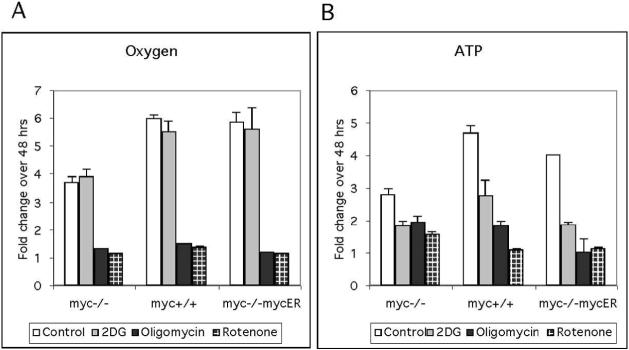
In many cell lines there is a direct relationship between growth and oxygen consumption (Wodnicka et al. 2000). As noted in our studies of mitochondrial function, increased oxygen consumption is associated with Myc-expression (Figure 1). Here we tested the hypothesis that rapid growth in myc-positive cells is correlated with and also dependent on O2 consumption. Using data generated from oxygen consumption over a 48 hr time-course (summarized in Fig 4A) we demonstrated that there is a correlation between O2 consumption and cell growth in myc-positive cells (Table 1), as indicated by a Pearson correlation coefficient R2 value approaching 1. To further differentiate between the two genotypes, we compared O2 consumption and cell growth in the presence of OxPhos or glycolytic inhibitors. As shown in figure 4A, 2-DG did not effect oxygen consumption, while there was a significant decrease in oxygen consumption for all cell lines in the presence of the OxPhos inhibitors oligomycin and rotenone. The correlation between growth and O2 consumption remained significant in both myc−/−and myc-positive cells treated with 2-DG (P<0.01, Table 1). In contrast, treatment with oligomycin or rotenone revealed a difference between genotypes. The R2 value for myc−/−cells dropped and was no longer significant (P>0.05). However, growth and oxygen consumption remained significantly correlated (P<0.01) for myc-positive cells treated with OxPhos inhibitors These data reflect the continued growth of myc−/−cells when respiration is inhibited, versus reduced growth in myc-positive cells to compensate for reduced respiration. We performed a similar correlation analysis between cellular ATP and growth in the presence of inhibitors (Table 1) using data generated from a 48 hr time-course (Fig. 4B). In contrast to the O2 data, all genotypes retained a significant correlation between ATP level and growth, irrespective of the inhibitor tested. Comparing the fold change in ATP levels after 48 h of inhibition, there were evident differences between genotypes (Figure 4B). The reduction in ATP in myc−/−cells treated with inhibitors (1.7-fold) was less than observed with myc-positive cells, perhaps accounting for the minimal effects on long term growth. For myc-positive cells, the effects of inhibitors on ATP production mirrored that seen for cell cycle and growth effects. 2-DG treatment reduced ATP levels by ∼ 2-fold, while OxPhos inhibitors had a more significant effect, reducing ATP levels by ∼4-fold. The observation that 2-DG treatment reduced ATP levels by ∼ 2-fold, yet did not impact proliferation suggests that ATP is produced in excess of requirements for cell proliferation in myc-positive cells. OxPhos inhibitors appear to have reduced ATP levels below a threshold required for rapid, Myc-dependent proliferation, consistent with a large contribution of oxidative respiration to ATP production in myc-positive cells.
Figure 4. The effect of metabolic inhibitors on oxygen consumption and ATP production.
These experiments employed myc−/−, myc+/+ and myc−/−mycER cultures and a serum deprivation-restimulation regimen. The glycolysis inhibitor 2-deoxyglucose, the OxPhos inhibitor oligomycin (Complex V), or the OxPhos inhibitor rotenone (Complex I) were added on serum stimulation, and incubation was continued for 48 h. Data are normalized to cell number and are expressed as the fold change relative to the values obtained at the time of drug addition (3 h). (A) Oxygen consumption was measured using oxygen biosensor plates (Becton Dickinson). (B) ATP levels were measured using the CellTiterGlo assay (Promega) The fold change in ATP production for myc-positive cells in the presence of OxPhos inhibitors was more significant (P<0.001) than for myc-/-cells. The results are representative of three independent experiments performed in triplicate.
Table 1.
Proliferation in the presence of Myc is highly correlated with oxygen consumption and ATP production.
| OXYGEN | ATP | ||||||
|---|---|---|---|---|---|---|---|
| R2 value | P value | R2 value | P value | ||||
| Myc−/− | Control | 0.79 | 0.001 ** | Myc−/− | Control | 0.66 | 0.007 ** |
| 2DG | 0.65 | 0.009 ** | 2DG | 0.62 | 0.01 * | ||
| OL | 0.28 | 0.13 | OL | 0.59 | 0.01 * | ||
| ROT | 0.38 | 0.07 | ROT | 0.85 | 0.0004 *** | ||
| myc+/+ | Control | 0.96 | 0.0004 ** | myc+/+ | Control | 0.92 | 0.0001 *** |
| 2DG | 0.85 | 0.007 ** | 2DG | 0.85 | 0.0004 *** | ||
| OL | 0.74 | 0.003 ** | OL | 0.73 | 0.003 ** | ||
| ROT | 0.76 | 0.002 ** | ROT | 0.89 | 0.004 ** | ||
| myc−/−mycER | Control | 0.98 | 0.0001 ** | myc−/−mycER | Control | 0.95 | 0.0001 *** |
| 2DG | 0.92 | 0.002 ** | 2DG | 0.66 | 0.008 ** | ||
| OL | 0.51 | 0.03 * | OL | 0.53 | 0.02 * | ||
| ROT | 0.84 | 0.001 ** | ROT | 0.81 | 0.01 * |
= significant
= very significant
=extremely significant
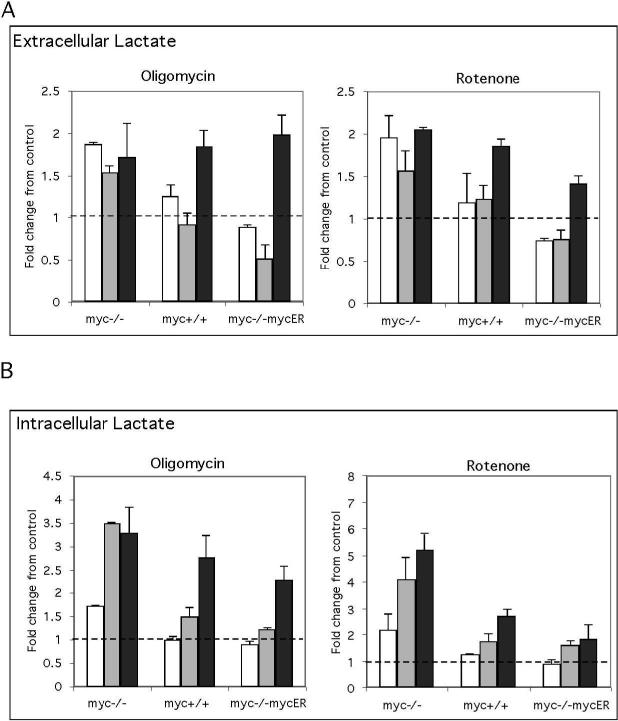
The oxygen consumption and cell growth correlation analysis suggests that myc-/cells maintain proliferation in the presence of OxPhos inhibitors. This could be achieved by switching to an alternate metabolic pathway or by increasing flux through an existing pathway, and we found that extracellular lactate levels in the myc−/−cells increased in the presence of OxPhos inhibitors (Figure 5A). We evaluated intracellular lactate levels at early time points after OxPhos inhibition to determine if there were differences between cell lines in the timing of this response. We found intracellular lactate in myc−/−cells increased 2−3 fold within the first 6 h. In contrast levels of intracellular lactate did not change in Myc positive cells until 16 h after OxPhos inhibition (Figure 5B). The increased capacity of myc−/−cells to rapidly increase lactate production may result from a pre-programming of glycolysis as the major energy source for cell division in these cells due to their inherent mitochondrial defects. This is further supported by the fact that cell cycle entry in myc−/−cells is most affected by the glycolysis inhibitor 2-DG while, in contrast, OxPhos inhibitors have the greatest effect on myc-positive cells (Figure 3B). Taken together, these results indicate that rapid cell cycle entry and growth of mycpositive cells is selectively dependent on mitochondrial metabolism. Furthermore, inhibiting mitochondrial function in these cells results in a slow growth phenotype that recapitulates myc−/−cells, as the growth rate for myc-positive cells in oligomycin or rotenone is similar to myc−/−cells in the absence of inhibitors (Figure 3C ).
Figure 5. Loss of Myc accelerates the Pasteur response to OxPhos inhibitors.
These experiments employed myc−/−, myc+/+ and myc−/−mycER cultures and a serum deprivationrestimulation regimen with addition of inhibitors on serum stimulation. Extracellular (A) and intracellular (B) lactate was measured at 3 h (white bars), 6 h (grey bars) and 16 h (black bars) after serum stimulation in the presence of oligomycin or rotenone using an Amplex red coupled assay. Data are expressed as fold changes relative to untreated controls. These results are representative of three independent experiments performed in triplicate.
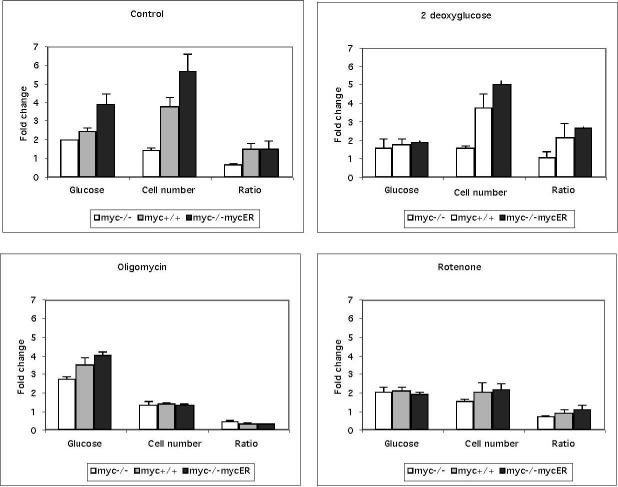
The above results suggest that the coupling of OxPhos to glycolysis is important for Myc-dependent cell proliferation. Therefore, we would also anticipate a significant effect of OxPhos inhibitors on the conversion of glucose to cellular biomass. We determined the total glucose consumed and the increase in cell number over a 48 h time period. The ratio of cell number to glucose consumption was used as a measure of metabolic capacity. It should be noted that we found no changes in cell size in these experiments (data not shown). Data from this analysis revealed a significant difference between genotypes when evaluated as glucose to cell number conversion in the presence of metabolic inhibitors (Figure 6). Under normal conditions, myc-positive cells produce more cells per glucose consumed than myc−/−cells, which is indicative of the greater efficiency of carbon metabolism in cells expressing Myc. In the presence of 2-DG both cell types show decreased glucose consumption and cell proliferation, but myc-positive cells maintain a higher cell number to glucose ratio, possibly by maximizing the use of alternate energy sources such as glutamine. In contrast, treatments with rotenone or oligomycin reduced the proliferation in myc-positive cells as well as the cell number to glucose ratio, whereas myc−/−cells continued to proliferate and maintained cell number to glucose ratio. The differential response of myc-positive cells to oligomycin and rotenone are likely linked to the induction of ROS by rotenone. These data provide further evidence that mitochondrial function is an important component of Myc-dependent proliferation following serum addition and in particular, suggest a link between regulation of mitochondrial respiration and Myc induced proliferation.
Figure 6. Oxphos inhibitors decrease glucose to cell number production in myc-positive cells.
Glucose consumption was measured using the Amplex red method (Molecular Probes) in the presence and absence of metabolic inhibitors over 48 hours and expressed as the fold change in glucose consumed. The change in cell number over 48 hours in the presence or absence of inhibitors was evaluated by computing the fold change in Hoechst 33342 fluorescence evaluated in a quantitative 96 well assay. Data to determine how well a cell converts glucose to increased cell number are expressed as the ratio of DNA content/Glucose. These data are representative of two independent experiments performed in triplicate.
Genes associated with mitochondrial function show a temporal increase in expression on induction of Myc in myc−/−cells
Many genes involved in mitochondrial function have been identified as Myc target genes (Morrish et al. 2003; Li et al. 2005; Zeller et al. 2006). To define the temporal increase in mitochondrial gene expression regulated by Myc in a myc−/−background, we evaluated microarray data from a large scale time-course experiment (Neretti and Sedivy, unpublished results). The data were evaluated for expression changes in genes linked to mitochondrial metabolism (Bai et al. 2007). We reasoned that the key processes required for Myc-dependent mitochondrial biogenesis would include the following: transcription and translation from mitochondrial DNA, transcription, translation and import of nuclear encoded proteins into the mitochondria, assembly of electron transport chain protein complexes, and production of mitochondrial membranes. In addition, we included metabolic pathways requiring the contribution of mitochondria, including purine and pyrimidine biosynthesis, lipid, glutamate, and folate/C1 metabolism, and the tricarboxylic acid cycle. Finally, we included genes that would protect against mitochondrial generation of ROS.
Transcription, translation and protein assembly
Genes involved in mitochondrial gene transcription including TFB1M, TOP1MT, TFAM, MRPL12 and POLG were all increased at multiple time points following induction of Myc (Supplemental data Figure S1, see Table S1 for gene descriptions). In addition a key co-factor associated with regulators of mitochondrial biogenesis PPRC1 (Vercauteren et al. 2006) and a series of mitochondrial ribosomal proteins MRPL16, MRPL38, MRPL42, MRPS12, MRPS7 were temporally up-regulated (Supplemental data Figure S1). Genes for protein import including TOMM34, TIMM9 and TIMM13 were increased in parallel with the mitochondrial chaperones PHB2, HSPD1 and TRAP1 and several electron transport complex assembly genes including BCS-1, SCO1, ATPAF1, ECSIT and LRPPRC (Supplemental Figure S2).
Mitochondrial metabolism
In agreement with prior microarray studies in this model (O'Connell et al. 2003), the SHMT1 (cytosolic) and SHMT2 (mitochondrial) genes for essential enzymes in folate/C1 metabolism regulating purine, pyrimidines, and S-adenosylmethionine biosynthesis, were increased. There was also a notable early increase in expression of the folate transporter SCL19A1 and other genes associated with folate/C1 metabolism including MTHFD1, DDX18 and GGH (Supplemental Figure S2). The key mitochondrial gene for purine metabolism, DHODH was induced along with increased expression of CAD, GART, ADSL, AK2 with roles in purine and pyrimidine metabolism (Supplemental Figure S3). There was also an increase in GLS expression, which is essential for CAD function (Supplemental Figure S4).
In our analysis of genes associated with the TCA cycle we found a decrease in expression of PDK2 and PC (Supplemental Figure S4). Genes from pathways that rely on the TCA cycle, including glutamine metabolism (GLS), and fatty acid metabolism MECR, ACSL1, AACS, ACAT1 were induced (Supplemental Figure S4). The coordinate induction of genes involved in fatty acid synthesis, including ASCL1 and MECR, a key mitochondrial gene for fatty acid elongation, in addition to genes controlling glycerophospholipid synthesis (AGAPT5, DGAT2 and LYPLA1) suggest increased in membrane lipid biosynthesis (Supplemental Table S1). Increased metabolic activity in the mitochondria would require increased transport of metabolites across mitochondrial membranes, and we found temporal increases in the glutamate/aspartate carrier SCL25A22 (Supplemental Figure S5), and a second reduced folate/thiamine carrier SLC19A2. Additionally, the mitochondrial deoxynucleotide carrier SLC25A19, the iron transporter SCL25A37, as well as the plasma membrane iron transporter TFRC (a Myc target gene O'Donnell et al. 2005), were induced by Myc at early time-points while the ATP/ADP transporter SCL25A5 was induced with delayed kinetics (Supplemental Figure S5).
As a consequence of increased oxidative metabolism expected from the above expression profile, we also observed increases in genes with known antioxidant function including SOD2, PRDX6, and TRAP1 (Supplemental Figure S6). These data, and our demonstration of increased mitochondrial function in response to serum stimulation, support the role of Myc in mitochondrial biogenesis and regulation of metabolic pathways that depend on mitochondrial biosynthetic activities.
Discussion
Addition of serum to quiescent cells rapidly stimulates cell cycle entry. The Myc oncogene is one of the first genes induced by serum (Dean et al. 1986) and initiates a cascade of transcriptional responses. Understanding Myc's role in the cell cycle is a major question in Myc biology and has driven the analysis of downstream gene targets (O'Connell et al. 2003; Li et al. 2005; Zeller et al. 2006). Use of Myc knockout models demonstrated Myc-dependent rapid upregulation of specific cyclins (Mateyak et al. 1999). However, efforts to reconstitute the Myc phenotype by expression of individual cell cycle genes have been unsuccessful (Berns et al. 2000) leading to the proposal that it is Myc's coordinate regulation of gene networks that enables rapid cell cycle entry (Nikiforov et al. 2002; Morrish and Hockenbery 2003). In particular, Myc's regulation of metabolic genes may play a significant role, as the bioenergetic and metabolic capacity of the cell act as checkpoints to rapid cell cycle entry and progression (Jones et al. 2005; Mandal et al. 2005; Liao et al. 2006). The current study addresses the contribution of Myc's regulation of mitochondrial function to Myc-dependent cell proliferation and additionally tested the possibility that this gene coordinately regulates both the metabolic and cell cycle genes required for cell cycle entry.
Our metabolic phenotyping experiments to evaluate sensitivity to metabolic inhibitors and our analysis of mitochondrial function confirm that high proliferative rates induced by Myc are linked to increased mitochondrial function. In addition we found that there is a specific requirement for OxPhos metabolism during cell cycle entry in mycpositive cells because Myc-dependent cell cycle entry, as well as cell growth and glucose to biomass conversion all decreased in the presence of OxPhos inhibitors. In contrast, these inhibitors had minimal effects on myc−/−cells. We additionally found a strong correlation between growth and oxygen consumption in the myc-positive cells. Overall these inhibitor studies suggest that mitochondria play a critical role in cell cycle entry induced by Myc. These findings are in keeping with results from recent screens for cell proliferation mutants in Drosophila which revealed several genes with mitochondrial functions for example cytochrome c maturation (CCHL) and ATP synthesis (ATP synthase delta) (Liao et al. 2006) and a recent RNAi screen in U2OS cells, which found that 10% of genes with cell cycle effects functioned in cell metabolism (Mukherji et al. 2006). These reports all point to a role for increased cellular metabolism in cell cycle entry. Myc, as one of the first transcription factors induced by serum, is well positioned to induce cell cycle genes as well as the metabolic gene transcriptome required for production of energy and metabolites required for DNA synthesis and cell signaling.
We previously demonstrated direct binding of Myc to the cytochrome c promoter by chromosomal immunoprecipitation, associated with an accelerated induction of cytochrome c by serum stimulation (Morrish et al. 2003). Here we analyzed a unique temporal genome-wide array for genes in functional categories related to transcription and translation of the mitochondrial genome, mitochondrial protein import, and assembly of mitochondrial respiratory complexes. We find that induction of Myc in myc−/−cells coordinately up-regulated several mitochondrial DNA transcriptional components of the core mitochondrial transcription machinery (Fig 7). Myc's temporal regulation of this class of genes should support increased transcription and translation from the mitochondrial genome. The additional increased temporal expression of chaperones and protein import genes in concert with genes regulating the assembly of respiratory complexes (Figure 7) demonstrates a role for Myc in regulation of respiratory complex assembly. While additional studies will be necessary to corroborate Myc's ability to regulate key aspects of mitochondrial biogenesis these data are the first to provide evidence of a single transcription factor, with a known role in regulation of cell cycle genes, that also regulates the temporal coordinate expression of genes required for respiratory complex assembly in addition to transcription and translation of the mitochondrial genome.
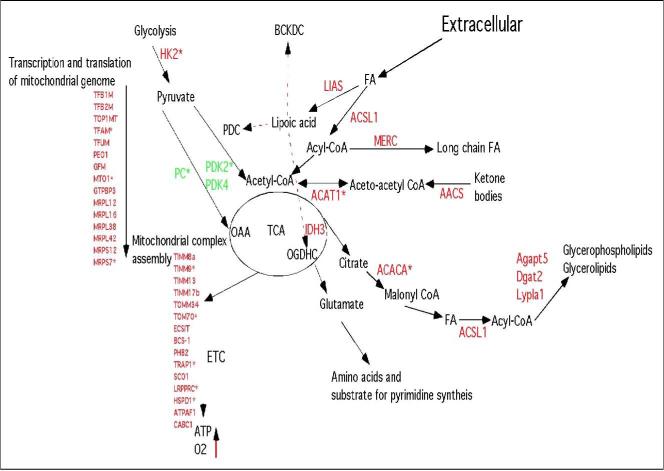
Figure 7. Regulation of mitochondrial function provides continuity of energy, reducing power and substrate generation to the metabolic networks required for cell cycle entry.
Pyruvate generated by glycolysis, after conversion to acetyl-CoA, is converted by the tricarboxylic acid (TCA) cycle to glutamate, a nodal substrate for amino acid, nucleotide and fatty acid biosynthesis. Acetyl-CoA generated from other sources also fuels the TCA. Activation of the TCA cycle, coupled to increased abundance of electron transport chain (ETC) complexes would stimulate cellular bioenergetics, leading to increased oxygen consumption and ATP generation. PDC, pyruvate dehydrogenase complex; BCKDC, branched chain ketoacid dehydrogenase; OGDH, oxoglutarate (alpha-ketoglutarate) dehydrogenase complex; FA, fatty acids; OAA, oxaloacetate. Expression analysis (Supplemental Table S1) revealed that the genes highlighted in red are Myc-inducible. The three genes shown in green are repressed by Myc. Genes with an asterix are known to be bound by Myc (data from c-Myc target database, www.myccancergene.org , Zeller et al. 2006 and Zhang et al. 2006).
Mitochondria are important, not only as a source of energy but also as an essential link for metabolite production in many metabolic pathways. Myc induction resulted in the temporal up-regulation of genes involved in C1 and folate metabolism, including mitochondrial isoform of SHMT, which together provide key metabolic intermediates for the synthesis of purines and pyrimidines. The additional temporal coordinate regulation of other key genes in these metabolic pathways, for example DHOD which is localized to mitochondria, illustrate how Myc's effects on mitochondrial function maintains the generation of metabolites required for DNA and RNA synthesis. These results also provide some insight into why the expression of SHMT2 resulted in partial rescue of the myc−/−phenotype (Nikiforov et al. 2002) as it is evident that maximal activity of folate/C1 metabolism requires co-induction of other genes.
We found that Myc induction resulted in decreased expression of the pyruvate dehydrogenase complex (PDHC) repressor pyruvate dehydrogenase kinase 2 and 4. This could increase entry of pyruvate into the TCA cycle and provide intermediates for amino acid and lipid biosynthesis as well as reducing equivalents for oxidative phosphorylation. The paradoxical increase in PDK1, recently shown to be activated by Myc (Kim et al., 2007), is confounding. However, recent work indicates both PDK2 and 4 are regulated by the PPARs delta/beta (Degenhart et al. 2008) and the drop in PPARs levels seen here may thereby lead to a concomitant drop in PDK2 and 4. The status of PDHC activity in these cells will require further study.
We also observed decreased expression of pyruvate carboxylase. These changes are similar to those observed on activation of PDHC by dichloroacetate in fibroblasts (Simpson et al. 2006). Additionally, while glutamine provided in the media is readily converted to glutamate the possible activation of the TCA cycle suggests that Myc expression may provide an additional de novo generated source of glutamate. However, further studies using 13C labeled substrates will be needed to examine Myc-induced metabolic pathway fluxes and these studies are in progress.
Interestingly, we found that expression of the mitochondrial NAD(+)-dependent deacetylase SIRT3, known to activate AceCS2 by deacetylation (Hallows et al. 2006), increased temporally on induction of Myc. Activation of AceCS2 would also increase acetyl CoA generation as a key nodal metabolite and substrate in many metabolic reactions. Notably the availability of acetyl-CoA is essential for the activity of histone acetyl transferases (HATs) (Takahashi et al. 2006). On addition of serum the promoter of the HAT gene GCN5, a Myc target gene, and key regulator of global chromatin remodeling (Knoepfler et al. 2006) has been shown to display a 5-to 10-fold increase in Myc binding. The expression of GCN5 is severely compromised in myc−/−cells resulting in hypoacetylation of H3 and H4 compared to wild type cells, while reintroduction of Myc into myc−/−cells restored histone acetylation to wildtype levels. We also observed the temporal induction of GCN5 (Suplemental Figure S4) and propose that Myc's ability to activate SIRT3 and other pathways that increase the production of acetyl-CoA may facilitate the activation of chromatin remodeling by GCN5, by supplying the acetyl-CoA substrate required for histone acetylation. The activation of these pathways would, in addition, increases the production of lipids required for membrane biosynthesis (Figure 7).
Finally, the increased mitochondrial activity ensuing from the changes described above would be expected to generate increased ROS, and the temporal increase in expression of SOD2 and PRDX6 observed would help to reduce oxidative stress. Additionally, the bi-functional genes LIAS and TRAP1 would also participate in the reduction of ROS generated from the mitochondria. We expected to observe an increase in genes involved in glutathione metabolism, based on recent data (Benassi et al. 2007) but neither of the glutamine-cysteine ligase (GCL) subunits were increased above our threshold. It is possible that is the increased generation of ROS in myc−/−cells (Figure 1) may activate glutathione synthesis, obscuring the ability to detect a Myc-induced response. This increased ROS may be linked to increases in β-oxidation of fatty acids as many of the genes down regulated by Myc are involved in β-oxidation (Supplemental Table S1) and the correlated decrease in peroxisome proliferator-activated receptor gamma coactivator 1 alpha, verb3 and estrogen receptor 1, regulators of lipid and steroid metabolism, suggest the activation of compensatory pathways for energy generation in the absence of Myc.
In summary, our results show that Myc activates mitochondrial function and initiates a transcriptional program that facilitates the coordinate metabolic activities required for rapid cell cycle entry. We propose that the ability to rapidly increase the metabolic capacity of the cell has implications for our understanding of how Myc deregulation leads to cancer. Increasing our understanding of how the metabolic events induced by Myc are modified in the presence of synergistic oncogene such as Bcl-2 may be an important avenue to developing early detection assays and novel therapies that target metabolic signatures in human cancers associated with Myc deregulation.
Material and methods
Cell culture
The myc-null HO15 (myc−/−), heterozygote HET15 (myc+/−), wild type TGR1 (myc+/+) and myc−/−MycER reconstituted fibroblast cell line KO-MycER used in this study have been previously described (O'Connell et al. 2003). Cells were grown in Dulbecco Modified Eagle medium (DMEM; Hyclone) supplemented with 10% calf serum (CS; Hyclone), at 37°C in an atmosphere of 5% CO2. To obtain quiescent cells, 80% confluent cultures were serum starved (0.25% CS) for 48−72 h. In serum re-addition studies cells were washed and harvested by trypsinization and plated in 10% CS. In studies of oxygen consumption, glucose consumption, ATP, lactate and pyruvate generation Myc was pre-activated for 24 hours, 2 days prior to serum starvation by addition of 100 nM 4hydroxytamoxifen (4-OHT;Sigma). On serum addition 4-OHT was added at 100 nM to all cells.
Metabolic inhibitors
All of the metabolic inhibitors used in this study were obtained from Sigma-Aldrich and dissolved in water (2-DG and rotenone), DMSO (antimycin A) or ethanol (oligomycin). The concentration of inhibitors used in our cell cycle inhibitor studies were selected to reduce metabolism but not induced cell death and are as follows; 2-deoxyglucose (5mM), oligomycin ( 1 pg/ul) and rotenone (25 pM).
Measurement of total cellular Oxygen consumption
Cells were plated at a density of 2 × 105 per 50 ul onto 50 ul of Cytotex 3 microcarrier beads (Sigma) (25 mg/ml) in a 96 well BD oxygen biosensor system (BD Biosciences). Fluorescence readings were taken at the times indicated using a Fluroskan Ascent reader (Thermo Electron Corp) at 480 nm excitation λ and 620 nm emission λ. The data were normalized according to the manufacturer's protocol (technical bulletin no. 448: BD Biosciences) and represented as normalized relative fluorescence units. The CellTiter Glo assay kit (Promega) was used to evaluate ATP. After 10 minutes incubation in the lysis mix, luminescence was measured in a 96 well plate luminometer reader (Packard, Top Count NXT microplate, Luminescence Counter) and expressed as LucLight fluorescence units.
Glucose, lactate and pyruvate
For analysis of extra-cellular glucose and lactate, cell culture media was harvested over the time course indicated. The media was diluted 1:500 before analysis using the Amplex red Glucose/Glucose oxidase kit (Molecular Probes/InVitrogen). Lactate was evaluated by substituting Lactate oxidase for glucose oxidase. Intracellular lactate and pyruvate were measured in cellular lysates generated with NP40 lysis buffer (50 mM Tris, 150 mM NaCL and 1% NP-40, all from Sigma) supplemented with Complete™ (Roche). The 50 ul lysate was diluted 1:4 in 1x phosphate buffer from the Amplex red kit and dispensed into round bottom 96 well plates and centrifuged at 3,000 rpm, 4°C for 10 minutes. A total of 50 ul of this clarified supernatant was then diluted 1:4 in 1x phosphate buffer pH 7.4 for lactate assays and the same buffer at pH 5.7 for pyruvate assays. Lactate was assayed as outlined above. For the pyruvate assay, pyruvate oxidase was substituted for lactate oxidase in the reaction and the co-factor Flavin adenine dinucleotide (0.0097 mM), MgSO4 (9.7mM), and thiamine pyrophosphate (0.2 mM) were also added. The pyruvate assay required incubation at 37°C rather than room temperature as recommended for lactate and glucose. For all assays the concentration of product was obtained by comparison with known standards (1−150 uM) evaluated in parallel with the cell extracts. Fluorescence was measured in a Fluroskan Ascent plate reader (Thermo Electron Corp) at 560 nm excitation λ and 580 nm emission λ.
Cell cycle analysis
Cell fixation, propidium iodide staining and cell cycle distribution was performed as outlined previously (Morrish et al. 2003)
Cell viability and proliferation HO33342 assay
After removal of media, cells were washed using 200 μl PBS per well and tapped dry on a tissue. Plates were stored at −20 °C prior to lysis with 100 μl of 0.01% SDS. After shaking for 30 min plates were then frozen at −70 °C. After thawing 100 μl of a HO33342 staining solution was added (2 μg/ml HO 33342, 10 mM Tris-HCl pH 7.4, 1 mM EDTA, 1 M NaCl) and plates were shaken for 1 hr in the dark at 37 °C. Fluorescence was measured in a Fluoroskan Ascent plate reader (Thermo Electron Corp) at 350 nm excitation λ and 460 nm emission λ.
Mitochondrial membrane potential, mass and reactive oxygen species
Cells were harvested, counted and 1 × 106 cells/ ml were stained with 200 nM tetramethylrhodamine methyl and ethyl esters (TMRM; Molecular probes) or 10 μM 5, 5«, 6, 6«-tetrachloro-1, 1«, 3, 3«-tetraethylbenzimidazol-carbocyanine iodide (JC1; Molecular Probes) for analysis of mitochondrial membrane potential. Controls consisted of unstained cells and cells treated with the uncoupler carbonyl cyanide mchlorophenylhydrazone (CCCP: 30 μM). Cells were stained for 30 minutes at 37 °C. Hydrogen peroxide and superoxide were measured with dichlorofluorescin diacetate (DCHF-DA; 5μM) and dihydroethidium (DHE; 5μM) respectively. Cells were stained for 30 minutes at 37 °C, washed × 2 with PBS and resuspended in PBS. Mass was evaluated using non-acridine orange 10 μM (NAO; Molecular Probes). All cells were evaluated using a BD Calibur flow cytometer and data were evaluated and graphed using CellQuest™ (Becton Dickinson).
Cell culture for array analysis and expression profiling
The cell line HOMycER12 was used (O'Connell et al. 2003). This cell line was derived from the c-myc−/−cell line HO15.19, and expresses a cDNA encoding a c-Myc -estrogen receptor (ER) fusion protein. MycER cells were kept in a constant state of proliferation and the MycER protein was activated by addition of 4-hydroxytamoxifen (OHT) to the medium, as described (O'Connell et al. 2003). Cells were collected at the following time (t) points after addition of OHT: 1, 2, 3, 4, 5, 6, 8, 10, 12, 16, 20 and 24 hours. A sample was also collected at the time of OHT addition (t=0 h). Negative controls were collected at 8 h, 16 h and 24 h following mock addition of OHT (ethanol vehicle). Total RNA was harvested from each sample, and was hybridized to GeneChip (r) Rat Exon 1.0 ST Arrays (Affymetrix). The entire process was repeated to generate three independent biological replicates.
Probeset filtering and normalization
The PLIER algorithm from the Affymetrix Power Tools (APT) software package was used to generate gene and exon expression scores. Probes were filtered and remapped according to the ENTREZG tables provided by the Microarray Lab of the Molecular and Behavioral Neurosience Institute, University of Michigan (Dai et al. 2005). Probes were remapped by blasting their sequences onto the rn4 genome and removing probes mapping to multiple locations in the genome. Genes were redefined as sets of at least three probes mapping to the same Entrez gene designation. These data will be deposited in the National Center for Biotechnology Information Gene expression Omnibus (GEO).
Statistical Analysis for microarrays
We first excluded from the analysis genes whose PLIER scores, averaged over the three biological replicates, did not exceed a value of 48 for all time points, (the 1st quartile of all the average scores over all replicates, which we considered below the noise threshold). We then applied a 3-way ANOVA with the following factors: time, treatment, and experiment (each biological replicate and its associated controls were considered as one independent experiment). The experiment factor was included since we have evidence that scores from the same experiment are correlated. For each gene, the time factor p value was calculated based on differential expression between any time point (t “greater or equal to” 1 h) and the t = 0 time point; the treatment factor p value was calculated based on differential expression between treated and control samples. The thresholds for these two sets of p values were set by requiring a false discovery rate (FDR) of 1% according to Story and Tibshirani (Storey and Tibshirani 2003); this corresponds to a threshold of 0.00716 for the time factor p value, and of 0.0134 for the treatment factor p value. The experiment factor p-value was not included in this filtering process. After this analysis, 4,186 genes were deemed significantly differentially expressed after c-Myc induction.
Statistical analysis for cell culture
Analysis of significant differences between means was evaluated using the standardized student t test using Microsoft Excel. Correlation analysis was performed using Instat (GraphPad softwear) by calculating the r squared value (coefficient of determination) from the Pearson correlation coefficient.
Acknowledgements
This work was funded by R01CA106650-02 to DH and supported in part by grants R01 GM41690 to J.M.S. and K25 AG028753 to N.N. from NIH.
Supplementary Material
References
- Andersson U, Scarpulla RC. Pgc-1-related coactivator, a novel, serum-inducible coactivator of nuclear respiratory factor 1-dependent transcription in mammalian cells. Mol Cell Biol. 2001;21(11):3738–3749. doi: 10.1128/MCB.21.11.3738-3749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvanitis C, Felsher DW. Conditional transgenic models define how MYC initiates and maintains tumorigenesis. Semin Cancer Biol. 2006;16(4):313–317. doi: 10.1016/j.semcancer.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Bai X, Wu J, Zhang Q, Alesci S, Manoli I, Blackman M, Chrousos G, Goldstein A, Rennert O, Su Y. Third-generation human mitochondria-focused cDNA microarray and its bioinformatic tools for analysis of gene expression. Biotechniques. 2007;42:365–375. doi: 10.2144/000112388. [DOI] [PubMed] [Google Scholar]
- Benassi B, Zupi G, Biroccio A. Gamma-glutamylcysteine synthetase mediates the c-Myc-dependent response to antineoplastic agents in melanoma cells. Mol Pharmacol. 2007;72:1015–1023. doi: 10.1124/mol.107.038687. [DOI] [PubMed] [Google Scholar]
- Berns K, Hijmans EM, Koh E, Daley GQ, Bernards R. A genetic screen to identify genes that rescue the slow growth phenotype of c-myc null fibroblasts. Oncogene. 2000;19(29):3330–3334. doi: 10.1038/sj.onc.1203639. [DOI] [PubMed] [Google Scholar]
- Bi M, Naczki C, Koritzinsky M, Fels D, Blais J, Hu N, Harding H, Novoa I, Varia M, Raleigh J, Scheuner D, Kaufman RJ, Bell J, Ron D, Wouters BG, Koumenis C. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. Embo J. 2005;24(19):3470–3481. doi: 10.1038/sj.emboj.7600777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, Watson SJ, Meng F. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M, Levine RA, Ran W, Kindy MS, Sonenshein GE, Campisi J. Regulation of c-myc transcription and mRNA abundance by serum growth factors and cell contact. J Biol Chem. 1986;261(20):9161–9166. [PubMed] [Google Scholar]
- Degenhardt T, Saramaki A, Malinen M, Rieck M, Vaisanen S, Huotari A, Herzig KH, Muller R, Carlberg C. Three members of the human dehydrogenase kinase gene family are direct targets of the peroxisome proliferator-activated receptor beta/delta. J Mol Biol. 2008;372(2):341–355. doi: 10.1016/j.jmb.2007.06.091. [DOI] [PubMed] [Google Scholar]
- Eilers M, Schirm S, Bishop JM. The MYC protein activates transcription of the alpha-prothymosin gene. Embo J. 1991;10(1):133–141. doi: 10.1002/j.1460-2075.1991.tb07929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9(6):425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- Gardner LB, Li Q, Park MS, Flanagan WM, Semenza GL, Dang CV. Hypoxia inhibits G1/S transition through regulation of p27 expression. J Biol Chem. 2001;276(11):7919–7926. doi: 10.1074/jbc.M010189200. [DOI] [PubMed] [Google Scholar]
- Hallows W, Lee S, Denu J. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig RP, Scacco S, Scarpulla RC. Sequential serum-dependent activation of CREB and NRF-1 leads to enhanced mitochondrial respiration through the induction of cytochrome c. J Biol Chem. 2000;275(17):13134–13141. doi: 10.1074/jbc.275.17.13134. [DOI] [PubMed] [Google Scholar]
- Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18(3):283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- Kim J, Gao P, Liu Y, Semenza G, Dang C. HIF-1 and Dysregulated c-Myc Cooperatively Induces VEGF and Metabolic Switches, HK2 and PDK1. Mol Cell Biol. 2007;27:7381–7393. doi: 10.1128/MCB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. Embo J. 2006;25(12):27232734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O'Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25(14):62256234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao TS, Call GB, Guptan P, Cespedes A, Marshall J, Yackle K, Owusu-Ansah E, Mandal S, Fang QA, Goodstein GL, Kim W, Banerjee U. An efficient genetic screen in Drosophila to identify nuclear-encoded genes with mitochondrial function. Genetics. 2006;174(1):525–533. doi: 10.1534/genetics.106.061705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal S, Guptan P, Owusu-Ansah E, Banerjee U. Mitochondrial regulation of cell cycle progression during development as revealed by the tenured mutation in Drosophila. Dev Cell. 2005;9(6):843–854. doi: 10.1016/j.devcel.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Mateyak MK, Obaya AJ, Adachi S, Sedivy JM. Phenotypes of c-Mycdeficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8(10):1039–1048. [PubMed] [Google Scholar]
- Mateyak MK, Obaya AJ, Sedivy JM. c-Myc regulates cyclin D-Cdk4 and -Cdk6 activity but affects cell cycle progression at multiple independent points. Mol Cell Biol. 1999;19(7):4672–4683. doi: 10.1128/mcb.19.7.4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrish F, Giedt C, Hockenbery D. c-MYC apoptotic function is mediated by NRF-1 target genes. Genes Dev. 2003;17(2):240–255. doi: 10.1101/gad.1032503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrish F, Hockenbery D. Myc's mastery of mitochondrial mischief. Cell Cycle. 2003;2:11–13. doi: 10.4161/cc.2.1.275. [DOI] [PubMed] [Google Scholar]
- Mukherji M, Bell R, Supekova L, Wang Y, Orth AP, Batalov S, Miraglia L, Huesken D, Lange J, Martin C, Sahasrabudhe S, Reinhardt M, Natt F, Hall J, Mickanin C, Labow M, Chanda SK, Cho CY, Schultz PG. Genome-wide functional analysis of human cell-cycle regulators. Proc Natl Acad Sci US A. 2006;103(40):14819–14824. doi: 10.1073/pnas.0604320103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforov MA, Chandriani S, O'Connell B, Petrenko O, Kotenko I, Beavis A, Sedivy JM, Cole MD. A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol Cell Biol. 2002;22(16):5793–5800. doi: 10.1128/MCB.22.16.5793-5800.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell BC, Cheung AF, Simkevich CP, Tam W, Ren X, Mateyak MK, Sedivy JM. A large scale genetic analysis of c-Myc-regulated gene expression patterns. J Biol Chem. 2003;278(14):12563–12573. doi: 10.1074/jbc.M210462200. [DOI] [PubMed] [Google Scholar]
- O'Donnell KA, Yu D, Zeller KI, Kim JW, Racke F, Thomas-Tikhonenko A, Dang CV. Activation of transferring receptor 1 by c-Myc enhances cellular proliferation and tumorigenesis. Mol Cell Biol. 2005;26(6):2373–2386. doi: 10.1128/MCB.26.6.2373-2386.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M, Xu Y, Wonsey D, Lee LA, Dang CV. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. 2000;275(29):21797–21800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- Pirity M, Blanck JK, Schreiber-Agus N. Lessons learned from Myc/Max/Mad knockout mice. Curr Top Microbiol Immunol. 2006;302:205–234. doi: 10.1007/3-540-32952-8_8. [DOI] [PubMed] [Google Scholar]
- Poyton RO, McEwen JE. Crosstalk between nuclear and mitochondrial genomes. Annu Rev Biochem. 1996;65:563–607. doi: 10.1146/annurev.bi.65.070196.003023. [DOI] [PubMed] [Google Scholar]
- Prathapam T, Tegen S, Oskarsson T, Trumpp A, Martin GS. Activated Src abrogates the Myc requirement for the G0/G1 transition but not for the G1/S transition. Proc Natl Acad Sci U S A. 2006;103(8):2695–2700. doi: 10.1073/pnas.0511186103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla RC. Nuclear control of respiratory gene expression in mammalian cells. J Cell Biochem. 2006;97(4):673–683. doi: 10.1002/jcb.20743. [DOI] [PubMed] [Google Scholar]
- Schorl C, Sedivy JM. Loss of protooncogene c-Myc function impedes G1 phase progression both before and after the restriction point. Mol Biol Cell. 2003;14(3):823–835. doi: 10.1091/mbc.E02-10-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shio Y, Suh KS, Lee H, Yuspa SH, Eisenman RN, Aebersold R. Quantitative proteomic analysis of myc-induced apoptosis: a direct role for Myc induction of he mitochondrial chloride ion channel, mtCLIC/CLIC4. J Biol Chem. 2006;281:2750–2756. doi: 10.1074/jbc.M509349200. [DOI] [PubMed] [Google Scholar]
- Simpson N, Han Z, Berendzen K, Sweeney C, Oca-Cossio J, Constantinidis I, Stacpoole P. Magnetic resonance spectroscopic investigation of mitochondrial fuel metabolism and energetics in cultured human fibroblasts: effects of pyruvate dehydrogenase complex deficiency and dichloroacetate. Mol Genet Metab. 2006;89:97–105. doi: 10.1016/j.ymgme.2006.04.015. [DOI] [PubMed] [Google Scholar]
- Storey J, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad SciU S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sweet S, Singh G. Accumulation of human promyelocytic leukemic (HL-60) cells at two energetic cell cycle checkpoints. Cancer Res. 1995;55(22):5164–5167. [PubMed] [Google Scholar]
- Sweet S, Singh G. Changes in mitochondrial mass, membrane potential, and cellular adenosine triphosphate content during the cell cycle of human leukemic (HL-60) cells. J Cell Physiol. 1999;180(1):91–96. doi: 10.1002/(SICI)1097-4652(199907)180:1<91::AID-JCP10>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Takahashi H, McCaffery J, Irizarry R, Boeke J. Nucleocytosolic acetylcoenzyme a synthetase is required for histone acetylation and global transcription. Mol Cell. 2006;23:207–217. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- Tamura K, Hua B, Adachi S, Guney I, Kawauchi J, Morioka M, Tamamori-Adachi M, Tanaka Y, Nakabeppu Y, Sunamori M, Sedivy JM, Kitajima S. Stress response gene ATF3 is a target of c-myc in serum-induced cell proliferation. Embo J. 2005;24(14):2590–2601. doi: 10.1038/sj.emboj.7600742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercauteren K, Pasko RA, Gleyzer N, Marino VM, Scarpulla RC. PGC-1-related coactivator: immediate early expression and characterization of a CREB/NRF-1 binding domain associated with cytochrome c promoter occupancy and respiratory growth. Mol Cell Biol. 2006;26(20):7409–7419. doi: 10.1128/MCB.00585-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodnicka M, Guarino RD, Hemperly JJ, Timmins MR, Stitt D, Pitner JB. Novel fluorescent technology platform for high throughput cytotoxicity and proliferation assays. J Biomol Screen. 2000;5(3):141–152. doi: 10.1177/108705710000500306. [DOI] [PubMed] [Google Scholar]
- Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, Ooi HS, Orlov YL, Shahab A, Yong HC, Fu Y, Weng Z, Kuznetsov VA, Sung WK, Ruan Y, Dang CV, Wei CL. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci U S A. 2006;103(47):17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gao P, Fukuda R, Kuman G, Krishnamachary B, Zeller KI, Dang CV, Semenza GL. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of CMYC activity. Cancer Cell. 2007;11(5):407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Zhang X, DeSalle LM, McMahon SB. Identification of novel targets of Myc whose transcription requires the essential MbII domain. Cell Cycle. 2006;5(3):238241. doi: 10.4161/cc.5.3.2409. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.