Abstract
Plastidial phosphorylase (Pho1) accounts for ∼96% of the total phosphorylase activity in developing rice (Oryza sativa) seeds. From mutant stocks induced by N-methyl-N-nitrosourea treatment, we identified plants with mutations in the Pho1 gene that are deficient in Pho1. Strikingly, the size of mature seeds and the starch content in these mutants showed considerable variation, ranging from shrunken to pseudonormal. The loss of Pho1 caused smaller starch granules to accumulate and modified the amylopectin structure. Variation in the morphological and biochemical phenotype of individual seeds was common to all 15 pho1-independent homozygous mutant lines studied, indicating that this phenotype was caused solely by the genetic defect. The phenotype of the pho1 mutation was temperature dependent. While the mutant plants grown at 30°C produced mainly plump seeds at maturity, most of the seeds from plants grown at 20°C were shrunken, with a significant proportion showing severe reduction in starch accumulation. These results strongly suggest that Pho1 plays a crucial role in starch biosynthesis in rice endosperm at low temperatures and that one or more other factors can complement the function of Pho1 at high temperatures.
INTRODUCTION
The current view for starch biosynthesis in higher plants is that amylopectin, the major component of starch, is synthesized by the action of ADP glucose pyrophosphorylase (AGPase), starch synthases (SS), starch branching enzymes (BE), and starch debranching enzymes (DBE) (Nakamura, 2002; Ball and Morell, 2003). Furthermore, there is evidence that disproportionating enzyme (Colleoni et al., 1999; Ball and Morell, 2003) and α-glucan phosphorylase (Schupp and Ziegler, 2004; Dauvillée et al., 2006) are also involved in this process.
Two types of starch phosphorylase (Pho) were observed in the following organs of higher plants: spinach (Spinacia oleracea) leaves (Steup and Schächtele, 1981), pea (Pisum sativum) cotyledons (van Berkel et al., 1991), potato (Solanum tuberosum) leaves (St-Pierre and Brisson, 1995), fava bean (Vicia faba) cotyledons (Buchner et al., 1996), banana (Musa acuminata) fruits (da Mota et al., 2002), and wheat (Triticum aestivum) endosperm (Schupp and Ziegler, 2004). The two Pho types differ from each other not only in structure and kinetic properties but also in their expression pattern and subcellular localization (Steup, 1990). The plastidial isoform, referred to as Pho1 or Pho-L, has an extra 80–amino acid insertion not present in Pho2 and possesses a high affinity toward low molecular weight linear malto-oligosaccharides (MOS) and amylose, whereas the cytosolic isoform, Pho2 or Pho-H, has a high affinity toward highly branched polyglucans, such as glycogen (Shimomura et al., 1982; Steup, 1988; Yu et al., 2001).
Both Pho types catalyze a reversible reaction where glucose 1-phosphate (G1P) is used as a substrate to add a glucose unit to the nonreducing end of the α-glucan chain with the release of Pi or is generated as a product in the presence of Pi in the reverse reaction. Although the enzyme can contribute to both degradation and synthesis of starch in plant tissues, biochemical data seem to favor a degradative role in starch metabolism in nonphotosynthetic tissues. The estimated physiological levels of this hexose-phosphate are much lower than Pho1's apparent affinity (Km) for this substrate, indicating that it would have suboptimal activity in the synthesis direction (Preiss and Levi, 1980; Steup, 1988; Preiss and Sivak, 1996). However, no direct evidence for a role of Pho1 in starch biosynthesis or degradation has been obtained. An Arabidopsis thaliana mutant deficient in the plastidial form of α-glucan phosphorylase (At PHS1, homolog of Pho1 in rice [Oryza sativa]) does not exhibit a significant change in the starch synthesis rate, in net starch levels, in starch structure and composition, or in starch degradation in green leaves (Zeeman et al., 2004). Furthermore, antisense repression of a Pho1 gene in potato decreases the detectable Pho activity in leaves but has no major impact on the accumulation of starch (Sonnewald et al., 1995).
On the other hand, circumstantial evidence suggests the involvement of Pho1 in starch biosynthesis in storage tissues. Pho1 enzyme activity and protein levels are coordinated with starch production in potato tuber (Kossmann et al., 1991; St-Pierre and Brisson, 1995), maize (Zea mays) endosperm (Ozbun et al., 1973), and wheat endosperm (Schupp and Ziegler, 2004). The exact role of Pho1 in the biosynthesis of polysaccharides in both photosynthetic and nonphotosynthetic tissues of higher plants is still controversial and a subject of many discussions.
Recently, Dauvillée et al. (2006) found that a mutation in STA4, which encodes one of the two plastidial Pho genes in the unicellular green alga Chlamydomonas reinhardtii, significantly reduces starch content and results in abnormally shaped granules with a modified amylopectin structure and a high amylose content. These results indicate that PhoB is required for normal starch biosynthesis in the alga. Interestingly, PhoB has a low affinity for MOS, whereas PhoA, the other plastidial Pho in C. reinhardtii, has a high affinity for MOS as does the higher plant Pho1 (Dauvillée et al., 2006).
Mutational analysis is a powerful tool to identify and clarify the function of a given enzyme. We have generated numerous rice mutants altered in endosperm starch by N-methyl-N-nitrosourea (MNU) treatment of fertilized egg cells (Satoh and Omura, 1981; Satoh et al., 2003a). In this study, we screened seed extracts for a loss of polypeptide bands on SDS-polyacrylamide gels, and we have succeeded in identifying multiple allelic lines lacking the 106-kD Pho1 protein in rice endosperm. Pho1 deficiency specifically resulted in the modification of amylopectin structure, which altered the physicochemical properties of starch in rice endosperm. More dramatically, a significant percentage of the pho1 seeds of the mutant were shrunken with severely reduced starch content, a phenotype readily apparent when seed development occurs at lower temperatures. These results indicate that Pho1 plays a critical role in starch synthesis in the rice endosperm, presumably not only during the maturation of amylopectin but also during its initial stages of α-glucan biogenesis. Based on our results, the putative role of Pho1 in synthesizing starch granules in rice endosperm is discussed.
RESULTS
Presence of Two Forms of α-Glucan Pho in Rice Endosperm
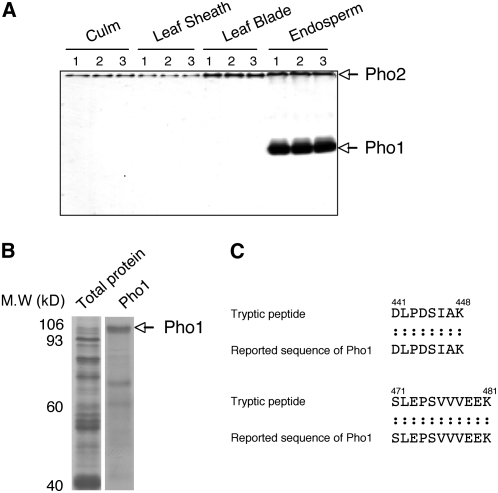
Pea cotyledons contain two Pho enzymes, Pho1 and Pho2, which differ in affinity for glycogen and in subcellular location. The amyloplast-localized Pho1 has a low affinity for glycogen, while the cytosolic Pho2 has a high affinity for this α-glucan (van Berkel et al., 1991). Native polyacrylamide gel electrophoresis in the presence of glycogen and subsequent zymogram analysis (Steup, 1990) reveals that developing rice endosperm contains two Pho activities with significant differences in electrophoretic mobility: a weak activity band located at the top of the polyacrylamide gel and a major activity band (Figure 1A) whose mobility was not significantly affected by the presence or absence of glycogen. Based on their electrophoretic mobilities, the results suggest that the strongly retarded and mobile activities are the cytosolic Pho2 and plastidial Pho1, respectively.
Figure 1.
Tissue-Specific Expression of Two Types of Phosphorylase in Rice.
(A) Activities of two starch Pho isoforms, Pho1 and Pho2, were detected by native-PAGE/enzymatic activity staining analysis in various tissues. The identification of Pho1 and Pho2 was based on our previous study (Yamanouchi and Nakamura, 1992). Lane 1, cv Kinmaze; lane 2, cv Taichung 65 (T65); lane 3, EM583 (a wx mutant derived from the MNU treatment of T65).
(B) SDS-PAGE analysis of Pho1 excised from the native-PAGE gel from rice developing endosperm.
(C) Primary sequences of a tryptic peptide from the Pho1 protein obtained by liquide chromatography–tandem mass spectrometry analysis and alignment with the unique insertion sequences of Pho1 reported in the database (NCBI nr).
The Pho2 activity was observed not only in endosperm but also in photosynthetic organs (culm, leaf sheath, and leaf blade), while a dominant Pho1 activity was detected in developing endosperm (Figure 1A). These findings are consistent with a previous report (Ohdan et al., 2005) that shows that the transcript levels of Pho1 are ∼6-fold higher than that of Pho2 in the rice endosperm, whereas Pho2 is 10-fold higher than Pho1 in the rice leaf blade.
Identification of the 106-kD Pho1 Protein by Mass Spectrometry
After detecting Pho1 activity by zymogram analysis (Figure 1A), the native polyacrylamide gels were stained with Coomassie blue. The corresponding Pho bands were excised, extracted, and then analyzed by SDS-PAGE. The Pho1 protein was detected as a band with a molecular mass of 106 kD (Figure 1B). Liquid chromatography–tandem mass spectrometry analysis of the tryptic peptide fragments derived from the 106-kD protein showed that the peptide sequences aligned with the rice Pho1 primary sequence consisting of 928 amino acids (National Center for Biotechnology Information [NCBI] BLAST of gil13195340) (Figure 1C; see Supplemental Figure 1 online). Moreover, the unique insertion sequence (80 amino acids from 439N to 518V), specific to the Pho1 isoform, was observed.
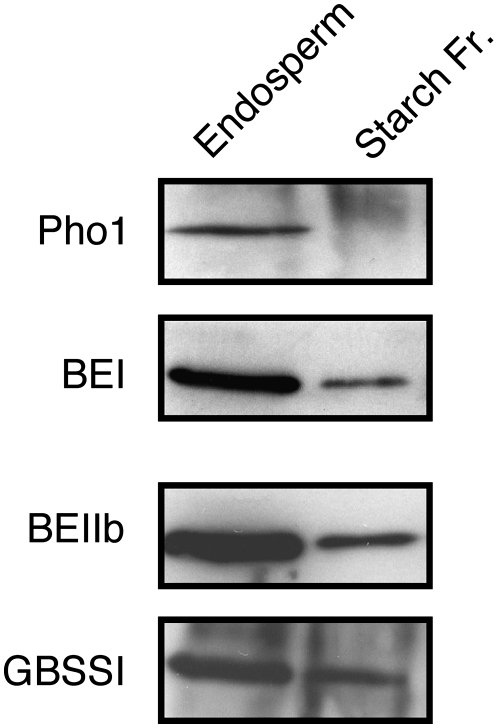
Pho1 Is Localized in the Stromal Compartment of Rice Endosperm Amyloplasts
Pho1 is localized in amyloplasts, the site of starch synthesis (Yu et al., 2001). Several starch biosynthetic enzymes, such as starch synthase I (SSI), are localized in the amyloplast stroma and are also associated with starch (Fujita et al., 2006). Unlike the amyloplast resident marker enzymes BEI and BEIIb, which were detected in both the stroma and starch fractions or the starch-localized GBSSI, only trace amounts of Pho1 were starch bound (Figure 2). Hence, Pho1 is restricted to the stroma compartment of the amyloplasts. A stroma location was also observed for maize endosperm Pho1 (Yu et al., 2001).
Figure 2.
Subcellular Localization of Pho1 in Developing Endosperm Cells of Rice.
Total endosperm proteins and proteins associated with starch granules of mid-developing seeds were resolved by SDS-PAGE and then subjected to immunoblotting for Pho1, BEI, BEIIb, and GBSSI. “Endosperm” denotes the total proteins extracted from the endosperm of developing seeds.
Screening and Isolation of Pho1-Deficient Mutant Lines
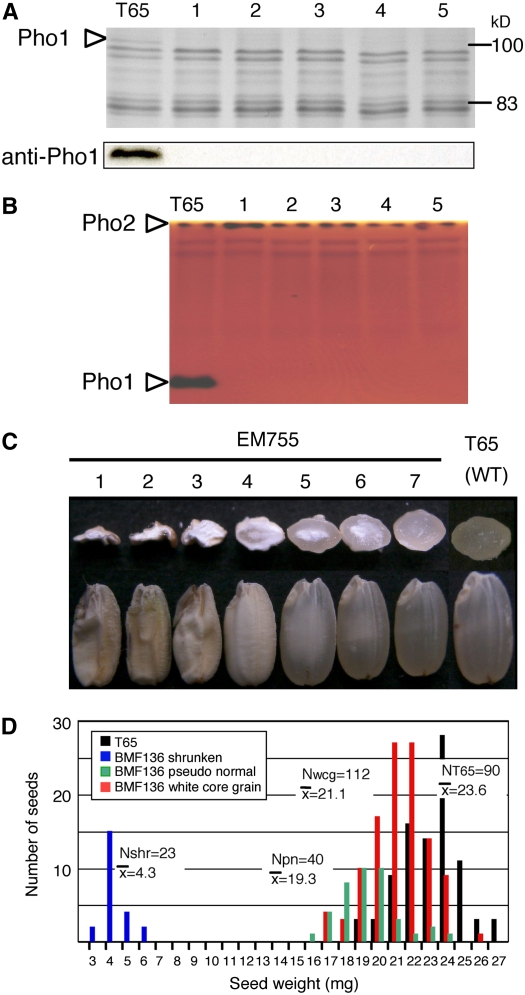
Mature seeds of >1700 mutant lines generated by MNU treatments of the Kinmaze and Taichung 65 (T65) japonica rice cultivars were screened for Pho1 mutants by SDS-PAGE analysis. Many mutant lines were identified that lacked the 106-kD polypeptide completely (Figure 3A) but did not exhibit any significant alterations in the expression of the other major endosperm proteins when assessed by SDS-PAGE. The complete loss of Pho1 was verified by protein gel blot analysis using anti-Pho1 (Figure 3A). Native-PAGE/activity staining analysis also showed the absence of the Pho1 activity band in the mutant lines, while the activity levels of Pho2 were comparative to that of the wild type (Figure 3B), indicating that expression of Pho1 and Pho2 are regulated independently.
Figure 3.
Effects of pho1 Mutation on Pho1 Content and Activity in Rice Endosperm and on Morphology of the Kernel.
(A) SDS-PAGE (top panel) and protein gel blot (bottom panel) profiles of Pho1 protein from the maturing endosperm of five allelic mutant lines induced by independent MNU treatment. Lane 1, EM755; lane 2, EM719; lane 3, EM640; lane 4, EM786; lane 5, EM876. The total proteins extracted from a single brown rice seed for each mutant line and T65 were resolved by SDS-PAGE and analyzed by immunoblotting using anti-Pho1.
(B) Zymogram depicting the Pho activities of wild-type T65 and the same five allelic mutant lines as in (A). Plastidial Pho1 activity moved down the gel, while the cytosolic Pho2 activity band is located at the top of the polyacrylamide gel. The activity responsible for the pair of lighter bands below Pho2 activity has not been identified.
(C) and (D) pho1-induced changes in grain morphology (C) and grain weight (D).
(C) Mature seeds from a single panicle of the pho1 mutant, EM755, grown under field conditions were analyzed. Views of a representative sample of sectioned (top row) and intact (bottom row) seeds are shown. Three distinct grain types were identified based on size and endosperm appearance severely shrunken (lanes 1 to 3), white-core grain (possessing a chalky endosperm core) (lanes 4 to 6), and pseudonormal (lane 7).
(D) Distribution of grain weight among the various seed phenotypes. The mean weight for each seed phenotype is shown. White-core grain and pseudonormal seeds were smaller than wild-type T65. Although exhibiting a vitreous appearance much like T65, pseudonormal seeds were ∼8% smaller than white-core seeds. Shrunken seeds weighed only ∼18% of wild-type T65. Nshr, number of shrunken seeds; Npn, number of pseudonormal; Nwcg, number of white-core grains; NT65, number of wild-type T65 seeds.
The total α-glucan Pho activity in the mutant endosperm (0.41 ± 0.03 nmoles/min/endosperm) was ∼3.6% that of the wild type (11.3 ± 0.12 nmoles/min/endosperm). Assuming that the activity of the cytosolic form is unaffected by the mutation, we estimated that Pho1 and Pho2 activities account for 96.4 and 3.6% of the total Pho activity, respectively. This estimation of the relative levels of these enzymes was supported by the intensities of their activity bands on zymograms (Figures 1A and 3B).
CDNAs (cDNAs) for Pho1 and Pho2 have been identified and isolated in developing rice seeds (Ohdan et al., 2005). Using this information, we generated sequence-specific primers and conducted real-time RT-PCR analysis to estimate Pho1 and Pho2 transcript levels. The analysis showed that Pho1 and Pho2 transcripts were present at 31,850 ± 1936 and 842 ± 164 copies per ng total RNA, respectively, in a pho1 mutant line (BMF136), whereas they occurred at 53,800 ± 15,300 and 1198 ± 236 copies per ng total RNA, respectively, in the wild type T65 (see Supplemental Figure 2 and Supplemental Table 1 online). The presence of Pho1 RNA in this mutant line indicates that the loss of Pho1 protein in the mutant was predominantly due to posttranscriptional regulation of the pho1 gene.
Grain Morphology of pho1 Mutants
More than 15 plastidial Pho1-deficient mutants were isolated by independent mutation treatments of the rice cultivars Kinmaze and T65 (representative examples of some of these are shown in Figure 3 and Supplemental Figure 3 online). Surprisingly, in the 15 lines examined, all produced seeds with marked variation in morphology, ranging from a pseudonormal phenotype, comparable in appearance but much lower in weight than the wild type, to a shrunken phenotype with extremely reduced starch levels. Examples of seed variability are depicted in Figure 3C for the mutant line EM755 (see also Figure 10A). Most of the seeds (65%) exhibited a white-core endosperm that varied in size. Approximately 23% were of the pseudonormal type, displaying a vitreous endosperm but with a grain weight of ∼8% less than seeds containing a white-core endosperm, which, in turn, were 10% lighter than wild-type seeds. A smaller percentage (13%) of the seeds were severely shrunken (Figure 3D). The shrunken seeds were randomly distributed along the panicle length (see Supplemental Figure 4 online), indicating that this extreme phenotype was not due to possible differences in nutrient partitioning along the panicle.
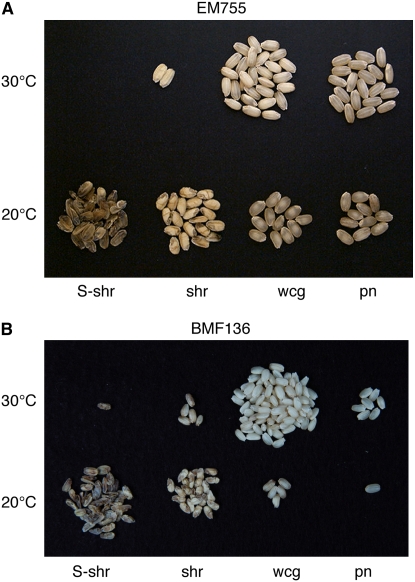
Figure 10.
Effects of Temperature on Kernel Morphology during Development of pho1 Mutant Seeds.
Mutant plants of EM755 (A) and BMF136 (B) were removed from the field plot at the maximum flowering stage and grown at temperatures of either 30°C (top rows) or 20°C (bottom rows) until they reached maturity. S-shr, severely shrunken grains; shr, shrunken grains; wcg, white-core endosperm grains; pn, pseudonormal grains.
This variability in seed morphology was observed in every generation examined annually since 2002. Moreover, seeds exhibited the same variation in morphology regardless of whether they were the progeny of a self-pollinated plant grown from a shrunken, white-core, or pseudonormal seed. As this polymorphism in grain phenotype was observed not only in all of the allelic pho1 mutant lines derived from several independent MNU treatments of cv T65 but also in those obtained from the cv Kinmaze, we conclude that this variation in seed morphology and, in turn, starch accumulation is caused by the pho1 mutation. Thus, Pho1 appears to be essential for normal starch biosynthesis.
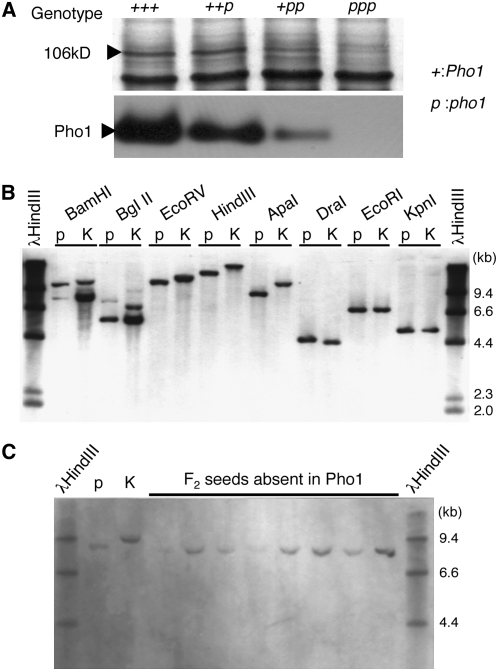
Gene Dosage Effects of pho1 Mutant Gene on the Activity Staining and Protein Gel Blot Band Patterns of Pho1
The 15 selected Pho1 mutants induced independently by MNU treatments of Kinmaze and T65 were crossed with each other and the F1 and F2 segregating populations were examined. All of the F1 seeds derived from the crosses between independent Pho1-deficient mutant lines and progeny from subsequent F2 populations were missing the Pho1 protein on SDS polyacylamide gels, indicating that they contain an allelic mutation at the same locus resulting in Pho1 deficiency. The segregation of F2 seeds derived from an initial cross with the wild-type parent fitted well with the expected 3:1 ratio of a single gene inheritance pattern (Table 1), indicating that this pho1 mutation is determined by a single gene, which we tentatively named pho1 (for plastidial starch phosphorylase1 missing mutation). Restriction fragment length polymorphism (RFLP) analysis using Pho1 cDNA as a probe showed no recombination between the Pho1 gene and pho1 mutation type (pho1) (Figures 4B and 4C), suggesting that the pho1 mutation is caused by the lesion of a structural gene encoding Pho1. The Pho1 gene was also mapped to the same position on chromosome 3 occupied by a BAC clone containing a putative Pho1 gene (see Supplemental Figure 5 online) as reported by the International Rice Genome Sequencing Project (2005).
Table 1.
Segregation Mode of Normal and pho1 Phenotypes and Their Grain Weight in F2 Derived from Crosses between pho1 Mutant Lines and the Parental Cultivar T65
| Segregation in F2
|
||||||
|---|---|---|---|---|---|---|
| Cross Combination | +
|
pho1
|
||||
| F1 | Normala | wcgb | shrc | Total | χ2(3:1) | |
| T65/EM755-1 | Normal | 69 | 21 | ND | 90 | 0.1 |
| (21.8 ± 1.3) | (20.1 ± 1.7) | |||||
| T65/EM755-2 | Normal | 63 | 19 | 4 | 86 | 0.1 |
| (21.8 ± 0.8) | (20.2 ± 1.1) | (4.1 ± 0.1) | ||||
| T65/EM640 | Normal | 61 | 23 | ND | 84 | 0.1 |
| (24.6 ± 1.5) | (23.0 ± 1.6) | |||||
Average grain weight (mg) is given in parentheses (means ± se).
Grains with a normal phenotype.
wcg, white-core grains.
shr, shrunken grains.
Figure 4.
Genetic Analysis of the pho1 Mutation in Rice.
(A) Gene dosage effects of pho1 mutation on protein amount (top) and activity (bottom) of plastidic α-glucan Pho in rice endosperm. “+” and “p” tentatively represent a wild-type and mutant gene, respectively. Since the endosperm has a gene dosage of 3, the genotypes of the wild type (T65), F1 derived from a cross between a T65 female parent and a EM755 male parent, F1 from a cross between a EM755 female parent and a T65 male parent, and a pho1 mutant (EM755) are represented as “+++,” “++p,” “+pp,” and “ppp”, respectively. Pho1 protein amounts were detected by SDS-PAGE of protein extracted from respective mature seeds. The activity of Pho1 was measured by native-PAGE/enzymatic activity staining analysis.
(B) RFLP analysis of the Pho1 gene. The Pho1 digestion products were detected by DNA gel blot analysis using DNA prepared from leaf blades of the pho1 mutant EM755 (p) and of an indica rice cultivar Kasalath (K).
(C) Segregation of an RFLP marker in homozygous pho1/pho1 F3 plants. DNA from pho1/pho1 F3 plants (p), which were selected by SDS-PAGE analysis, was digested with ApaI. K represents DNA from the Kasalath cultivar that underwent the same digestion analysis.
When the EM755 pho1 mutant line was crossed in a reciprocal manner with a wild-type cultivar, T65, the clear dosage gene effects of Pho1 were observed not only at the enzyme activity level but also at the protein level (Figure 4A). The expression level of Pho1 protein increased linearly with an increase in the number of dominant Pho1 genes. These results suggest that the lack of Pho1 is caused by a nonsense mutation in the Pho1 gene.
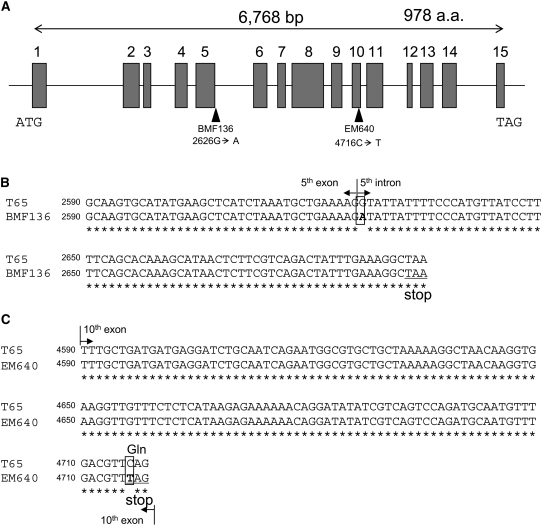
Analysis of Nucleotide Sequences of the Pho1 Gene from the pho1 Mutants
Genomic DNA sequences of the Pho1 gene in two pho1 mutant lines, BMF136 (a near isogenic line for the pho1 mutation derived from a cross between T65 and EM755) and EM640, as well as of the parent cultivar T65, were determined to identify the mutation points in the two pho1 mutant lines. The rice gene encoding Pho1 spans 6768 bp and consists of 15 exons and 14 introns (Figure 5A). MNU almost exclusively induces G:C to A:T transitions in mammalian genes (Engelbergs et al., 2000; Cooks et al., 2003). A similar tendency of mutagenesis by MNU was found in the treatment of the fertilized egg cell in rice, in which most of the mutations were observed to be nucleotide transitions of G:C to A:T by TILLING analysis (Suzuki et al., 2008). Comparison of the nucleotide sequences showed that both pho1 mutants contained single nucleotide substitutions that would potentially reduce Pho1 expression by altering synthesis of the intact 106-kD protein. BMF136 contained a G-to-A transition at nucleotide position 2626 located at the 5′ end of the fifth intron (Figure 5B). In this instance, the highly conserved border sequence at the splice site was disrupted, resulting in a loss of intron splicing (Brown, 1996). Retention of the intron would result in a frame shift and the insertion of a premature stop codon 66 nucleotides downstream of the mutation site resulting in a truncated polypeptide. A similar MNU-induced nonsense mutation near the splicing site was observed in the rice SSIIIa gene (Fujita et al., 2007). In EM640, a C-to-T transition occurred at nucleotide 4716, rendering a codon change from CAG (Gln) to a TAG stop codon in the tenth exon (Figure 5C). The reduction of gene expression by nonsense mutation mediated mRNA decay is well documented (Maquat, 2004) and is likely the basis for the absence of 106-kD Pho1 mRNA in EM640. These results demonstrate that Pho1 is the structural gene coding for the 106-kD plastidial Pho and is responsible for the abnormal accumulation of starch.
Figure 5.
Schematic Representation of Mutations in the Pho1 Gene of BMF136 and EM640.
(A) Schematic representation of the Pho1 gene and the locations of single nucleotide substitutions in pho1 mutant lines BMF136 and EM640. ATG and TAG indicate the initiation and termination codons, respectively. Gray boxes, lines, and arrowheads indicate exons, introns, and mutation sites, respectively. a.a., amino acid.
(B) Alignment of the Pho1 gene from T65 with the pho1 gene from the BMF136 mutant. The splice site border sequence between the fifth exon and intron (at nucleotide 2626) was disrupted by a G-to-A mutation, which results in intron retension and generates a stop codon 66 nucleotides downstream of the mutation site.
(C) Alignment of the Pho1 gene from T65 with the pho1 gene from the EM640 mutant. In EM640, a C-to-T transition occurred at nucleotide 4716 in the tenth exon, and this resulted in a codon change from CAG (Gln) to a stop codon (TAG).
The pho1 Mutations Have No Significant Effects on the Activities of the Major Starch Biosynthetic Enzymes
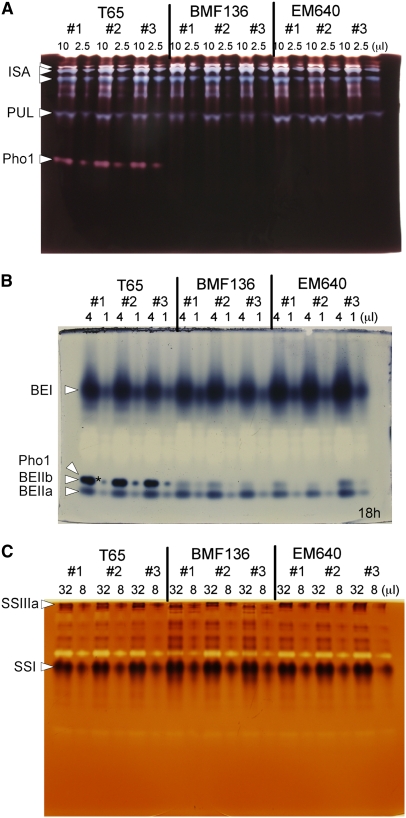
To examine possible pleiotropic effects of the pho1 mutation on other starch synthesizing enzymes, zymogram analyses were performed on two pho1 mutant lines, BMF136 and EM640, and the wild-type T65. No significant differences in activities of DBE isozymes (isoamylase and pullulanase) (Figure 6A), BE isoforms (BEI, BEIIa, and BEIIb) (Figure 6B), and SS isoforms (SSI and SSIIIa) (Figure 6C) were found between the pho1 mutants and their wild-type parental cultivar T65. AGPase activity was also found not to be significantly different in the mutant (4.14 ± 0.33 nmoles/min/endosperm) than in the wild type (3.28 ± 0.25 nmoles/min/endosperm). Therefore, mutation of the Pho1 gene encoding the plastidic Pho showed no obvious pleiotropic effects on the activity levels of the other major starch biosynthetic enzymes.
Figure 6.
Effects of pho1 Mutations on Activities of Starch Biosynthetic Enzymes.
Native-PAGE/activity staining analysis was performed using enzymes from three developing endosperms of pho1 mutants BMF136 and EM640 and of their wild-type parent, T65. The numbers above the lanes indicate the volume (μL) of crude enzyme extract applied to each gel lane. The activity bands for isoamylase, pullulanase, and Pho1 (A), for branching enzymes, BEI, BEIIa, and BEIIb, and for Pho1 (B), and for SSI and SSIIIa (C) are indicated with arrowheads. Note that the activity bands for BEIIb and Pho1 are not resolved and overlap on the polyacrylamide gel as denoted by the asterisk in (B), as previously described (Yamanouchi and Nakamura, 1992). Note that the staining intensity of this band is mainly due to Pho1 activity, which is absent in BMF136 and EM640.
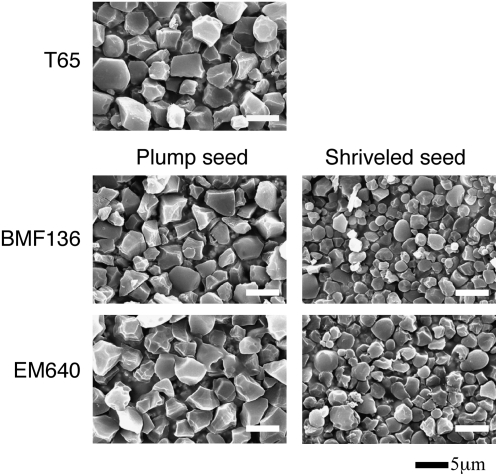
The pho1 Mutations Altered the Morphological Properties of Starch Granules in Rice Endosperm
Scanning electron microscopy revealed that the pho1 mutation resulted in modification in starch granule morphology (Figure 7). Starch granules from plump pho1 seeds from pho1 mutant lines BMF136 and EM640 were slightly smaller than wild-type seeds, and some granules were more spherical than the irregular polyhedron-shaped granules typical of wild-type starch grains. Starch granules from shrunken seeds were markedly smaller than those from wild-type seeds, with the larger granules having an irregular spherical shape instead of the normal irregular polyhedron shape evident for wild-type large starch granules. The x-ray diffraction patterns of endosperm starch granules were similar between the pho1 mutants and the wild-type T65 (see Supplemental Figure 6 online).
Figure 7.
Effects of pho1 Mutation on Starch Granules as Viewed by Scanning Electron Microscopy.
Starch granules were prepared from plump or shrunken seeds from the pho1 mutant lines BMF136 and EM640 and their wild-type parental cultivar, T65. Bars = 5 μm.
Effects of the pho1 Mutation on the Fine Structure of Amylopectin in the Mature Endosperm
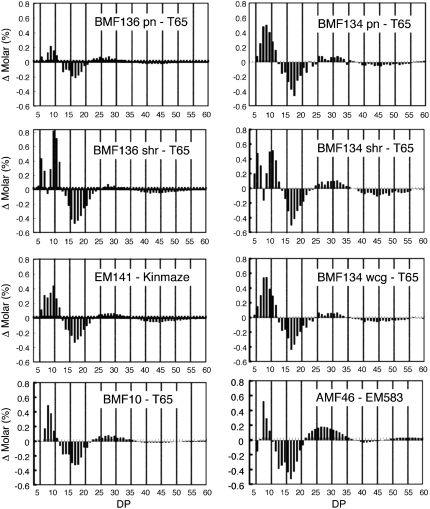
To examine the structural changes of amylopectin in the endosperm of pho1 mutants, the chain length distribution of amylopectin isoamylolysates was determined using high-resolution capillary electrophoresis. Figure 8 presents the changes in chain length profiles of amylopectins between three selected pho1 mutant lines and their wild-type parent, T65 or Kinmaze. The pho1 mutants had a higher proportion of short chains with a degree of polymerization (DP) ≤ 11 and a lower proportion of intermediate chains with a DP of 13 to 21. Interestingly, in most instances, differences in chain length distribution between the pho1 and parental wild-type starches showed a sinusoidal pattern, which was strongly attenuated as the chain length increased. Although the changes in chain length distribution were consistently observed in the pho1 mutants, the changes were relatively small. This indicates that Pho1 is not a major activity but simply a modifier in the elongation of amylopectin, a process performed by SS, BE, and DBE activities.
Figure 8.
The Change in Chain Length Distribution of Amylopectin in Rice Endosperm between Various pho1 Mutants and Their Wild-Type Parents (T65 or Kinmaze) as Determined by the APTS-Capillary Electrophoresis Method.
The change in the molar amounts (y axis) of specific size chains (x axis) between various pho1 mutants and their wild-type parent, T65 or Kinmaze, are depicted. For pho1 mutants BMF136 and 134, differences in chain length distribution for pseudonormal (pn), shrunken (sh), and white-core grain (wcg for BMF 134 only) from wild-type T65 are shown. BMF136 and BMF134 are near isogenic lines derived from crosses between the parent cultivar, T65, and EM755 and EM719, respectively. BMF10 is a near isogenic line from a cross between cv Kinmaze and EM141. AMF46 is an amylose-free pho1 double mutant line (pho1/pho1 wx/wx) derived from a cross between a waxy mutant (wx, EM583) and pho1 mutant EM755.
In spite of large differences in seed weight (Figure 3), the chain length distribution pattern of amylopectin was nearly the same for the pseudonormal seed and the shrunken and white-core seed of the pho1 mutant lines BMF134 and BMF136. In the range of DP ≤ 25, amylopectin chain length differences between the pho1 mutants and T65 were somewhat more significant in the shrunken seed than the pseudonormal seed (Figure 8). Collectively, these results suggest that Pho1 operates at two distinct phases of starch biosynthesis, one phase consisting of starch initiation and a second phase encompassing starch elongation.
The pho1 Mutation Changes the Gelatinization Properties of Starch in Rice Endosperm
To determine if mutation in the Pho1 gene affects the physicochemical properties of starch, the thermal properties of starch from wild-type and pho1 plump white-core and shrunken kernels were analyzed by differential scanning calorimetry (DSC) (Table 2). Onset gelatinization temperatures (To) and enthalpy changes (ΔH) as well as peak (Tp) and conclusion (Tc) temperature were significantly lower in the white-core endosperm pho1 seeds than in the wild type (T65). The To value of starch from shrunken seeds was much lower than that from mutant seeds with plump kernels, but shrunken seeds had the highest Tp and lowest ΔH values. The lower amount of energy required to gelatinize pho1 starch is consistent with its increased levels of smaller chains relative to the wild type (Figure 8).
Table 2.
Effects of pho1 Mutation on the Gelatinization Properties of Starch in Rice as Determined by DSC
| To (°C)a | Tp (°C)b | Tc (°C)c | ΔH (mJ/mg)d | ||
|---|---|---|---|---|---|
| T65 | 51.7 ± 0.3 | 59.6 ± 0.2 | 72.6 ± 0.5 | 8.7 ± 0.3 | |
| BMF136 | wcge | 47.3 ± 0.3 | 56.6 ± 0.2 | 69.8 ± 0.4 | 6.8 ± 0.3 |
| BMF136 | shrf | 39.8 ± 1.2 | 60.5 ± 0.1 | 70.0 ± 1.0 | 4.2 ± 0.8 |
Onset temperature.
Peak temperature.
Conclusion temperature.
Gelatinization enthalpy of starch.
wcg, white-core grains.
shr, shrunken grains.
Characterization of the Chain Elongation Reaction Catalyzed by Pho1 and SSIIa from Rice
The chain elongation properties of rice Pho1 were compared with those of rice SSIIa by analyzing their reaction products using MOS of DP4, DP6, or DP7 as primers. In this in vitro analysis, purified preparations of recombinant rice enzymes of Pho1 (rPho1) and SSIIa (rSSIIa) expressed in Escherichia coli were used. Regardless of the initial primer used in the reaction, the two enzymes showed different product distributions to each other. While rPho1 produced a broad distribution of MOS products of increasing size, rSSIIa showed a much narrower distribution of MOS products (see Supplemental Figure 7 online).
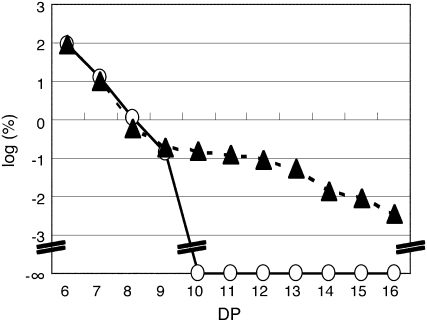
To precisely compare the action pattern of products generated by rPho1 and rSSIIa, the mol % of MOS products generated by each of these enzymes when ∼13% of the substrate maltohexaose was metabolized was plotted on a logarithmic scale (Figure 9). Strikingly, rPho1 synthesized MOS with DP values of at least 16, while rSSIIa only produced MOS with DP values of 7 to 9. The results indicate that Pho1 can synthesize much longer linear glucans than SSIIa.
Figure 9.
Comparison of Chain Elongation Reaction Products of rPho1 and rSSIIa Using MOS as Primers.
Specific MOS products were generated by rPho1 (35 ng protein, closed triangles) in 5 min and by rSSIIa (10.8 μg protein, open circles) in 60 min, using maltohexaose as a substrate. The percentage of product formed for each MOS was plotted on a logarithmic scale. Note that under these conditions similar amounts of maltohexaose were metabolized by rPho1 and rSSIIa (12.9 and 13.5%, respectively) and that no substantial amounts of MOS with DP ≥10 were synthesized by rSSIIa.
Starch Accumulation in the Endosperm of the pho1 Mutant Was Affected by Temperature
As mentioned, the pho1 mutation mediates significant polymorphic variation in grain morphology, starch structure, and starch content among seeds within the same panicles of homozygous pho1 plants (Figure 3C). In addition, this polymorphism in seed morphology and weight and in starch modifications was stable from one generation to the next. There was a tendency, however, for seeds derived from late flowering panicles to have a lower starch content than those borne by earlier flowering panicles. As most of the rice plants flowered in early September with temperatures gradually decreasing during the month, we postulated that the pho1-induced seed polymorphism was temperature dependent.
To test this hypothesis, wild-type and pho1 homozygous plants were transferred from the paddy field to the phytotron immediately after flowering and grown at 20, 25, or 30°C (Table 3). After maturation, the morphology and weight of the harvested seeds were compared. Mature seeds of the wild-type T65 exhibited little variation in grain weight, regardless of the temperature condition (Table 3). Hence, starch accumulation in the endosperm of wild-type rice is not significantly influenced by these temperature conditions. While seeds of the pho1 mutants BMF136 and EM755 displayed the usual polymorphism in seed morphology seen in field-grown plants, the frequency of the various grain phenotypes and extent of starch accumulation differed markedly depending on the growth temperature (Figure 10, Table 3). At 30°C, nearly all (94 to 96%) of the seeds produced from mutant plants contained near normal starch levels, of which the bulk were of the white-core endosperm type (Table 3). Only 4 to 6% of the seeds were of the shrunken type, which accumulated <10 mg of starch. Similar to plants grown in the field, these shrunken seeds had a chalky endosperm (Figure 10). Chalky indicates that the seeds were not vitreous (glass-like) in appearance. The white-core endosperm contains a chalky core (see sectioned seeds in Figure 3C). The frequency of shrunken endosperm grains increased significantly when the growth temperature was lowered, with a corresponding decrease in the number of seeds displaying the pseudonormal and white-core endosperm seed phenotypes. When grown at 25°C, the frequency of shrunken seeds produced by EM755 or BMF136 increased to 35 to 39% of the harvested seeds (Table 3). The frequency of shrunken seeds and extent of inhibition of starch accumulation was more pronounced when seed development occurred at 20°C than at higher temperatures (Figure 10). More than two-thirds of the total seeds of EM755 or BMF136 showed a shrunken or severely shrunken phenotype, with the latter type predominating (Table 3). The severely shrunken seeds were less than half the weight (5 mg) of shrunken seeds and accumulated less than one-twentieth of the amount of starch that the wild type did. Unlike the chalky endosperm of shrunken seeds, the severely shrunken seeds were transparent. Interestingly, the seed weight distributions displayed by severely shrunken and shrunken seeds did not overlap but were markedly discontinuous (Table 3). This marked discontinuity in weight distribution by severely shrunken and shrunken seeds suggests that normal starch accumulation requires at least two Pho1-dependent events.
Table 3.
Frequency of Four Grain Phenotypes in pho1 Mutant Seeds Developed under Three Different Temperature Conditions
| Exp. 1 (EM755)
| ||||||
|---|---|---|---|---|---|---|
| 30°C
|
25°C
|
20°C
|
||||
| Type | No. (%) | G.W. (mg) | No. (%) | G.W. (mg) | No. (%) | G.W. (mg) |
| T65 | 20 (100) | 22.8 ± 1.2 | 20 (100) | 22.3 ± 1.1 | 20 (100) | 24.8 ± 0.8 |
| EM755 | ||||||
| pn | 20 (39) | 20.2 ± 1.2 | 16 (18) | 22.0 ± 1.3 | 12 (16) | 21.4 ± 1.2 |
| wcg | 29 (57) | 19.6 ± 2.8 | 42 (47) | 23.8 ± 1.1 | 12 (16) | 20.6 ± 1.0 |
| shr | 2 (4) | 10.8 ± 0.2 | 31 (35) | 5.6 ± 0.9 | 23 (32) | 9.5 ± 2.5 |
| S-shr | 0 (0) | 0 (0) | 26 (36) | 4.4 ± 0.7 | ||
| Total | 51 (100) | 89 (100) | 73 (100) | |||
| Exp. 2 (BMF136)
| ||||||
| 30°C
|
25°C
|
20°C
|
||||
| Type | No. (%) | G.W. (mg) | No. (%) | G.W. (mg) | No. (%) | G.W. (mg) |
| T65 | 30 (100) | 22.4 ± 1.8 | 30 (100) | 22.5 ± 2.1 | 30 (100) | 22.9 ± 1.7 |
| BMF136 | ||||||
| pn | 18 (17) | 20.9 ± 3.4 | 26 (24) | 22.1 ± 1.1 | 2 (3) | 19.5 ± 0.3 |
| wcg | 82 (77) | 18.4 ± 3.7 | 40 (37) | 22.7 ± 2.5 | 15 (19) | 20.5 ± 1.3 |
| shr | 6 (6) | 10.0 ± 1.2 | 41 (39) | 6.6 ± 1.5 | 24 (31) | 7.9 ± 1.4 |
| S-shr | 0 (0) | 0 (0) | 36 (47) | 3.6 ± 1.0 | ||
| Total | 106 (100) | 107 (100) | 77 (100) | |||
G.W., grain weight; pn, pseudonormal grain; wcg, white-core endosperm grain; shr, shrunken grain; S-shr, severely shrunken grain.
To confirm the effect of pho1 mutation on the grain filling phenotypes under low temperature, a similar experiment was performed using a near isogenic line, BMF136. The same phenomena were observed in this experiment (Figure 10B, Table 3, Exp. 2), indicating that grain filling was severely reduced in the pho1 mutant seeds relative to the wild type, especially when they were developed under low temperature conditions.
Overall, our results indicate that the plastidic Pho1 is essential for normal starch synthesis and accumulation and that the wide variation in starch accumulation in pho1 seeds grown in the field is caused by growth temperature conditions.
DISCUSSION
We have identified and studied 15 independent rice endosperm mutants that lack the Pho1 protein due to a lesion in the Pho1 gene (Figures 1 to 5). We showed that Pho1 plays a crucial role in starch synthesis in plastids of nonphotosynthetic storage tissue of higher plants. Although the shrunken4 mutant of maize was reported to lack Pho activity, the exact genetic lesion responsible for the shrunken phenotype remains unidentified (Tsai and Nelson, 1969; Burr and Nelson, 1973). Our study describes a novel rice grain phenotype, distinct from other cereal starch mutants, that implicates Pho1 in starch biosynthesis in the rice endosperm. The phenotype of the rice pho1 mutant was not predictable from previous studies of leaf starch metabolism in the Arabidopsis pho1 mutant (phs1) (Zeeman et al., 2004), indicating that Pho1 probably plays different roles in photosynthetic and nonphotosynthetic organs.
It is particularly interesting that pho1 grain phenotype can be classified into two types: a shrunken endosperm type containing less than one-fifth of the wild type starch levels that predominated at 20°C and a white-core/pseudonormal endosperm type containing nearly normal starch levels, which predominated at 30°C (Table 3). The starch level in the shrunken endosperm of the pho1 mutant seed (4.2 ± 0.1 mg/grain) was lower than that detected in the rice endosperm shr2 allele (5.6 ± 1.9 mg/grain), which codes for a defective AGPase subunit (see also Satoh et al., 2003a; Ohdan et al., 2005). Since AGPase provides the substrate for starch synthesis, the more severe reduction in starch levels by pho1 than that seen for shr2 supports an essential role for Pho1 activity for maximum starch biosynthesis. In addition, Pho1 had a higher transcript level during the early stages of seed development than did AGPase (Ohdan et al., 2005). In the potato tuber, Pho1 gene expression was also maximal during the early developmental stages of starch accumulating tissues (Mori et al., 1991). The early temporal expression of Pho1 during seed development together with the starch deficient phenotype of the shrunken seeds indicate that pho1 is essential for one or more early steps of starch biosynthesis in the rice endosperm.
In this study, the mode of elongation of MOS of DP6 by the rice endosperm proteins Pho1 and SSIIa was compared using recombinant enzymes (rPho1 and rSSIIa). The in vitro elongation experiments shown in Figure 9 indicate that Pho1 has a significantly higher capacity to synthesize long linear glucans from small MOS than SSIIa does. Remarkably, potato tuber Pho1 can synthesize amylose with molecular weights of up to the order of 106 in the presence of G1P and maltopentaose (Kitamura et al., 1982). These results are consistent with the idea that Pho1 has the potential to play an important role in synthesizing linear glucans, which can serve as linear substrates for BE to form branched glucans in the starch initiation process (Borovsky et al., 1976). It has been suggested that Pho1 preferentially acts on linear glucans, while Pho2 acts preferentially on branched glucans (Shimomura et al., 1982; Steup, 1988). In addition, the catalytic activity of rPho1 from rice is significantly higher (75 μmoles G1P/mg protein/min) toward MOS than rSSIIa is (24 nmoles ADPglucose/mg protein/min). Therefore, these results support a role for Pho1 in extending small MOS, whereas rice SSIIa is unlikely to be involved in this process.
Our genetic and biochemical results indicate that Pho1 activity is a major factor in starch biosynthesis of the maturing rice endosperm. Although the exact role is not known, several possibilities exist. First, although past kinetic analysis of this enzyme suggested otherwise (Preiss et al., 1980), Pho1 might play a crucial role in starch granule formation at a very early stage of the grain filling process (Tsai and Nelson., 1969). Based on the very high Km for G1P and the estimated low concentrations of this substrate in the plastid, Pho1 was suggested to play mainly a degradative role in starch metabolism (Preiss and Sivak, 1996). However, recent kinetic studies of Pho1 support a role for this enzyme in starch biosynthesis (S.-K. Hwang, H. Satoh, and T.W. Okita, unpublished data). Incorporation of 14C-G1P into starch was only partially affected by Pi. Even under physiological G1P substrate levels (0.2 mM), Pho1 was still able to carry out the biosynthetic reaction, although at low rates, in the presence of 50-fold excess of Pi (see Supplemental Table 2 online). Hence, under conditions that would favor the degradation of starch, Pho1 preferentially carries out biosynthesis. In addition to the direct involvement of Pho1 in starch biosynthesis in the endosperm, the possibility that Pho1 plays an important role in influencing endosperm development or in partitioning carbon to the endosperm for starch synthesis cannot be ruled out.
Second, Pho1 might play some crucial role in starch biosynthesis by forming a functional protein–protein complex with other carbohydrate metabolizing enzymes such as BE or SS isoforms. Tetlow et al. (2004) showed that Pho1, BEIIb, and BEI form a protein complex in wheat amyloplast that is dependent on protein phosphorylation under in vivo conditions. The activity levels of SS, BE, and DBE isoforms and their mobilities in the native-PAGE gels were not affected by the pho1 mutation of rice (Figure 6). Thus, it is unlikely that the protein–protein interactions between Pho1 and these other enzymes play a major role in the phenotype of pho1 mutants, although further studies are necessary to assess the role of starch biosynthetic enzyme complexes containing Pho1 in starch synthesis.
This study showed that the extent of starch accumulation as measured by seed weight varied considerably depending on the growth temperature. At 20°C, the bulk of the seeds produced were shrunken or severely shrunken while at 30°C, the majority of the seeds accumulated near normal levels of starch (Table 3). This variation of the grain morphology ranging from pseudonormal/white-core types to heavily shrunken types caused by a reduction of starch production in the endosperm is not due to the difference in their genetic background because this polymorphic condition was stably inherited regardless of the seed morphological type. Likewise, the grain variability was not due to nutrient availability. The shrunken seeds in pho1 mutants were positioned randomly on the panicles (see Supplemental Figure 4 online), indicating that this phenotype was not due to possible differences in assimilate unloading within this reproductive organ. To explain the above temperature-dependent phenomenon, we postulate the existence of a second unidentified factor, factor X, that can complement the function of Pho1 but whose ability is temperature dependent. Since we did not observe any marked pleiotropic effects on the levels of activity of AGPase, SS, BE, and DBE isoforms in the pho1 mutant (Figure 6), we assume that the overlapping role of Pho1 and factor X can be separated from the normal starch biosynthetic processes operated by AGPase, SS, BE, and DBE, as illustrated in Figure 11. The temperature-dependent phenotypic changes found in the amount of starch production in the seed can be explained by postulating that the amounts or activity of factor X are higher at elevated temperatures but are greatly reduced at lower temperatures during the seed development, whereas Pho1 is not significantly affected by temperature. In the absence of Pho1, as in the pho1 mutant, factor X cannot satisfactorily substitute the function of Pho1 at lower temperatures; therefore, many seeds are unable to accumulate normal levels of starch, while at higher temperatures, its activity is able to nearly restore starch biosynthesis capacity in the endosperm. Further studies are needed to characterize the function of Pho1 and to identify factor X using a wide range of materials and methodologies.
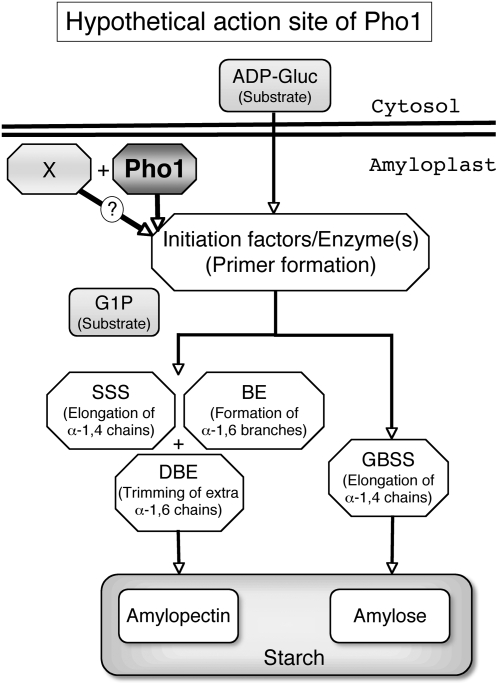
Figure 11.
Model Representing the Possible Role of Pho1 in Starch Biosynthesis in the Rice Endosperm.
In this model, Pho1 is suggested to play a major role in one or more events of starch initiation. One possible role for Pho1 is to extend the chain length of the initial primer so that it is acted upon by SSs. This involvement of Pho1 in the starch initiation process would account for the shrunken phenotype and loss of starch in pho1 mutants grown at 20°C. A hypothetical X factor is also proposed that can partially complement the function of Pho1 in starch initiation at 30°C. The X factor is suggested to have low activity or be present at low amounts at 20°C but to be fully active at 30°C. Hence, at 20°C, the loss of Pho1 and low net activity of X factor results in a high incidence of shrunken seeds, while at higher temperature the X factor can partially complement Pho1 role in starch initiation leading to the production of a high percentage of seeds that accumulate near normal starch levels.
Factor X may be the elusive amylogenin. In mammalian and yeast cells, glycogenin with autoglucosylation ability serves as a primer for the glycogen synthases (Cheng et al., 1995; Whelan, 1998). Glycogenin is capable of accepting up to eight glucose residues covalently bound to the hydroxyl group of a Tyr residue, using UDPglucose as a glucosyl donor (Lomako et al., 1992, 1993). An analogous protein with autoglycosylation ability was reported in some plant tissues, such as the potato tuber (Lavintman and Cardini, 1973; Lavintman et al., 1974), carrots (Daucus carota) (Quentmeier et al., 1987), and maize endosperm (Rothschild and Tandecarz, 1994). The analogous protein purified from potato tuber has a molecular mass of 38 kD (Ardila and Tandecarz, 1992), similar to that of glycogenin, which has a molecular mass of 37 kD (Smythe and Cohen, 1991). Chatterjee et al. (2005) reported that reducing the expression of glycogenin-like protein leads to a reduction of starch accumulation in Arabidopsis leaves, suggesting the possible involvement of glycogenin in starch biosynthesis in leaves. Thus, one should not exclude the possibility that glycogenin-like protein is a primer for starch biosynthesis.
In summary, our results show that the loss of Pho1 has varying and significant impacts on starch biosynthesis in rice endosperm. First, this mutation dramatically decreases starch accumulation in the endosperm, especially when development occurs under low temperature conditions. Second, the mutation markedly affects the fine structure of amylopectin even when the mutant seeds develop normally.
We established that the plastidial α-glucan Pho (Pho1) plays an important role in starch biosynthesis in the endosperm of rice and propose that Pho1 plays an important role in the glucan initiation process by synthesizing glucan primers with long DP values. Although direct evidence to support the hypothesis is needed, our study provides evidence that starch biosynthesis in higher plants is composed of two distinct phases: the glucan initiation process and the starch amplification process. Moreover, the two processes appear to be regulated by different mechanisms.
METHODS
Plant Materials
The endosperm mutant lines used in this experiment were stocked at the Plant Genetics Laboratory of Institute of Genetic Resources, Kyushu University, Japan. These mutant lines were produced by treating fertilized egg cells of japonica rice cultivars (Oryza sativa cv Kinmaze and T65) with MNU as reported by Satoh and Omura (1979). Rice plants were grown at an experimental field of Kyushu University Farm under natural conditions. Mature seeds were harvested and dried fully under natural conditions in a greenhouse and stored in the stockroom controlled at 4°C until they were used. Developing seeds and fresh plant materials such as leaves, leaf sheaths, and culms were stored at −80°C until used.
Screening of the pho1 Mutants
Screening of mutants for Pho1 deficiency was performed by SDS-PAGE analysis on the developing and mature seeds of 1764 endosperm mutant lines derived from MNU treatment of fertilized egg cells of japonica rice cultivars, cv Kinmaze (500 lines) and cv T65 (1264 lines). SDS-PAGE of the crude enzyme extract and of the protein extract and protein gel blotting were performed as described previously (Nishi et al., 2001).
For screening of Pho1-deficient mutants, the total proteins were extracted from one mature brown rice seed from each mutant line. The seed was crushed by pliers, and the tissue was placed into a micro test tube (1.5 mL) and homogenized with 500 μL of a buffer containing 8 M urea, 4% SDS, 5% 2-mercapthoetanol (2ME), and 0.125 M Tris-HCl, pH 6.8. The homogenates were shaken at 100 rpm for at least 18 h and centrifuged at 12,000g for 10 min at 10°C. The supernatant referred to as the protein extract was applied to SDS-PAGE analysis. Twenty microliters of the supernatant was loaded onto a gel containing 10% acrylamide/0.035% BIS and analyzed by SDS-PAGE. After electrophoresis, the gel was stained with Coomassie Brilliant Blue R 300, and the presence or absence of the 106-kD protein band was examined. The loss of Pho1 activity in the developing endosperm of isolated mutants deficient in 106-kD protein was confirmed by native-PAGE/activity staining for Pho. Ten microliters of the crude enzyme extract prepared in 50 mM HEPES-NaOH, pH 7.4, 2 mM MgCl2, 50 mM 2ME, and 12.5% (v/v) glycerol was applied onto the native-PAGE gel containing 0.8% (w/v) oyster glycogen (G8751; Sigma-Aldrich). After native-PAGE, the gel was incubated in the reaction mixture containing 50 mM HEPES-NaOH, 50 mM G1P, and 2.5 mM AMP adjusted at pH 7.4 at 30°C for 6 h and then stained with 2% I2/0.2% KI solution.
Preparation of Near Isogenic Lines of the pho1 Mutants
Near isogenic lines (BMF) for several Pho1-deficient mutants were selected for the further analysis. The BMF lines were derived from a cross between a female parent of the original cultivar and a male parent of the mutant line to eliminate the potential cytoplasmic mutations that were caused by mutations of intracellular organelles containing their own DNA, such as mitochondria or amyloplasts, which are transmitted maternally to the next generation. These crossings were repeated at least three times, and the BMF lines were selected from self-pollinated offspring exhibiting a similar phenotype to that of the original cultivar (wild type) except for the pho1 mutation. BMF134 and BMF136 (BC2F4) derived from crosses between T65 and EM 719, and T65 and EM755, respectively, and T65 were used. BMF10 (BC2F4) was derived from a cross between Kinmaze and EM141. EM640, EM755, EM719, EM786, EM876, EM141, and EM583 (wx) were used as additional materials. AMF46 is an amylose-free Pho1-deficient mutant line derived from a cross between EM755 and BMF159 (BC2F4 from a cross between T65 and a waxy mutant EM583 induced by MNU treatment of T65).
Effects of Temperature on the pho1 Phenotype during Endosperm Development
Rice plants were carefully removed from the experimental field at the maximum flowering stage and transplanted into plastic pots. After removing spikelets that have already pollinated, the plants were transferred to phytotron rooms (in the greenhouse) controlled at temperatures of 20, 25, or 30°C located at the Biotron Institute of Kyushu University. They were self-pollinated in their respective rooms and developed to maturity. After harvesting, seeds were carefully removed from the hull and the grain was weighed. Starch content in the endosperm was measured by the glucoamylase method as reported previously (Nakamura et al., 1989).
Genetic Analysis of Pho1-Deficient Mutant EM755
Dosage Effect
The endosperm gene dosage series was generated by self-crosses or reciprocal crosses within or between homozygous Pho1/Pho1 (T65) and pho1/pho1 (EM755) parents. Endosperms containing 3, 2, 1, and 0 copies of the Pho1 gene were those from the self-pollinated T65, T65 (female) × EM755 (male), EM755 (female) × T65 (male), and the self-pollinated pho1 mutant strain EM755, respectively.
The dosage effects of the Pho1 gene were analyzed by SDS-PAGE and the native-PAGE/activity staining methods.
Linkage Analysis
RFLP analysis of the Pho1 gene was performed by DNA gel blotting analysis as described previously (Satoh et al., 2003b) using a fragment of cDNA amplified by PCR (primers: forward, 5′-TGCAGTGCAGATGAATGACA-3′; reverse, 5′- CGCATGGTGCTTAATGTTTG-3′) as a probe. DNA was extracted from leaves of the indica rice cv Kasalath, EM755, and F2 plants derived from a cross between Kasalath and EM755. Two grams of fresh leaves from each strain were freeze-dried for >12 h. The leaves were powdered using a Multi Beads Shocker (Yasui Kikai) and the DNA extracted by the CTAB method of Murray and Thompson (1980). Three micrograms each of DNA was digested by BamHI, BglII, EcoRV, HindIII, ApaI, DraI, EcoRI, or KpnI (TAKARA BIO) and analyzed for RFLP. Linkage between the Pho1 gene and Pho1 deficiency were analyzed by RFLP using F2 and F3 plants derived from a cross between Kasalath and EM755.
Determination of Mutation Sites in the Pho1 Gene of BMF136 and EM640
To establish the mutation sites in the Pho1 gene of the pho1 mutant lines, the nucleotide sequences of the genomic DNA from two mutant lines, BMF136 and EM640, were determined. Genomic DNA was prepared from seedling leaves (200 mg each) of cv T65 and the mutant lines BMF136 and EM640 by the CTAB method of Murray and Thompson (1980). The genomic DNA sequences were determined using the ABI Dye Terminator Ready Reaction kits and an automated ABI 3130 DNA sequencer (Applied Biosystems). Primers to analyze the DNA sequence were designed by Primer3 Input (primer3_www.cgi v 0.2; http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). Primers used in this analysis are listed in Supplemental Table 2 online. Templates for the sequence were prepared using LA Taq polymerase (TAKARA BIO) following the protocol. DNA sequence analysis was performed using EditView1.0.1 and Auto Assembler 2.1. Wild-type and mutant sequences were aligned using ClustalW (http://clustalw.ddbj.nig.ac.jp/top-e.html) at the website default values.
Measurement of Transcript Levels of Pho1 and Pho2 Genes by Quantitative Real-Time RT-PCR Analysis
Transcript levels of Pho1 and Pho2 genes were determined by quantitative real-time PCR methods using oligonucleotide primer pairs and DNA amplification conditions as previously described by Ohdan et al. (2005). Total RNAs from seeds of cv T65 and its pho1 mutant BMF65 were extracted with EASYPrep RNA (TaKaRa) according to the manufacturer's instructions. The extracts, including 5 μg total RNAs, were treated with 5 units of DNase I (Amplification Grade; Invitrogen) to completely remove contaminating genomic DNA. Two micrograms of seed total RNA was used for first-strand cDNA synthesis with the iScript cDNA synthesis kit (Bio-Rad). An aliquot of the first-strand cDNA mixture corresponding to 5 ng of total RNA served as the template for quantitative real-time RT-PCR analysis with the Quantitect SYBR Green PCR kit (Qiagen). Reactions were performed on an iCycler (Bio-Rad) according to the manufacturer's protocols. The gene-specific primers used for quantitative PCR are listed in Supplemental Table 1 online. To optimize PCR conditions for each primer set, annealing temperature, PCR efficiency, and standard curve were examined. The specificity of the PCR amplification was checked with a melt curve analysis (from 55 to 94°C) following the final cycle of the PCR. To verify the specificity of each primer set, their amplification products were cloned in the pGEM T-Easy vector and sequenced in an ABI PRISM 3100 genetic analyzer (Applied Biosystems).
Determination of mRNA Copy Number
The copy number of mRNA was determined following the procedure recommended by Applied Biosystems (http://www.appliedbiosystems.com/support/apptech/). In this procedure, the mass of a single plasmid template was calculated, and this mass was equated to one copy of the target gene sequence. The purified plasmid template was then quantified and serially diluted in TE to obtain plasmid solutions that differed in several orders of magnitude. Aliquots were used as templates for quantitative real-time PCR. Data were plotted to generate the standard curve. Plotting the values obtained from any sample against this standard curve yielded the approximate copy number of the target gene in the sample according to the methods described by Bustin (2000) and Gachon et al. (2004). In this study, we equated the gene sequence copy number to mRNA copy number.
Fractionation of the Amyloplast
The amyloplast preparation was fractionated from developing rice endosperm using a modified method described by Tetlow et al. (2003). Briefly, 200 developing rice seeds at the mid-milking stage with their pericarps and embryos removed were soaked in 2 mL of buffer A containing 0.8 M sorbitol, 50 mM HEPES-NaOH, pH 7.5, 1 mM EDTA, 1 mM KCl, 2 mM MgCl2, 2 mM DTT, and 1 g L−1 BSA for 1 h on ice. The plasmolyzed rice seeds were sliced carefully into fine tips with a razor blade in a glass Petri dish with an additional 750 μL of buffer A. The resulting suspension was filtered through four layers of miracloth, centrifuged at 100g for 10 min at 4°C, and the supernatant was used as the cytosolic fraction. The precipitate was washed carefully by the buffer A two times and then used as the amyloplast fraction. The amyloplast fraction was then suspended in 1.0 mL of buffer B containing 5 mL Tricine-HCl, pH 7.8, 5 mM NaCl, and 1 mM MgCl2 and then sonicated to disrupt the amyloplast membrane. After centrifugation at 12,000g for 15 min at 4°C, the supernatant and the pellet were used as the stroma plus membrane fraction and the starch fraction, respectively. The stroma membrane fraction was treated with 1 mL of 20% trichloroacetic acid, gently mixed, and then placed on ice for 15 min. After centrifugation at 14,000g for 15 min at 4°C, the supernatant was removed. The resulting precipitate was washed with 1 mL of ethanol by centrifugation at 14,000g for 15 min at 4°C. The procedure was repeated twice. The proteins in the amyloplast faction and starch fraction were dissolved into 2 mL each of SDS sample buffer containing 8 M urea, and those of the stroma and membrane fraction were dissolved in 1 mL of the buffer, while a developing seed at the mid-milking stage free of pericarp and embryo was homogenized in a 1.5 mL microtube with 300 μL of the same sample buffer and used as a control. After centrifugation, 15 μL each of the supernatant was applied to a gel containing 10% acrylamide/0.035% BIS for SDS-PAGE. Protein gel blot analysis was performed after transferring proteins from the SDS-PAGE gel to PVDF membrane (Amersham-Pharmacia) with polyclonal antibodies raised against the respective proteins as described by Takemoto et al. (2002).
Preparation of Enzyme Extract
For assay of enzyme activities, 1 g (fresh weight) of endosperms at the late milking stage was removed from the embryo and pericarp and was homogenized with 10 mL of an extraction buffer containing 50 mM HEPES-NaOH, pH 7.4, 4 mM MgCl2, 50 mM 2ME, and 12.5% glycerol. The homogenate was centrifuged at 10,000g for 10 min at 4°C. The supernatant referred to as the soluble enzyme extract was used for enzyme assay.
For assay of Pho activities in various tissues of rice, the developing rice seed at the late-milky-ripe stage, the leaf blade of the flag leaf, the leaf sheath of the flag leaf, and the second internode (stem) were used. Five developing rice seeds (60 mg) at the late-milky-ripe stage with their pericarp and embryo tissues removed were homogenized with 1.5 mL of the extraction buffer (GS) containing 50 mM HEPES, 2 mM MgCl2, 50 mM 2ME, and 12.5% glycerol adjusted to pH 7.4 and then centrifuged. For analysis of Pho activity in leaf, leaf sheath, and stem tissue, 500 mg of the respective tissues were frozen in liquid nitrogen and then powdered by Multi Beads Shocker (Yasui Kikai). The powdered samples were homogenized in 1.5 mL of GS buffer, and the supernatants were collected for enzyme assays.
Assay of AGPase
The activity of AGPase was determined by the method described by Nishi et al. (2001).
Zymogram Analysis of Starch Synthesizing Enzymes
A single endosperm at the late-milky-ripe stage from BMF136, EM640, and T65 was homogenized with 10 volumes (on a fresh weight basis) of cold grinding solution (GS) containing 50 mM HEPES-NaOH, pH 7.4, 2 mM MgCl2, 50 mM 2ME, and 12.5% (v/v) glycerol. The homogenate was centrifuged at 20,000g at 4°C for 10 min, and the supernatant was used for native-PAGE.
Native-PAGE gels for assay of enzymes were prepared with 130 mM Tris-HCl, pH 6.8, 0.027% TEMED, and 0.08% ammonium persulfate as the stacking gels and with 375 mM Tris-HCl, pH 8.8, 0.05% TEMED, and 0.05% ammonium persulfate as the resolving gels. The concentration of acrylamide in the resolving gels was varied depending on the enzymes to be analyzed, as described below. Native-PAGE/activity staining analyses of Pho activity staining was performed on a 5% (w/v) acrylamide slab gel containing 0.8% (w/v) oyster glycogen (Sigma-Aldrich) with the reaction mixture including 50 mM HEPES-NaOH, 50 mM G1P, 2.5 mM AMP, and 20% glycerol adjusted to pH 7.4. Native-PAGE/activity staining analyses of DBE and BE were performed on 5% acrylamide slab gels, including 0.4% (w/v) potato amylopectin (Sigma-Aldrich) and no glucan, respectively, using the methods of Fujita et al. (1999) and Yamanouchi and Nakamura (1992), respectively. SS activity staining was performed on a 7.5% (w/v) acrylamide slab gel containing 0.8% (w/v) oyster glycogen (G8751; Sigma-Aldrich) according to Nishi et al. (2001) with the modification that 0.5 M citrate was included in the reaction mixture. The reactions were run at 30°C for 2, 18, and 17 h for assay of DBE, BE, and SS, respectively.
Cloning of the Region Encoding the Mature Pho1 and SSIIa of Rice Endosperm
The rice Pho1 transit peptide sequence was predicted by ChloroP 1.1 Server (http://www.cbs.dtu.dk/services/ChloroP/). The putative transit peptide was encoded by the first 153 nucleotide bases from the A nucleotide of the ATG start codon. First-strand cDNA was synthesized with total RNA from developing rice grains in the japonica rice cultivar Nipponbare, and the region encoding the mature Pho1 (nucleotide bases 154 to 2856) was amplified by PCR using the sense primer 5′-ATGCAGAATTCCTAGGGCAGGATGACAGGCTTGATGTC-3′ containing the EcoRI site paired with the antisense primer 5′-ATTATGTTAACTAGCGTGGCGAGCGATCGGGGCGT-3′. The incorporation of the EcoRI site in the sense primer was needed for cloning into the expression vector pET-32b. The single-band PCR product was gel purified and cloned in pET-32b and introduced into Escherichia coli strain DH5α. It was verified by sequencing. The resulting Pho1 gene in pET-32b was then transformed into the expression host E. coli strain AD494 (DE3) pLysS (Novagen). The region coding for the mature SSIIa in an indica rice cultivar IR36 was cloned into pET-32b, as described previously (Nakamura et al., 2005).
Induction and Purification of Recombinant Plastidic Pho and SSIIa of Rice Endosperm Expressed in E. coli
The transformed E. coli AD494 (DE3) pLysS cells mentioned above were grown at 25°C to OD600 of ∼0.6 in Luria-Bertani medium containing 100 μg of carbenicillin, 15 μg of kanamycin, and 34 μg of chloramphenicol per milliliter of the medium. The cells were then induced by the addition of 1 mM isopropylthio-β-galactoside and cultured overnight at 25°C. The cells were collected by centrifugation at 20,000g at 4°C for 5 min and frozen at −80°C until used. The frozen cells were thawed, resuspended in 200 mL of imidazole-HCl, pH 7.4, 50 mM 2ME, 8 mM MgCl2, and 12.5% glycerol, disrupted by sonication on ice in an Ultrasonic Disruptor model UD201 (TOMY) set at a power output level of 3 and 30% duty for 10 bursts, and centrifuged at 20,000g at 4°C for 5 min. The supernatant, which was designated as the crude enzyme extract, was collected and applied to a HitrapQ HP anion exchange column (5 mL; Amersham Biosciences) that had been equilibrated with Medium A, containing 50 mM imidazole, pH 7.4, 8 mM MgCl2, and 1 mM DTT. The protein was eluted with a linear gradient of 0.5 M NaCl for 30 min at a flow rate of 2 mL/min. The peak fraction of Pho1 activity eluted at the NaCl concentration of ∼0.367 to 0.433 M was concentrated to 0.6 mL by a Centricon 50 centrifugal concentrator (Millipore) and added to the same volume of 2 M ammonium sulfate. The mixture was applied to an Ether-5PW column (7.5 mm in diameter × 75 mm in length; Tosoh), which had been equilibrated with Medium A containing 2 M ammonium sulfate. The proteins were eluted with a descending ammonium sulfate linear gradient (2 to 0 M) for 60 min at a flow rate of 1 mL/min. The Pho1 activity was eluted at the ammonium sulfate concentration of ∼0.67 M. The Pho1 activity fractions were collected and concentrated to ∼0.7 mL using a Centricon 50 centrifugal concentrator. As the Pho1 activity profile was the same as that of the UV curve of the Ether-5PW chromatography, and the pooled fraction was shown to contain only a single protein band by SDS-PAGE, the fraction was designated as the purified recombinant Pho1 from rice (rPho1). The recombinant rice SSIIa was also purified in the same way as above, and the purified SSIIa was designated as rSSIIa. The partially purified preparation was stored at −80°C until used for characterization of the enzyme. The protein amount was determined by the method of Bradford (1976).
Analysis of Linear MOS Produced by Enzymatic Reactions of Plastidic Pho and SSIIa in the Presence of MOS as Primers
The enzymatic reaction of rPho1 was performed with 0.2 mL of the buffer solution containing 50 mM HEPES-NaOH buffer, pH 7.0, 5 mM G1P, 0.5 mM MOS, and the purified rPho1 (0.35 or 0.70 μg protein) in a tube. The reaction was run for 5 to 10 min at 30°C and terminated by heating the tube in boiling water for 2 min. The enzymatic reaction of rSSIIa was performed with 0.2 mL of the buffer solution containing 100 mM Bicine-NaOH buffer, pH 7.5, 500 mM citrate, 2 mM ADPglucose, 0.5 mM MOS, and the purified rSSIIa (5.4 or 10.8 μg protein) in a tube. The reaction was run for 60 min at 30°C and terminated by heating the tube in boiling water for 2 min. The analysis of the products was performed by measuring the APTS-labeled α-1,4-glucans using a P/ACE System 5000 high-resolution capillary electrophoresis equipped with a laser-induced fluorescence detector (Beckman Instruments) according to the method by O'Shea and Morell (1996) as described previously (Nakamura et al., 2002).
Chain Length Profile of Amylopectin
The chain length distribution analysis of isoamylorysates of amylopectin was performed by measuring the APTS-labeled α-1,4-glucans using a P/ACE System 5000 high-resolution capillary electrophoresis equipped with a laser-induced fluorescence detector (Beckman Instruments) according to the method by O'Shea and Morell (1996) as described previously (Nakamura et al., 2002).
Morphological and Physicochemical Properties of Starches
Analyses of scanning electron microscopy (JSM-56000LV; JEOL) and x-ray diffraction pattern of starches were performed as described by Kubo et al. (2005). DSC of starches was measured as described by Nakamura et al. (2002).
Measurement of Carbohydrates
The starch content was enzymatically measured as described previously (Nakamura et al., 1989).
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative, GenBank/EMBL, or DDBJ (http://getentry.ddbj.nig.ac.jp/top-e.html) databases under the following accession numbers: AB441692 (pho1, Taichung65), AB441693 (pho1-1, BMF136), and AB441694 (pho1-2, EM640).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Amino Acid Sequences of the 106-kD Protein Fragments Analyzed by LC-MS/MS.
Supplemental Figure 2. Transcript Levels of Pho1 and Pho2 Genes Determined by Real-Time RT-PCR.
Supplemental Figure 3. Effects of phol Mutations on the Morphology of Rice Kernels Grown under Field Conditions.
Supplemental Figure 4. Distribution of Shrunken Seeds on the Panicles of a pho1 Mutant Plant.
Supplemental Figure 5. Location of the Pho1 gene on Chromosome 3 in Rice.
Supplemental Figure 6. X-Ray Diffraction Pattern of Starch in the Endosperm of T65 and a pho1 Mutant Line BMF136.
Supplemental Figure 7. Chain Elongation Reaction of rPho1 and rSSIIa Using Malto-Oligosaccharides as Primers.
Supplemental Table 1. Primers of Pho Genes for Real-Time RT-PCR.
Supplemental Table 2. The Effect of Phosphate on the Biosynthetic Activity of Rice Starch Phosphorylase1 (Pho1).
Supplemental Table 3. Primer List for Sequencing Analysis of the Pho1 Gene.
Supplementary Material
Acknowledgments
This research was partly funded by the Japan Society for the Promotion of Science, by the Bio-oriented Technology Research Advancement Institution, by the Core Research for Evolutional Science and Technology, and by the U.S. Department of Energy Grant DE-FG02-96ER20216.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Hikaru Satoh (hsatoh@agr.kyushu-u.ac.jp).
Online version contains Web-only data.
References
- Ardila, F.J., and Tandecarz, J.S. (1992). Potato tuber UDP-Glucose:Protein transglucosylase catalyzes its own glucosylation. Plant Physiol. 99 1342–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball, S.G., and Morell, M.K. (2003). From bacterial glycogen to starch: Understanding the biogenesis of the plant starch granule. Annu. Rev. Plant Biol. 54 207–233. [DOI] [PubMed] [Google Scholar]
- Borovsky, D., Smith, E.E., and Whelan, W.J. (1976). On the mechanism of amylose branching by potato Q-enzyme. Eur. J. Biochem. 62 307–312. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- Brown, J.W. (1996). Arabidopsis intron mutations and pre-mRNA splicing. Plant J. 10 771–780. [DOI] [PubMed] [Google Scholar]
- Buchner, P., Borisjuk, L., and Wobus, U. (1996). Glucan phosphorylases in Vicia faba L.: Cloning, structural analysis and expression patterns of cytosolic and plastidic forms in relation to starch. Planta 199 64–73. [DOI] [PubMed] [Google Scholar]
- Burr, B., and Nelson, O.E. (1973). The phosphorylases of developing maize seeds. Ann. N. Y. Acad. Sci. 210 129–138. [DOI] [PubMed] [Google Scholar]
- Bustin, S.A. (2000). Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25 169–193. [DOI] [PubMed] [Google Scholar]
- Chatterjee, M., Berbezy, P., Vyas, D., Coates, S., and Barsby, T. (2005). Reduced expression of a protein homologous to glycogenin leads to reduction of starch content in Arabidopsis leaves. Plant Sci. 168 501–509. [Google Scholar]
- Cheng, C., Mu, J., Farkas, I., Huang, D., Goebl, M.G., and Roach, P.J. (1995). Requirement of the self-glucosylating initiator proteins Glg1p and Glg2p for glycogen accumulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 15 6632–6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleoni, C., Dauvillée, D., Mouille, G., Morell, M., Samuel, M., Slomiany, M.C., Liénard, L., Wattebled, F., d'Hulst, C., and Ball, S. (1999). Biochemical characterization of the Chlamydomonas reinhardtii α-1,4 glucanotransferase supports a direct function in amylopectin biosynthesis. Plant Physiol. 120 1005–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooks, M.S., Evans, M.D., Dizdaroglu, M., and Lunec, J. (2003). Oxidative DNA damage: Mechanisms, mutation, and disease. FASEB J. 17 1195–1214. [DOI] [PubMed] [Google Scholar]
- da Mota, R.V., Cordenunsi, B.R., do Nascimento, J.R.O., Purgatto, E., Rosseto, M.R.M., and Lajolo, F.M. (2002). Activity and expression of banana starch phosphorylases during fruit development and ripening. Planta 216 325–333. [DOI] [PubMed] [Google Scholar]
- Dauvillée, D., et al. (2006). Plastidial phosphorylase is required for normal starch synthesis in Chlamydomonas reinhardtii. Plant J. 48 274–285. [DOI] [PubMed] [Google Scholar]
- Engelbergs, J., Thomale, J., and Rajewsky, M.F. (2000). Role of DNA repair in carcinogen-induced ras mutation. Mutat. Res. 450 139–153. [DOI] [PubMed] [Google Scholar]
- Fujita, N., Kubo, A., Francisco, P.B., Jr., Nakakita, M., Harada, K., Minaka, N., and Nakamura, Y. (1999). Purification, characterization, and cDNA structure of isoamylase from developing endosperm of rice. Planta 208 283–293. [DOI] [PubMed] [Google Scholar]
- Fujita, N., Yoshida, M., Asakura, N., Ohdan, T., Miyao, A., Hirochika, H., and Y. Nakamura, Y. (2006). Function and characterization of starch synthase I using mutants in rice. Plant Physiol. 140 1070–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, N., et al. (2007). Characterization of SSIIIa-deficient mutants of rice (Oryza sativa L.,). The function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol. 144 2009–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon, C., Mingam, A., and Charrier, B. (2004). Real-time PCR: What relevance to plant studies. J. Exp. Bot. 55 1445–1454. [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005). The map-based sequence of the rice genome. Nature 436 793–800. [DOI] [PubMed] [Google Scholar]
- Kitamura, S., Yunokawa, H., Mitsuie, S., and Kuge, T. (1982). Study on polysaccharides by the fluorescence method. II. Micro-Brownian motion and conformational change of amylase in aqueous solution. Polym. J. 14 93–99. [Google Scholar]
- Kossmann, J., Visser, R.G.F., Müller-Röber, B., Willmitzer, L., and Sonnewald, U. (1991). Cloning and expression analysis of a potato cDNA that encodes branching enzyme: Evidence for co-expression of starch biosynthetic genes. Mol. Gen. Genet. 230 39–44. [DOI] [PubMed] [Google Scholar]
- Kubo, A., Rahman, S., Utsumi, Y., Li, Z., Mukai, Y., Yamamoto, M., Ugaki, M., Harada, K., Satoh, H., Konik-Rose, C., Morell, M., and Nakamura, Y. (2005). Complementation of sugary-1 phenotype in rice endosperm with the wheat Isoamylase1 gene supports a direct role for Isoamylase1 in amylopectin biosynthesis. Plant Physiol. 137 43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavintman, N., and Cardini, C.E. (1973). Particulate UDP-glucose: Protein transglucosylase from potato tuber. FEBS Lett. 29 43–46. [DOI] [PubMed] [Google Scholar]
- Lavintman, N., Tandecarz, J., Carceller, M., Mendiara, S., and Cardini, C.E. (1974). Role of uridine diphosphate glucose in the biosynthesis of starch. Mechanism of formation and enlargement of a glucoproteic acceptor. Eur. J. Biochem. 50 145–155. [DOI] [PubMed] [Google Scholar]
- Lomako, J., Lomako, W.M., and Whelan, W.J. (1992). The substrate specificity of isoamylase and the preparation of apo-glycogenin. Carbohydr. Res. 227 331–338. [DOI] [PubMed] [Google Scholar]
- Lomako, J., Lomako, W.M., Whelan, W.J., Dombro, R.S., Neary, J.T., and Norenberg, M.D. (1993). Glycogen synthesis in the astrocyte: from glycogenin to proglycogen to glycogen. FASEB J. 7 1386–1393. [DOI] [PubMed] [Google Scholar]
- Maquat, L.E. (2004). Nonsense-mediated mRNA decay: Splicing, translation and mRNA dynamics. Nat. Rev. Mol. Cell Biol. 5 89–99. [DOI] [PubMed] [Google Scholar]
- Mori, H., Tanizawa, K., and Fukui, T. (1991). Potato tuber type H phosphorylase isozyme. Molecular cloning, nucleotide sequence, and expression of a full-length cDNA in Escherichia coli. J. Biol. Chem. 266 18446–18453. [PubMed] [Google Scholar]
- Murray, M.G., and Thompson, W.F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, Y. (2002). Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: Rice endosperm as a model tissue. Plant Cell Physiol. 43 718–725. [DOI] [PubMed] [Google Scholar]
- Nakamura, Y., Francisco, P.B., Jr., Hosaka, Y., Sato, A., Sawada, T., Kubo, A., and Fujita, N. (2005). Essential amino acids of starch synthase IIa differentiate amylopectin structure and starch quality between japonica and indica rice varieties. Plant Mol. Biol. 58 213–227. [DOI] [PubMed] [Google Scholar]
- Nakamura, Y., Sakurai, A., Inaba, Y., Kimura, K., Iwasawa, N., and Nagamine, T. (2002). The fine structure of amylopectin in endosperm from Asian cultivated rice can be largely classified into two classes. Starch 54 117–131. [Google Scholar]
- Nakamura, Y., Yuki, K., Park, S.-Y., and Ohya, T. (1989). Carbohydrate metabolism in the developing endosperm of rice grains. Plant Cell Physiol. 30 833–839. [Google Scholar]
- Nishi, A., Nakamura, Y., Tanaka, N., and Satoh, H. (2001). Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiol. 127 459–472. [PMC free article] [PubMed] [Google Scholar]
- Ohdan, T., Francisco, P.B., Jr., Sawada, T., Hirose, T., Terao, T., Satoh, H., and Nakamura, Y. (2005). Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J. Exp. Bot. 56 3229–3244. [DOI] [PubMed] [Google Scholar]
- O'Shea, M.G., and Morell, M.K. (1996). High resolution slab gel electrophoresis of 8-amino-1,3,6-pyrenetrisulfonic acid (APTS) tagged oligosaccharides using a DNA sequencer. Electrophoresis 17 681–686. [DOI] [PubMed] [Google Scholar]
- Ozbun, J.L., Hawker, J.S., Greenberg, E., Lammel, C., and Preiss, J. (1973). Starch synthetase, phosphorylase, ADPglucose pyrophosphorylase, and UDPglucose pyrophosphorylase in developing maize kernels. Plant Physiol. 51 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss, J., and Levi, C. (1980). Starch biosynthesis and degradation. In The Biochemistry of Plants, Vol. 3, J. Preiss, ed (New York: Academic Press), pp. 371–423.
- Preiss, J., Okita, T.W., and Greenberg, E. (1980). Characterization of the spinach leaf phosphorylases. Plant Physiol. 66 864–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss, J., and Sivak, M. (1996). Starch synthesis in sinks and sources. In Photoassimilate Distribution in Plants and Crops: Source-Sink Relationships, E. Zamski and A.A. Schaffer, eds (New York: Marcel Dekker), pp. 139–168.
- Quentmeier, H., Ingold, E., and Seitz, H.U. (1987). Purification of an autocatalytic protein-glycosylating enzyme from cell suspensions of Daucus carota L. Planta 171 483–488. [DOI] [PubMed] [Google Scholar]
- Rothschild, A., and Tandecarz, J.S. (1994). UDP-glucose:protein transglucosylase in developing maize endosperm. Plant Sci. 97 119–127. [Google Scholar]
- Satoh, H., Nishi, A., Fujita, N., Kubo, A., Nakamura, Y., Kawasaki, T., and Okita, T. (2003. a). Isolation and characterization of starch mutants in rice. J. Appl. Glycosci. (1999) 50 225–230. [Google Scholar]
- Satoh, H., Nishi, A., Yamashita, K., Takemoto, Y., Tanaka, Y., Hosaka, Y., Sakurai, A., Fujita, N., and Nakamura, Y. (2003. b). Starch-branching enzyme I-deficient mutation specifically affects the structure and properties of starch in rice endosperm. Plant Physiol. 133 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh, H., and Omura, T. (1979). Induction of mutation by the treatment of fertilized egg cell with N-methyl-N-nitrosourea in rice. J. Fac. Agric. Kyushu Univ. 24 165–174. [Google Scholar]
- Satoh, H., and Omura, T. (1981). New endosperm mutations induced by chemical mutagens in rice, Oryza sativa L. Japan J. Breed. 31 316–326. [Google Scholar]
- Schupp, N., and Ziegler, P. (2004). The relation of starch phosphorylases to starch metabolism in wheat. Plant Cell Physiol. 45 1471–1484. [DOI] [PubMed] [Google Scholar]
- Shimomura, S., Nagai, M., and Fukui, T. (1982). Comparative glucan specificities of two types of spinach leaf phosphorylase. J. Biochem. 91 703–717. [DOI] [PubMed] [Google Scholar]
- Smythe, C., and Cohen, P. (1991). The discovery of glycogenin and the priming mechanism for glycogen biogenesis. Eur. J. Biochem. 200 625–631. [DOI] [PubMed] [Google Scholar]
- Sonnewald, U., Basner, A., Greve, B., and Steup, M. (1995). A second L-type isozyme of potato glucan phosphorylase: Cloning, antisense inhibition and expression analysis. Plant Mol. Biol. 27 567–576. [DOI] [PubMed] [Google Scholar]
- St-Pierre, B., and Brisson, N. (1995). Induction of the plastidic starch-phosphorylase gene in potato storage sink tissue. Planta 195 339–344. [Google Scholar]
- Steup, M. (1988). Starch degradation. In Biochemistry of Plants, Vol. 14, J. Preiss, ed (San Diego, CA: Academic Press), pp. 255–296.
- Steup, M. (1990). Starch degrading enzymes. In Methods in Plant Biochemistry, Vol. 3, P.J. Lea, ed (London: Academic Press), pp. 103–128.
- Steup, M., and Schächtele, C. (1981). Mode of glucan degradation by purified phosphorylase forms from spinach leaves. Planta 153 351–361. [DOI] [PubMed] [Google Scholar]
- Suzuki, T., Eiguchi, M., Kumamaru, T., Satoh, H., Matsusaka, H., Moriguchi, K., Nagato, Y., and Kurata, N. (2008). MNU-induced mutant pools and high performance TILLING enable finding of any gene mutation in rice. Mol. Genet. Genomics 279 213–223. [DOI] [PubMed] [Google Scholar]
- Takemoto, Y., Coughlan, S.J., Okita, T.W., Satoh, H., Ogawa, M., and Kumamaru, T. (2002). Protein disulfide isomerase is essential for the localization of prolamin and glutelin storage proteins to separate intracellular compartments. Plant Physiol. 128 1212–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetlow, I.J., Davies, E.J., Vardy, K.A., Bowsher, C.G., Burrell, M.M., and Emes, M.J. (2003). Subcellular localization of ADPglucose pyrophosphorylase in developing wheat endosperm and analysis of the properties of a plastidial isoform. J. Exp. Bot. 54 715–725. [DOI] [PubMed] [Google Scholar]
- Tetlow, I.J., Wait, R., Lu, Z., Akkasaeng, R., Bowsher, C.G., Esposito, S., Kosar-Hashemi, B., Morell, M.K., and Emes, M.J. (2004). Protein phosphorylation in amyloplasts regulates starch branching enzyme activity and protein-protein interactions. Plant Cell 16 694–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, C.Y., and Nelson, O.E. (1969). Mutations at the shrunken-4 locus in maize that produce three altered phosphorylases. Genetics 61 813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berkel, J., Conrads-Strauch, J., and Steup, M. (1991). Glucan-phosphorylase forms in cotyledons of Pisum sativum L.: Localization, developmental change, in-vitro translation, and processing. Planta 185 432–439. [DOI] [PubMed] [Google Scholar]
- Whelan, W.J. (1998). Pride and prejudice: The discovery of the primer for glycogen synthesis. Protein Sci. 7 2038–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanouchi, H., and Nakamura, Y. (1992). Organ specificity of isoforms of starch branching enzyme (Q-enzyme) in rice. Plant Cell Physiol. 33 985–991. [Google Scholar]
- Yu, Y., Mu, H.H., Wassermanm, B.P., and Carman, G.M. (2001). Identification of the maize amyloplast stromal 112-kD protein as a plastidic starch phosphorylase. Plant Physiol. 125 351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman, S.C., Thorneycroft, D., Schupp, N., Chapple, A., Weck, M., Dunstan, H., Haldimann, P., Bechtold, N., Smith, A.M., and Smith, S.M. (2004). Plastidial α-glucan phosphorylase is not required for starch degradation in Arabidopsis leaves but has a role in the tolerance of abiotic stress. Plant Physiol. 135 849–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.