Abstract
We demonstrate that XopD, a type III effector from Xanthomonas campestris pathovar vesicatoria (Xcv), suppresses symptom production during the late stages of infection in susceptible tomato (Solanum lycopersicum) leaves. XopD-dependent delay of tissue degeneration correlates with reduced chlorophyll loss, reduced salicylic acid levels, and changes in the mRNA abundance of senescence- and defense-associated genes despite high pathogen titers. Subsequent structure-function analyses led to the discovery that XopD is a DNA binding protein that alters host transcription. XopD contains a putative helix-loop-helix domain required for DNA binding and two conserved ERF-associated amphiphilic motifs required to repress salicylic acid– and jasmonic acid–induced gene transcription in planta. Taken together, these data reveal that XopD is a unique virulence factor in Xcv that alters host transcription, promotes pathogen multiplication, and delays the onset of leaf chlorosis and necrosis.
INTRODUCTION
Bacterial pathogens of plants and animals use the type III secretion (T3S) system to introduce protein substrates (effectors) that alter host signaling and maintain host cell viability (Alfano and Collmer, 2004; Troisfontaines and Cornelis, 2005). The suppression of eukaryotic immune responses by T3S effectors is a shared theme of many pathogens (Abramovitch et al., 2006a; Coburn et al., 2007). However, little is known about other host processes affected by these proteins during host–microbe interactions.
In plants, it is known that T3S effectors inhibit both basal immunity and resistance (R) protein–activated innate immunity (Chisholm et al., 2006). Insight into Pseudomonas syringae effector function reveals that distinct biochemical activities are used to perturb the activity and/or stability of host proteins involved in defense signaling. T3S-dependent suppressors of host basal defense responses include HopAI1, a phosphothreonine lyase that inhibits the phosphorylation of mitogen-activated protein kinases, MPK3 and MPK6 (Zhang et al., 2007); HopU1, a mono-ADP-ribosyltransferase that modifies Gly-rich RNA binding protein GRP7 (Fu et al., 2007); and HopM1, an inhibitor of vesicle trafficking via proteasome-dependent degradation of At MIN7 (Arabidopsis thaliana HopM interactor 7), an adenosine diphosphate ribosylation factor guanine nucleotide exchange factor (Nomura et al., 2006). Suppressors of host R protein–dependent responses include AvrRpt2, a Cys protease that degrades RIN4 interfering with RPM1-dependent defense signaling (Axtell et al., 2003; Mackey et al., 2003) and the AvrPtoB E3 ligase, a general suppressor of programmed cell death (Abramovitch et al., 2006b; Rosebrock et al., 2007).
In contrast with P. syringae, much less is known about the diverse biochemical roles of T3S effectors in Xanthomonas pathogenesis. What is known is that the AvrBs3/PthA effector family is highly conserved in xanthomonads, being essential for both strain virulence and symptom production (Schornack et al., 2006). These effectors share features with eukaryotic transcription factors, possessing an acidic activation domain and nuclear localization signals at the C terminus. This suggests that the AvrBs3/PthA effectors might alter plant transcription during infection. Consistent with these structural features, AvrBs3, AvrXa27, PthXo1, PthXo6, and PthXo7 activate host transcription leading to disease susceptibility and/or defense responses (Gu et al., 2005; Yang et al., 2006; Kay et al., 2007; Römer et al., 2007; Sugio et al., 2007). Interestingly, AvrBs3 binds directly to plant promoters, and its binding specificity is in part mediated by its central variable repeats (Kay et al., 2007). The conservation and diversity of AvrBs3/PthA effectors in the genus Xanthomonas indicates that host transcription is a prime target for this group of plant pathogens.
Our laboratory is interested in elucidating how T3S effectors from Xanthomonas campestris pathovar vesicatoria (Xcv) alter plant physiology to elicit bacterial spot disease on tomato (Solanum lycopersicum). In previous work, we discovered that the XopD effector (545 amino acids) encodes a Cys protease that cleaves tomato small ubiquitin-related modifier (SUMO) precursors and removes SUMO from SUMO-conjugated proteins (Hotson et al., 2003). After delivery into plant cells, XopD localizes to subnuclear foci, indicating that the host targets are likely nuclear sumoylated protein(s). The crystal structure of the XopD C terminus (amino acids 335 to 520) confirmed that XopD shares structural similarity with the yeast ubiquitin-like protease ULP1 and has provided insight into which residues impart plant SUMO substrate specificity (Chosed et al., 2007). The N terminus of XopD is required for trafficking to the plant nucleus; however, its role in regulating effector specificity was unclear.
Here, we show that the full-length XopD protein suppresses disease symptom development in tomato leaves, enabling Xcv to successfully multiply within the tissue. XopD-dependent delay in tissue collapse correlates with reduced chlorophyll loss, reduced salicylic acid (SA) levels, and modulation of host mRNA levels encoding senescence- and defense-associated genes. Furthermore, expression of XopD in planta is sufficient to repress SA and jasmonic acid–induced gene transcription. Suppression of disease symptoms and modulation of host transcription required XopD protease activity, DNA binding activity, and two conserved transcriptional repressor motifs. Together, these data show that XopD is a unique virulence factor that promotes Xcv growth and suppresses host defense and pathogen-induced cell death responses triggered in diseased tissue.
RESULTS
XopD Is Required for Maximal Pathogen Growth in Planta and to Delay Leaf Chlorosis and Necrosis
XopD (545 amino acids) is a modular T3S effector protein that is translocated into the plant nucleus during Xcv infection (Hotson et al., 2003). The N-terminal domain of XopD is required for targeting the effector to subnuclear foci, and the C-terminal domain encodes a Cys protease that cleaves SUMO-conjugated proteins. These biochemical features suggest that XopD may play an important virulence role in the plant nucleus during infection. Unfortunately, phenotypes associated with XopD action inside plant cells have remained elusive.
In attempt to identify XopD-dependent phenotypes in planta, we performed mutational analysis of the protein to identify domains capable of eliciting symptoms in the nonhost Nicotiana benthamiana. Agrobacterium tumefaciens–mediated transient expression of full-length XopD fused to the HA epitope [XopD(wt)-HA] in N. benthamiana leaves resulted in tissue chlorosis (yellowing) and necrosis (cell death) by 5 to 7 d after inoculation (DAI) (see Supplemental Figure 1 online). The timing of tissue necrosis is slow compared with the rapid, localized hypersensitive cell death response characteristic of R protein–mediated defenses in resistant hosts. Therefore, the necrotic phenotype observed in N. benthamiana leaves likely reflects cell death due to XopD accumulation and cytotoxicity. Overexpression of a XopD protease mutant [XopD(C470A)-HA] did not elicit leaf necrosis, indicating that protease activity is required for symptom production. Similarly, overexpression of the N-terminal domain (XopD1-242-HA) or the C-terminal domain (XopD243-545-HA) failed to trigger leaf necrosis. All mutant proteins were expressed in N. benthamiana leaves (see Supplemental Figure 1 online). These data show that the N-terminal domain and an active C-terminal domain are required to trigger leaf chlorosis and necrosis in N. benthamiana, highlighting the importance of both domains inside plant cells.
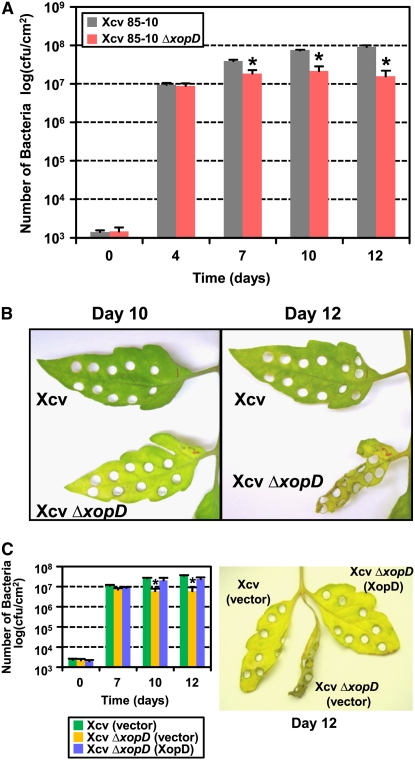
Next, we engineered a xopD null mutant in Xcv strain 85-10 to assess the role of XopD in Xcv growth and symptom production in tomato. The entire XopD open reading frame (amino acids 1 to 545) was deleted by homologous recombination, creating the Xcv ΔxopD mutant strain. Susceptible VF36 tomato leaves were completely infiltrated using a needleless syringe with a 105 colony-forming units (cfu)/mL suspension of wild-type Xcv and Xcv ΔxopD. Both strains grew equally well in the tomato leaves until 4 DAI. Xcv ΔxopD multiplication declined at 7 DAI, whereas wild-type Xcv multiplication continued to increase through day 12 (Figure 1A). This demonstrates that XopD is required for maximal Xcv growth in susceptible tomato leaves at the late stages of infection.
Figure 1.
XopD Is Required for Maximal Xcv Growth and to Delay the Development of Disease Symptoms in Infected Tomato Leaves.
(A) Growth of Xcv (gray bars) and Xcv ΔxopD (red bars) strains in susceptible VF36 tomato. Leaves were hand-inoculated with a 105 cfu/mL suspension of bacteria. Data points represent mean log10 cfu per cm2 ± sd of three independent experiments. Error bars indicate sd of three independent experiments. The asterisk above the bars indicates statistically significant (t test, P < 0.05) differences between the bacterial numbers for Xcv and Xcv ΔxopD.
(B) Phenotype of tomato leaves sampled in (A). Hole punches were used to quantify bacterial numbers depicted in (A). Leaves were photographed at 10 and 12 DAI. Similar phenotypes were observed in three independent experiments.
(C) Expression of XopD in Xcv ΔxopD complements reduced pathogen growth and suppresses tissue necrosis in infected leaves. Susceptible VF36 tomato leaves were hand-inoculated with a 105 cfu/mL suspension of bacteria: Xcv (vector) containing pDSK519 (green bars), Xcv ΔxopD (vector) containing pDSK519 (yellow bars), and Xcv ΔxopD (XopD) containing pDSK519(xopD promoter-xopD(wt)-HA) (blue bars). Pathogen growth is shown in the left panel. Data points represent mean log10 cfu per cm2 ± sd of three independent experiments. The asterisk above the bars indicates statistically significant (t test, P < 0.05) differences between the bacterial numbers of Xcv (vector) and Xcv ΔxopD (vector). Leaf symptoms are shown in the right panel. Hole punches were used to quantify bacterial numbers depicted in the left panel. Leaves were photographed at 12 DAI. Similar phenotypes were observed in three independent experiments.
Unexpectedly, tomato leaves inoculated with Xcv ΔxopD became chlorotic by 10 DAI despite reduced bacterial growth (Figure 1B, left panel). By contrast, leaves inoculated with Xcv sustaining a higher titer of bacteria were relatively green at the same time point. By 12 DAI, leaves inoculated with Xcv ΔxopD exhibited severe chlorosis and tissue necrosis (Figure 1B, right panel), typical of the disease symptoms observed in leaves at the late stages of infection just prior to death. At 12 DAI, leaves inoculated with Xcv exhibited chlorosis but no significant tissue necrosis. No noticeable difference in the size or number of bacterial lesions (i.e., spots associated with bacterial spot disease in tomato) was observed for tissue inoculated with Xcv or Xcv ΔxopD. The XopD-dependent increase in Xcv growth and suppression of tissue chlorosis and necrosis was also observed in tomato cultivars Pearson (see Supplemental Figure 2 online) and Moneymaker (Figures 3B and 3C), two other susceptible tomato cultivars, with similar kinetics. These leaf phenotypes indicate that XopD action generally inhibits the onset of chlorosis and necrosis in susceptible tomato leaves. We herein define these XopD-specific phenotypes as “a delay in disease symptom development at the late stages of Xcv infection (i.e., 10 to 12 DAI).” Collectively, these data indicate that XopD plays a distinct role during Xcv pathogenesis. It promotes Xcv growth while suppressing symptom development (Figures 1A and 1B). XopD-dependent virulence is thus defined as the ability to promote maximal bacterial multiplication prior to organ death.
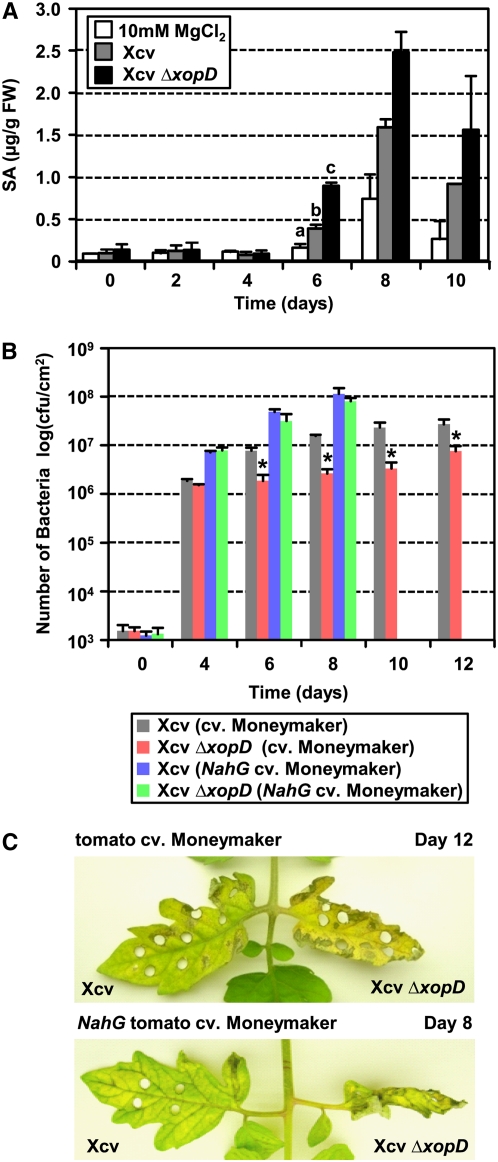
Figure 3.
XopD Reduces SA Levels during Infection.
(A) XopD reduces SA levels in leaves inoculated with Xcv. Susceptible VF36 tomato leaves were hand-inoculated with buffer (white bars) or a 105 cfu/mL suspension of Xcv (gray bars) or Xcv ΔxopD (black bars). SA levels (i.e., pools of free SA and existing methyl salicylate) in infected tissue were quantified for 10 d. μg/g FW = μg of free and endogenous methyl salicylate per gram of fresh weight. Error bars indicate sd for two biological samples. Different letters above the day 6 bars indicate statistically significant (one-way analysis of variance and Tukey's HSD test, P < 0.05) differences between the samples.
(B) Xcv ΔxopD growth is less restricted in NahG Moneymaker tomato leaves. Growth of Xcv (blue bars) and Xcv ΔxopD (green bars) strains in transgenic NahG tomato and that of Xcv (gray bars) and Xcv ΔxopD (red bars) strains in the wild-type Moneymaker tomato leaves are indicated. Leaves were hand-inoculated with a 105 cfu/mL suspension of bacteria. Data points represent mean log10 cfu per cm2 ± sd of three independent experiments. The asterisk above the bars indicates statistically significant (t test, P < 0.05) differences between the bacterial numbers of Xcv and Xcv ΔxopD in the wild-type Moneymaker tomato leaves. NahG leaves were not analyzed beyond 8 DAI because they were fully necrotic (see [C]) and died by 9 DAI.
(C) XopD delays disease symptom production in wild-type Moneymaker and NahG tomato leaves inoculated with Xcv. Phenotype of leaves analyzed in (B) at 12 DAI (wild-type Moneymaker tomato) and 8 DAI (NahG Moneymaker tomato).
Mutant Xcv ΔxopD strains expressing wild-type XopD from a broad host plasmid were complemented for bacterial growth and symptom development in susceptible VF36 tomato leaves (Figure 1C), indicating that the ΔxopD mutant phenotype was specifically due to the loss of XopD function. Thus, in susceptible tomato leaves, XopD action promotes Xcv multiplication but slows the rate of disease symptom development. These data suggest that XopD may directly or indirectly interfere with pathogen-induced leaf senescence.
XopD Suppresses Chlorophyll Loss and Alters Senescence- and Defense-Associated mRNA Levels during Infection
Pathogen infection is known to accelerate the initiation and progression of leaf senescence (Gan, 2007). Leaf senescence is defined as the age-dependent deterioration process at the organ level that is associated with programmed cell death (Lim et al., 2007a). Based on the Xcv phenotypes observed in tomato, we hypothesized that XopD may be suppressing premature leaf senescence and/or defense responses triggered during bacterial colonization. To address this, we analyzed molecular markers that reflect different stages of leaf senescence to determine (1) when senescence- and defense-associated markers are altered during Xcv infection and (2) the role of XopD in affecting the abundance of these markers during bacterial multiplication.
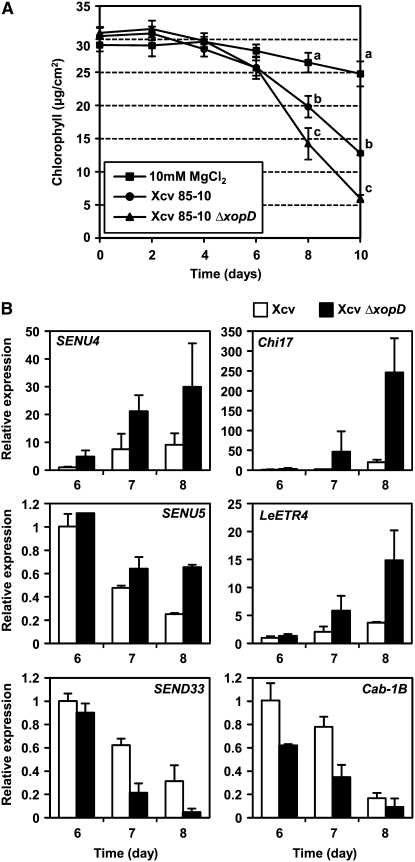
First, we monitored chlorophyll levels because chlorophyll degradation is an integral part of tissue senescence that reflects physiological and biochemical changes associated with nutrient recycling (Gan, 2007). Tomato leaves infiltrated with 10 mM MgCl2 slowly lost chlorophyll over the course of 10 d, demonstrating the rate of chlorophyll degradation in uninfected tissue. Tomato leaves inoculated with Xcv ΔxopD lost significantly more chlorophyll at 8 DAI compared with leaves inoculated with wild-type Xcv (Figure 2A). This is consistent with the observation that tomato leaves infected with Xcv ΔxopD are more chlorotic than leaves infected with wild-type Xcv at 10 DAI (Figure 1B), even though they contain fewer bacteria (Figure 1A). These data indicate that both wild-type Xcv and Xcv ΔxopD initiate premature chlorophyll breakdown as early as 6 DAI. However, the rate of pigment loss between 8 and 10 DAI was significantly faster in leaves infected with Xcv ΔxopD (Figure 1A). Thus, XopD suppresses the rate of chlorophyll degradation delaying the appearance of chlorosis in the highly infected leaf tissue.
Figure 2.
XopD Suppresses Senescence-Associated Responses during Infection.
(A) XopD delays chlorophyll degradation in leaf tissue inoculated with Xcv. Susceptible VF36 tomato leaves were hand-inoculated with 10 mM MgCl2 buffer (square) or a 105 cfu/mL suspension of Xcv (circles) or Xcv ΔxopD (triangles). Total chlorophyll (μg/cm2) was extracted from the inoculated tissue. Error bars indicate sd. Different letters at each time point indicate statistically significant (one-way analysis of variance and Tukey's HSD test, P < 0.05) differences between the samples.
(B) XopD alters the mRNA abundance of senescence- and defense-associated genes. Total RNA was isolated from tomato leaves hand-inoculated with105 cfu/mL of Xcv (white rectangles) or Xcv ΔxopD (black rectangles), respectively. Quantitative real-time RT-PCR was performed for four classes of tomato genes: (1) senescence/upregulated (SENU5); (2) senescence and defense/upregulated (SENU4); (3) defense/upregulated (Chi17); and (4) senescence/downregulated (SEND33 and Cab-1B) as well as the ethylene receptor gene ETR4. Relative expression levels at 6, 7, and 8 DAI are shown. Actin expression was used to normalize the expression value in each sample, and relative expression values were determined against the average value of the sample infected with wild-type Xcv at 6 DAI. The averages of the two independent experiments are shown. Error bars indicate sd.
Second, we used the same diseased tissue to monitor mRNA levels for senescence- and defense-associated genes. Our objective was to determine if XopD specifically or generally affects the transcription and/or abundance of genes whose expression levels significantly change during age- and pathogen-induced senescence. We assayed four classes of tomato genes: (1) senescence-associated/upregulated (SENU2, SENU3, and SENU5); (2) senescence- and defense-associated/upregulated (SENU4); (3) defense-associated/upregulated (Chi17); and (4) senescence-associated/downregulated (SEND33 and Cab-1B). SENU2 and SENU3 encode two different tomato Cys proteases (Drake et al., 1996). SENU5 encodes a NAC transcription factor that is highly expressed at the advanced stages of tomato leaf senescence (John et al., 1997; Olsen et al., 2005). SENU4 encodes a pathogenesis-related protein 1b1, and its mRNA levels increase in response to aging and SA (John et al., 1997; Block et al., 2005). Chi17 encodes chitinase (Danhash et al., 1993). SEND33 and Cab-1B encode two chloroplast proteins (ferredoxin and chlorophyll binding proteins, respectively). SEND33 and Cab-1B mRNA levels decrease at the late stages of senescence (John et al., 1995, 1997).
We monitored the impact of Xcv and Xcv ΔxopD infection on the mRNA abundance of the aforementioned genes between 6 and 8 DAI when significant changes in pathogen titers are clearly evident in leaves (Figure 1A). XopD-dependent changes in mRNA abundance were clearly detected by 7 DAI (Figure 2B), days before the appearance of tissue chlorosis at 10 DAI (Figure 1B). For example, SENU4 mRNA levels were higher in leaves infected with Xcv ΔxopD compared with leaves infected with wild-type Xcv (Figure 2B). Similar trends were observed for SENU5 and Chi17 mRNA levels, although the relative mRNA expression level varied in magnitude and timing between 6 and 8 DAI (Figure 2B). These data show that XopD action reduces mRNA levels for senescence- and defense-associated genes that are upregulated in infected tomato leaves, possibly by suppressing gene transcription or affecting mRNA stability. By contrast, SEND33 and Cab-1B mRNA levels were higher in leaves infected with Xcv compared with Xcv ΔxopD (Figure 2B). This indicates that XopD action can also increase the mRNA level of some senescence downregulated genes. An increase in mRNA level could occur if XopD action increased transcription or inhibited mRNA degradation.
XopD, on the other hand, did not alter the mRNA levels of SENU2 or SENU3 (see Supplemental Figure 3 online). SENU2 and SENU3 mRNAs are most abundant at the late stages of leaf senescence when tissue degeneration is clearly visible (Drake et al., 1996). These data show that XopD action is specifically affecting the abundance of some but not all senescence-associated mRNAs during Xcv infection.
We also monitored mRNA abundance for two ethylene receptors, ETR2 and ETR4. ETR4 is upregulated by Xcv and ethylene and negatively regulates tissue degeneration triggered by high ethylene levels (Ciardi et al., 2000). By contrast, ETR2 mRNA levels are not affected by Xcv or ethylene treatment. By 7 DAI, leaves infected with wild-type Xcv had less ETR4 mRNA compared with leaves infected with Xcv ΔxopD (Figure 2B), whereas ETR2 mRNA levels were similar (see Supplemental Figure 3 online). Reducing ETR4 mRNA levels in the cell would be expected to increase leaf sensitivity to ethylene in terms of symptom development; however, this was not observed (Figure 1B). Collectively, these data indicate that XopD is directly or indirectly opposing changes in mRNA levels that occur in the host in response to Xcv ΔxopD infection.
XopD Reduces SA Levels in Leaves Infected with Xcv
SA is a hormone that regulates the transcription of some plant genes during organ senescence (Morris et al., 2000; Buchanan-Wollaston et al., 2005). Considering that Xcv infection induces SA levels (O'Donnell et al., 2003; Block et al., 2005) and XopD action delays leaf chlorosis and necrosis (Figure 1B), we hypothesized that XopD action might be affecting SA levels in the diseased tissue. To test this, we estimated the SA pool (i.e., free SA plus existing methyl salicylate) in susceptible VF36 leaves inoculated with Xcv over the course of 10 d (Figure 3A). SA pools remained similar from 0 to 4 DAI for all infections. By 6 DAI, leaves inoculated with Xcv ΔxopD had roughly sixfold more SA than leaves inoculated with buffer alone and threefold more than leaves inoculated with Xcv (Figure 3A). Xcv and Xcv ΔxopD bacterial titers at this time point were similar. The significant reduction in the pool of SA in leaves inoculated with Xcv at 6 DAI thus indicates that XopD action leads to the suppresson of SA biosynthesis and/or accumulation. This result also demonstrates that XopD action does not completely abolish SA synthesis but rather dampens the magnitude and timing of SA accumulation in infected leaves.
SA Is Required to Inhibit Xcv Growth
We next hypothesized that reduced Xcv ΔxopD growth in planta could be due to high SA levels in tissue. To address this, we quantified Xcv growth in susceptible, transgenic tomato cultivar Moneymaker expressing NahG, a bacterial gene that encodes salicylate hydroxylase, which cleaves SA into catechol (Delaney et al., 1994). As expected, Xcv ΔxopD multiplication was significantly reduced in Moneymaker leaves compared with wild-type Xcv (Figure 3B). Leaf chlorosis and necrosis were also delayed in a XopD-dependent manner (Figure 3C), consistent with the phenotypes observed in VF36 tomato plants. By contrast, wild-type Xcv and mutant Xcv ΔxopD strains grew similarly in Moneymaker NahG leaves (Figure 3B); however, the rate of bacterial multiplication was much faster in this line than in wild-type Moneymaker leaves. Tissue degeneration was still delayed in a XopD-dependent manner (Figure 3C), although the overall time to death was more rapid in the infected NahG line presumably due to the higher titer of bacteria (Figure 3B). Leaves inoculated with Xcv ΔxopD were fully necrotic by 8 DAI, while leaves inoculated with wild-type Xcv were just beginning to exhibit chlorosis (Figure 3C). These data indicate that SA in tissue infected with Xcv inhibits pathogen growth and does not substantially promote leaf symptom development.
XopD Binds DNA and DNA Binding Is Required for Virulence in Tomato Plants and Tissue Necrosis in N. benthamiana
We next explored whether XopD might influence host gene transcription by directly binding to DNA. This seemed likely since XopD is localized to subnuclear foci in N. benthamiana (Hotson et al., 2003). BLASTP (Altschul et al., 1997) was used to inspect XopD's full-length protein sequence to identify short, nearly exact matches characteristic of DNA binding domains (DBDs). We identified three regions within the N terminus that share sequence similarity with regions of known or predicted DNA binding proteins (see Supplemental Figure 4A online), indicating that XopD may contain a DBD (Figure 4A). Motif 1 (44 to 58 amino acids) is similar to the homeodomain of the Xenopus laevis VENT-2 transcription factor. Motif 2 (121 to 155 amino acids) is similar to the Azotobacter vinelandii IclR family of transcriptional regulatory proteins. Motif 3 (149 to 159 amino acids) is similar to the Vibrio cholerae DNA polymerase III subunit epsilon. Furthermore, analysis of XopD's structure using the HNN secondary structure prediction method (Combet et al., 2000) identified several putative α-helices. All three motifs are located in regions of XopD predicted to form α-helices (see Supplemental Figure 4A online), consistent with the structural nature of some DNA binding proteins (Harrison, 1991).
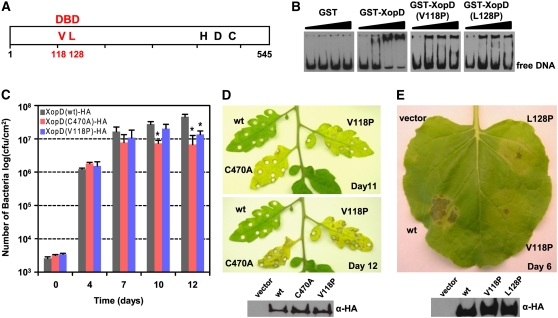
Figure 4.
XopD Binds DNA and DNA Binding Is Required for Virulence in Tomato and Tissue Necrosis in N. benthamiana.
(A) Schematic of the putative DNA binding domain of XopD protein (open rectangle). In red, the N-terminal DBD containing critical residues, V118 and L128 (VL), for DNA binding activity; and in black, the C-terminal SUMO protease domain containing catalytic core residues His, Asp, and Cys (HDC).
(B) In vitro XopD DNA binding activity. Arabidopsis PDF1.2 promoter probes were used for EMSA by being incubated with an increasing concentration (0, 0.06, 0.12, and 0.25 μM) of purified GST, GST-XopD, GST-XopD(V118A), and GST-XopD(L128P) and then visualized by chemiluminescece.
(C) XopD DBD is required for maximal Xcv growth in tomato. Leaves were hand-inoculated with a 1 × 105 cfu/mL suspension of Xcv ΔxopD expressing XopD(wt)-HA (gray bars), XopD(C470A)-HA (red bars), or XopD(V118P)-HA (blue bars). Data points represent mean log10 cfu per cm2 ± sd of three independent experiments. The asterisk above the bars indicates statistically significant (t test, P < 0.05) differences between the bacterial numbers of Xcv ΔxopD expressing XopD(wt)-HA and the XopD mutant proteins.
(D) XopD protease activity and DBD are required to delay disease symptoms in VF36 leaves inoculated with Xcv (top panel). Representative phenotypes of tomato leaves inoculated in (C) after 11 and 12 d: wt, XopD(wt)-HA; C470A, XopD(C470A)-HA; V118P, XopD(V118P)-HA. Immunoblot analysis (bottom panel) of vector and protein expressed in Xcv. Xcv strains described in (C) were grown in XVM2 medium for 6 h with appropriate antibiotics. Total cellular lysate (∼5 × 107 cells) was examined by protein gel blot analysis using HA antisera.
(E) Phenotype of XopD DNA binding mutants in N. benthamiana (top panel). Leaf was infiltrated with a suspension (OD600 = 0.5) of A. tumefaciens expressing the following: vector, vector control; wt, XopD(wt)-HA; V118P, XopD(V118P)-HA; and L128P, XopD(L128P)-HA. The leaf was photographed 6 DAI. Immunoblot analysis (bottom panel) of proteins transiently expressed in N. benthamiana leaves as shown in the top panel. Total protein (∼50μg) was extracted from tissue at 48 h after inoculation and then examined by protein gel blot analysis using HA antisera. These experiments were repeated three times with similar results.
To determine if XopD directly binds DNA, in vitro electrophoretic mobility shift assays (EMSAs) were performed using labeled plant DNA and recombinantly purified glutathione S-transferase (GST) or GST-XopD fusion protein (see Supplemental Figure 5 online). We examined XopD binding to the promoter region of PDF1.2 and PR1, two Arabidopsis genes whose mRNA expression is upregulated by SA and bacterial infection (Penninckx et al., 1996; Kunkel and Brooks, 2002). As a control, we also monitored binding to the Arabidopsis actin promoter ACT2. GST protein incubated with PDF1.2 promoter did not alter DNA mobility. By contrast, GST-XopD bound strongly to the PDF1.2 promoter, resulting in a DNA mobility shift (Figure 4B). Similar results were obtained when the PR1 promoter was used as the DNA probe (see Supplemental Figure 4B online).
XopD DNA binding activity was found to be nonspecific under the conditions tested in vitro. This was initially evident when strong binding of XopD to the ACT2 promoter was observed (see Supplemental Figure 4B online). In competition assays, XopD binding to both PR1 and PDF1.2 promoter probes was inhibited by increasing the concentration of nonspecific DNA, poly(dI·dC) (see Supplemental Figure 4C online). Furthermore, random binding site selection assays failed to identify a XopD binding consensus sequence. The sequence of 42 enriched DNA 16-mers indicated that there was not an obvious XopD DNA binding motif, apart from all sequences being GC rich.
Eight point mutants spanning the three identified motifs were constructed to identify XopD regions required to bind DNA (see Supplemental Figure 4A online). Six point mutants were stably expressed but did not affect DNA binding. By contrast, the binding of GST-XopD(V118P) and GST-XopD(L128P) (Figure 4A) to the PDF1.2 promoter DNA probe was reduced compared with GST-XopD (Figure 4B, right panels). Similar results were obtained in EMSA studies using the PR1 and ACT2 probes (see Supplemental Figure 4B online). To ensure that the Pro substitutions in this region did not grossly affect XopD's protein conformation and its known characteristics, we monitored XopD protein stability, localization, and protease activity. GST-XopD(V118P) and GST-XopD(L128P) expression was similar to GST-XopD (see Supplemental Figure 5 online). Agrobacterium-mediated transient transformation showed that enhanced yellow fluorescent protein (EYFP)-XopD(V118P) and EYFP-XopD(L128P) were localized to subnuclear foci in N. benthamiana (see Supplemental Figure 6 online) and possessed SUMO isopeptidase activity (see Supplemental Figure 7 online) similar to wild-type EYFP-XopD. Together, these studies indicate that the V118P and L128P mutations did not globally affect XopD stability, localization, or protease activity. Therefore, only the putative helix-loop-helix region spanning amino acids 113 to 131 in XopD appears to participate in DNA binding, compared with the other regions studied.
Next, we assessed whether or not the putative helix-loop-helix region spanning amino acids 113 to 131 is important for XopD-dependent bacterial growth in tomato. Susceptible VF36 tomato leaves were inoculated with different Xcv ΔxopD strains carrying a plasmid containing wild-type XopD [XopD(wt)-HA], the DBD mutant [XopD(V118P)-HA], or the protease mutant [XopD(C470A)-HA]. Multiplication of Xcv ΔxopD expressing XopD(C470A) in tomato leaves was significantly reduced at 10 and 12 DAI relative to Xcv ΔxopD expressing XopD-HA (Figure 4C). Growth of Xcv ΔxopD expressing XopD(V118P)-HA was partially impaired, and its titer was only significant different from that of Xcv ΔxopD expressing XopD(wt)-HA at 12 DAI. As observed with the Xcv ΔxopD null mutant, the onset and severity of disease symptoms in leaves inoculated with Xcv ΔxopD expressing XopD(V118P)-HA or XopD(C470A) were inversely related to the titer of bacteria at 12 DAI (Figure 4D). There was no difference in the expression of XopD(V118P)-HA and XopD(C470A)-HA in Xcv ΔxopD compared with that of wild-type XopD-HA (Figure 4D). We also found that Agrobacterium-mediated transient expression of XopD(V118P)-HA and XopD(L128P)-HA in N. benthamiana failed to elicit strong tissue necrosis by 6 DAI (Figure 4E). These mutant proteins were expressed as well as wild-type XopD-HA in planta (Figure 4E), indicating that the point mutations were not affecting protein stability in N. benthamiana. Thus, this region in XopD is required for XopD DNA binding activity, for maximal Xcv virulence in tomato, and to promote severe tissue necrosis in N. benthamiana.
XopD Contains Two EAR Motifs Required for Virulence in Tomato and Tissue Necrosis in N. benthamiana
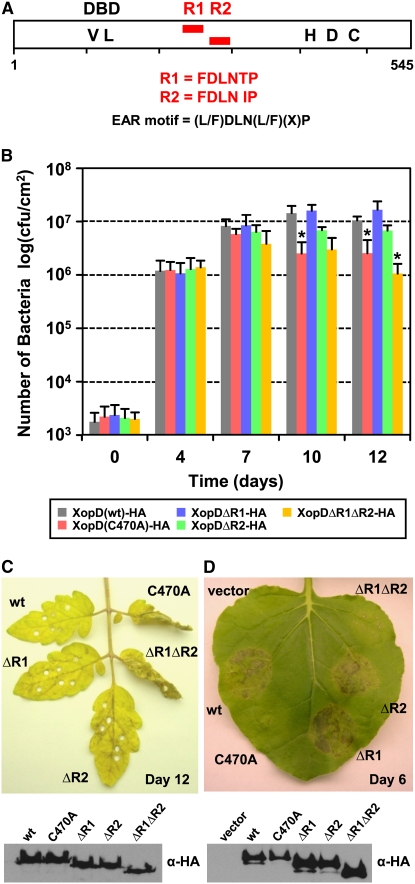
The lack of DNA binding specificity suggested to us that effector specificity must be determined by another XopD domain or by additional DNA–protein or protein–protein interactions within the host nucleus. Interestingly, visual inspection of XopD's sequence revealed two tandemly repeated EAR (ERF-associated amphiphilic repression) motifs [L/FDLNL/F(x)P] (Ohta et al., 2001), which we designated R1 (amino acids 244 to 249) and R2 (amino acids 284 to 289) (Figure 5A). Plant transcription factors containing highly conserved EAR motifs often function as repressors to negatively regulate gene transcription induced during defense and stress responses (Kazan, 2006). To determine if the EAR motifs in XopD are essential for function, the growth and phenotype of Xcv strains containing mutant XopD proteins lacking one (ΔR1 and ΔR2) or both (ΔR1ΔR2) motifs were analyzed.
Figure 5.
XopD Has Two EAR Motifs Required for Virulence in Tomato and Tissue Necrosis in N. benthamiana.
(A) Schematic view of XopD protein. The open rectangle represents XopD protein, and two EAR motifs, R1 and R2, are represented by red rectangles. The motifs are located between the N-terminal DBD and C-terminal SUMO protease domain containing catalytic core residues His, Asp, and Cys (HDC).
(B) XopD EAR motifs are required for maximal Xcv growth in tomato. Susceptible VF36 tomato leaves were hand-inoculated with a 1 × 105 cfu/mL suspension of Xcv ΔxopD expressing XopD(wt)-HA (gray bars), XopD(C470A)-HA (red bars), XopDΔR1-HA (blue bars), XopDΔR2-HA (green bars), or XopDΔR1ΔR2-HA (yellow bars). Data points represent mean log10 cfu per cm2 ± sd of three independent experiments. The asterisk above the bars indicates statistically significant (t test, P < 0.05) differences between the bacterial numbers of Xcv ΔxopD expressing XopD(wt)-HA and the XopD mutant proteins.
(C) XopD EAR motifs are required to suppress disease symptoms in tomato (top panel). Representative phenotype of VF36 tomato leaves inoculated in (B) after 12 d: wt, XopD(wt)-HA; C470A, XopD(C470A)-HA; ΔR1, XopDΔR1-HA; ΔR2, XopDΔR2-HA; ΔR1ΔR2, XopDΔR1ΔR2-HA. Immunoblot analysis (bottom panel) of proteins expressed in the bacteria. Xcv strains described in (B) were grown in XVM2 medium for 6 h with appropriate antibiotics. Total cellular lysate (∼5 × 107 cells) was examined by protein gel blot analysis using HA antisera.
(D) XopDΔR1ΔR2-HA does not elicit necrosis in N. benthamiana (top panel). Leaf was infiltrated with the A. tumefaciens strains expressing the following: vector, vector control; wt, XopD(wt)-HA; C470A, XopD(C470A)-HA; ΔR1, XopDΔR1-HA; ΔR2, XopDΔR2-HA; ΔR1ΔR2, XopDΔR1ΔR2-HA. The phenotype was photographed 6 DAI. Immunoblot analysis (bottom panel) of proteins transiently expressed in N. benthamiana leaves as shown in top panel. Total protein (∼50μg) was extracted from tissue at 48 h after inoculation and then examined by protein gel blot analysis using HA antisera. These experiments were repeated three times with similar results.
Xcv ΔxopD strains expressing XopDΔR1-HA or XopDΔR2-HA grew similarly to Xcv ΔxopD expressing XopD(wt)-HA, reaching ∼1 × 107 cfu/cm2 of leaf tissue at 12 DAI (Figure 5B). However, growth of Xcv ΔxopD expressing XopDΔR1ΔR2-HA was reduced 10-fold. The XopD(C470A)-HA protease deficient strain grew slightly better than the XopDΔR1ΔR2-HA mutant but significantly less than XopD(wt)-HA at 12 DAI (Figure 5B). Deletion of one or both EAR motifs did not affect XopD nuclear localization (see Supplemental Figure 6 online) or protease activity (see Supplemental Figure 7 online), indicating that the mutations did not drastically alter protein structure.
Consistent with their impact on Xcv multiplication, the EAR motifs are required for XopD-dependent suppression of symptom development (Figure 5C). Mutant XopD proteins containing either R1 or R2 suppressed tissue chlorosis and necrosis triggered in leaves infected with Xcv ΔxopD (Figure 5C), indicating that the EAR motifs are functionally redundant. However, we observed that wild-type XopD containing both EAR motifs was able to suppress symptoms elicited by Xcv ΔxopD slightly longer than XopD containing R1 or R2. XopD protease activity was also required to maximally suppress Xcv ΔxopD-elicited symptoms (Figure 5C). The expression of each mutant protein in Xcv ΔxopD was similar to that of wild-type XopD-HA (Figure 5C).
In N. benthamiana, both repressor motifs and protease activity were required for XopD-dependent leaf necrosis. Agrobacterium-mediated transient expression of mutant XopD protein lacking both EAR motifs did not elicit symptoms in N. benthamiana, despite its overexpression (Figure 5D), while overexpression of XopD containing either R1 or R2 triggered tissue necrosis. Taken together, these data show that both EAR motifs and protease activity are required for XopD-dependent virulence in tomato and symptom production in N. benthamiana.
XopD Represses Plant Gene Expression
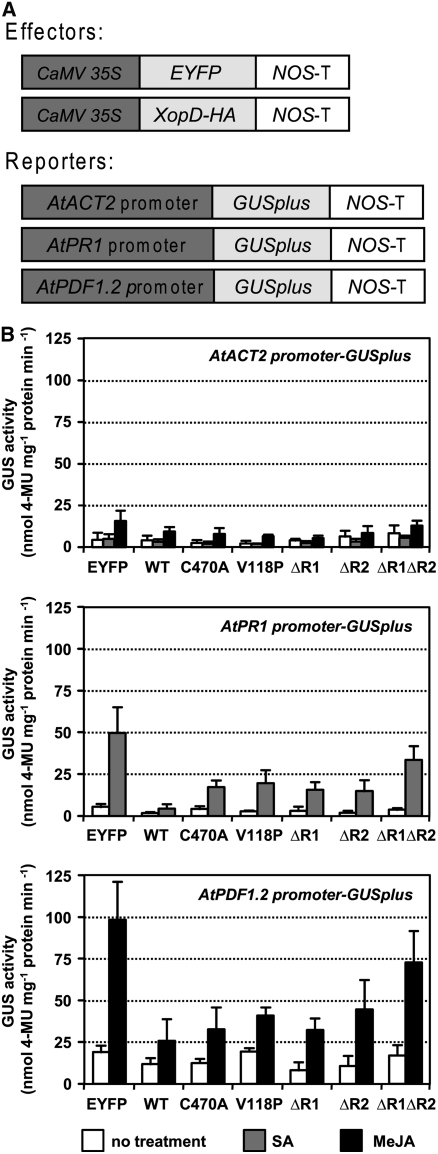
We next determined if Agrobacterium-mediated transient expression of XopD in N. benthamiana is sufficient to alter the transcription of reporter genes in response to hormone treatment. Three Arabidopsis promoters (PR1, PDF1.2, and ACT2) were used in a heterologous expression study in N. benthamiana. The promoters were fused to GUSplus (a modified Staphylococcus β-glucuronidase [GUS] gene and a more sensitive reporter gene than Escherichia coli GUS gene; http://www.cambia.org) and served as reporters of transcription (Figure 6A). EYFP, XopD-HA, and select XopD mutants served as putative effectors of transcription. The effectors were coexpressed with the reporters to determine if they could alter reporter transcription (Figure 6A). Reporters and effectors were transiently coexpressed in N. benthamiana leaves for 18 h. Leaves were then treated with buffer, SA, or methyl jasmonate for 12 h. GUS activity was quantified to determine the level of transcription at each promoter in response to each putative effector.
Figure 6.
XopD Represses Plant Defense Gene Transcription.
(A) Schematic of transcription effector and reporter constructs coexpressed in N. benthamiana. Effectors (EYFP, XopD-HA, and mutant XopD-HA derivatives) were constitutively expressed using the cauliflower mosaic virus 35S promoter. The GUSplus reporter was fused to the ACT2, PR1, and PDF1.2 promoters. The nopaline synthase terminator (NOS-T) was used in all constructs.
(B) XopD EAR motifs are required for repression of PR1 and PDF1.2 transcription. N. benthamiana leaves inoculated with each A. tumefaciens strain for 18 h were treated with buffer, SA (2 mM), or methyl jasmonate (MeJA; 100 μM). Samples were collected 12 h later and GUS activity (nmol of 4-methylumbelliferone [4-MU] mg−1 protein min−1) quantified. For each reporter, the following effectors were tested: EYFP, enhanced yellow fluorescent protein; WT, XopD-HA; C470A, XopD(C470A)-HA; V118P, XopD(V118P)-HA; ΔR1, XopDΔR1-HA; ΔR2, XopDΔR2-HA; ΔR1ΔR2, XopDΔR1ΔR2-HA. Bars represent buffer (white), SA (gray), and methyl jasmonate (black) treatment. Error bars indicate sd of three independent experiments.
Transcription at the PR1 and PDF1.2 promoters was induced by SA or methyl jasmonate treatment, whereas transcription from the ACT2 promoter was relatively unaffected (Figure 6B). In leaves carrying the ACT2 promoter-GUSplus reporter construct, low GUS activity was consistently detected under all conditions tested despite the effector being expressed or the hormone treatment (i.e., SA or methyl jasmonate; Figure 6B, top panel). Despite the fact that XopD can nonspecifically bind to the ACT2 promoter (Figure 4B), XopD-dependent repression of transcription to background levels was not detected. For the PR1 reporter, high GUS activity was detected in SA-treated leaves expressing the PR1promoter-GUSplus reporter and the EYFP effector (Figure 6B, middle panel). Yet, when XopD-HA served as the effector, PR1 transcription was strongly repressed. Each of the XopD mutants tested was less effective than XopD in reducing PR1 transcription, indicating that protease activity, DNA binding activity, and the repressor motifs all contribute to maximal transcriptional repression. Alleviation of XopD repressor activity was only observed when both R1 and R2 were deleted, indicating that the motifs are also functionally redundant for this phenotype. Similar trends were detected in plants coexpressing the XopD effector and the PDF1.2 promoter-GUSplus reporter (Figure 6B, bottom panel). These results demonstrate that XopD expression in planta is sufficient to repress SA- and jasmonic acid–induced gene transcription.
DISCUSSION
In this work, we discovered that Xcv employs a T3S-dependent virulence factor to suppress tissue degeneration in diseased leaves. When comparing wild-type Xcv infections to those of Xcv ΔxopD, we observed that pathogen titer in susceptible tomato leaves does not correlate with the onset of disease symptoms (Figures 1A and 1B). Our data suggest that XopD functions inside the plant nucleus to alter basal defense and cell death responses triggered in the host during Xcv pathogenesis. These responses are likely triggered in the host tissue to eliminate the infected organ and restrict the spread of the pathogen. We speculate that XopD might function at the later stages of Xcv infection to suppress abrupt physiological changes in the host (e.g., hormone signaling, defense signaling, and cell death) to sustain the bacterial population within the infected tissue. The importance of XopD-dependent phenotypes at 6 to 7 DAI suggests that it might be secreted later than other Xcv type III effector proteins. Or, a threshold concentration of XopD must be reached to alter plant physiology. These ideas stem from the striking observation that XopD function in tomato leaves correlates with plant tolerance (i.e., the ability of the host to cope with bacterial colonization). Thus, XopD is a T3S effector that functions as a virulence factor by contributing to Xcv multiplication but also functions as a tolerance-promoting factor by suppressing leaf symptom development.
Effector-mediated suppression of disease symptoms was first noted by López-Solanilla et al. (2004) while studying Pseudomonas–tomato interactions. They showed that HopN1 (previously referred to as HopPtoN) action inside susceptible tomato leaves reduces the number of macroscopic lesions observed 12 DAI even though P. syringae pathovar tomato (Pst) DC3000 and Pst DC3000 ΔhopN1 titers were similar. The mechanisms by which HopN1 suppresses symptom production were not investigated. However, it was shown that HopN1 has Cys protease activity in vitro characteristic of papain-like enzymes, suggesting that it might cleave specific host targets associated with defense (López-Solanilla et al., 2004).
Although HopN1 and XopD encode Cys proteases, they each have distinct enzyme activities (Hotson et al., 2003; López-Solanilla et al., 2004). HopN1 belongs to the CA clan (proteases in this clan are characterized by a Cys nucleophile and a catalytic core that is comprised of three amino acid residues, C/H/D or N; the peptidase database MEROPS, http://merops.sanger.ac.uk/), whereas XopD belongs to the CE clan (proteases in this clan are characterized by a Cys nucleophile and a catalytic core that is comprised of three amino acid residues, H/D or E/C). This suggests that the targets and mechanisms by which these effectors alter tomato signaling to suppress symptom development are likely different. XopD-dependent phenotypes observed for Xcv infection are however distinct from those of HopN1 during Pst DC3000 infection. XopD alters the kinetics of leaf chlorosis and necrosis without affecting the number or rate of appearance of necrotic lesions. Moreover, XopD promotes pathogen multiplication and suppresses pathogen-induced senescence in leaves.
Currently, we do not know if XopD virulence function is important for Xcv colonization in susceptible pepper (Capsicum annuum). We were unable to determine if XopD promotes Xcv growth in susceptible Early Cal Wonder pepper leaves because leaves inoculated with Xcv typically abscised between 7 and 8 DAI, the time period when the titers of Xcv and Xcv ΔxopD are significantly different in susceptible tomato leaves (Figure 1A). A previous study showed that Xcv mutant strains lacking the protease domain of XopD (amino acids 311 to 545) produce typical symptoms in pepper leaves (i.e., water soaking in susceptible lines or the hypersensitive response in resistant lines) when compared with wild-type Xcv strains (Noël et al., 2002). These data suggest that XopD might not be required for virulence in pepper, or perhaps another effector playing a functionally redundant role in pepper might mask XopD action.
SA is implicated in the development of normal leaf senescence and the inhibition of pathogen growth (Delaney et al., 1994; Morris et al., 2000). In tomato, we found that high SA pools were not required for tissue necrosis in leaves inoculated with Xcv. Other senescence promoting factors, yet to be determined, are likely influencing these physiological changes. Based on our studies with the NahG tomato plants, the primary role for SA in this interaction appears to be to limit Xcv growth, consistent with previous findings (O'Donnell et al., 2003). Our work also shows that XopD can breach SA-dependent defense by 6 DAI (Figure 3A). XopD-dependent reduction in the SA pool in infected leaves at 6 DAI presumably enables wild-type Xcv to multiply in the stressed organ, whereas mutant strains lacking XopD cannot.
How XopD breaches host defenses is still not clear. However, the discovery of XopD-dependent virulence phenotypes and additional biochemical activities in planta provides new insight to potential host targets and pathways affected by XopD. The fact that XopD alters the abundance of distinct mRNAs during the late stages of disease progression in leaves argues that the effector has specific host targets, whose functions may intersect both defense and senescence signaling pathways. XopD-specific suppression of SENU5, encoding a NAC transcription factor associated with leaf senescence, suggests that transcription factors and/or regulators of transcription may be direct targets of XopD.
Our structure-function studies of XopD performed in vitro and in planta reveal that XopD might be mimicking the function of some plant transcriptional regulators. XopD-dependent modulation of senescence- and defense-associated mRNA levels during Xcv infection provided evidence that XopD is altering host gene transcription (Figure 2B). We then discovered that XopD possesses DNA binding activity and transcriptional repression activity. XopD appears to be a nonspecific DNA binding protein that uses a helix-loop-helix domain between amino acids 113 and 131 to maximally bind DNA (Figure 4B). We were unable to identify a defined DNA binding target sequence for XopD, other than stretches of GCs. This is intriguing because some plant transcription factors bind to GCC-boxes in promoters in response to senescence and stress, both abiotic and biotic (Fujimoto et al., 2000).
The identification of two conserved EAR motifs in XopD spanning the DNA binding domain and the C-terminal protease domain ultimately provided a clue that XopD might be capable of repressing host gene transcription. In plants, EAR motifs are found on a number of transcriptional regulators and they are sufficient to repress transcription (Ohta et al., 2001). It is not clear how EAR repressors work, but the pathways that they negatively regulate ultimately control their own transcription, indicating that such repressors tightly regulate signaling by a feedback mechanism. For example, NIMIN1 negatively regulates SA-dependent defense genes (e.g., PR1) in Arabidopsis by interacting with the transcription factor NPR1 (Weigel et al., 2005; Kazan, 2006).
Our work shows that the XopD EAR motifs are required to repress plant gene transcription, to promote maximal Xcv multiplication, and to delay the onset of disease symptoms infected tomato leaves (Figures 5 and 6). Protease activity and DNA binding activity only partially contributed to transcriptional repression in N. benthamiana. Our phenotypic studies show that a significant effect on pathogen virulence was only apparent when both EAR motifs were deleted from XopD or when protease activity was inactivated (Figures 4C and 5B), whereas DNA binding mutants significantly affected pathogen growth only at 12 DAI (Figure 4C). Together, these data suggest that repressor and protease activities contribute more to XopD virulence than DNA binding activity.
A summary of all the phenotypes for XopD mutants examined in this study is presented in Table 1. Collectively, the domain structure and distinct activities associated with these domains indicates that XopD is a modular T3S effector protein. XopD's function in the plant nucleus is thus more complex than expected. The effector likely partakes in multiple protein–DNA and protein–protein interactions to alter host transcription.
Table 1.
Summary of the Phenotypes of XopD Mutant Proteins
| Protein Tested | Virulence (S. lycopersicum) | Symptom Development (S. lycopersicum) | Cell Death (N. benthamiana) | SUMO Protease Activity (N. benthamiana) | DNA Binding (in Vitro) | Nuclear Localization (N. benthamiana) |
|---|---|---|---|---|---|---|
| XopD(wt) | Normal | Normal | Yes | Yes | Yes | Yes |
| XopD(C470A) | Reduced | Earlier than the wild type | No | No | Yes | Yes |
| XopD(V118P) | Reduced | Earlier than the wild type but later than XopD(C470A) and XopDΔR1ΔR2 | Delayed | Yes | No | Yes |
| XopDΔR1 | Normal | Normal | Yes | Yes | Yes | Yes |
| XopDΔR2 | Normal | Normal | Yes | Yes | Yes | Yes |
| XopDΔR1ΔR2 | Reduced | Earlier than wild type | No | Yes | Yes | Yes |
How then does XopD repress host transcription? Multiple mechanisms can be envisioned based on the central importance of all three XopD activities in controlling eukaryotic transcription (see Supplemental Figure 8 online for working model). For instance, DNA binding domains provide specific or nonspecific access to chromatin and transcriptional units. EAR motifs play a dominant role in the repression of hormone-induced transcription in plants (Ohta et al., 2001; Tiwari et al., 2004) and are emerging as important negative regulators of defense gene expression (Kazan, 2006). Moreover, SUMO posttranslational modification, opposed to monoubiquitination, plays a central role in transcriptional repression (Gill, 2004). Based on this information, XopD may work in the following manner. (1) XopD could confer repression by dimerizing with a DNA-bound transcriptional activator via the two EAR domains, thereby directly affecting gene transcription. (2) XopD could desumoylate a transcriptional regulator, altering its activity or stability, thereby indirectly regulating gene expression. (3) XopD could work at the level of chromatin remodeling and thus affect both the repression and activation of gene transcription.
We currently favor the model that XopD may be functioning at the level of chromatin-remodeling based on the finding that ERF7, an Arabidopsis EAR-containing protein, appears to be altering chromatin status by recruiting the corepressor SIN3 and histone deacetylase HDA19 to promoters containing GCC boxes (Song et al., 2005; Weigel et al., 2005; Kazan, 2006). Also, the overexpression of ORE7, nuclear chromatin remodeling, AT-hook DNA binding motif prolongs leaf longevity in Arabidopsis (Lim et al., 2007b).
Based on our findings, we speculate that XopD alters host transcription to reverse rapid changes in host gene expression in response to Xcv colonization. The central importance of the EAR motifs and protease activity in XopD virulence indicate that these biochemical activities might play a major role in substrate specificity. For example, the XopD EAR motifs and protease domain might engage in distinct interactions with chromatin factors or the transcriptional machinery. XopD nonspecific DNA binding activity might enable XopD to increase the affinity of these interactions and/or to access distinct promoter sites to modulate the expression of several host genes. Future work will be aimed at addressing the mechanism of XopD transcriptional repression and identifying genes and pathways affected by XopD during the course of Xcv infection.
In conclusion, we have shown that XopD functions as a transcriptional repressor, resulting in the suppression of host responses at the late stages of infection. XopD represses SA-induced defense responses that limit Xcv growth and counteracts cellular events orchestrated in diseased leaves. By prolonging leaf viability, we hypothesize that the pathogen is not only sustaining its niche but ensuring that enough resources are available for maximal community growth prior to organ death and pathogen dissemination.
METHODS
Xanthomonas Strains and Growth Conditions
Xanthomonas campestris pathovar vesicatoria strain 85-10 (originally obtained from Brian Staskawicz) was used for this study. Wild-type and mutant Xcv 85-10 strains were grown on nutrient yeast glycerol agar (NYGA) (Turner et al., 1984) at 28°C. For protein expression analysis, strains were grown in Xanthomonas vesicatoria minimal media 2 (XVM2) liquid broth (Noël et al., 2002).
Construction of the Xcv 85-10 ΔxopD Null Mutant
Standard methods were used for DNA cloning, restriction mapping, and gel electrophoresis (Sambrook et al., 1989). All primer information is listed in Supplemental Table 1 online. The Xcv xopD deletion mutant was generated by insertion of the spectinomycin (Sp)-resistant gene into wild-type Xcv strain 85-10 by marker exchange. The 1.5-kb upstream and 1.2-kb downstream regions of xopD were PCR amplified using Xcv genomic DNA as template and cloned into pENTR/D-TOPO (Invitrogen), creating pENTR/D-TOPO(ΔxopD). PCR primer sets JG76/JG78 and JG79/JG80 were used to amplify the upstream and downstream region of xopD, respectively. The 1.5-kb Sp cassette was cloned into the BamHI site between the upstream and downstream region of xopD in pENTR/D-TOPO(ΔxopD), creating pENTR/D-TOPO(ΔxopD∷Sp). The 4.2-kb ΔxopD∷Sp insert in pENTR/D-TOPO(ΔxopD∷Sp) was recombined into suicide vector pLVC18-Rfc (courtesy of C. Morales and B. Staskawicz) using the Gateway system, generating pLVC18-Rfc(ΔxopD∷Sp). The plasmid was introduced into Xcv 85-10 by triparental mating. Xcv exconjugants were selected by growth on NYGA media containing 100 μg/mL rifampicin (Rif), 50 μg/mL Sp, and 10 μg/mL tetracycline (Tet). Xcv strains (Rifr, Spr, and Tets) were then analyzed by PCR to confirm that homologous recombination occurred at the xopD locus.
XopD Mutagenesis
Site-directed mutagenesis of xopD was performed using the PCR-mediated megaprimer method (Barettino et al., 1994). In the first PCR amplification, pEG202-nls(xopD) was used as a template, and MB160 primer and individual mutagenic primers were used (see Supplemental Table 2 online). The PCR products were isolated from 1.5% agarose gels, purified using the QIAEX II gel extraction kit (Qiagen), and resuspended in water for use in subsequent PCRs as megaprimers. In the second PCR amplification, each megaprimer was used with primer DH29 and the pCRII(xopD1-242-HA) template. The respective PCR products were cloned into pCRII and sequenced. The BamHI-StuI fragment for each construct was subcloned into pGEX-5X-3(xopD) and pESC-URA(xopD-HA). For transient expression of the XopD mutants in Nicotiana benthamiana, the BamHI-NheI fragment of pESC-URA (mutant xopD-HA) was subcloned into the BamHI-XbaI site of pEZRK-LCY plasmid.
Bacterial Growth Curves
To monitor Xcv growth in planta, Solanum lycopersicum cv VF36 leaves were hand-inoculated by complete infiltration of the leaf tissue with a 1 × 105 cfu/mL suspension of bacteria in 10 mM MgCl2 using a needleless syringe. Leaves of the same age on the same branch were used for each experimental test. Plants were kept under 16 h light/day at 28°C. Under these conditions, small black bacterial lesions or spots generally appeared by 6 DAI. Four leaf discs (0.5 cm2) per treatment per time point were ground in 10 mM MgCl2 and diluted and spotted onto NYGA plates in triplicate to determine bacterial load. Three biological replicates (i.e., three plants) were used, and the experiment was repeated at least three times. The average bacterial titer ± sd for the three experiments is reported.
Chlorophyll Quantification
Total chlorophyll in leaves was determined as described (Arnon, 1949) with slight modification. VF36 tomato leaves were hand-inoculated with a needleless syringe with 10 mM MgCl2 or a 1 × 105 cfu/mL suspension of Xcv or Xcv ΔxopD in 10 mM MgCl2. Four leaf discs (1 cm2) from three biological replicates were pooled, ground in 1 mL of 80% acetone, and then centrifuged at 18,000 g for 1 min. Optical density of the supernatant at 645 and 663 nm was determined to calculate total chlorophyll content. The experiment was repeated three times, and the average chlorophyll content ± sd is reported.
RNA Isolation and Quantitative RT-PCR
Total RNA was isolated from leaves hand-inoculated with a needleless syringe containing a 1 × 105 cfu/mL suspension of Xcv using Trizol reagent (Invitrogen) according to the manufacturer's instructions. Five micrograms of RNA were used for cDNA synthesis. Quantitative RT-PCR was performed using the cDNA and gene-specific primers (see Supplemental Table 1 online). Each cDNA was amplified by real-time PCR using HotStart-IT SYBR Green qPCR Master Mix (USB) and the ABI 7300 PCR system (Applied Biosystems). Actin expression was used to normalize the expression value in each sample, and relative expression values were determined against the average value of wild-type Xcv sample at 6 DAI. Experiments were repeated at least twice.
SA Quantification
Extraction, methylation, and analysis of the combined free SA and existing methyl salicylate pools via a chemical ionization–gas chromatography/mass spectrometry method were performed as previously described (Schmelz et al., 2004). Approximately 150 mg of infected leaf material was extracted with three biological replicates analyzed.
Transient Protein Expression in N. benthamiana
Agrobacterium tumefaciens strain C58C1 pCH32 (Tai et al., 1999) was used for transient protein expression in planta. Strains were grown overnight at 28°C on Luria agar medium containing 100 μg/mL Rif, 5 μg/mL Tet, and 35 μg/mL kanamycin. Bacteria were incubated in induction media (10 mM MES, pH 5.6, 10 mM MgCl2, and 150 μM acetosyringone; Acros Organics) 2 h before inoculation. N. benthamiana leaves were hand-inoculated with a bacterial suspension (OD600 = 0.5) in induction media. Plants were incubated at room temperature under continuous low light for 2 to 4 d.
Protein Extraction and Immunoblot Analysis
Protein pellets were washed in 1 M Tris and then resuspended in 8 M urea sample buffer. Protein was extracted from plant cells as previously described (Mudgett and Staskawicz, 1999). Proteins separated by SDS-PAGE and then transferred to nitrocellulose were detected by ECL chemiluminescence (Amersham) using HA sera (Covance) and horseradish peroxidase–conjugated secondary antibodies (Bio-Rad).
GST-XopD Protein Expression and Purification
GST-XopD1-545, GST-XopD(V118P), and GST-XopD(L128P) were expressed in Escherichia coli BL21 Star (DE3) cells (Invitrogen) and purified by GST affinity chromatography (Hotson et al., 2003). Cells were grown in 2× yeast extract/tryptone media to OD600 = 0.6 to 0.8 and then induced with 400 μM isopropylthio-β-galactoside (Roche) for 4 h at room temperature. Cells were lysed in PBS, pH 8.0, 1% Triton X-100, 0.1% β-mercaptoethanol, and 1 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich) with a sonicator (Branson). Protein bound to Glutathione Sepharose 4B beads (Amersham Biosciences) was eluted with 10 mM reduced glutathione. Purified proteins were analyzed by SDS-PAGE and quantified using the Coomassie protein assay reagent kit (Pierce).
XopD Gel Mobility Shift Assay
DNA binding assays were done using the Lightshift Chemiluminescent EMSA kit as described (Pierce). Plant promoters were amplified with the appropriate primer pair using Arabidopsis thaliana genomic DNA template, and then sequences were confirmed: 320-bp PR1 promoter with primers JG242/JG131, 313-bp PDF1.2 promoter with primers JG240/JG133, and 319-bp ACT2 promoter with primers JG163/JG312. DNA labeled with biotin using the Biotin 3′ end DNA labeling kit (Pierce) was incubated with purified GST or GST-XopD in binding buffer (10 mM Tris, pH 7.5, 50 mM KCl, 5 mM MgCl2, 5% glycerol, and 1 mM DTT) for 30 min at 28°C. The final concentration of each protein in the reaction was 0, 0.06, 0.12, and 0.25 μM. Mixtures were separated on a 6% polyacrylamide gel in 0.5× Tris-Borate-EDTA buffer and transferred to nitrocellulose membranes (Amersham Biosciences) to detect DNA by chemiluminescence.
Random DNA Binding Site Selection
Random binding site selection was performed using the method described previously with some modification (Okuno et al., 2001). Double-stranded oligonucleotide containing random 16 bases was prepared by PCR using primer sets JG107, JG108, and JG109. The DNA was incubated with the GST-XopD protein, and the binding mixture was separated by PAGE, and the DNA of the complex was eluted from the gel. The recovered DNA was PCR amplified with the primer set JG108/JG109, and the PCR product was used for a subsequent round of selection. The DNA from the sixth round of selection was cloned into pCR-BluntII vector (Invitrogen) for sequencing. The MDscan algorithm (http://ai.stanford.edu/∼xsliu/MDscan/) was used for sequence analysis.
Plant GUS Constructs
For GUS reporter assays in N. benthamiana, a 1-kb EcoRI-HindIII promoter fragment from the Arabidopsis PR1 and PDF1.2 genes was PCR amplified using primer pairs JG130/JG131 and JG132/JG133, respectively. PCR products were cloned into pCRII, creating pCRII(PR1 promoter) and pCRII(PDF1.2 promoter) and sequence confirmed. To make two AvrII sites in pBluescriptII SK+, PCR primers JG118 and JG119 were used to amplify the entire vector. Ligation of this DNA introduced one AvrII site upstream of the SacI site in the vector. Using this plasmid as template, PCR was repeated using primers JG120 and JG121. Ligation generated a second AvrII site downstream of the KpnI site, creating pBSII-AvrII. A 2.3-kb XhoI-GUSplus-Nos Terminator-KpnI fragment was amplified using PCR primers JG114, JG115, and pCAMBIA1305.1 (http://www.cambia.org) as a template and then cloned into the XhoI-KpnI sites of pBSII-AvrII, creating pBSII-AvrII(GUSplus). The EcoRI-HindIII PR1 and PDF1.2 promoter fragments were subcloned into pBSII-AvrII(GUSplus), creating pBSII-AvrII(PR1 promoter-GUSplus) and pBSII-AvrII(PR1 promoter-GUSplus). Finally, the 3.3-kb AvrII fragment of the PR1 promoter- or PDF1.2 promoter-GUSplus-NOS terminator regions was subcloned into the single SpeI site in pEZRK-LCY (a binary vector containing EYFP; a gift from David Ehrhardt) or pEZRK-LCY(xopD-HA), creating pEZRK-LCY(PR1 promoter-GUSplus), pEZRK-LCY(xopD-HA + PR1-promoter-GUSplus), pEZRK-LCY(PDF1.2 promoter-GUSplus), and pEZRK-LCY(xopD-HA + PDF1.2 promoter-GUSplus).
Plant GUS Assays
GUS reporter assays in N. benthamiana were performed as described, with modification (Yang et al., 2000). Agrobacteria cells (OD600 = 0.3) harboring reporter plasmids pEZRK-LCY(PR1 or PDF1.2 promoter-GUSplus) or pEZRK-LCY(xopD-HA + PR1 or PDF1.2 promoter-GUSplus) were inoculated into leaves as described above. For chemical treatment, leaves were sprayed with 2 mM SA (Sigma-Aldrich) or 100 μM methyl jasmonate (Sigma-Aldrich) 18 h after agroinfiltration. After 12 h, leaf discs (0.75 cm2) were ground in 300 μL of GUS extraction buffer (50 mM sodium phosphate, pH 7.0, 10 mM EDTA, 0.1% sodium lauryl sarcosine, 0.1% Triton X-100, and 10 mM β-mercaptoethanol) and centrifuged at 16,000 g for 10 min at 4°C. Fifty microliters of supernatant was mixed with 450 μL of GUS extraction buffer with 1 mM 4-methylumbelliferyl-d-glucuronide (Sigma-Aldrich). The mixture was incubated at 37°C for 1 h. Total protein concentration was quantified using the Coomassie protein assay reagent kit, and GUS activity was determined using the Gilford Fluoro IV spectrofluorometer (Ciba Corning Diagnostics).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AM039952 (xopD), M21444 (SENU2), Z48736 (SENU3), Y08804 (SENU4), Z75524 (SENU5), Z75520 (SEND33), M14443 (Cab-1B), Z15139 (Chi17), AF043085 (ETR2), AY600438 (ETR4), U60480 (Actin), At3g18780 (At ACT2), At2g14610 (At PR1), and At5g44420 (At PDF1.2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. XopD Causes Tissue Necrosis in N. benthamiana.
Supplemental Figure 2. XopD Is Required for Xcv Multiplication and to Delay Disease Symptom Production in Tomato Cultivar Pearson.
Supplemental Figure 3. XopD Does Not Affect the mRNA Levels of SENU2, SENU3, or ETR2 Genes during Xcv Infection in Tomato.
Supplemental Figure 4. XopD Is a Nonspecific DNA Binding Protein.
Supplemental Figure 5. Purified Recombinant Proteins Used for EMSA.
Supplemental Figure 6. EYFP-XopD Mutant Proteins Are Localized to Nuclear Foci in Planta.
Supplemental Figure 7. All EYFP-XopD Mutant Proteins Have SUMO Protease Activity in Planta, Except for EYFP-XopD(C470A).
Supplemental Figure 8. Possible Repression Model Depicting XopD-Dependent Action within the Plant Nucleus.
Supplemental Table 1. List of Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Dominique Bergmann, Or Gozani, Sharon Long, and Charles Yanofsky for instrument use, lab members for intellectual discussion, Christopher Aakre for manuscript comments, and Harry Klee for reagents, for setting up our collaboration, and for critical feedback. J.-G.K. was supported in part by Korea Research Foundation Grant KRF-2004-037-C00254. M.B.M. was supported by National Institutes of Health Grant 1R01 GM-068886, the Terman Fellowship, and the Hellman's Scholar Fellowship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Mary Beth Mudgett (mudgett@stanford.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abramovitch, R.B., Anderson, J.C., and Martin, G.B. (2006. a). Bacterial elicitation and evasion of plant innate immunity. Nat. Rev. Mol. Cell Biol. 7 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramovitch, R.B., Janjusevic, R., Stebbins, C.E., and Martin, G.B. (2006. b). Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. Proc. Natl. Acad. Sci. USA 103 2851–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano, J.R., and Collmer, A. (2004). Type III secretion system effector proteins: Double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42 385–414. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon, D.I. (1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell, M.J., Chisholm, S.T., Dahlbeck, D., and Staskawicz, B.J. (2003). Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Mol. Microbiol. 49 1537–1546. [DOI] [PubMed] [Google Scholar]
- Barettino, D., Feigenbutz, M., Valcarcel, R., and Stunnenberg, H.G. (1994). Improved method for PCR-mediated site-directed mutagenesis. Nucleic Acids Res. 22 541–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block, A., Schmelz, E., O'Donnell, P.J., Jones, J.B., and Klee, H.J. (2005). Systemic acquired tolerance to virulent bacterial pathogens in tomato. Plant Physiol. 138 1481–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston, V., Page, T., Harrison, E., Breeze, E., Lim, P.O., Nam, H.G., Lin, J.F., Wu, S.H., Swidzinski, J., Ishizaki, K., and Leaver, C.J. (2005). Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 42 567–585. [DOI] [PubMed] [Google Scholar]
- Chisholm, S.T., Coaker, G., Day, B., and Staskawicz, B.J. (2006). Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124 803–814. [DOI] [PubMed] [Google Scholar]
- Chosed, R., Tomchick, D.R., Brautigam, C.A., Mukherjee, S., Negi, V.S., Machius, M., and Orth, K. (2007). Structural analysis of Xanthomonas XopD provides insights into substrate specificity of ubiquitin-like protein proteases. J. Biol. Chem. 282 6773–6782. [DOI] [PubMed] [Google Scholar]
- Ciardi, J.A., Tieman, D.M., Lund, S.T., Jones, J.B., Stall, R.E., and Klee, H.J. (2000). Response to Xanthomonas campestris pv. vesicatoria in tomato involves regulation of ethylene receptor gene expression. Plant Physiol. 123 81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn, B., Sekirov, I., and Finlay, B.B. (2007). Type III secretion systems and disease. Clin. Microbiol. Rev. 20 535–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combet, C., Blanchet, C., Geourjon, C., and Deleage, G. (2000). NPS@: Network protein sequence analysis. Trends Biochem. Sci. 25 147–150. [DOI] [PubMed] [Google Scholar]
- Danhash, N., Wagemakers, C.A.M., Kan, J.A.L., and Wit, P.J.G.M. (1993). Molecular characterization of four chitinase cDNAs obtained from Cladosporium fulvum-infected tomato. Plant Mol. Biol. 22 1017–1029. [DOI] [PubMed] [Google Scholar]
- Delaney, T.P., Uknes, S., Vernooij, B., Friedrich, L., Weymann, K., Negrotto, D., Gaffney, T., Gut-Rella, M., Kessmann, H., Ward, E., and Ryals, J. (1994). A central role of salicylic acid in plant disease resistance. Science 266 1247–1250. [DOI] [PubMed] [Google Scholar]
- Drake, R., John, I., Farrell, A., Cooper, W., Schuch, W., and Grierson, D. (1996). Isolation and analysis of cDNAs encoding tomato cysteine proteases expressed during leaf senescence. Plant Mol. Biol. 30 755–767. [DOI] [PubMed] [Google Scholar]
- Fu, Z.Q., Guo, M., Jeong, B.R., Tian, F., Elthon, T.E., Cerny, R.L., Staiger, D., and Alfano, J.R. (2007). A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature 447 284–288. [DOI] [PubMed] [Google Scholar]
- Fujimoto, S.Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan, S. (2007). Senescence Processes in Plants. (Oxford, UK: Blackwell Publishing).
- Gill, G. (2004). SUMO and ubiquitin in the nucleus: Different functions, similar mechanisms? Genes Dev. 18 2046–2059. [DOI] [PubMed] [Google Scholar]
- Gu, K., Yang, B., Tian, D., Wu, L., Wang, D., Sreekala, C., Yang, F., Chu, Z., Wang, G.L., White, F.F., and Yin, Z. (2005). R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 435 1122–1125. [DOI] [PubMed] [Google Scholar]
- Harrison, S.C. (1991). A structural taxonomy of DNA-binding domains. Nature 353 715–719. [DOI] [PubMed] [Google Scholar]
- Hotson, A., Chosed, R., Shu, H., Orth, K., and Mudgett, M.B. (2003). Xanthomonas type III effector XopD targets SUMO-conjugated proteins in planta. Mol. Microbiol. 50 377–389. [DOI] [PubMed] [Google Scholar]
- John, I., Drake, R., Farrell, A., Cooper, W., Lee, P., Horton, P., and Grierson, D. (1995). Delayed leaf senescence in ethylene-deficient ACC-oxidase antisense tomato plants: Molecular and physiological analysis. Plant J. 7 483–490. [Google Scholar]
- John, I., Hackett, R., Cooper, W., Drake, R., Farrell, A., and Grierson, D. (1997). Cloning and characterization of tomato leaf senescence-related cDNAs. Plant Mol. Biol. 33 641–651. [DOI] [PubMed] [Google Scholar]
- Kay, S., Hahn, S., Marois, E., Hause, G., and Bonas, U. (2007). A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science 318 648–651. [DOI] [PubMed] [Google Scholar]
- Kazan, K. (2006). Negative regulation of defence and stress genes by EAR-motif-containing repressors. Trends Plant Sci. 11 109–112. [DOI] [PubMed] [Google Scholar]
- Kunkel, B.N., and Brooks, D.M. (2002). Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5 325–331. [DOI] [PubMed] [Google Scholar]
- Lim, P.O., Kim, H.J., and Nam, H.G. (2007. a). Leaf senescence. Annu. Rev. Plant Biol. 58 115–136. [DOI] [PubMed] [Google Scholar]
- Lim, P.O., Kim, Y., Breeze, E., Koo, J.C., Woo, H.R., Ryu, J.S., Park, D.H., Beynon, J., Tabrett, A., Buchanan-Wollaston, V., and Nam, H.G. (2007. b). Overexpression of a chromatin architecture-controlling AT-hook protein extends leaf longevity and increases the post-harvest storage life of plants. Plant J. 52 1140–1153. [DOI] [PubMed] [Google Scholar]
- López-Solanilla, E., Bronstein, P.A., Schneider, A.R., and Collmer, A. (2004). HopPtoN is a Pseudomonas syringae Hrp (type III secretion system) cysteine protease effector that suppresses pathogen-induced necrosis associated with both compatible and incompatible plant interactions. Mol. Microbiol. 54 353–365. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Belkhadir, Y., Alonso, J.M., Ecker, J.R., and Dangl, J.L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112 379–389. [DOI] [PubMed] [Google Scholar]
- Morris, K., MacKerness, S.A., Page, T., John, C.F., Murphy, A.M., Carr, J.P., and Buchanan-Wollaston, V. (2000). Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J. 23 677–685. [DOI] [PubMed] [Google Scholar]
- Mudgett, M.B., and Staskawicz, B.J. (1999). Characterization of the Pseudomonas syringae pv. tomato AvrRpt2 protein: Demonstration of secretion and processing during bacterial pathogenesis. Mol. Microbiol. 32 927–941. [DOI] [PubMed] [Google Scholar]
- Noël, L., Thieme, F., Nennstiel, D., and Bonas, U. (2002). Two novel type III-secreted proteins of Xanthomonas campestris pv. vesicatoria are encoded within the hrp pathogenicity island. J. Bacteriol. 184 1340–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, K., Debroy, S., Lee, Y.H., Pumplin, N., Jones, J., and He, S.Y. (2006). A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313 220–223. [DOI] [PubMed] [Google Scholar]
- O'Donnell, P.J., Schmelz, E., Block, A., Miersch, O., Wasternack, C., Jones, J.B., and Klee, H.J. (2003). Multiple hormones act sequentially to mediate a susceptible tomato pathogen defense response. Plant Physiol. 133 1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, M., Matsui, K., Hiratsu, K., Shinshi, H., and Ohme-Takagi, M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno, M., Arimoto, E., Ikenobu, Y., Nishihara, T., and Imagawa, M. (2001). Dual DNA-binding specificity of peroxisome-proliferator-activated receptor γ controlled by heterodimer formation with retinoid X receptor α. Biochem. J. 353 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, A.N., Ernst, H.A., Leggio, L.L., and Skriver, K. (2005). NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 10 79–87. [DOI] [PubMed] [Google Scholar]
- Penninckx, I.A., Eggermont, K., Terras, F.R., Thomma, B.P., De Samblanx, G.W., Buchala, A., Métraux, J.P., Manners, J.M., and Broekaert, W.F. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8 2309–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römer, P., Hahn, S., Jordan, T., Strauss, T., Bonas, U., and Lahaye, T. (2007). Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science 318 645–648. [DOI] [PubMed] [Google Scholar]
- Rosebrock, T.R., Zeng, L., Brady, J.J., Abramovitch, R.B., Xiao, F., and Martin, G.B. (2007). A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 448 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schmelz, E.A., Engelberth, J., Tumlinson, J.H., Block, A., and Alborn, H.T. (2004). The use of vapor phase extraction in metabolic profiling of phytohormones and other metabolites. Plant J. 39 790–808. [DOI] [PubMed] [Google Scholar]
- Schornack, S., Meyer, A., Römer, P., Jordan, T., and Lahaye, T. (2006). Gene-for-gene-mediated recognition of nuclear-targeted AvrBs3-like bacterial effector proteins. J. Plant Physiol. 163 256–272. [DOI] [PubMed] [Google Scholar]
- Song, C.P., Agarwal, M., Ohta, M., Guo, Y., Halfter, U., Wang, P., and Zhu, J.K. (2005). Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17 2384–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio, A., Yang, B., Zhu, T., and White, F.F. (2007). Two type III effector genes of Xanthomonas oryzae pv. oryzae control the induction of the host genes OsTFIIAγ1 and OsTFX1 during bacterial blight of rice. Proc. Natl. Acad. Sci. USA 104 10720–10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai, T., Dahlbeck, D., Stall, R.E., Peleman, J., and Staskawicz, B.J. (1999). High-resolution genetic and physical mapping of the region containing the Bs2 resistance gene of pepper. Theor. Appl. Genet. 99 1201–1206. [Google Scholar]
- Tiwari, S.B., Hagen, G., and Guilfoyle, T.J. (2004). Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troisfontaines, P., and Cornelis, G.R. (2005). Type III secretion: More systems than you think. Physiology (Bethesda). 20 326–339. [DOI] [PubMed] [Google Scholar]
- Turner, P., Barber, C., and Daniels, M. (1984). Behaviour of the transposons Tn5 and Tn7 in Xanthomonas campestris pv. campestris. Mol. Gen. Genet. 195 101–107. [Google Scholar]
- Weigel, R.R., Pfitzner, U.M., and Gatz, C. (2005). Interaction of NIMIN1 with NPR1 modulates PR gene expression in Arabidopsis. Plant Cell 17 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, B., Sugio, A., and White, F.F. (2006). Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. USA 103 10503–10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., Li, R., and Qi, M. (2000). In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J. 22 543–551. [DOI] [PubMed] [Google Scholar]
- Zhang, J., et al. (2007). A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1 175–185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.