Abstract
In prokaryotes, PurU (10-formyl tetrahydrofolate [THF] deformylase) metabolizes 10-formyl THF to formate and THF for purine and Gly biosyntheses. The Arabidopsis thaliana genome contains two putative purU genes, At4g17360 and At5g47435. Knocking out these genes simultaneously results in plants that are smaller and paler than the wild type. These double knockout (dKO) mutant plants show a 70-fold increase in Gly levels and accumulate elevated levels of 5- and 10-formyl THF. Embryo development in dKO mutants arrests between heart and early bent cotyledon stages. Mature seeds are shriveled, accumulate low amounts of lipids, and fail to germinate. However, the dKO mutant is only conditionally lethal and is rescued by growth under nonphotorespiratory conditions. In addition, culturing dKO siliques in the presence of sucrose restores normal embryo development and seed viability, suggesting that the seed and embryo development phenotypes are a result of a maternal effect. Our findings are consistent with the involvement of At4g17360 and At5g47435 proteins in photorespiration, which is to prevent excessive accumulation of 5-formyl THF, a potent inhibitor of the Gly decarboxylase/Ser hydroxymethyltransferase complex. Supporting this role, deletion of the At2g38660 gene that encodes the bifunctional 5,10-methylene THF dehydrogenase/5,10-methenyl THF cyclohydrolase that acts upstream of 5-formyl THF formation restored the wild-type phenotype in dKO plants.
INTRODUCTION
Folates act as donors of one-carbon (C1) units in all organisms. These C1 units exist at various oxidation states and are attached to nitrogen 5 and/or to nitrogen 10 of the pteridine ring of folates (Cossins and Chen, 1997). In plants, tetrahydrofolate (THF) is activated with formate in an ATP-dependent manner by 10-formyl THF synthetase (SYN; EC 6.3.4.4) to yield one of the most oxidized folates, 10-formyl THF. Subsequently, 10-formyl THF can be converted through other folate intermediates to the most reduced activated folate, 5-methyl THF (see Supplemental Figure 1A online; Chen et. al., 1997). Activated folates are essential in the biosynthesis of purines, formylmethionyl-tRNA, and thymidylate and in the metabolism of several amino acids, including Met, Gly, and Ser (Cossins and Chen, 1997).
In Escherichia coli, purine biosynthesis involves PurN, a folate-dependent transformylase (EC 2.1.2.2), which catalyzes the conversion of glycinamide-5′-phosphoribonucleotide (GAR) to N-formylglycinamide-5′-phosphoribonucleotide by transferring the formyl group from 10-formyl THF and releasing THF (see Supplemental Figure 1B online; Dev and Harvey, 1978; Smith and Daum, 1987; Inglese et al., 1990a). PurN homologs have also been characterized in Arabidopsis thaliana and soybean (Glycine max; Schnorr et al., 1994, 1996; Boldt and Zrenner, 2003). Under aerobic conditions, E. coli can also hydrolyze 10-formyl THF via one of the folate interconversion enzymes, 10-formyl THF deformylase (10-FDF, PurU; EC 3.5.1.10), which releases THF and formate (Nagy et al., 1993). Formate produced by PurU can be used by PurT, a folate-independent transformylase, to convert GAR to N-formylglycinamide-5′-phosphoribonucleotide (Nygaard and Smith, 1993; Marolewski et al., 1994). THF generated by PurN and PurU is used as a cofactor in Gly and 5,10-methylene THF biosyntheses from Ser by Ser hydroxymethyltransferase (SHMT; EC 2.1.2.1). In this reaction, Ser serves as an alternate donor of C1 units as its carbon 3 is incorporated into 5,10-methylene THF in bacteria, animals, and probably plants (Cossins, 2000; Engel et al., 2007).
In photosynthetic tissues, the reaction catalyzed by SHMT is driven in the opposite direction by the buildup of Gly. Thus, photorespiratory Gly and 5,10-methylene THF are converted to Ser and THF in plant mitochondria. THF is recycled back to Gly decarboxylase (GDC; EC 1.4.4.2, 2.1.2.10 and 1.8.1.4), which is involved in 5,10-methylene-THF formation from Gly and THF (see Supplemental Figure 1C online; Ogren, 1984; Douce and Neuburger, 1999). In bacteria and plants, 5,10-methylene-THF can be reversibly oxidized to 10-formyl THF by the bifunctional 5,10-methylene-THF dehydrogenase/5,10-methenyl-THF cyclohydrolase (DHC; EC 1.5.1.5/3.5.4.9) (Dev and Harvey, 1978; D'Ari and Rabinowitz, 1991; Chen et al., 1997). The gene encoding DHC has been cloned from pea (Pisum sativum) and its sequence confirmed the bifunctional character of this protein (Chen et al., 1999).
Two putative PurU homologs (At4g17360 and At5g47435) and four loci encoding DHC (At2g38660, At3g12290, At4g00600, and At4g00620) were identified in the Arabidopsis genome (Hanson et al., 2000). Based on available online software described below, both Arabidopsis PurU homologs and one DHC (At2g38660) were predicted to be mitochondrial proteins. We have identified T-DNA insertion mutants in these three genes and generated double and triple knockout mutant plants. Here, we report on the characterization of these mutants, showing that At4g17360 and At5g47435 proteins are crucial for photorespiration and that all three proteins are components of the photorespiratory THF cycle.
RESULTS
Identification and Characterization of Putative 10-FDF Genes
A BLAST search analysis using the E. coli PurU sequence (Nagy et al., 1993) against The Arabidopsis Information Resource database revealed the presence of two putative PurU genes, At4g17360 and At5g47435, encoding proteins with 30 and 31% amino acid sequence identity to E. coli PurU. The E. coli PurU protein contains 10 out of 11 conserved amino acid residues present in the predicted catalytic and folate binding domains of the transformylase E. coli PurN (Almassy et al., 1992; Nagy et al., 1995). In At4g17360 and At5g47435 corresponding proteins, nine residues out of the 11 are preserved in these conserved domains, including Asn-234 and -229, His-237 and -231, and Asp-272 and -267 (Figure 1A). These amino acid residues correspond to Asn-106, His-108, and Asp-144 of the E. coli PurN and are crucial for folate binding and catalysis (Inglese et al., 1990b; Almassy et al., 1992). The predicted GAR binding site in PurN involves eight residues (Almassy et al., 1992), of which only Ile-107 is preserved in E. coli PurU and both Arabidopsis proteins, indicating the absence of this site. This observation is consistent with PurU being a deformylase and not a transformylase. Both E. coli PurU and Arabidopsis proteins also contain an ACT domain, an amino acid binding domain that is present in many enzymes involved in amino acid metabolism and functions as a means of allosteric regulation (Chipman and Shaanan, 2001). While some residues are conserved between E. coli and Arabidopsis deformylases, there also are some differences. In E. coli PurU, 14 of 22 consensus ACT residues are conserved, while in Arabidopsis, the putative deformylases contain 16 conserved consensus ACT residues (Figure 1B).
Figure 1.
Sequence Similarities between E. coli and Putative Arabidopsis Deformylases.
(A) Pairwise alignments of PurN and PurU from E. coli and At4g17360 and At5g47435. Amino acid residues important for enzyme activity in PurN are highlighted in black. PurN Asn-106, His-108, and Asp-144 are crucial for catalysis (*), while Ser-110 and His-137 are thought to be less important (*#) (Almassy et al., 1992). The folate binding site includes Met-89, Ileu-91, Leu-118, and Leu-143 ($).
(B) The ACT domains of E. coli and Arabidopsis deformylases. The top row shows the consensus sequence of the ACT domain (Chipman and Shaanan, 2001). Small letters denote the following amino acid groups: b, big (FILMVWYKREQ); c, charged (DEHKR); h, hydrophobic (ACFILMVWY); l, branched (ILV); p, polar (DEHKNQRST); s, small (ACSTDNVGP). Conserved residues are highlighted in black.
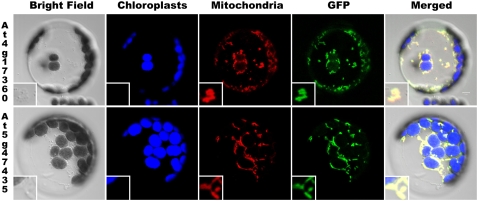
Based on the results of several computer-based prediction programs compiled in the Arabidopsis Mitochondrial Protein Database (http://www.ampdb.bcs.uwa.edu.au/) (Heazlewood and Millar, 2005), both At4g17360 and At5g47435 gene products contain a putative N-terminal mitochondrial targeting sequence. However, no proteomics data for the two proteins are available in the Subcellular Proteomics Database to support this prediction (Heazlewood et al., 2007). Therefore, we have confirmed the predicted mitochondrial localization of At4g17360 and At5g47435 proteins in Arabidopsis protoplasts experimentally using green fluorescent protein (GFP) fusion constructs (Figure 2). Merging GFP and MitoTracker fluorescence images has demonstrated that both proteins colocalize with mitochondria.
Figure 2.
Localization of At4g17360 and At5g47435 Proteins to Arabidopsis Mitochondria.
GFP fused with 5′-untranslated region (UTR) and the 5′-region of the coding sequence of each gene was transiently expressed in Arabidopsis leaf protoplasts and colocalized to mitochondria stained with MitoTracker. Bright-field, chloroplast autofluorescence, MitoTracker Red, GFP, and all four merged images are presented from left to right.
At4g17360 and At5g47435 Genes Are Expressed in Various Organs
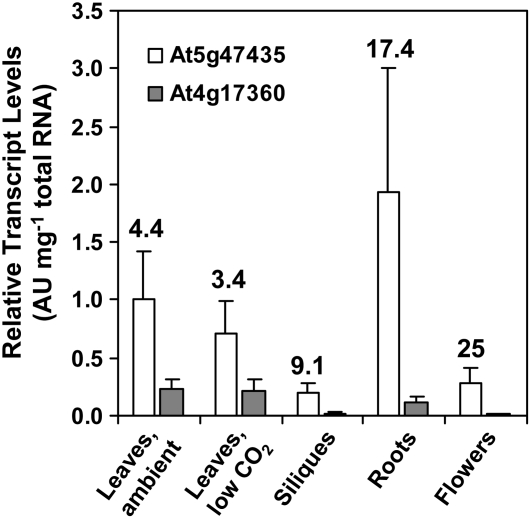
To assess sites of At4g17360 and At5g47435 gene expression, we performed TaqMan real-time quantitative RT-PCR with total RNA isolated from rosettes, siliques, flowers, and roots of wild-type Arabidopsis plants (Figure 3). Because of possible involvement of At4g17360 and At5g47435 gene products in photorespiration, a set of plants was also subjected to low CO2 levels. No statistically significant differences in At4g17360 and At5g47435 transcript levels were found between plants grown in ambient air and low CO2 conditions. Our RT-PCR data presented in Figure 3 are consistent with the publicly available microarray results in the Genevestigator (Zimmermann et al., 2004) and Weigel World (Schmid et al., 2005) databases. Both genes are expressed in all organs tested and show similar expression profiles. At5g47435 is expressed at least at 3.5-fold higher levels than At4g17360, depending on the organ. The highest expression of both genes is found in all types of leaves (rosette leaves of various age, cauline leaves, and cotyledons), followed by roots, seeds, and flowers (Zimmermann et al., 2004; Schmid et al., 2005).
Figure 3.
Expression Profiles of At4g17360 and At5g47435 in Various Tissues.
mRNAs were reverse transcribed using oligo(dT16) and gene-specific primers, and the resulting cDNAs were used as a template in TaqMan real-time PCR. Results are presented as an average of normalized relative transcript levels ± sd of three to four biological replicates. Numbers represent the ratios of relative At5g47435 to At4g17360 transcript levels.
Redundancy of Arabidopsis At4g17360 and At5g47435 Genes
A reverse genetics approach was taken to study the roles of At4g17360 and At5g47435 proteins in Arabidopsis. T-DNA insertion mutants (lines Garlic_169F06 and Salk_062946) were identified in the Torrey Mesa Research Institute and the Salk Institute mutant seed collection databases, respectively. Both lines have a single T-DNA insertion as determined by DNA gel blot analysis (see Supplemental Figure 3 online for Garlic_169F06). As confirmed by sequencing the flanking regions of the T-DNA, the Salk_062946 line has a T-DNA insertion in the fifth intron of the At4g17360 gene, while Garlic_169F06 in the first exon of the At5g47435 gene. RT-PCR showed no detectable At4g17360 or At5g47435 transcript in the corresponding single homozygous 10-FDF insertion mutants, and no compensation for the lack of expression of either gene was observed (data not shown). The single homozygous 10-FDF mutant plants were indistinguishable from the wild type in appearance. To determine whether this was due to At4g17360 and At5g47435 proteins being functionally redundant, we generated double homozygous 10-FDF mutant plants by crossing Salk_062946 and Garlic_169F06 mutant plants. For all subsequent experiments, F2 or F3 generations of the double homozygous mutant plants were used. These plants, subsequently referred to as double knockout (dKO) plants, showed a range of phenotypes described below.
Embryos of dKO Mutant Seeds Show Delayed Development and an Albino Phenotype
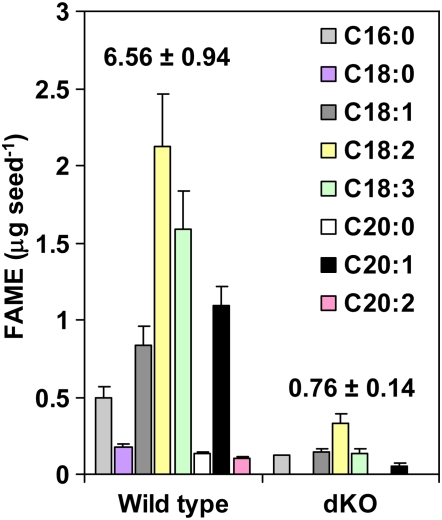
Compared with their single-gene mutant parents, double homozygous 10-FDF mutant (dKO) plants were smaller and paler than wild-type plants (Figure 4A). Flowering of the dKO mutant plants was delayed by ∼2 weeks and they produced shriveled nonviable seeds carrying colorless embryos (Figures 4B to 4D; see Supplemental Figure 4 online). These mutant embryos accumulated low amounts of oil, not exceeding 20% of the wild-type levels (Figure 5). Fatty acids present in wild-type seeds at low levels (stearic [C18:0], eicosanoic [C20:0], and eicosadienoic [C20:2]) were not detected in seeds of the dKO mutant. Only palmitic (C16:0), oleic (C18:1), linoleic (C18:2), linolenic (C18:3), and eicosenoic (C20:1) acids were detected in dKO seeds.
Figure 4.
Visible Phenotypes of dKO Leaves and Seeds.
(A) Three-week-old wild-type and dKO plants.
(B) Thirteen-day-old seeds.
(C) Mature dry seeds.
(D) Dissected 13-d-old embryos.
Figure 5.
Seed Lipid Levels and Composition in Wild-Type and dKO Seeds Grown at Ambient CO2 Conditions.
Fatty acid methyl esters (FAMEs) were analyzed by a gas chromatograph equipped with a flame ionization detector on seeds obtained from four different plants for each genotype. Numbers above the bars represent the total seed FAME levels presented as the mean ± sd of four biological replicates.
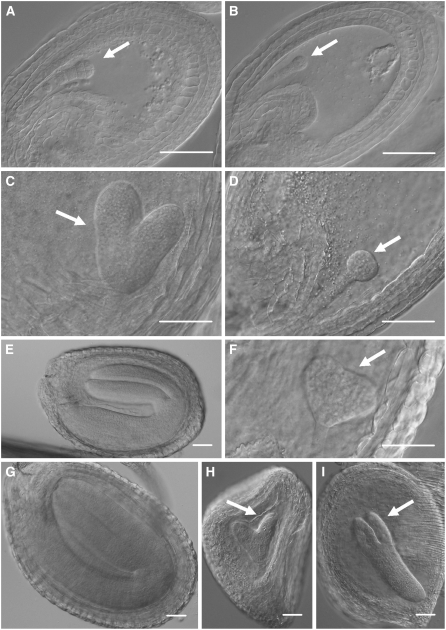
Further analysis of developing seeds revealed that although the initial seed size was similar between wild-type and dKO mutant plants, the development of dKO embryos, but not homozygous single mutant embryos (see Supplemental Figure 2 online), was severely delayed compared with wild-type embryos (Figure 6). At 2 d after flowering (DAF), both wild-type and dKO embryos were at the early globular stage (Figures 6A and 6B). While wild-type embryos were already at the heart stage at 5 DAF, dKO embryos were only at the late globular stage (Figures 6C and 6D). At 9 DAF, wild-type embryos were at the bent cotyledon developmental stage, whereas dKO embryos were still at the triangular stage (Figures 6E and 6F). At 13 DAF, the wild type was at the final developmental stage, but most dKO embryos were at the heart stage, though some were approaching torpedo stage (Figures 6G to 6I). At this stage, a majority of the dKO seeds, but not the wild type, began to brown and dry out, resulting in a shriveled appearance and nonviable mature seeds. In rare instances, dKO embryos approached the early bent cotyledon stage before browning and drying and remained white (Figure 4D), whereas wild-type embryos at this stage are green, indicating that the dKO albino phenotype cannot be explained by delays in embryo development alone.
Figure 6.
Nomarski Images Showing Different Developmental Stages of Wild-Type and dKO Embryo Development.
(A), (C), (E), and (G) Wild-type seeds.
(B), (D), (F), (H), and (I) dKO seeds.
(A) and (B) Two DAF.
(C) and (D) Five DAF.
(E) and (F) Eight DAF.
(G) to (I) Thirteen DAF.
White arrows point to the embryos at early developmental stages. Bars = 50 μm.
Leaves of dKO Plants Accumulate Photorespiratory Gly and Are Rescued at High CO2 Conditions
The seed and embryo phenotypes of the dKO mutant were only observed when the mother plant was homozygous dKO. The presence of a single copy of either At4g17360 or At5g47435 (segregation lines from crosses between the two homozygous parents) was sufficient to prevent these phenotypes. These observations indicate the occurrence of a maternal effect during seed development whereby the healthy plant provides the seeds with unknown factors that are absent, or present at inadequate levels, in the mutant plants. We therefore used metabolite profiling to probe changes in the relative levels of major metabolites in leaves. The pale phenotype of dKO leaves indicated that primary metabolism and/or photosynthesis was likely to be affected. Comparing metabolite profiles of the wild type and dKO grown in ambient air showed substantial differences in amino acid, sugar, and carboxylic acid levels (Tables 1 and 2).
Table 1.
Amino Acid Levels in 4-Week-Old Plants Grown in Ambient Air or High CO2 Conditions
| AA | Wild Type | dKO Mutant | shm1-1 Mutant |
|---|---|---|---|
| Gly | 0.37 ± 0.04 | 26.2 ± 6.1** | 38.5 ± 2.8** |
| Gly↑ | 0.32 ± 0.07 | 0.46 ± 0.09* | 0.44 ± 0.17* |
| Ser | 2.06 ± 0.30 | 1.66 ± 0.38 | 5.60 ± 0.57** |
| Ser↑ | 1.97 ± 0.29 | 2.96 ± 0.68 | 1.47 ± 0.47 |
| Asp | 2.12 ± 0.14 | 0.61 ± 0.26** | 0.43 ± 0.08** |
| Glu | 2.83 ± 0.10 | 1.60 ± 0.42** | 0.68 ± 0.59* |
| Asn | 0.43 ± 0.05 | 0.28 ± 0.06* | 3.98 ± 0.89* |
| Gln | 5.94 ± 0.75 | 2.31 ± 0.41** | 13.5 ± 1.1** |
| His | 0.047 ± 0.005 | 0.17 ± 0.07* | 1.21 ± 0.08** |
| Thr | 0.95 ± 0.13 | 0.77 ± 0.07 | 1.61 ± 0.07** |
| Arg | 0.064 ± 0.005 | 0.10 ± 0.02* | 1.77 ± 0.40* |
| Ala | 1.20 ± 0.04 | 0.39 ± 0.09** | 0.36 ± 0.01** |
| Pro | 0.40 ± 0.10 | 0.56 ± 0.18 | 0.98 ± 0.58 |
| Tyr | 0.014 ± 0.003 | 0.056 ± 0.012** | 0.20 ± 0.04* |
| Val | 0.10 ± 0.01 | 0.14 ± 0.01** | 0.43 ± 0.06** |
| Met | 0.026 ± 0.003 | 0.011 ± 0.001** | 0.058 ± 0.008* |
| Ileu | 0.028 ± 0.001 | 0.058 ± 0.005** | 0.21 ± 0.07* |
| Leu | 0.035 ± 0.002 | 0.083 ± 0.010** | 0.21 ± 0.05* |
| Lys | 0.025 ± 0.001 | 0.073 ± 0.008** | 0.18 ± 0.04* |
| Phe | 0.036 ± 0.005 | 0.12 ± 0.03** | 0.64 ± 0.21* |
Amino acid (AA) levels were analyzed in plant leaf extracts (four biological replicates) by HPLC and fluorescence detection. Gly and Ser levels in plants grown at high CO2 levels (↑) were analyzed by quantitative GC-MS analysis using deuteriated Gly and Ser as internal standards, while the levels of amino acids in plants grown in ambient CO2 conditions were analyzed by HPLC. The levels are expressed as an average ± sd in nmol mg−1 dry weight. Levels of amino acids in dKO and/or shm1-1 mutants that increased more than threefold or that were below 40% of the wild type are in bold. Asterisks represent statistically significant Student's t test values when mutants were compared with the wild type (P < 0.05* or P < 0.01**).
Table 2.
Relative Levels of Major Selected Metabolites in Leaves of dKO and shm1-1 Mutants
| Metabolite | dKO Mutant | shm1-1 Mutant |
|---|---|---|
| Glycerol | 0.67 ± 0.04* | 1.13 ± 0.24 |
| Succinate | 0.90 ± 0.15 | 0.67 ± 0.09 |
| Fumarate | 0.50 ± 0.08* | 0.39 ± 0.13** |
| Malate | 1.01 ± 0.16 | 5.40 ± 1.29** |
| Fructose 1 | 2.82 ± 0.45* | 0.60 ± 0.10** |
| Fructose 2 | 2.79 ± 0.51* | 0.76 ± 0.11 |
| Glucose 1 | 1.59 ± 0.33 | 0.50 ± 0.18** |
| Glucose 2 | 1.55 ± 0.35 | 0.40 ± 0.14** |
| Inositol | 0.38 ± 0.00** | 0.14 ± 0.02** |
| Sucrose | 0.21 ± 0.07* | 0.008 ± 0.002* |
| α-Tocopherol | 0.77 ± 0.36 | 6.97 ± 4.32* |
Semiquantitative GC-MS analyses of derivatized polar and nonpolar metabolites were performed on four biological replicates for each genotype. Glucose and fructose yield two trimethylsilyl derivatives, which are labeled as glucose 1 and 2, and fructose 1 and 2. Wild-type levels of each metabolite were set to 1.00, and levels of the corresponding metabolites in dKO and shm1-1 mutants are presented as values ± sd relative to the wild type. Levels of metabolites that showed statistically significant changes (P < 0.05* or P < 0.01**) compared with the wild type are indicated in bold. Semiquantitative means that only relative levels of metabolites were obtained, which is typical for MS-based metabolite profiling when nonlabeled nonauthentic internal standards are used. For example, metabolite 1 in one sample can be compared with metabolite 1 in another, but metabolite 1 levels cannot be compared with metabolite 2 even within the same sample due to differences in derivatization and ionization efficiencies of different metabolites.
The most dramatic difference between wild-type and dKO leaves was an ∼70-fold increase in Gly levels in dKO plants (Table 1). Accumulation of high leaf Gly levels as well as photobleaching of leaves has previously been observed in photorespiratory mutants deficient in GDC and SHMT activities (e.g., Arabidopsis shm1-1 and shm1-2 mutants) (Somerville and Ogren, 1981; Ogren, 1984; Somerville, 2001; Voll et al., 2006). To test whether the accumulation of high Gly levels in the dKO mutant (Table 1) was of photorespiratory origin, we analyzed Gly and Ser levels both in plants grown under elevated CO2 concentrations, which suppress photorespiration (Ogren, 1984) and in high CO2–grown plants transferred to ambient air for different periods of time.
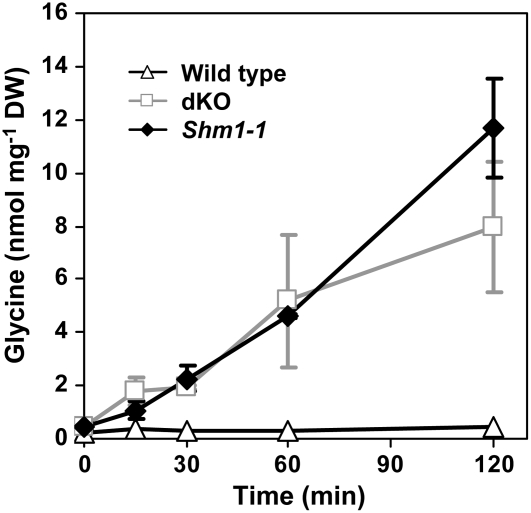
Leaf Gly levels in dKO and shm1-1 mutant plants grown in nonphotorespiratory conditions returned to near normal; Gly levels were elevated only by ∼60% relative to the corresponding wild type, while Ser levels were similar (Table 1). A rapid, linear (r2 > 0.95) accumulation of Gly was observed within a few minutes after exposing dKO and shm1-1 mutant to ambient air, while wild-type Gly levels remained low and stable during the course of the experiment (∼0.3 nmol mg−1 dry weight) (Figure 7). Leaves of dKO and shm1-1 plants accumulated ∼4.6 and 4.3 nmol of Gly per milligram of dry weight within the first hour, respectively.
Figure 7.
Time Course of Gly Accumulation in Arabidopsis Leaves.
Plants were pregrown at high CO2 conditions for 4 weeks and transferred to ambient air. Tissue was harvested at the indicated time points, and leaf extracts were analyzed by quantitative GC-MS. Gly levels are presented as an average ± sd of five biological replicates. DW, dry weight.
Levels of Amino Acids and Sugars Are Altered in dKO Mutant Plants
Levels of Ala, Asp, and Glu, amino acids that can donate their amino groups to photorespiratory glyoxylate instead of Ser (Liepman and Olsen, 2003), were depleted in both dKO and shm1-1 mutant leaves (Table 1). Levels of Ala and Asp in the leaves of dKO mutant decreased to ∼30%, while those of Gln, Glu, and Asn to 39, 56, and 66% of the wild-type levels, respectively. In the shm1-1 mutant leaves, levels of these amino acids varied between 20 and 30% of wild-type levels, though Gln and Asn levels were 2.3- and 9.3-fold higher than in the wild type. Arg and Lys levels were elevated by <3-fold in dKO leaves, whereas they increased 7- and 27-fold, respectively, in the shm1-1 mutant. This trend was also observed for the levels of some other amino acids. While the levels of His, Tyr, Val, Ile, Leu, and Phe increased between 2- and 4-fold in dKO, there was a 4- to 26-fold increase in the levels of these amino acids in the shm1-1 mutant leaves (Table 1).
Gas chromatography–mass spectrometry (GC-MS) profiling of metabolites in leaves of 4-week-old plants grown at ambient CO2 levels also revealed significant changes in sugar and carboxylic acid levels in dKO and shm1-1 mutants when compared with the wild type (Table 2). Sucrose levels in dKO and shm1-1 mutants grown in ambient air dropped to 20 and 0.8%, respectively, of wild-type levels. Fructose and glucose levels were slightly elevated in dKO but decreased in shm1-1 mutant leaves. Inositol levels decreased to 38 and 14% of wild-type levels in dKO and shm1-1 mutant leaves, respectively. In dKO leaves, fumarate levels were half those in the wild type. In the shm1-1 mutant, fumarate levels decreased to 40% of wild-type levels, while malate levels increased 5.4-fold relative to the wild type. For nonpolar metabolites, only α-tocopherol levels were significantly elevated (∼7-fold) in shm1-1 mutant leaves but not in dKO leaves (Table 2). The shm1-1 mutant used for the analysis showed severe yellowing of leaves due to photorespiration. An increase in α-tocopherol levels is known to occur during stress and senescence (Munne-Bosch and Alegre, 2002), and stress can result in protein degradation and the subsequent dramatic increase in the levels of most amino acids in shm1-1 mutant.
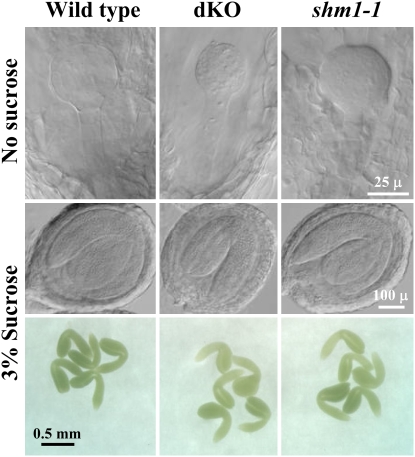
Sucrose Restores Normal Development of dKO and shm1-1 Mutant Embryos in Tissue Cultures
During embryo development, maternal source tissues provide nutrients for the sink tissues. In photorespiratory mutants, carbon fixation and recovery is inefficient (Somerville and Ogren, 1980, 1981), which could lead to low pools of carbon sources available for developing embryos. We hypothesized that the low availability of sucrose, the transport form of sugar in phloem, is causing the impaired embryo development and albino dKO phenotype. To test this hypothesis, excised and surface-sterilized siliques carrying embryos at the globular stage were allowed to grow in the presence or absence of sucrose on the solid media. We also used the Arabidopsis shm1-1 mutant as a photorespiratory control. After 7 d of culturing, none of the embryos including the wild type were able to develop in the absence of sucrose and were arrested at the late globular stage (Figure 8). None of these seeds were able to germinate. On the other hand, all siliques cultured in the presence of sucrose for 7 d carried many green seeds with embryos at the bent cotyledon stage (Figure 8), which were able to germinate upon reaching maturity. The germination efficiencies of wild-type, dKO, and shm1-1 mutant seeds that originated from these cultured siliques were 92, 93, and 94%, respectively. Therefore, supplementation of sucrose to the cultured siliques restored normal development of dKO and shm1-1 mutant embryos.
Figure 8.
Sucrose Supplementation to Excised Siliques Saves Embryo Development in Photorespiratory Mutants.
Excised surface-sterilized wild-type, dKO, and shm1-1 mutant siliques carrying globular embryos were cultured in the presence or absence of 3% sucrose on Murashige and Skoog medium for 7 d. Seeds were cleared in Hoyer's solution for Nomarski microscopy, or the embryos were dissected directly from the seeds.
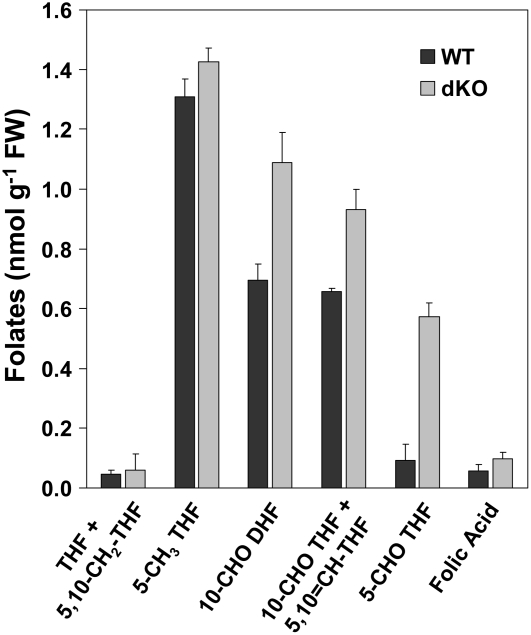
Leaf Folate Levels Are Elevated in the dKO Mutant
Because Arabidopsis PurU homologs are potentially involved in folate and formate metabolism, we analyzed their levels in dKO and wild-type leaves to investigate their function in vivo. While formate levels were similar in wild-type and dKO mutant plants (166 ± 1 and 173 ± 41 nmol g−1 fresh weight, respectively), there were significant differences in folate levels and composition (Figure 9). 5-Formyl THF, 10-formyl+5,10-methenyl-THF, and 10-formyl dihydrofolate/10-formyl THF levels were significantly (P ≤ 0.01) higher in dKO (6-fold, 30%, and 30%, respectively) than in wild-type leaves, while there was no change in THF+5,10-methylene-THF, 5-methyl THF, and folic acid levels. As a result, total folate levels nearly doubled in dKO mutant leaves relative to the wild type (4.18 ± 0.16 and 2.86 ± 0.13 nmol g−1 fresh weight, respectively). The procedures used for folate analyses do not distinguish between 10-formyl-THF and 5,10-methenyl-THF or between THF and 5,10-methylene-THF (Quinlivan et al., 2006). In addition, 10-formyl dihydrofolate is an oxidation product of 10-formyl-THF and does not occur naturally in plants, but it can be used to estimate 10-formyl THF levels.
Figure 9.
Leaf Folate Levels and Composition in Wild-Type and dKO Mutant Plants.
Folates were extracted from 5-week-old wild-type and dKO plants grown in ambient air. The bars represent folate levels as an average ± sd of four biological replicates. FW, fresh weight.
Photorespiratory Phenotypes in the dKO Mutant Are a Result of the Inhibition of the GDC/SHMT Complex
5-Formyl THF, a competitive inhibitor of folate-dependent enzymes, is generated either from 5,10-methenyl THF by SHMT in the presence of Gly (Schirch and Ropp, 1967; Grimshaw et al., 1984; Stover and Schirch, 1990, 1991; Holmes and Appling, 2002) or spontaneously in mildly acidic environments (Baggott, 2000). This inhibitor is metabolized by 5-formyl THF cycloligase (5-FCL) in Arabidopsis mitochondria (Roje et al., 2002). Moderately increased levels of 5-formyl THF result in mild photorespiratory phenotypes in the 5-FCL mutant as a consequence of inhibiting the GDC/SHMT complex in Arabidopsis (Goyer et al., 2005). In the case of the dKO mutant, it was not clear whether the elevated levels of 5-formyl THF were the sole cause for these phenotypes. Folate deformylases provide a way to regulate the THF/C1 activated THF ratio in microorganisms (Nagy et al., 1995), and THF availability likely limits the GDC activity in mitochondria (Rebeille et al., 1994). Deformylases, along with DHC, could provide a way to recycle THF trapped in the C1 form that is continuously produced from Gly and THF by GDC.
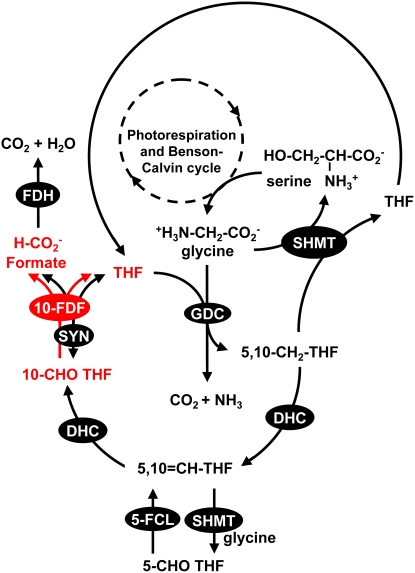
To distinguish between the two proposed roles of At4g17360 and At5g47435 proteins in photorespiration, we generated triple homozygous mutant plants by crossing the dKO mutant with a mutant affected in the DHC. Based on the proposed pathway delineated in Figure 10 (the THF cycle; see Discussion for more details), blocking DHC disrupts both THF recycling and the formation of the inhibitor 5-formyl THF, while blocking 10-FDF disrupts only the THF cycle downstream of 5-formyl THF biosynthesis. If THF recycling is important, the homozygous DHC mutant should also show some photorespiratory problems. Otherwise, all photorespiratory phenotypes caused by the accumulation of 5-formyl THF should be rescued in the dKO mutant in the DHC mutant background.
Figure 10.
The Proposed Operation of a Mitochondrial THF Cycle.
The reactions of photorespiration that result in the generation of Gly are omitted for clarity. Multiple reaction steps are represented by dashed arrows. The THF cycle includes folate interconversion enzymes DHC and 10-FDF. FDH, formate dehydrogenase.
Only dKO and shm1-1 mutants show visible phenotypes (see Supplemental Figure 4 online). By contrast, both homozygous dhc and triple homozygous dhc/dKO (triple) mutants showed no photorespiratory phenotypes. These mutants were indistinguishable from the wild type based on inspection as well as metabolite profiles (see Supplemental Figures 4 and 5 online). Both dhc and triple mutants were viable and produced normal green embryos (see Supplemental Figure 4B online). We used GC-MS metabolite profiling and Principal Component Analysis (PCA) on correlations of >50 major metabolites to assess differences in central carbon and nitrogen metabolism between wild-type and mutant plants. PCA revealed that both mutants clustered with the wild type, indicating that their metabolite profiles were similar to the wild type. Metabolite profiles of dKO and shm-1-1 mutants did differ noticeably from the wild type, which was reflected in PCA by separation of these mutants based on Principal Component 1 (see Supplemental Figure 5 online). These results suggest that the photorespiratory phenotypes in the dKO mutant are caused entirely by 5-formyl THF accumulation and subsequent inhibition of the GDC/SHMT complex.
DISCUSSION
The Phenotypes of the Arabidopsis dKO Mutant
Embryogenesis is essential for plant propagation and involves a coordinated interaction of developmental and metabolic processes. Developing Arabidopsis embryos are metabolically highly active, and mutations in central carbon metabolism or its regulation and signaling affect embryo development (Focks and Benning, 1998; Eastmond et al., 2002; Baud et al., 2003; Cernac and Benning, 2004; Baud and Graham, 2006). We have used a reverse genetic approach to study the roles of At4g17360 and At5g47435 proteins, putative 10-FDFs, in Arabidopsis. Pale Arabidopsis dKO mutant plants produced nonviable seeds carrying albino embryos. Embryo development was delayed and arrested typically at the heart or torpedo stages, resulting in the production of shriveled seeds accumulating low lipid levels. All these phenotypes were overcome by suppressing photorespiration under high CO2.
The embryo-defective phenotype was clearly a result of a maternal effect because homozygous dKO seeds that developed on segregating plants in ambient air were green and viable. Therefore, the presence of At4g17360 and At5g47435 in seeds and embryos does not seem to be required due to nearly absent photorespiratory fluxes in seeds. This is because CO2 levels in seeds are very high, most likely due to the low permeability of seed coat and silique walls to CO2. Seed CO2 levels in a related species (Brassica napus) were found to be 10-fold higher than the Km for CO2 of ribulose-1,5-bisphosphate carboxylase/oxygenase (Goffman et al., 2004), the enzyme that initiates photorespiration via the competition between O2 and CO2 for ribulose-1,5-bisphosphate, favoring carboxylation over oxygenation. This scenario differs from leaves, where CO2 levels are below Km, and the oxygenation reaction takes place quite efficiently, leading to high photorespiratory fluxes.
Developing embryos require maternally provided nutrients and energy for their growth. Sugars and amino acids represent carbon and nitrogen sources transported through phloem to the embryos (Hirner et al., 1998; Schwender and Ohlrogge, 2002). The occurrence of a maternal effect and low levels of sucrose in leaves of dKO and shm1-1 photorespiratory mutants indicated that these mutants did not receive an adequate supply of sucrose for embryo development. In photorespiratory mutants, slow recycling of photorespiratory carbon through the Benson-Calvin cycle results in a decrease of carbon availability for sugar synthesis (Somerville and Ogren, 1980, 1981). While the levels of glucose were similar, sucrose levels decreased to 20% of the wild type in dKO mutant leaves and were below 1% in the shm1-1 mutant, suggesting that sucrose metabolism was affected in these photorespiratory mutants. Therefore, a defect in photorespiration may affect sucrose availability for these developing embryos and other sink tissues.
We tested this hypothesis by culturing wild-type and photorespiratory mutant siliques with or without sucrose. Regardless of genotype, none of the embryos were able to develop in the absence of sucrose, indicating that the siliques themselves were not able to act as a carbon source for developing embryos. On the other hand, sucrose supplementation restored the development and viability of dKO and shm1-1 mutant embryos. These results collectively indicate that the seed and embryo-defective phenotypes observed in dKO and shm1-1 mutants are caused by inefficient sucrose supply by mother plants due to blocked photorespiration in leaves. Due to low availability of carbon for sucrose synthesis in photorespiratory mutants (Somerville and Ogren, 1980, 1981), this maternal affect may be a general phenomenon in mutants affected in photorespiration.
Functions of At4g17360 and At5g47435 Proteins in Photorespiration
In E. coli, PurU encodes a 10-FDF involved in purine biosynthesis (in conjunction with PurT), while providing THF for Gly biosynthesis (Nagy et al., 1993, 1995; Nygaard and Smith, 1993). In Arabidopsis, knocking out both At4g17360 and At5g47435 genes resulted in the presence of several visible and metabolic phenotypes typical for photorespiratory mutants, including reduced plant size, yellowish leaves, accumulation of Gly, and depletion of sucrose and amino donors. Only photorespiratory phenotypes, which were completely eliminated in high CO2 conditions, were observed in the dKO mutant grown in ambient air. Therefore, in Arabidopsis leaves, At4g17360 and At5g47435 are required to maintain photorespiratory fluxes but probably are not needed for purine biosynthesis. This view is also supported by the absence of an apparent PurT homolog in Arabidopsis that could attach formate released by PurU to GAR (Boldt and Zrenner, 2003). However, the possibility of At4g17360 and At5g47435 proteins complementing for PurN has not been investigated.
Arabidopsis At4g17360 and At5g47435 proteins show 50% overall similarity to E. coli PurU, and both contain conserved amino acids representing the predicted folate binding and catalytic sites that are present in the bacterial PurN and PurU proteins (Almassy et al., 1992; Nagy et al., 1995). However, all attempts to demonstrate the deformylase activity of At4g17360 and At5g47435 proteins expressed in E. coli and yeast and assays with wild-type and dKO mutant mitochondria as well as the complementation of the E. coli purU purN double mutant have failed (data not shown). This can be explained by the inability to obtain soluble At4g17360 and At5g47435 proteins for the enzyme assays, the recalcitrant nature of deformylases (Nagy et al., 1993), and by the differences in the regulation of deformylase activities between E. coli and Arabidopsis. Requirement for differential regulation is evident as bacterial SHMT-dependent Gly biosynthesis consumes THF, while photorespiratory SHMT generates THF as the reaction proceeds in the opposite direction compared with E. coli. Both E. coli PurU and Arabidopsis deformylases contain similar, but not identical, ACT domains at the N terminus, through which the regulation by amino acids occurs (Chipman and Shaanan, 2001). In fact, E. coli PurU activity is promoted by Met and inhibited by Gly (Nagy et al., 1993). Differences in the ACT domain sequences between these deformylases indicate that the allosteric regulation of these enzymes may be distinct between the two organisms, likely reflecting functional differences.
Arabidopsis At4g17360 and At5g47435 proteins influence folate levels and composition. In photorespiration, GDC and SHMT are the only known enzymes using folates as cofactors. A proposed mechanism for the involvement of At4g17360 and At5g47435 as putative folate deformylases in photorespiration is shown in Figure 10. A similar, but not a cyclic, route involving folate interconversion enzymes has previously been postulated to produce formate in plants (Hourton-Cabassa et al., 1998; Hanson et al., 2000; Goyer et al., 2005). (Photorespiration involves a series of cycles to metabolize toxic 2-phosphoglycolate and to prevent both the depletion of intermediates and cofactors and the accumulation of products due to large fluxes.) In the THF cycle proposed here, 5,10-methylene THF produced by GDC is not always used by SHMT to make Ser but can also be converted to 10-formyl THF by DHC (Cossins and Chen, 1997; Hanson et al., 2000). Subsequently, 10-FDF metabolizes 10-formyl THF to formate and THF, which can be reused by GDC. Formate either is oxidized by mitochondrial formate dehydrogenase to CO2 or can reenter one-carbon metabolism in other compartments via 10-formyl THF SYN (Hanson and Roje, 2001). Although SYN is encoded by a single gene in Arabidopsis (Hanson et al., 2000), in pea, its activity has been demonstrated in cytosol, mitochondria, and chloroplasts (Chen et al., 1997). This ATP-dependent enzyme condenses formate and THF to 10-formyl THF, but it is potentially reversible with an equilibrium constant Keq being between 20 and 61 (Himes and Rabinowitz, 1962). Under photorespiratory conditions (ambient CO2 levels) the concentrations of THF and 10-formyl THF in plant mitochondria likely favor the formation of formate and THF. Thus mitochondrial SYN could perform a function redundant to 10-FDF. However, the dKO mutant shows various photorespiratory phenotypes, which are associated with specific functions of At4g17360 and At5g47435 and are clearly not complemented by the possible presence of SYN in Arabidopsis mitochondria.
Accumulation of 5-Formyl THF Causes Photorespiratory Phenotypes in dKO
Based on the model in Figure 10, the THF cycle is disrupted in dKO mutant leaves downstream of the production of 5-formyl THF, which is an inhibitor of the mitochondrial GDC/SHMT complex in Arabidopsis. The degree of the severity of phenotypes in two folate interconversion mutants grown in ambient air coincides with the leaf levels of 5-formyl THF but not 10-formyl THF, the presumed substrate of 10-FDF. In the leaves of the 5-FCL mutant, 5-formyl THF levels were twofold higher than in the wild type, leading to mild photorespiratory phenotypes: a 20-fold increase in Gly levels and moderate growth retardation (Goyer et al., 2005). The phenotypes of the dKO mutant (pale infertile plants with up to a 70-fold increase in Gly levels) were much more severe than in the 5-FCL mutant, and there was a sixfold increase in 5-formyl THF levels in dKO leaves relative to the wild type.
Considering that 5-FCL is the only enzyme known to metabolize 5-formyl THF, it was seemingly incongruous to observe that the phenotypes of the dKO mutant were more severe than those of the 5-FCL mutant. The position of these two enzymes and other enzymes in the cycle provides a plausible explanation. The reaction catalyzed by 5-FCL is a branch in the proposed THF cycle, while 10-FDF is a part of the cycle ultimately leading to THF and formate/CO2 (Figure 10). Unlike 10-FDF, blocking 5-FCL does not result in the complete disruption of the cycle, and the C1 units that were not incorporated into 5-formyl THF by SHMT can still flow through the cycle and be released as CO2 in the 5-FCL mutant. In the dKO mutant, however, the accumulation of 5-formyl THF is more extreme because the cycle is interrupted. According to the model, 5-FCL activity alone cannot prevent the buildup of 5-formyl THF in dKO leaves as the 5,10-methenyl THF produced by 5-FCL can only be converted either to 10-formyl THF (a dead end because 10-FDF is disrupted) or back to 5-formyl THF by SHMT.
The positive trend between the levels of 5-formyl THF and the severity of the mutant phenotypes suggested that this inhibitor of the GDC/SHMT complex may be a direct cause of the photorespiratory phenotypes. Alternatively, the mitochondrial THF cycle could provide a way to regenerate THF from 5,10-methylene THF that is continuously produced from Gly and THF by GDC. The presence of photorespiratory phenotypes in the DHC mutant would indicate that THF recycling is important. No such phenotypes were observed in this mutant. In addition, all the phenotypic effects observed in dKO mutant plants were eliminated in the DHC mutant background. These results with the triple mutant suggest that all the phenotypes in the dKO mutant are associated with a large accumulation of 5-formyl THF and the subsequent inhibition of the GDC/SHMT complex, rather than with THF recycling. These observations point to a specific function of At4g17360 and At5g47435 proteins in the prevention of 5-formyl THF accumulation and provide evidence of a connection between the function of these proteins and other components of the THF cycle and the GDC/SHMT complex.
Knocking out mitochondrial DHC had no effect on plant performance. 10-Formyl THF produced by DHC is used in the biosynthesis of purines and formylmethionyl-tRNA, while 5,10-methenyl THF and 5-methyl THF donate methyl groups to thymidylate and Met, respectively (Coffin and Cossins, 1986; Neuburger et al., 1996; Clandinin and Cossins, 1974; Roje et al., 1999; Cossins, 2000; Boldt and Zrener; 2003). In the dhc mutant mitochondria, 10-formyl THF and 5-methyl THF can be synthesized in other ways (Figure 10; see Supplemental Figure 1A online). However, thymidylate synthesis requires 5,10-methenyl THF, the intermediate of the DHC reactions. Cytosolic pathways may provide either the folate and/or thymidylate that can be transported to mitochondria to fulfill the demands of DNA replication. Our results are consistent with the observations made with yeast mutants affected in the mitochondrial trifunctional C1–THF synthase (MSI1; which carries the SYN activity and DHC functions), which also does not seem to be essential for normal cell growth (Shannon and Rabinowitz, 1988). Overall, it appears that the redundant one-carbon metabolism pathways can compensate for each other and ensure normal operation of folate-dependent enzymes in different compartments.
METHODS
Constructs
Arabidopsis thaliana AI997177 and AI997164 cDNAs (Genome Systems) encoding two putative 10-FDFs (At4g17360 and At5g47435) were subcloned into the pGEMT-Easy vector (Promega). Clone AI997164 contained an additional “A,” which led to a frame shift. Therefore, the corresponding At5g47435 cDNA was amplified by RT-PCR before subcloning into pGEMT-Easy. The following primers (all primer sequences are in the 5′ → 3′ direction) were used for the amplification: CACCATGGTACGAAGAGTCTCCACCAC and GAATACGACAGTCCTTTGTGTCCCA for At4g17360 and CACCATGGTACGACGAATCACCGAGAGAG and GAATACAACAGTCTTGTTTGTACCA for At5g47435. For GFP fusions, expression of the full-length At4g17360 and At5g47435 cDNAs interfered with the stability of GFP fluorescence, so truncated versions of the two proteins containing the first 120 and 115 amino acids were fused with GFP. To ensure proper translation and targeting, the native 5′-UTRs (59 and 64 bp upstream of ATG for At4g17360 and At5g47435, respectively) were also included. These 5′-UTRs were amplified from genomic DNA with the following primers: XbaIfw1 (TGCTCTAGAATGTTTATAACGACGG) and rv1 (AAGCCGGGAATGAGAAGAGGT) for At4g17360 and XbaIfw2 (TGCTCTAGATTGGTCGGTCTCTCTCG) and rv2 (CTGACAATGGAAGACATGGACGC) for At5g47435. The N-terminal parts of the proteins were amplified from previously isolated full-length cDNAs with the primers BamHIrv1 (CGGGATCCTGAGCTCAAAGCACT) and fw1 (ACCTCTTCTCATTCCCGGCTT) for At4g17360 and BamHIrv2 (CGGGATCCTGAATTCAAGGCACCGTA) and fw2 (GCGTCCATGTCTTCCATTGTCAG) for At5g47435. The final PCR products were amplified from the two PCR products (5′-UTRs and N termini) using the flanking primers XbaIfw1 and BamHIrv1 for At4g17360, and XbaIfw2 and BamHIrv2 for At5g47435. These PCR products were ligated with the XbaI-BamHI–digested GFP reporter plasmid p35-SGFP(S65T), a generous gift of Andreas Nubenfuhr (University of Tennessee, Knoxville, TN).
Plant Material and Growth Conditions
T-DNA insertion mutants of Arabidopsis thaliana for each of the two genes encoding putative 10-FDFs were identified in the Syngenta (Garlic_169F06) and the Salk Institute (Salk_062946) mutant collection databases. The presence of T-DNA was confirmed by PCR using the following primers: At5g47435 gene-specific primers (GAATATGGACAGAACCACGGT and CTTCATCCACTTGGCTTCTC), T-DNA–specific primer LB3 (GCATCTGAATTTCATAACCAATC), At4g17360 gene-specific primers (TCACGTGAGAGACATGTCAGAGA and GAGGTTTCTCCAGAGGCATGG), and T-DNA–specific primer LBb1 (GCGTGGACCGCTTGCTGCAACT). dKO mutant plants were generated by crossing Salk_062946 and Garlic_169F06 homozygous mutants. Heterozygotes for both genes were screened by PCR and self-pollinated. Progeny were screened for homozygous mutants for both genes. Wild-type plants were obtained from segregating double heterozygous 10-FDF mutants.
The At2g38660 gene encodes DHC, which is predicted to be localized to mitochondria (TargetP; Emanuelsson et al., 2007). Two lines, Salk_142776 and Salk_143478, were obtained from the Salk Institute. The following primers were used for genotyping the Salk_142776 mutant line: GAACATAAGTTTGCGAGTCTCTTTG and TAGCTGAGTGAAAGCTTATGATAG. Salk_142776 has a T-DNA inserted in the 5′-UTR of the gene and showed low levels of wild-type At2g38660 transcript as demonstrated by RT-PCR using primers DHCF1, CTGTTTCCTTTGAGACTGAACAG, and DHCR1, TAATGTGCTCTGGATCCTTTGTAA (see Supplemental Figure 6A online). Primers DHCF (TCAGATTGGATTCAAGCTATGG) and DHCR (ACGGAATTCTTGTGCAACTTC) were used for genotyping the Salk_143478 line, in which the T-DNA insertion occurred in the second intron of At2g38660. This insertion resulted in a complete loss of the At2g38660 transcript (see Supplemental Figure 6A online). Therefore, Salk_143478 was used for subsequent experiments. The triple homozygous mutant was generated by crossing dKO and Salk_143478 homozygous mutant lines and PCR screening the F2 plants for a plant homozygous for all three genes. Progeny of a candidate triple homozygous mutant plant were all confirmed to be homozygous for all three T-DNA insertions (13 plants, an example of genotyping from different experiments using progeny of one of the 13 plants in addition to the wild type, dKO, and dhc mutant is shown in Supplemental Figures 6B and 6C online).
The SHMT mutant (shm1-1) originally isolated by Somerville and Ogren (1981) was kindly provided by Andreas Weber (Michigan State University, East Lansing, MI) (Voll et al., 2006) and was used as a photorespiratory control in some experiments. Due to its failure to grow in ambient CO2 levels, the shm1-1 mutant was pregrown at high CO2 and transferred to ambient air for at least 3 d before experiments were performed. All plants were grown in a controlled growth chamber environment at a 12-h photoperiod, 20/18°C light/dark cycle, photosynthetic photon flux density 80 ± 20 nmol m−2 s−1 in either ambient air (CO2 levels ∼320 ppm) or high CO2 (4100 ± 200 ppm) or low CO2 (130 ± 3 ppm). High CO2 conditions were obtained by mixing air with CO2, while low CO2 levels were generated by passing air through soda lime. CO2 levels were monitored with a Vernier CO2 sensor (Vernier Software and Technologies).
Subcellular Localization of At4g17360 and At5g47435 Gene Products
Subcellular localization of both proteins was demonstrated by colocalization analyses of the GFP fluorescence with MitoTracker Red CMXRos (Molecular Probes) in Arabidopsis protoplasts transiently expressing the GFP fusions of At4g17360 or At5g47435 native 5′-UTR and the corresponding N-terminal parts. Protoplasts from 3-week-old Arabidopsis leaves were prepared and transformed as described previously (Bauer et al., 2004). After transformation, protoplasts were incubated for 12 to 14 h under constant light. MitoTracker Red was used for specific staining of mitochondria according to the manufacturer's recommendations. Protoplasts were incubated with 500 nM MitoTracker for 15 min. Confocal imaging was performed using an inverted Zeiss LSM 210 laser scanning microscope equipped with a ×63 oil immersion objective (Carl Zeiss). GFP fluorescence was excited with the argon laser at 488 nm and detected between 505 and 530 nm. MitoTracker was excited with the green helium neon laser at 543 nm and detected between 560 and 615 nm, while far-red autofluorescence of chlorophyll was captured between 643 and 753 nm. All images were processed with LSM 5 Image Browser (Carl Zeiss), Paint Pro Shop 11 (Corel Corporation), and Adobe Photoshop 5.0 software simultaneously.
Real-Time Quantitative RT-PCR
Wild-type Arabidopsis plants were pregrown in ambient air and then either kept in ambient air or transferred to low CO2 conditions for 7 d. Three types of tissues were analyzed: 4-week-old rosettes and 9-d-old siliques from soil-grown plants and 4-week-old roots obtained from hydroponically grown plants (Gibeaut et al., 1997). Leaves of dKO mutant plants were used as a negative control (the threshold cycle [CT] numbers for At4g17360 and At5g47435 were 40, demonstrating the lack of the transcript for both genes and the specificity of the TaqMan gene expression assays). Each type of sample was represented by three to four biological replicates. Tissue was harvested 5 to 6 h after the beginning of the light cycle. Total RNA was extracted by the modified hot borate method (Wan and Wilkins, 1994). DNA was removed by treatment with RQ1 RNase-free DNase (Promega). Total RNA was quantified using a Ribo-green RNA quantitation kit (Promega). The quantitative TaqMan real-time PCR was performed in two steps. First, mRNA was reverse transcribed to cDNA using 10 μg of total RNA, oligo(dT16) primer, and the TaqMan reverse transcription reagents (Applied Biosystems). Second, cDNA corresponding to 2.25 μg of RNA was used in TaqMan real-time PCR, 40 cycles (ABI Prism 7900HT sequence detection system; Applied Biosystems). TaqMan gene expression assays for At4g17360, At5g47435, and control At5g60390 (elongation factor EF-1α, used for data normalization) were used according to the manufacturer's recommendations (Applied Biosystems). The CT numbers for each sample were obtained from the logarithmic phase of the amplification plots. To quantify steady state transcript levels in various organs, standard curves were constructed in triplicate for At4g17360 and At5g47435 genes using serial dilutions of purified individual PCR products corresponding to full-length cDNAs (from 10−5 to 1 pg).
Microscopy
Flowers were tagged at anthesis with colored cotton thread loops, and siliques of different developmental stages were harvested at specified times (2, 5, 9, and 13 DAF). Seeds were cleared in Hoyer's solution and mounted for microscopy as described (Liu and Meinke, 1998). Nomarski images of developing wild-type and dKO mutant seeds were obtained on an Olympus BH2 microscope equipped with a Nikon Coolpix 4500 camera with an Optem Coolpix adapter.
Tissue Culture
Wild-type, dKO, and shm1-1 mutant siliques with embryos at the globular stage (fifth silique from the top) were collected and surface-sterilized in 15% (v/v) bleach for 10 min followed by rinsing in sterile water. Siliques were cultured vertically in solid Murashige and Skoog medium with or without 3% (w/v) sucrose as a carbon source. Seven days later, some siliques were used to dissect embryos, while others were cleared in Hoyer's solution, and Nomarski microscopy of embryos was performed as described above. The remaining siliques were allowed to mature for 10 additional days and the seeds were dried, stratified, and tested for germination (n > 70).
Seed Lipid Analysis
Ten mature seeds per line were crushed by a brief vortexing with a 4-mm glass bead, and lipids were hydrolyzed and derivatized with 0.5 mL of 1 n methanolic HCl for 3 h at 80°C. Heptadecanoic acid (C17:0) was used as a standard to quantify the FAME content extracted with 0.5 mL hexane. One microliter of the hexane phase was injected directly into an Agilent 6890N series gas chromatograph equipped with a 30-m DB-23 column (0.25 mm × 0.25-μm film thickness) and interfaced with a flame ionization detector (Agilent Technologies).
Metabolite Profiling
Plants were grown at ambient or high CO2 conditions. Extractions and semiquantitative GC-MS analyses of polar and nonpolar metabolites were performed on four to five biological replicates as described previously (Duran et al., 2003; Goyer et al., 2005). Briefly, leaf samples were rapidly frozen in liquid nitrogen, lyophilized, weighed, disrupted with glass beads, and extracted with chloroform:water 1:1 (v/v). The chloroform phase containing nonpolar metabolites was dried under a stream of nitrogen, and FAMEs were prepared by derivatization in 1 n methanolic HCl at 50°C for 5 h. Samples were dried under a stream of nitrogen gas, and trimethylsilyl derivatives of glycerol and carbohydrates hydrolyzed from triacylglycerols and sugar lipids were prepared. For the analysis of polar metabolites, the aqueous phase was dried and trimethylsilyl derivatives were prepared. One microliter of derivatized samples was separated on an Agilent 6890 series gas chromatograph equipped with a 60-m DB-5 MS column (0.25 mm × 0.25 μm) and analyzed in scan and positive electron ionization mode using an Agilent 5973 series quadrupole mass spectrometer (Agilent Technologies). Heptadecanoic acid, phenanthrene, and ribitol were used as internal standards.
Amino Acid Analyses
Two types of amino acid analyses were performed. First, comprehensive HPLC amino acid analysis was performed on samples from four biological replicates of the wild type, dKO, and shm1-1 mutants (Goyer et al., 2005). γ-Aminobutyric acid was used as an internal standard, and amino acids were derivatized with AccQ-Fluor reagent (6-aminoquinolyl-N-hydroxysuccinimidylcarbamate; Waters). Second, a short quantitative GC-MS method was developed to follow changes in Ser and Gly levels in wild-type, purU dKO, and shm1-1 mutant leaves. Samples (five biological replicates) were spiked with [15N, 2,2-D2]-Gly and [2,3,3-D3]-l-Ser (Cambridge Isotope Laboratories) before extractions. Amino acids were derivatized in 50 μL of N-methyl-N-(tert-butyldimethylsilyl)-trifluoroacetamide containing 1% (v/v) tert-butyldimethylchlorosilane (Pierce)/pyridine (1:1 by volume) at 50°C for 1 h. Amino acid-tert-butyldimethylsilyl derivatives were analyzed using the same GC-MS instrumentation as described above. Fragments having mass-to-charge (m/z) ratios of 218 and 248, and 288 and 390 (and the corresponding M + 3 ions for the labeled internal standards) were used to quantify the levels of Gly and Ser, respectively.
Folate and Formate Analyses
Deglutamylated folates were analyzed in leaves (2 g) of the wild type and purU dKO mutant grown in ambient air by HPLC and electrochemical detection in extracts purified by affinity chromatography as described previously (Goyer et al., 2005). For the determination of formate levels, formate was extracted under alkaline conditions as described (Li et al., 2003) with modifications. Briefly, 0.6 to 0.8 g of leaves (3- to 4-week-old plants) was frozen in liquid N2 and subjected to grinding in 1 mL of freshly prepared 100 mM KOH (Sigma-Aldrich; low carbonate content) containing 100 nmol 13C-formate as an internal standard using polypropylene mesh bags (Pierce) as an alternative to mortar and pestle. The homogenate was centrifuged, and 50 μL of 1 M sulfuric acid was added per 0.5 mL of sample and kept on ice for 10 min. After centrifugation, 0.5 mL of supernate was subjected to transmethylation and solid phase microextraction using 70-μm Carboxen/polydimethylsiloxane fiber assemblies (Supelco) as described (Lee et al., 1999). Methyl formate analysis was performed on an Agilent GC-MS instrument (same as above). The separation was achieved on a 30-m DBwax column (0.25 mm × 0.25-μm film thickness). Mass spectra data were collected from m/z 50 to 100 in the positive ion mode at electron energy of 70 eV. Formate levels were calculated based on the recovery of the internal standard (m/z 60 and 61 for methyl formate, retention time 0.75 min). 13C-Formate recoveries in plant samples varied between 45 and 65% of these of the internal standard alone.
Statistical Analysis
Plants were pregrown for 3 weeks at high CO2 and transferred to ambient air for 2 weeks. Relative levels of >50 major metabolites in the wild type, dKO, shm1-1, dhc, and triple mutants were quantified by integrating areas of fragments specific for given compounds using Enhanced MSD ChemStation software (Agilent Technologies). Relative metabolite levels of individual samples were used in PCA on correlations using JMP 7.0.1 software (SAS Institute).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative and GenBank/EMBL databases. Accession numbers for the E. coli K12 PurU and PurN protein sequences are NP415748 and NP416995, respectively. Arabidopsis sequences are as follows: 10-FDFs At4g17360 and At5g47435 (corresponding mutant lines Salk_062946 [CS860076] and Garlic_169F06 [CS860077], respectively); GDC encoded by eight loci (four different subunits; Bauwe and Kolukisaoglu, 2003) (P, At2g26080 and At4g33010; H, At1g32470, At2g35120, and At2g35370; L, At3g17240 and At1g48030; and T, At1g11860); photorespiratory SHMT At4g37930 (CS8010 mutant; Voll et al., 2006); DHC in mitochondria At2g38660 (corresponding mutant lines Salk_143478 [CS860078] and Salk_142776, respectively); 5-FCL At5g13050 (Syngenta mutant line 28D07; Goyer et al., 2005); 10-formyl THF SYN At1g50480; and 10-formyl THF-dependent phosphoribosylglycinamide formyltransferase PurN At1g31220. The germplasm IDs for dKO and the triple homozygous mutants are CS860079 and CS860080, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Pathways Involving Folate Interconversions.
Supplemental Figure 2. Parallel Comparison of Embryo Development in Wild-Type, dKO, and Corresponding Single Homozygous Mutant Lines.
Supplemental Figure 3. DNA Gel Blot Analysis of Wild-Type and Garlic_169F06 Single Mutant Plants.
Supplemental Figure 4. Parallel Comparison of Phenotypes in Wild-Type and Mutant Plants.
Supplemental Figure 5. Principal Component Analysis on Correlations of Major Metabolites in Wild-Type and Mutant Plants.
Supplemental Figure 6. RT-PCR and Genotyping Results.
Supplementary Material
Acknowledgments
We thank Daniel Jones and Beverly Chamberlin at the Michigan State University (MSU) Mass Spectroscopy Facility, Joseph Leykam at the MSU Molecular Structure Facility, Annette Thelen at the MSU Research Technology Support Facility, and Melinda Frame at the MSU Center for Advanced Microscopy for providing technical and instrumental support. We also thank Frederica Brandizzi for help with GFP image processing. This work was supported by National Science Foundation grants to Y.S.-H., A.D.H., and J.F.G.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Eva Collakova (collakov@msu.edu) and Andrew D. Hanson (adha@ufl.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Almassy, R.J., Janson, C.A., Kan, C.C., and Hostomska, Z. (1992). Structures of apo and complexed Escherichia coli glycinamide ribonucleotide transformylase. Proc. Natl. Acad. Sci. USA 89 6114–6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggott, E.J. (2000). Hydrolysis of 5,10-methenyltetrahydrofolate to 5-formyltetrahydrofolate at pH 2.5 to 4.5. Biochemistry 39 14647–14653. [DOI] [PubMed] [Google Scholar]
- Baud, S., and Graham, I.A. (2006). A spatiotemporal analysis of enzymatic activities associated with carbon metabolism in wild type and mutant embryos of Arabidopsis using in situ histochemistry. Plant J. 46 155–169. [DOI] [PubMed] [Google Scholar]
- Baud, S., Guyon, V., Kronenberger, J., Wuilleme, S., Miquel, M., Caboche, M., Lepiniec, L., and Rochat, C. (2003). Multifunctional acetyl-CoA carboxylase 1 is essential for very long chain fatty acid elongation and embryo development in Arabidopsis. Plant J. 33 75–86. [DOI] [PubMed] [Google Scholar]
- Bauer, M., Dietrich, C., Nowak, K., Sierralta, W.D., and Papenbrock, J. (2004). Intracellular localization of Arabidopsis sulfurtransferases. Plant Physiol. 135 916–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauwe, H., and Kolukisaoglu, U. (2003). Genetic manipulation of glycine decarboxylation. J. Exp. Bot. 54 1523–1535. [DOI] [PubMed] [Google Scholar]
- Boldt, R., and Zrenner, R. (2003). Purine and pyrimidine biosynthesis in higher plants. Physiol. Plant. 117 297–304. [DOI] [PubMed] [Google Scholar]
- Cernac, A., and Benning, C. (2004). WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J. 40 575–585. [DOI] [PubMed] [Google Scholar]
- Chen, L.F., Chan, S.Y., and Cossins, E.A. (1997). Distribution of folate derivatives and enzymes for synthesis of 10-formyltetrahydrofolate in cytosolic and mitochondrial fractions of pea leaves. Plant Physiol. 115 299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L.F., Nargang, F.E., and Cossins, E.A. (1999). Isolation and sequencing of a plant cDNA encoding a bifunctional methylenetetrahydrofolate dehydrogenase: methenyltetrahydrofolate cyclohydrolase protein. Pteridines 10 171–177. [Google Scholar]
- Chipman, D.M., and Shaanan, B. (2001). The ACT domain family. Curr. Opin. Struct. Biol. 11 694–700. [DOI] [PubMed] [Google Scholar]
- Clandinin, M.T., and Cossins, E.A. (1974). Methionine biosynthesis in isolated pea mitochondria. Phytochemistry 13 585–591. [Google Scholar]
- Coffin, J.W., and Cossins, E.A. (1986). Mitochondrial folates and methionyl-tRNA transformylase activity during germination and early growth of seeds. Phytochemistry 25 2481–2487. [Google Scholar]
- Cossins, E.A. (2000). The fascinating world of folate and one-carbon metabolism. Can. J. Bot. 78 691–708. [Google Scholar]
- Cossins, E.A., and Chen, L.F. (1997). Folates and one-carbon metabolism in plants and fungi. Phytochemistry 45 437–452. [DOI] [PubMed] [Google Scholar]
- D'Ari, L., and Rabinowitz, J.C. (1991). Purification, characterization, cloning, and amino acid sequence of the bifunctional enzyme 5,10-methylenetetrahydrofolate dehydrogenase/5,10-methenyltetrahydrofolate cyclohydrolase from Escherichia coli. J. Biol. Chem. 266 23953–23958. [PubMed] [Google Scholar]
- Dev, I.K., and Harvey, R.J. (1978). N10-Formyltetrahydrofolate is formyl donor for glycinamide ribotide transformylase in Escherichia coli. J. Biol. Chem. 253 4242–4244. [PubMed] [Google Scholar]
- Douce, R., and Neuburger, M. (1999). Biochemical dissection of photorespiration. Curr. Opin. Plant Biol. 2 214–222. [DOI] [PubMed] [Google Scholar]
- Duran, A.L., Yang, J., Wang, L.J., and Sumner, L.W. (2003). Metabolomics spectral formatting, alignment and conversion tools (MSFACTs). Bioinformatics 19 2283–2293. [DOI] [PubMed] [Google Scholar]
- Eastmond, P.J., van Dijken, A.J.H., Spielman, M., Kerr, A., Tissier, A.F., Dickinson, H.G., Jones, J.D.G., Smeekens, S.C., and Graham, I.A. (2002). Trehalose-6-phosphate synthase 1, which catalyses the first step in trehalose synthesis, is essential for Arabidopsis embryo maturation. Plant J. 29 225–235. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Brunak, S., von Heijne, G., and Nielsen, H. (2007). Locating proteins in the cell using TargetP, SignalP, and related tools. Nat. Protocols 2 953–971. [DOI] [PubMed] [Google Scholar]
- Engel, N., van den Daele, K., Kolukisaoglu, U., Morgenthal, K., Weckwerth, W., Parnik, T., Keerberg, O., and Bauwe, H. (2007). Deletion of glycine decarboxylase in Arabidopsis is lethal under nonphotorespiratory conditions. Plant Physiol. 144 1328–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focks, N., and Benning, C. (1998). Wrinkled1: A novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 118 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibeaut, D.M., Hulett, J., Cramer, G.R., and Seemann, J.R. (1997). Maximal biomass of Arabidopsis thaliana using a simple, low-maintenance hydroponic method and favorable environmental conditions. Plant Physiol. 115 317–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffman, F.D., Ruckle, M., Ohlrogge, J., and Shachar-Hill, Y. (2004). Carbon dioxide concentrations are very high in developing oilseeds. Plant Physiol. Biochem. 42 703–708. [DOI] [PubMed] [Google Scholar]
- Goyer, A., Collakova, E., de la Garza, R.D., Quinlivan, E.P., Williamson, J., Gregory, J.F., Shachar-Hill, Y., and Hanson, A.D. (2005). 5-Formyltetrahydrofolate is an inhibitory but well tolerated metabolite in Arabidopsis leaves. J. Biol. Chem. 280 26137–26142. [DOI] [PubMed] [Google Scholar]
- Grimshaw, C.E., Henderson, G.B., Soppe, G.G., Hansen, G., Mathur, E.J., and Huennekens, F.M. (1984). Purification and properties of 5,10-methenyltetrahydrofolate synthetase from Lactobacillus casei. J. Biol. Chem. 259 2728–2733. [PubMed] [Google Scholar]
- Hanson, A.D., Gage, D.A., and Shachar-Hill, Y. (2000). Plant one-carbon metabolism and its engineering. Trends Plant Sci. 5 206–213. [DOI] [PubMed] [Google Scholar]
- Hanson, A.D., and Roje, S. (2001). One-carbon metabolism in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52 119–137. [DOI] [PubMed] [Google Scholar]
- Heazlewood, J.L., and Millar, A.H. (2005). AMPDB: The Arabidopsis Mitochondrial Protein Database. Nucleic Acids Res. 33 D605–D610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood, J.L., Verboom, R.E., Tonti-Filippini, J., Small, I., and Millar, A.H. (2007). SUBA: The Arabidopsis Subcellular Database. Nucleic Acids Res. 35 D213–D218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himes, R.H., and Rabinowitz, J.B. (1962). Formyltetrahydrofolate synthetase. 3. Studies on mechanism of reaction. J. Biol. Chem. 237 2903–2914. [PubMed] [Google Scholar]
- Hirner, B., Fischer, W.N., Rentsch, D., Kwart, M., and Frommer, W.B. (1998). Developmental control of H+/amino acid permease gene expression during seed development of Arabidopsis. Plant J. 14 535–544. [DOI] [PubMed] [Google Scholar]
- Holmes, W.B., and Appling, D.R. (2002). Cloning and characterization of methenyltetrahydrofolate synthetase from Saccharomyces cerevisiae. J. Biol. Chem. 277 20205–20213. [DOI] [PubMed] [Google Scholar]
- Hourton-Cabassa, C., Ambard-Bretteville, F., Moreau, F., de Virville, J.D., Remy, R., and des Francs-Small, C.C. (1998). Stress induction of mitochondrial formate dehydrogenase in potato leaves. Plant Physiol. 116 627–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglese, J., Johnson, D.L., Shiau, A., Smith, J.M., and Benkovic, S.J. (1990. a). Subcloning, characterization, and affinity labeling of Escherichia coli glycinamide ribonucleotide transformylase. Biochemistry 29 1436–1443. [DOI] [PubMed] [Google Scholar]
- Inglese, J., Smith, J.M., and Benkovic, S.J. (1990. b). Active-site mapping and site-specific mutagenesis of glycinamide ribonucleotide transformylase from Escherichia coli. Biochemistry 29 6678–6687. [DOI] [PubMed] [Google Scholar]
- Lee, X.P., Kumazawa, T., Kondo, K., Sato, K., and Suzuki, O. (1999). Analysis of methanol or formic acid in body fluids by headspace solid-phase microextraction and capillary gas chromatography. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 734 155–162. [DOI] [PubMed] [Google Scholar]
- Li, R., Moore, M., and King, J. (2003). Investigating the regulation of one-carbon metabolism in Arabidopsis thaliana. Plant Cell Physiol. 44 233–241. [DOI] [PubMed] [Google Scholar]
- Liepman, A.H., and Olsen, L.J. (2003). Alanine aminotransferase homologs catalyze the glutamate: glyoxylate aminotransferase reaction in peroxisomes of Arabidopsis. Plant Physiol. 131 215–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C.M., and Meinke, D.W. (1998). The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. Plant J. 16 21–31. [DOI] [PubMed] [Google Scholar]
- Marolewski, A., Smith, J.M., and Benkovic, S.J. (1994). Cloning and characterization of a new purine biosynthetic enzyme. A non-folate glycinamide ribonucleotide transformylase from Escherichia coli. Biochemistry 33 2531–2537. [DOI] [PubMed] [Google Scholar]
- Munne-Bosch, S., and Alegre, L. (2002). The function of tocopherols and tocotrienols in plants. Crit. Rev. Plant Sci. 21 31–57. [Google Scholar]
- Nagy, P.L., Marolewski, A., Benkovic, S.J., and Zalkin, H. (1995). Formyltetrahydrofolate hydrolase, a regulatory enzyme that functions to balance pools of tetrahydrofolate and one-carbon tetrahydrofolate adducts in Escherichia coli. J. Bacteriol. 177 1292–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy, P.L., McCorkle, G.M., and Zalkin, H. (1993). PurU, a source of formate for PurT-dependent phosphoribosyl-N-formylglycinamide synthesis. J. Bacteriol. 175 7066–7073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuburger, M., Rebeille, F., Jourdain, A., Nakamura, S., and Douce, R. (1996). Mitochondria are a major site for folate and thymidylate synthesis in plants. J. Biol. Chem. 271 9466–9472. [DOI] [PubMed] [Google Scholar]
- Nygaard, P., and Smith, J.M. (1993). Evidence for a novel glycinamide ribonucleotide transformylase in Escherichia coli. J. Bacteriol. 175 3591–3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogren, W.L. (1984). Photorespiration - Pathways, regulation, and modification. Annu. Rev. Plant Physiol. Plant Mol. Biol. 35 415–442. [Google Scholar]
- Quinlivan, E.P., Hanson, A.D., and Gregory, J.F. (2006). The analysis of folate and its metabolic precursors in biological samples. Anal. Biochem. 348 163–184. [DOI] [PubMed] [Google Scholar]
- Rebeille, F., Neuburger, M., and Douce, R. (1994). Interaction between glycine decarboxylase, serine hydroxymethyltransferase and tetrahydrofolate polyglutamates in pea leaf mitochondria. Biochem. J. 302 223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roje, S., Janave, M.T., Ziemak, M.J., and Hanson, A.D. (2002). Cloning and characterization of mitochondrial 5-formyltetrahydrofolate cycloligase from higher plants. J. Biol. Chem. 277 42748–42754. [DOI] [PubMed] [Google Scholar]
- Roje, S., Wang, H., McNeil, S.D., Raymond, R.K., Appling, D.R., Shachar-Hill, Y., Bohnert, H.J., and Hanson, A.D. (1999). Isolation, characterization, and functional expression of cDNAs encoding NADH-dependent methylenetetrahydrofolate reductase from higher plants. J. Biol. Chem. 274 36089–36096. [DOI] [PubMed] [Google Scholar]
- Schirch, L.V., and Ropp, M. (1967). Serine transhydroxymethylase. Affinity of tetrahydrofolate compounds for enzyme and enzyme-glycine complex. Biochemistry 6 253–257. [DOI] [PubMed] [Google Scholar]
- Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Scholkopf, B., Weigel, D., and Lohmann, J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37 501–506. [DOI] [PubMed] [Google Scholar]
- Schnorr, K.M., Laloue, M., and Hirel, B. (1996). Isolation of cDNAs encoding two purine biosynthetic enzymes of soybean and expression of the corresponding transcripts in roots and root nodules. Plant Mol. Biol. 32 751–757. [DOI] [PubMed] [Google Scholar]
- Schnorr, K.M., Nygaard, P., and Laloue, M. (1994). Molecular characterization of Arabidopsis thaliana cDNAs encoding 3 purine biosynthetic enzymes. Plant J. 6 113–121. [DOI] [PubMed] [Google Scholar]
- Schwender, J., and Ohlrogge, J.B. (2002). Probing in vivo metabolism by stable isotope labeling of storage lipids and proteins in developing Brassica napus embryos. Plant Physiol. 130 347–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, K.W., and Rabinowitz, J.C. (1988). Isolation and characterization of the Saccharomyces cerevisiae MIS1 gene encoding mitochondrial C1-tetrahydrofolate synthase. J. Biol. Chem. 263 7717–7725. [PubMed] [Google Scholar]
- Smith, J.M., and Daum, H.A. (1987). Identification and nucleotide sequence of a gene encoding 5′-phosphoribosylglycinamide transformylase in Escherichia coli K12. J. Biol. Chem. 262 10565–10569. [PubMed] [Google Scholar]
- Somerville, C.R. (2001). An early Arabidopsis demonstration. Resolving a few issues concerning photorespiration. Plant Physiol. 125 20–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville, C.R., and Ogren, W.L. (1980). Inhibition of photosynthesis in Arabidopsis mutants lacking leaf glutamate synthase activity. Nature 286 257–259. [Google Scholar]
- Somerville, C.R., and Ogren, W.L. (1981). Photorespiration deficient mutants of Arabidopsis thaliana lacking mitochondrial serine transhydroxymethylase activity. Plant Physiol. 67 666–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover, P., and Schirch, V. (1990). Serine hydroxymethyltransferase catalyzes the hydrolysis of 5,10-methenyltetrahydrofolate to 5-formyltetrahydrofolate. J. Biol. Chem. 265 14227–14233. [PubMed] [Google Scholar]
- Stover, P., and Schirch, V. (1991). 5-Formyltetrahydrofolate polyglutamates are slow tight-binding inhibitors of serine hydroxymethyltransferase. J. Biol. Chem. 266 1543–1550. [PubMed] [Google Scholar]
- Voll, L.M., Jamai, A., Renne, P., Voll, H., McClung, C.R., and Weber, A.P.M. (2006). The photorespiratory Arabidopsis shm1 mutant is deficient in SHM1. Plant Physiol. 140 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, C.Y., and Wilkins, T.A. (1994). A modified hot borate method significantly enhances the yield of high quality RNA from cotton (Gossypium Hirsutum L). Anal. Biochem. 223 7–12. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.