The plant hormone and signaling molecule auxin is a key player during pattern formation, organogenesis, and various physiological processes. Recent discoveries in auxin biology point toward an auxin pathway with a higher complexity than previously anticipated. This prompted us to review this constantly growing field and to put these novel and exciting findings into a broader developmental and evolutionary context.
Auxin signaling can be divided broadly into three layers that contribute to its complexity: the spatio-temporal pattern of its biosynthesis, its directional transport, and cell- or tissue-specific responses. In Arabidopsis thaliana, auxin is synthesized via several pathways in embryos, leaves, and roots (reviewed in Woodward and Bartel, 2005). The active transport of auxin, leading to the establishment of auxin maxima and gradients, is mediated by specific proteins: PIN-FORMED (PIN) efflux carriers, AUXIN1/LIKE AUX1 (AUX1/LAX) influx carriers, and MULTIDRUG RESISTANCE/P-GLYCOPROTEIN transporters (reviewed in Vieten et al., 2007). Upon reaching specific threshold concentrations, auxin induces specific responses that trigger diverse developmental and physiological effects (reviewed in Teale et al., 2006).
The abundant interlinkages between these layers and the variety of protein complexes involved in auxin-related processes contribute to the overall complexity of the auxin pathway. In addition, there are numerous inputs, intrinsic and environmental cues, that feed into this pathway, and this is quite frequently mediated by other hormonal pathways. Although the auxin pathway is being unraveled bit by bit, we will illustrate that there is still a long way to go before it will be fully understood.
THE MOLECULAR BASIS OF AUXIN SIGNALING
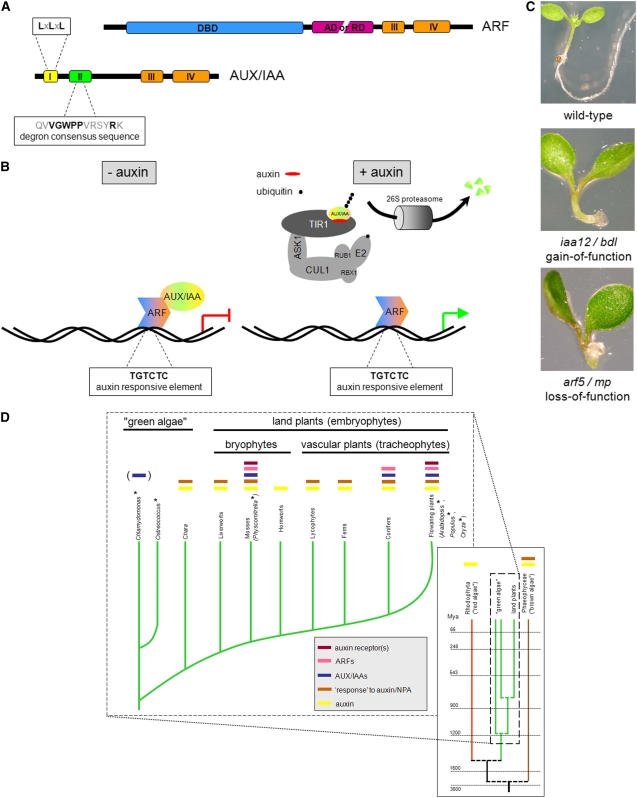
To start, we briefly introduce the essentials of auxin signaling. Intracellular auxin is perceived by the TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX PROTEIN1-3 (TIR1/AFB1-3) receptors, thereby triggering the degradation of AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) proteins. These AUX/IAAs, under low auxin concentrations, form dimers with AUXIN RESPONSE FACTOR (ARF) transcription factors, thereby blocking the activity of at least the activating ARFs. Once freed from the AUX/IAAs, these ARFs regulate the expression of auxin-responsive genes (Figures 1A and 1B). It should be noted that most likely there are auxin-induced responses that require additional signaling components (reviewed in Teale et al., 2006).
Figure 1.
Basics and Evolution of Auxin Signaling and Response.
(A) Schematic representation of important domains and motifs of ARFs and AUX/IAAs. LxLxL resembles the repressive motif in ETHYLENE RESPONSE FACTOR–associated amphiphilic repression domains. DBD, B3 DNA binding domain; AD, activation domain; RD, repression domain.
(B) Simplified scheme of auxin-responsive gene regulation through an activating ARF in the absence or presence of auxin. ASK1, ARABIDOPSIS SKP1-LIKE; CUL1, CULLIN1; E2, a conjugating enzyme; RBX1, RING-BOX PROTEIN1; RUB1, RELATED TO UBIQUITIN1.
(C) Identical phenotypes of gain-of-function AUX/IAA (iaa12/bdl) and loss-of-function ARF (arf5/mp) auxin response mutants.
(D) Phylogenetic scheme highlighting the evolution of auxin signaling and response, based on physiological and genomic data. The colored bars above the branches point out positive evidence for the indicated feature. It should be noted that only from mosses onward the available genome sequence has been analyzed in depth, and likewise the observed auxin responses display biological significance. Years taken from Yoon et al. (2004). *, fully sequenced genome; (), unconfirmed finding; NPA, naphthylphthalamic acid (an auxin transport inhibitor).
The 29 members of the Arabidopsis AUX/IAA family usually contain four conserved domains (Figure 1A) and display strong genetic redundancy, since even triple loss-of-function mutants have no apparent abnormal phenotype (Overvoorde et al., 2005). By contrast, when AUX/IAA proteins are stabilized through a specific amino acid exchange in the degron sequence within domain II (Figure 1A), auxin responses are disrupted, and this often results in dramatic phenotypes, as exemplified by solitary root (slr)/iaa14 (Fukaki et al., 2002) or bodenlos (bdl)/iaa12 (Hamann et al., 2002) (Figure 1C).
Interestingly, one clade within the AUX/IAA protein family contains proteins that lack all or some of domain II and are stable relative to other AUX/IAAs. Overexpression of some of these noncanonical AUX/IAAs causes strong auxin-related aberrant phenotypes, such as malformed vasculature of cotyledons, collapse of the root apical meristem, and defects in gravitropic response (Sato and Yamamoto, 2008).
In Arabidopsis, there are 23 ARF genes encoding proteins that contain a B3 DNA binding domain, a repression or activation domain, and two domains that share high similarity with domains III and IV of AUX/IAAs (Figure 1A). ARFs bind to the auxin-responsive element TGTCTC, thereby regulating the expression of auxin-responsive genes. Domains III and IV mediate the dimerization of AUX/IAAs and ARFs, and this specific interaction blocks the activity of at least activating ARFs (Figure 1B; reviewed in Guilfoyle and Hagen, 2007). Therefore, arf loss-of-function mutations, such as arf7 arf19 (Okushima et al., 2005; Wilmoth et al., 2005) or monopteros (mp)/arf5 (Hardtke and Berleth, 1998), often result in phenotypes similar to those caused by the stabilized AUX/IAA interaction partner (Figure 1C).
In the past few years, it was shown that TIR1 and some close homologs act as auxin receptors in Arabidopsis. TIR1 is an integral component of the SKP1/CULLIN/F-BOX PROTEIN (SCF)TIR1 complex that eventually mediates the ubiquitination of AUX/IAAs and thereby destines them for 26S proteasome-dependent degradation (reviewed in Abel, 2007). AUX/IAAs are bound by TIR1 in the TIR1 pocket via their domain II, and auxin acts as a molecular glue to enhance this interaction (Figure 1B; Tan et al., 2007).
EVOLUTIONARY PERSPECTIVES ON AUXIN SIGNALING
During evolution of land plants, developmental mechanisms arose that resulted in coordinated multicellular growth. It is thought that this may well have been achieved, at least in part, through the evolution of the auxin pathway (Rensing et al., 2008). The current knowledge of this evolutionary issue is summarized in Figure 1D.
An indication of an ancient evolutionary origin of auxin-related mechanisms might be suggested by the as yet unconfirmed finding of at least two AUX/IAAs in Chlamydomonas reinhardtii (Palenik et al., 2007). If confirmed, this finding might suggest that single-celled green algae that diverged from the land plant lineage some 1 billion years ago already possessed specific components of the auxin pathway. However, this conclusion remains in doubt as several researchers have been unable to convincingly reproduce these results (S. Lau and I. De Smet, unpublished data). In addition, the function of such proteins in Chlamydomonas is unknown, and Ostreococcus, a single-celled member of a different class of green algae, appears to have lost the respective genes (Palenik et al., 2007).
For morphologically more complex green algae, such as Chara contraria, there is no sequence information available, but their generative cells respond to auxin treatment with the depolymerization of microtubules, which is reminiscent of what happens in vascular plants (Jin et al., 2007). Red and brown algae also appear to contain and/or respond to auxin (reviewed in Cooke et al., 2002). For instance, in Fucus distichus, a model brown alga, the embryo is affected by auxin and by auxin transport inhibitors (Sun et al., 2004). However, it should be noted that these physiological responses and the presumptive presence of some parts of the auxin pathway might not reflect a real endogenous role for auxin in these organisms.
While the function and importance of auxin in the case of these diverse and polyphyletic algal lineages is not well understood, there are numerous indications of its emerging role in land plants. For example, several physiological experiments revealed the occurrence of a basic auxin metabolism, an auxin-dependent apical dominance, and polar auxin transport within the bryophytes (liverworts, hornworts, and mosses) and pteridophytes (lycophytes, horsetails, and ferns) (reviewed in Cooke et al., 2002). Moreover, compelling evidence for the existence of at least a basic auxin pathway in mosses can be drawn from the sequenced genome of Physcomitrella patens and the first analysis of its content. All major players of the higher plant auxin pathway, such as biosynthesis proteins (YUCCAs), receptors (TIR1/AFBs), transporters (PINs and AUX1/LAXs), and transcriptional regulators (AUX/IAAs and ARFs), are encoded in the Physcomitrella genome (Rensing et al., 2008). In agreement with these genomic data, it was recently shown that a small auxin-related molecule, which specifically affects the TIR1-mediated response to auxin in Arabidopsis, also antagonizes auxin response in Physcomitrella (Hayashi et al., 2008).
Vascular plants are distinguished from Physcomitrella by a higher developmental complexity, and this is accompanied by an expansion of the genetic machinery of the auxin pathway in those species (Goldfarb et al., 2003; Jain et al., 2006; Kalluri et al., 2007; Rensing et al., 2008). Whereas the Physcomitrella genome contains 55 auxin-related genes representing 0.14% of all protein coding loci, the corresponding numbers in, for example, Arabidopsis are 174 and 0.65%, respectively (Rensing et al., 2008). Remarkably, a strikingly low fraction of auxin-related genes in Physcomitrella encodes AUX/IAAs compared with vascular plants (Rensing et al., 2008). This suggests that enrichment of auxin-related gene families has occurred within vascular plant genomes, for example, via preferential preservation of these genes after duplication events.
An extended number of genes enables a higher developmental complexity via different nonexclusive means: changes in the expression pattern and evolution of new or different functions of the corresponding gene products. In the case of the auxin pathway, the relative contributions of these mechanisms to the assumed increased complexity have been thoroughly investigated for the AUX/IAAs in Arabidopsis. Several promoter swapping experiments for AUX/IAAs suggest a predominant role for gene-specific expression patterns compared with the contributions of protein specificities. Differences in protein specificity nonetheless confer a certain degree of distinctiveness to the investigated AUX/IAAs, since protein-specific differences in function were noted when their expression was driven by the same promoter (Weijers et al., 2005; Muto et al., 2007).
Both altered expression and function appear to have arisen since the last common ancestor of Arabidopsis and Physcomitrella and may have played a significant role in the evolution of greater morphological complexity in vascular plants. The increased number of possible combinations of auxin signaling components in vascular plants, with each combination potentially giving rise to another output, enhances the diversity or at least the fine-tuning of auxin responses. Still, it remains to be elucidated whether, for instance, the enormous number of theoretically possible AUX/IAA–ARF interaction pairs is of any biological relevance.
REGULATION OF AUX/IAAs AND ARFs
Cell- or tissue-specific auxin responses depend on mechanisms that bring about specific control of transcript and/or protein levels and the subsequent differential interpretation of auxin maxima and gradients through specific combinations of auxin signaling components.
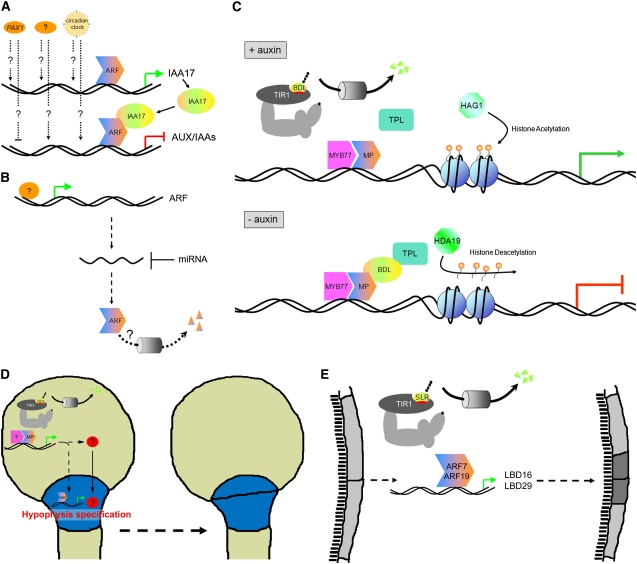
As indicated by their designation, most AUX/IAAs are primary auxin response genes. Whereas degradation of AUX/IAA proteins has been extensively studied (see above), very little detailed information is available on the transcriptional regulation of AUX/IAA genes apart from their auxin inducibility, which depends on the cell or tissue type, the actual auxin concentration, and the duration of exposure to certain auxin levels (Paponov et al., 2008). For example, it has been shown that PARTIAL SUPPRESSOR OF AXR3-1 (PAX1), an uncharacterized locus, positively regulates the expression of AUXIN RESISTANT3 (AXR3)/IAA17. While PAX1 also affects the expression of other AUX/IAAs, it remains to be demonstrated if this transcriptional regulation happens via a direct mechanism or through a feedback loop involving AXR3/IAA17 (Figure 2A; Tanimoto et al., 2007). Recently it was shown that the circadian clock also regulates the transcription of components of the auxin pathway and that plant growth in response to exogenous auxin is gated by the circadian clock (Figure 2A). Interestingly, the circadian expression patterns of the AUX/IAAs and ARFs that are clock regulated show an antiphasic relationship (Covington and Harmer, 2007).
Figure 2.
Upstream Regulation and Downstream Events of Auxin Signaling and Response.
(A) Transcriptional regulation of AUX/IAAs. PAX1 regulation happens via a direct mechanism or through a feedback loop involving AXR3/IAA17. Dotted arrows indicate an unclear regulatory mechanism.
(B) Regulation of the levels of ARF transcripts and proteins. Dotted arrow indicates an unclear regulatory mechanism.
(C) Scheme of putative interactions between AUX/IAAs (IAA12/BDL), ARFs (ARF5/MP), a corepressor (TPL), a coactivator (MYB77), and proteins involved in histone acetylation (HAG1) and deacetylation (HDA19) in the presence and absence of auxin. Orange/yellow balloons indicate acetyl residues.
(D) Model for hypophysis specification during embryogenesis, highlighting the role of IAA12/BDL and ARF5/MP in the embryo proper and an additional auxin response in combination with an MP-dependent putative mobile signal in the hypophysis. Hatched lines indicate MP-mediated auxin responses; red disc with a question mark indicates putative mobile signal.
(E) Model for the asymmetric cell division of pericycle cells during lateral root initiation, focusing on SLR/IAA14, ARF7, ARF19, and their downstream targets LBD16 and LBD29.
In addition, a means of modulating or impeding auxin responses is via the negative posttranscriptional regulation of the expression of ARFs through small RNAs (Figure 2B; reviewed in Teale et al., 2006) For example, transcript levels of ARF8 are regulated by miR167, and this regulation was shown to play a role in auxin-mediated lateral root development (Gifford et al., 2008).
Another mechanism of fine-tuning auxin responses could be the (developmentally) controlled degradation of ARFs. This topic has not received much attention, but a recent report describes the degradation properties of ARF1, which appears to be degraded in a pathway distinct from the way AUX/IAAs are degraded (Salmon et al., 2008). These researchers showed that the degradation of ARF1 at moderate rates is proteasome dependent but does not require a CULLIN1-based SCF complex and is not affected by auxin (Figure 2B; Salmon et al., 2008). Provided that the degradation of ARF proteins is regulated in a developmentally relevant manner, this opens up intriguing new possibilities for an even more subtle regulation of auxin responses. A possible role of targeted ARF degradation could be the prevention of (specific) auxin responses in certain tissues, which could be especially important during the restriction of the expression domain of ARFs.
COMBINATORIAL CONTROL OF AUXIN SIGNALING
With respect to events occurring during auxin responses, most attention has been paid to the interaction of AUX/IAAs and ARFs. Undoubtedly, AUX/IAA–ARF interaction pairs are principal regulators of auxin-responsive gene expression, but recent studies have revealed that other proteins are also recruited to this core unit.
One protein that is able to act as a coactivator with ARFs is MYB DOMAIN PROTEIN77 (MYB77). It was recently shown that MYB77 interacts in vitro with several supposedly activating and repressing ARFs via their C termini and with IAA19 to promote auxin-responsive gene expression (Shin et al., 2007). In the case of ARF7, this was confirmed in planta, and based on the lateral root phenotype of the arf7 myb77 double mutant, it was demonstrated that ARF7 and MYB77 act in a synergistic manner (Shin et al., 2007). While MYB transcription factors had not been implicated previously in auxin signaling, there were indications that the expression of some MYB family members is itself regulated by auxin (Kranz et al., 1998). Because both auxin response elements and putative MYB binding motifs are present in close proximity in promoters that are potentially regulated by ARFs and MYB77, it is feasible that ARFs and MYB77 could interact at those promoters and combinatorially regulate gene expression (Figure 2C). However, since MYB77 directly binds to the ARF dimerization domain, it is possible that these MYB binding sites are not required and that MYB77 is recruited to promoters in a similar manner as AUX/IAAs (Shin et al., 2007).
MAINTENANCE OF DEVELOPMENTAL DECISIONS
In the temperature-sensitive topless-1 (tpl-1) mutant, which acts as a dominant negative for the whole TOPLESS RELATED family, the shoot pole is transformed into a second root pole (Long et al., 2006). TPL was proposed to be a transcriptional corepressor based on the properties of its domains as well as on genetic interactions with HISTONE DEACETYLASE19 (HDA19) and HISTONE ACETYLTRANSFERASE OF THE GNAT FAMILY1 (HAG1) (Long et al., 2006). Recent data demonstrated that tpl-1 can suppress all phenotypic abnormalities caused by the stabilizing mutation in the AUX/IAA gene BDL. A better understanding of the mechanisms underlying this observation comes from the finding that TPL acts in a complex together with BDL and MP. TPL binds to domain I of BDL, specifically to a motif resembling the ETHYLENE RESPONSE FACTOR–associated amphiphilic repression motif in this domain, which generally is required for repression (Figures 1A and 2C; Szemenyei et al., 2008).
These results are consistent with a report that PICKLE, a putative chromatin remodeling factor thought to function in cooperation with a histone deacetylase, is required for the SLR/IAA14-mediated suppression of lateral root initiation. In addition, application of a histone deacetylase inhibitor was found to suppress the lateral root defects seen in the slr mutant (Fukaki et al., 2006).
The involvement of histone modifications in auxin response could be plausibly explained via auxin-dependent changes in the histone acetylation status and that, for instance, in the tpl-1 mutant, this becomes unstable leading to aberrant developmental programs. This mechanism of stabilizing auxin-induced developmental decisions might be regarded as a counterpart of the well-studied developmental control exerted by Polycomb group complexes in plants and animals (reviewed in Schwartz and Pirrotta, 2007).
WHAT IS HAPPENING DOWNSTREAM OF AUX/IAAs AND ARFs?
For auxin maxima or auxin gradients to entail any consequences, they must trigger physiologically or developmentally relevant processes. Numerous fundamental processes in plants are known to be auxin dependent, including the establishment of the axis of polarity in the embryo, the specification of the hypophysis, the phenomenon of phyllotaxis, and the initiation of lateral roots (reviewed in De Smet and Jürgens, 2007), and various physiological responses, such as gravitropism (reviewed in Vieten et al., 2007) and shade avoidance (Tao et al., 2008). In addition, a large number of genes exhibits auxin-induced changes of expression and/or contains auxin-responsive elements in the promoter region (Nemhauser et al., 2004; Paponov et al., 2008), making them good target candidates for ARF action. In spite of seemingly favorable prerequisites, precise descriptions of specific auxin-dependent events that depend on the activity of ARFs have remained elusive, but the first important steps are being taken.
The best-characterized ARF-dependent developmental pathway during embryogenesis is the one that leads to the establishment of the root pole via the proper specification of the hypophysis. The involvement of BDL/IAA12 and MP/ARF5 in this process has been clearly demonstrated (reviewed in De Smet and Jürgens, 2007). Intriguingly, both BDL and MP are not expressed in the hypophysis, but immediately above that cell, in the embryo proper. Hence, it has been postulated that a MP-dependent signal must be conveyed to the hypophysis from the above cells, be it (solely) auxin or (additionally) something else. Because auxin application does not overcome the root defect of the bdl mutant, it is likely that an auxin-independent signal is required (Weijers et al., 2006). It is interesting to note that the overexpression of MP, which is sufficient to overcome the bdl phenotype and which principally could cause a continuous auxin response, does not give rise to developmental defects in the embryo but rather leads to normal seedlings (Weijers et al., 2006). The reason for this outcome is currently unclear, but a plausible explanation is that specific cofactors, as for instance the above-described MYB77, are required in combination with MP/ARF5 to execute the correct auxin responses (Figure 2D).
Lately, crucial progress has been made in the understanding of the downstream events of the auxin pathway during lateral root initiation and development. In the root, periodic auxin responses can be observed during priming of pericycle cells for lateral root initiation in the basal meristem. The phasing of ∼15 h, which does not coincide with the circadian rhythm, correlates with the formation of consecutive lateral roots. The recurrence of the auxin response may be caused (at least in part) by periodic gravitropism-induced fluctuations in auxin redistribution within the root apex (De Smet et al., 2007). Apart from these observations, approximately half of the predominant expression patterns along the longitudinal root axis, including those of several genes of the auxin pathway, showed expression changes that fluctuate over developmental time (Brady et al., 2007).
ARF7 and ARF19, which appear to be able to interact with IAA3, IAA14, IAA19, and IAA28 (Tatematsu et al., 2004; Fukaki et al., 2005; Weijers et al., 2005; Okushima et al., 2007), are proposed to induce the expression of LATERAL ORGAN BOUNDARIES DOMAIN PROTEIN16 (LBD16) and LBD29 and thereby promote the formation of lateral roots. Importantly, ARF7 binds in vitro to LBD promoter fragments containing auxin-responsive elements (Figure 2E; Okushima et al., 2007). These data from Arabidopsis support the earlier finding of a similar relationship in rice (Oryza sativa; Inukai et al., 2005).
Another possible target for ARF transcription factors during lateral root development is PUCHI, which contains transcriptionally relevant auxin-responsive elements in its promoter region. PUCHI encodes a putative APETALA2/ETHYLENE-RESPONSIVE ELEMENT BINDING PROTEIN transcription factor and is involved in the regulation of lateral root morphogenesis via control of the pattern of cell divisions during the early stages of primordium development, most likely in combination with other position-specific factors (Hirota et al., 2007).
Auxin-related events within the root apex can be regarded as a paradigm for the interlinked actions of the different components of the auxin pathway. Their integration finally leads to a self-sustained auxin gradient (Grieneisen et al., 2007) being superimposed on a matrix of differentially susceptible cells, which is brought about by the PLETHORA (PLT) proteins (Galinha et al., 2007). The distributions of PLT proteins can be regarded as readout of the auxin gradient, since the correct expression of PLT genes depends on auxin distribution and response and at least indirectly on MP/ARF5 and NONPHOTOTROPIC HYPOCOTYL4/ARF7. Distinct PLT protein levels are eventually translated into distinct cellular responses. The different PLT levels appear to regulate processes as diverse as the acquisition of stem cell identity, the mitotic activity of stem cell daughters, and cell differentiation (Galinha et al., 2007). This provides a quantitative readout of auxin distribution, in contrast with the earlier described on/off situations. However, the relevant mechanisms underlying this output remain unclear.
CRUCIAL ROLE FOR AUXIN BIOSYNTHESIS
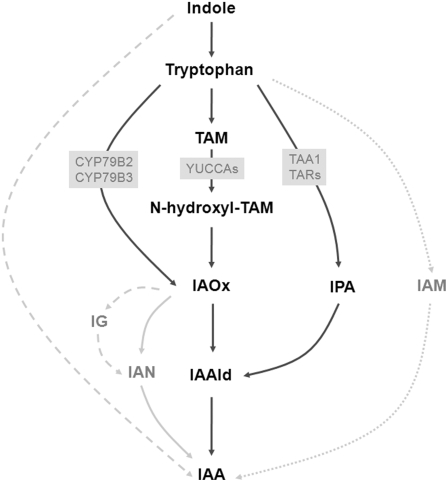
As described above, various layers of regulation are prevalent within the auxin pathway, but a pivotal prerequisite for the auxin pathway to function at all is the production of auxin itself by the plant, and the by far most important naturally occurring auxin is IAA. IAA biosynthesis is generally assumed to be accomplished via the multibranched Trp-dependent and the (scarcely investigated) Trp-independent pathway. Traditionally, the different branches of the Trp-dependent pathway are distinguished by more or less branch-specific intermediates, namely, indole-3-pyruvic acid (IPA), indole-3-acetamide (IAM), tryptamine (TAM), and indole-3-acetaldoxime (IAOx) (Figure 3). The IAM pathway, however, can probably be employed only by several pathogenic bacteria (reviewed in Woodward and Bartel, 2005).
Figure 3.
Proposed Pathways of IAA Biosynthesis in Arabidopsis.
The simplified scheme depicts the putative main pathways (black), some additional branches (gray), important precursors and intermediates, and the enzymes that are discussed in the text (gray boxes). The IAM pathway is most likely bacteria specific (dotted lines), and from IAOx via IG to IAN there are several intermediates that are not mentioned (hatched lines). CYP, cytochrome P450; IG, indole-3-methylglucosinolate; IAN, indole-3-acetonitrile; IAAld, indole-3-acetaldehyde.
The cytochrome P450s CYP79B2 and CYP79B3 have long been implicated in the biosynthesis of auxin, converting Trp into IAOx (Figure 3; reviewed in Woodward and Bartel, 2005). Recent insights have been gained with respect to the conversion of TAM into N-hydroxyl-TAM by the YUCCA family of flavin monooxygenases (Cheng et al., 2006, 2007; Kim et al., 2007) and of Trp into IPA by TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1 (TAA1) and its four homologs TRYPTOPHAN AMINOTRANSFERASE RELATED1 (TAR1) to TAR4 (Stepanova et al., 2008; Tao et al., 2008) (Figure 3).
The overexpression of the YUCCAs tested so far yielded essentially the same phenotype, whereas particular knockout combinations of the differentially expressed YUCCAs led to more specific defects. Corroborating the functional relevance of the differential expression of the various YUCCAs, different YUCCA knockout combinations led to a dampened auxin response only in those tissues where these particular YUCCAs are normally expressed (Cheng et al., 2006). The earliest developmental defect that was observed, eventually giving rise to an mp-like phenotype, is displayed by yuc1 yuc4 yuc10 yuc11 quadruple mutants (Cheng et al., 2007). These four YUCCAs are all expressed in the embryo, and YUC1 and YUC4 expression occurs in the apical part of the embryo (Cheng et al., 2007), the site that has long been proposed to be the major site of auxin production during embryogenesis.
Like the YUCCAs, TAA1 and its homologs that have been investigated until now are also expressed in discrete cell populations. For example, in roots, the expression of TAA1 is strongest in the quiescent center, whereas TAR2 is expressed in the provasculature. Knocking out both genes leads to the differentiation of the root meristematic cells and eventually to root growth arrest. In early embryogenesis, TAA1 is expressed apically and from the heart stage onwards in the apical and root meristematic regions. The importance of this embryonic expression of TAA1, and most likely also its homologs TAR1 and TAR2, for the production of auxin during embryogenesis can be inferred from the typical auxin-related mp-like phenotype in the taa1 tar1 tar2 triple mutant (Stepanova et al., 2008).
Notably, the yuc1 yuc4 yuc10 yuc11 quadruple and taa1 tar1 tar2 triple mutants extend the guild of mutants in the auxin pathway that show an impaired embryonic development and finally fail to initiate a primary root, joining the tir1 afb1 afb2 afb3 (Dharmasiri et al., 2005), pin1 pin3 pin4 (Friml et al., 2003), bdl (Hamann et al., 2002), and mp mutants (Hardtke and Berleth, 1998). The phenotypic equivalence of all these mutants emphasizes the importance of each level of the auxin pathway for a proper embryonic development. Therefore, both YUCCA-dependent and TAA1/TAR-dependent auxin biosynthesis contribute to the adequate supply of auxin during embryogenesis. Since there is no obvious genetic redundancy between the two pathways, it is unclear why mutants in the YUCCA-dependent and the TAA1/TAR-dependent auxin biosynthesis pathways display the same phenotype as mutants in auxin transport, perception, and response. It might be speculated that both pathways are coupled, thereby leading to especially low auxin levels when one pathway is knocked out. Mutant combinations between both pathways and/or analysis of enzyme activity when one pathway is knocked out could provide an answer to this intriguing question.
Although recent analyses have indicated that auxin transport is sufficient to generate and maintain auxin maxima and gradients (Grieneisen et al., 2007), these novel data strongly suggest the indispensability of site-specific auxin biosynthesis for establishing, maintaining, and/or enforcing auxin maxima. In addition to revealing the identity of important auxin biosynthesis proteins, these findings highlight the fundamental role of the spatio-temporal regulation of auxin biosynthesis gene expression in Arabidopsis.
CONCLUSIONS AND FUTURE PERSPECTIVES
To date, the simple molecule auxin continues to be the brightest star of plant development and of many physiological processes. The distribution of auxin and consequently the occurrence of auxin maxima and gradients within the plant appear to be the result of the interplay of auxin biosynthesis and transport. The subsequent different auxin responses depend on a tight regulation of gene expression, transcript abundance, and stability of proteins of the key components of the auxin signaling machinery. How all these regulatory processes are precisely controlled by the plant to generate specific outputs is being revealed step by step. The manner in which cells preserve an auxin-mediated developmental decision might be revealed soon, and several recently identified key players, such as those involved in histone modifications, are the ideal tools for this purpose. Additionally, the INDOLE-3-BUTYRIC ACID RESPONSE5 phosphatase was recently suggested to regulate auxin response in an entirely novel way (Strader et al., 2008), thereby opening up a completely new line of auxin research.
It appears that some sort of responsiveness to auxin evolved early in the lineage leading to land plants and seemingly several times independently in other lineages. However, the biological significance of these observations and whether, for example, red and brown algae have comparable auxin sensing and signaling mechanisms as those present in members of the land plant lineage are unclear. Rapidly expanding knowledge of plant genomes and the availability of new chemical compounds that specifically interfere with auxin responses will most likely shed more light on these questions in the near future.
Acknowledgments
We thank Dolf Weijers, Ute Voβ, three anonymous referees, and Nancy Eckardt for comments, suggestions, and editing. This work was supported by the European Molecular Biology Organization (EMBO-ALTF 108-2006) and the Deutsche Forschungsgemeinschaft (SFB 446)
References
- Abel, S. (2007). Auxin is surfacing. ACS Chem. Biol. 2 380–384. [DOI] [PubMed] [Google Scholar]
- Brady, S.M., Orlando, D.A., Lee, J.Y., Wang, J.Y., Koch, J., Dinneny, J.R., Mace, D., Ohler, U., and Benfey, P.N. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318 801–806. [DOI] [PubMed] [Google Scholar]
- Cheng, Y., Dai, X., and Zhao, Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20 1790–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y., Dai, X., and Zhao, Y. (2007). Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19 2430–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke, T.J., Poli, D., Sztein, A.E., and Cohen, J.D. (2002). Evolutionary patterns in auxin action. Plant Mol. Biol. 49 319–338. [PubMed] [Google Scholar]
- Covington, M.F., and Harmer, S.L. (2007). The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol. 5 e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet, I., and Jürgens, G. (2007). Patterning the axis in plants - Auxin in control. Curr. Opin. Genet. Dev. 17 337–343. [DOI] [PubMed] [Google Scholar]
- De Smet, I., et al. (2007). Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134 681–690. [DOI] [PubMed] [Google Scholar]
- Dharmasiri, N., Dharmasiri, S., Weijers, D., Lechner, E., Yamada, M., Hobbie, L., Ehrismann, J.S., Jürgens, G., and Estelle, M. (2005). Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9 109–119. [DOI] [PubMed] [Google Scholar]
- Friml, J., Vieten, A., Sauer, M., Weijers, D., Schwarz, H., Hamann, T., Offringa, R., and Jürgens, G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426 147–153. [DOI] [PubMed] [Google Scholar]
- Fukaki, H., Nakao, Y., Okushima, Y., Theologis, A., and Tasaka, M. (2005). Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J. 44 382–395. [DOI] [PubMed] [Google Scholar]
- Fukaki, H., Tameda, S., Masuda, H., and Tasaka, M. (2002). Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 29 153–168. [DOI] [PubMed] [Google Scholar]
- Fukaki, H., Taniguchi, N., and Tasaka, M. (2006). PICKLE is required for SOLITARY-ROOT/IAA14-mediated repression of ARF7 and ARF19 activity during Arabidopsis lateral root initiation. Plant J. 48 380–389. [DOI] [PubMed] [Google Scholar]
- Galinha, C., Hofhuis, H., Luijten, M., Willemsen, V., Blilou, I., Heidstra, R., and Scheres, B. (2007). PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449 1053–1057. [DOI] [PubMed] [Google Scholar]
- Gifford, M.L., Dean, A., Gutierrez, R.A., Coruzzi, G.M., and Birnbaum, K.D. (2008). Cell-specific nitrogen responses mediate developmental plasticity. Proc. Natl. Acad. Sci. USA 105 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb, B., Lanz-Garcia, C., Lian, Z., and Whetten, R. (2003). Aux/IAA gene family is conserved in the gymnosperm, loblolly pine (Pinus taeda). Tree Physiol. 23 1181–1192. [DOI] [PubMed] [Google Scholar]
- Grieneisen, V.A., Xu, J., Maree, A.F., Hogeweg, P., and Scheres, B. (2007). Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449 1008–1013. [DOI] [PubMed] [Google Scholar]
- Guilfoyle, T.J., and Hagen, G. (2007). Auxin response factors. Curr. Opin. Plant Biol. 10 453–460. [DOI] [PubMed] [Google Scholar]
- Hamann, T., Benkova, E., Bäurle, I., Kientz, M., and Jürgens, G. (2002). The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 16 1610–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke, C.S., and Berleth, T. (1998). The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, K., Tan, X., Zheng, N., Hatate, T., Kimura, Y., Kepinski, S., and Nozaki, H. (2008). Small-molecule agonists and antagonists of F-box protein-substrate interactions in auxin perception and signaling. Proc. Natl. Acad. Sci. USA 105 5632–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota, A., Kato, T., Fukaki, H., Aida, M., and Tasaka, M. (2007). The auxin-regulated AP2/EREBP gene PUCHI is required for morphogenesis in the early lateral root primordium of Arabidopsis. Plant Cell 19 2156–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai, Y., Sakamoto, T., Ueguchi-Tanaka, M., Shibata, Y., Gomi, K., Umemura, I., Hasegawa, Y., Ashikari, M., Kitano, H., and Matsuoka, M. (2005). Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 17 1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, M., Kaur, N., Garg, R., Thakur, J.K., Tyagi, A.K., and Khurana, J.P. (2006). Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct. Integr. Genomics 6 47–59. [DOI] [PubMed] [Google Scholar]
- Jin, Q., Scherp, P., Heimann, K., and Hasenstein, K.H. (2007). Auxin and cytoskeletal organization in algae. . Cell Biol. Int. 32 542–545. [DOI] [PubMed] [Google Scholar]
- Kalluri, U.C., Difazio, S.P., Brunner, A.M., and Tuskan, G.A. (2007). Genome-wide analysis of Aux/IAA and ARF gene families in Populus trichocarpa. BMC Plant Biol. 7 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.I., et al. (2007). yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiol. 145 722–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz, H.D., et al. (1998). Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 16 263–276. [DOI] [PubMed] [Google Scholar]
- Long, J.A., Ohno, C., Smith, Z.R., and Meyerowitz, E.M. (2006). TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312 1520–1523. [DOI] [PubMed] [Google Scholar]
- Muto, H., Watahiki, M.K., Nakamoto, D., Kinjo, M., and Yamamoto, K.T. (2007). Specificity and similarity of functions of the Aux/IAA genes in auxin signaling of Arabidopsis revealed by promoter-exchange experiments among MSG2/IAA19, AXR2/IAA7, and SLR/IAA14. Plant Physiol. 144 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser, J.L., Mockler, T.C., and Chory, J. (2004). Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2 E258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima, Y., Fukaki, H., Onoda, M., Theologis, A., and Tasaka, M. (2007). ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima, Y., et al. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 17 444–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde, P.J., et al. (2005). Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell 17 3282–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palenik, B., et al. (2007). The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl. Acad. Sci. USA 104 7705–7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paponov, I.A., Paponov, M.T., Teale, W., Menges, M., Chakrabortee, S., Murray, J.A.H., and Palme, K. (2008). Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol. Plant 1 321–337. [DOI] [PubMed] [Google Scholar]
- Rensing, S.A., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319 64–69. [DOI] [PubMed] [Google Scholar]
- Salmon, J., Ramos, J., and Callis, J. (2008). Degradation of the auxin response factor ARF1. Plant J. 54 118–128. [DOI] [PubMed] [Google Scholar]
- Sato, A., and Yamamoto, K.T. (2008). Overexpression of the non-canonical Aux/IAA genes causes auxin-related aberrant phenotypes in Arabidopsis. . Physiol. Plant. 133 397–405. [DOI] [PubMed] [Google Scholar]
- Schwartz, Y.B., and Pirrotta, V. (2007). Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 8 9–22. [DOI] [PubMed] [Google Scholar]
- Shin, R., Burch, A.Y., Huppert, K.A., Tiwari, S.B., Murphy, A.S., Guilfoyle, T.J., and Schachtman, D.P. (2007). The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 19 2440–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova, A.N., Robertson-Hoyt, J., Yun, J., Benavente, L.M., Xie, D.Y., Dolezal, K., Schlereth, A., Jürgens, G., and Alonso, J.M. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133 177–191. [DOI] [PubMed] [Google Scholar]
- Strader, L.C., Monroe-Augustus, M., and Bartel, B. (2008). The IBR5 phosphatase promotes Arabidopsis auxin responses through a novel mechanism distinct from TIR1-mediated repressor degradation. BMC Plant Biol. 8 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H., Basu, S., Brady, S.R., Luciano, R.L., and Muday, G.K. (2004). Interactions between auxin transport and the actin cytoskeleton in developmental polarity of Fucus distichus embryos in response to light and gravity. Plant Physiol. 135 266–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei, H., Hannon, M., and Long, J.A. (2008). TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319 1384–1386. [DOI] [PubMed] [Google Scholar]
- Tan, X., Calderon-Villalobos, L.I., Sharon, M., Zheng, C., Robinson, C.V., Estelle, M., and Zheng, N. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446 640–645. [DOI] [PubMed] [Google Scholar]
- Tanimoto, M., Jowett, J., Stirnberg, P., Rouse, D., and Leyser, O. (2007). pax1-1 partially suppresses gain-of-function mutations in Arabidopsis AXR3/IAA17. BMC Plant Biol. 7 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, Y., et al. (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133 164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu, K., Kumagai, S., Muto, H., Sato, A., Watahiki, M.K., Harper, R.M., Liscum, E., and Yamamoto, K.T. (2004). MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16 379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teale, W.D., Paponov, I.A., and Palme, K. (2006). Auxin in action: Signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 7 847–859. [DOI] [PubMed] [Google Scholar]
- Vieten, A., Sauer, M., Brewer, P.B., and Friml, J. (2007). Molecular and cellular aspects of auxin-transport-mediated development. Trends Plant Sci. 12 160–168. [DOI] [PubMed] [Google Scholar]
- Weijers, D., Benkova, E., Jäger, K.E., Schlereth, A., Hamann, T., Kientz, M., Wilmoth, J.C., Reed, J.W., and Jürgens, G. (2005). Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J. 24 1874–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers, D., Schlereth, A., Ehrismann, J.S., Schwank, G., Kientz, M., and Jürgens, G. (2006). Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev. Cell 10 265–270. [DOI] [PubMed] [Google Scholar]
- Wilmoth, J.C., Wang, S., Tiwari, S.B., Joshi, A.D., Hagen, G., Guilfoyle, T.J., Alonso, J.M., Ecker, J.R., and Reed, J.W. (2005). NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 43 118–130. [DOI] [PubMed] [Google Scholar]
- Woodward, A.W., and Bartel, B. (2005). Auxin: Regulation, action, and interaction. Ann. Bot. (Lond.) 95 707–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, H.S., Hackett, J.D., Ciniglia, C., Pinto, G., and Bhattacharya, D. (2004). A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 21 809–818. [DOI] [PubMed] [Google Scholar]