Abstract
Although calmodulin (CaM) is known to play multiple regulatory roles in eukaryotes, its direct function as transcriptional regulator is unknown. Furthermore, the physiological functions of CaM are largely unknown in plants. Here, we show that one of the four Arabidopsis thaliana CaM isoforms, CAM7, is a transcriptional regulator that directly interacts with the promoters of light-inducible genes and promotes photomorphogenesis. CAM7 overexpression causes hyperphotomorphogenic growth and an increase in the expression of light-inducible genes. Mutations in CAM7 produce no visible effects on photomorphogenic growth, indicating likely redundant gene functions. However, cam7 mutants display reduced expression of light-inducible genes, and cam7 hy5 double mutants show an enhancement of the hy5 phenotype. Moreover, overexpression of CAM7 can partly suppress the hy5 phenotype, indicating that the two factors work together to control light-induced seedling development. The mutational and transgenic studies, together with physiological analyses, illustrate the concerted function of CAM7 and HY5 basic leucine zipper transcription factor in Arabidopsis seedling development.
INTRODUCTION
Calmodulin (CaM) is ubiquitous in eukaryotes and is a highly conserved Ca2+ binding protein that plays multiple regulatory functions responding to a wide variety of stimuli (Berridge et al., 2000; Hepler, 2005). CaM has a common helix-loop-helix structure, the EF-hand, which is known to perform its regulatory function by modulating the activity of specific CaM binding proteins. CaM regulation of basic-helix-loop-helix transcription factors has been reported, where CaM inhibits the DNA–protein interactions by competing with the DNA binding domains of the basic-helix-loop-helix proteins (Corneliussen et al., 1994). Interestingly, recent studies have shown that some proteins with EF-hands have the ability to directly interact with DNA. For example, the human DRE antagonist modulator (DREAM) has four EF-hands and specifically interacts with the DNA DRE element (Carrion et al., 1999; Gilchrist et al., 2001; Craig et al., 2002). Various studies have shown that Ca2+/CaM is involved in multiple signaling pathways in plants (Miller and Sanders, 1987; Braam and Davis, 1990; Knight et al., 1991; Szymanski et al., 1996; Yang and Poovaiah, 2002; Yoo et al., 2004). The Arabidopsis thaliana genome contains seven CAM genes that encode only four protein isoforms: CAM1/CAM4, CAM2/CAM3/CAM5, CAM6, and CAM7. The CAM7 protein sequence shows the most similarity to consensus among all the members of the family, but all the CAM isoforms are very highly conserved. CAM1/CAM4 differs from CAM7 by four amino acids, whereas CAM2/3/5 and CAM6 differ from CAM7 by a single amino acid substitution (McCormack et al., 2005).
Arabidopsis seedlings grow with two distinct developmental patterns in the presence and absence of light (Nagy and Schaefer, 2002; Chen et al., 2004; Huq and Quail, 2005). The dark-grown seedlings exhibit elongated hypocotyls and closed cotyledons with apical hooks, designated as skotomorphogenic growth. When exposed to light, seedlings grow with a short hypocotyl and open and expanded cotyledons, known as photomorphogenic growth. The expression of light-inducible genes, which remains suppressed in the dark, is strongly induced during photomorphogenesis. A complex molecular network operates to sense the dark–light transitions and regulate the seedling morphology and gene expression accordingly (Jiao et al., 2007). The basic leucine zipper transcription factor, Long Hypocotyl 5 (HY5), plays an important role in the transition from skotomorphogenesis to photomorphogenesis. The loss-of-function mutants of HY5 display partial photomorphogenic growth at various wavelengths of light with reduced expression of light-regulated genes (Oyama et al., 1997; Ang et al., 1998; Chattopadhyay et al., 1998). The abundance of HY5 protein has been correlated with the extent of photomorphogenic growth (Osterlund et al., 2000). Recently, genome-wide promoter target studies have revealed that there are >3000 chromosomal sites in the Arabidopsis genome that have putative HY5 binding targets (Lee et al., 2007)
The homeostasis of Ca2+ has been shown to be associated with blue/UV-A light–induced gene expression (Long and Jenkins, 1998). A recent genetic study using SHORT UNDER BLUE LIGHT1 (SUB1) has suggested the possible involvement of local Ca2+ concentration change in phytochrome- and cryptochrome-mediated light signaling (Guo et al., 2001). Biochemical and pharmacological studies have revealed three branched pathways of light-induced gene expression. In one of these pathways, CaM has been shown to be involved in the regulation CAB gene expression (Neuhaus et al., 1993, 1997; Bowler et al., 1994). All these studies suggest that Ca2+/CaM is involved in light-mediated seedling development and gene expression. However, the molecular and physiological function of CaM or structurally related Ca2+ binding protein, which interprets and specifically transduces the information into appropriate cellular responses, remains largely unknown (Veitia, 2005).
The activity of a CAB1 minimal promoter containing an essential Z-box light-responsive element (LRE) is controlled by HY5 (Yadav et al., 2002). Recently, two Z-box binding transcription factors, ZBF1/MYC2 and ZBF2/GBF1, have been identified from a ligand binding screen and shown to function in cryptochrome-mediated blue light signaling (Yadav et al., 2005; Mallappa et al., 2006). In this study, we demonstrate the functional relevance of ZBF3, encoding CAM7, in light-mediated seedling development and gene expression.
RESULTS
CAM7 Specifically Binds to the Z-/G-Box of Light-Regulated Promoters
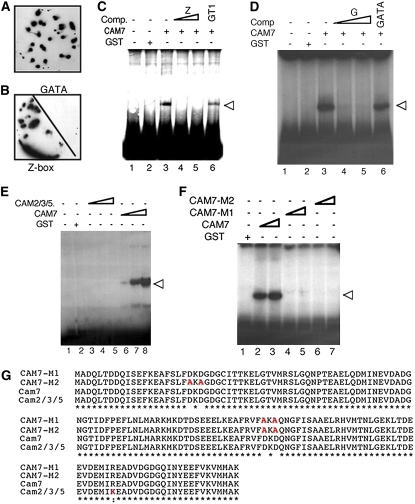
We had identified and cloned ZBF3/CAM7, which was represented by three independent cDNA clones in a ligand binding screen (Yadav et al., 2005). The DNA binding analyses, which examined binding of labeled probe DNA sequences to proteins immobilized on nylon membranes, revealed that ZBF3/CAM7 was able to specifically bind to the Z-box LRE (Figures 1A and 1B). To further examine the results obtained from these analyses, we performed electrophoretic mobility shift assays (EMSAs) using CAB1 minimal promoter containing an essential Z-box and purified glutathione S-transferase-CAM7 (GST-CAM7) fusion protein. As shown in Figure 1C, GST-CAM7 was able to bind to the Z-box of CAB1 minimal promoter. Excess unlabeled Z-box DNA, but not a nonspecific competitor (GT1 LRE; Chattopadhyay et al., 1998), was able to compete for the binding activity of GST-CAM7. Since recent studies have suggested that the Z- and G-box LREs are functionally equivalent (Yadav et al., 2005; Mallappa et al., 2006), we also investigated the binding ability of CAM7 to the G-box. As shown in Figure 1D, CAM7 could specifically bind to the essential G-box of RBCS-1A minimal promoter. Taken together, these results suggest that CAM7 specifically binds to the Z-/G-box of light-regulated CAB1 and RBCS-1A minimal promoters.
Figure 1.
CAM7 Binds to the Essential Z-/G-Box of CAB1 or RBCS-1A Minimal Promoter.
(A) Identification of CAM7 in a ligand binding (protein/DNA gel blot) screen. The blotted nylon membrane (containing protein-expressing plaques from the tertiary screen for proteins that bind to the Z-box) was probed with the radioactively labeled Z-box LRE (Yadav et al., 2005).
(B) The specificity of interaction of CAM7 to the Z-box. The blotted nylon membrane was cut into two halves and probed with the Z-box or GATA LRE (Yadav et al., 2002).
(C) EMSAs showing GST-CAM7 (CAM7) specifically binds to the Z-box of 189-bp CAB1 minimal promoter (Yadav et al., 2005). Approximately 200 ng of recombinant protein was added (lanes 3 to 6) to radioactively labeled CAB1 promoter fragment. Approximately 500 ng GST protein was added in lane 2. The triangle indicates the increased amount of unlabeled Z-box DNA added (50 and 100 molar excess in lanes 4 and 5, respectively) to the reaction as competitor (Comp.). In lane 6, 100 molar excess GT1 LRE (Yadav et al., 2002) was added. The presence of CAM7 or GST protein is indicated by plus signs in their respective rows. The minus signs indicate the absence of competitors CAM7 or GST. The arrowhead indicates the protein–DNA complex.
(D) EMSA showing GST-CAM7 (CAM7) protein specifically binds to the essential G-box of 196-bp RBCS-1A minimal promoter (Chattopadhyay et al., 1998). For experimental detail, see (C). In this case, the unlabeled competitor DNA is G-box LRE.
(E) EMSA of CAM2/3/5 to the CAB1 minimal promoter. Approximately 200 ng, 1 μg, and 3 μg (lanes 3 to 5), and 100, 200, and 300 ng (lanes 6 to 8) of recombinant proteins were added to radioactively labeled CAB1 minimal promoter. For experimental detail, see (C).
(F) EMSAs showing that CAM7, but not CAM7-M1 and CAM7-M2, is able to bind to CAB1 minimal promoter. Approximately 200, 300, 200, 500, 200, and 500 ng (lanes 2 to 7, respectively) of recombinant proteins were added to radioactively labeled CAB1 minimal promoter. For experimental detail, see (C).
(G) Amino acid sequences of CAM7, CAM2/3/5, and site-directed mutagenesis products of CAM7 (CAM7-M1 and CAM7-M2) are shown. The amino acid substitutions are shown in red.
It has been postulated that substitution of amino acid in the EF-hand could contribute to select the target specificity of CaM (McCormack et al., 2005). CAMs have highly conserved amino acid sequences, and the amino acid sequence of the CAM2/3/5 isoform differs from CAM7 by a single amino acid substitution (Figure 1G). To determine whether CAM2/3/5 was also able to interact with the Z-box, EMSAs were performed using purified GST-CAM3 fusion protein and CAB1 minimal promoter as probe. However, no DNA–protein complex was detected; thereby, these results suggest that CAM2/3/5 is unable to bind to the CAB1 minimal promoter (Figure 1E). To further test this observation, we generated mutated versions of CAM7, CAM7-M1 and CAM7-M2, by site-directed mutagenesis. Whereas two Asp residues of CAM7 were substituted by Ala in CAM7-M1, four Asp residues were substituted by Ala in CAM7-M2 protein (Figure 1G). We used purified GST-CAM7-M1 or GST-CAM7-M2 fusion proteins in EMSAs (see Supplemental Figure 1 online). None of these mutated versions of CAM7 was also able to bind to the CAB1 minimal promoter (Figure 1F). Taken together, these results suggest that CAM7 is likely to be a unique member of the CAM gene family that directly binds to the Z-/G-box of light-regulated promoters.
Overexpression of CAM7 Leads to Hyperphotomorphogenic Growth Irrespective of Light Qualities
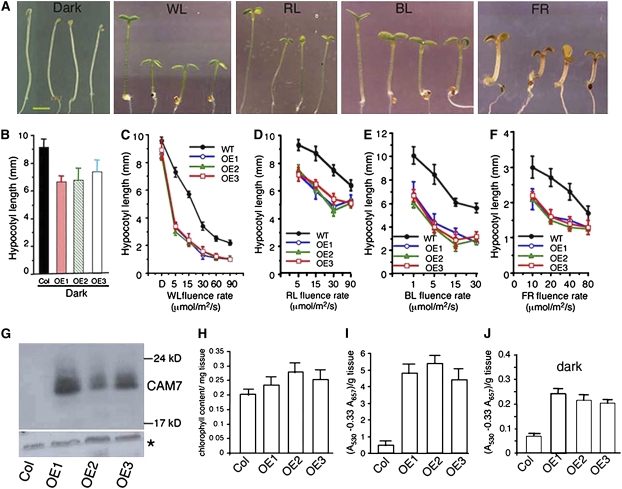
To investigate the physiological function of CAM7 in light-mediated seedling development, 27 Arabidopsis transgenic lines overexpressing CAM7 fused to three copies of c-Myc epitope were generated. The c-Myc epitope was fused to either the C- or N-terminal end of the CAM7 protein, and the proteins showed high levels of accumulation in the transgenic lines (Figure 2G). The transgenic seedlings exhibited short hypocotyl phenotype at various wavelengths of light, including red (RL), far-red (FR), and blue light (BL) (Figure 2A). Measurements of hypocotyl length revealed that the enhanced inhibition of hypocotyl elongation was more evident at lower fluence rates especially in RL- or FR-grown seedlings (Figures 2C to 2F). Strikingly, the transgenic seedlings displayed a weak photomorphogenic growth with shorter hypocotyl and partly opened cotyledons without apical hooks in the darkness (Figures 2A and 2B). The overexpresser transgenic seedlings also showed higher levels of chlorophyll in light and of anthocyanin in both dark and light growth conditions (Figures 2H to 2J). Taken together, these results suggest that overexpression of CAM7 induces a partial photomorphogenic development in the dark and also promotes photomorphogenic growth in various wavelengths of light.
Figure 2.
CAM7 Promotes Photomorphogenic Growth.
(A) The visible phenotypes of the seedlings grown in constant dark, WL (15 μmol/m2/s), RL (30 μmol/m2/s), BL (20 μmol/m2/s), or FR (40 μmol/m2/s) are shown. In each panel, 6-d-old wild-type (Columbia [Col]) and CAM7 overexpresser transgenic seedlings (OE1, OE2, and OE3 in Col background) are shown from left to right, respectively. OE1 and OE2 contain CAM7 with c-Myc tagged at the N-terminal end, whereas OE3 contains CAM7 with c-Myc tagged at the C-terminal end. Bar = 1 mm.
(B) to (F) Quantification of hypocotyl length of 6-d-old seedlings grown in constant dark or at various fluences of WL, RL, BL, or FR. Approximately 25 to 30 seedlings were used for the measurement of hypocotyl length. The error bars indicate sd.
(G) Immunoblot (using anti-c-Myc antibodies) of 20 μg of total protein prepared from wild-type (Col) or overexpresser transgenic plants. The asterisk in the bottom panel shows a cross-reacting band in the same gel as loading control.
(H) The level of total chlorophyll content in 6-d-old wild-type (Col) or transgenic seedlings grown in WL (30 μmol/m2/s) is shown.
(I) and (J) Accumulation of anthocyanin in 6-d-old wild-type or transgenic seedlings grown in WL (30 μmol/m2/s) or dark, respectively. Approximately 30 to 40 seedlings were used for the measurement of chlorophyll or anthocyanin accumulation. The error bars indicate sd.
CAM7 Interacts with CAB1 Minimal Promoter in Vivo and Promotes Light-Induced Gene Expression
To determine whether CAM7 was able to promote the transcriptional activity of light-regulated genes, we performed RNA gel blot analysis using transgenic seedlings grown in constant dark or light. The expression of CAB was strongly elevated in transgenic seedlings compared with the wild type in white light (WL) (Figures 3A and 3E). Whereas very little expression, as expected, of CAB was detected in wild-type background in the dark, the expression was strikingly elevated in transgenic seedlings. To determine the light-controlled expression of the CAB or RBCS gene, 5-d-old dark-grown seedlings were transferred to WL for various time points. The level of expression was further elevated in transgenic seedlings compared with wild-type background (Figure 3B). Taken together, these results provide evidence that CAM7 acts as a positive regulator of CAB and RBCS gene expression.
Figure 3.
CAM7 Interacts with CAB1 Promoter in Vivo and Promotes Light-Regulated Gene Expression.
(A) The RNA gel blot shows the level of CAB1 gene expression in 6-d-old wild-type (Col) and CAM7 overexpresser transgenic seedlings (OE1, OE2, and OE3) grown in dark or WL (30 μmol/m2/s). Ten micrograms of total RNA was loaded onto each lane. 18S rRNA has been shown as loading control. The numbers indicate the relative mRNA levels. To quantify the RNA gel blot data, the intensity of each band was quantified by the Fluor-S-MultiImager (Bio-Rad), and ratios of CAB1 versus its corresponding rRNA band were determined and plotted (Fluor-S-MultiImager; Bio-Rad).
(B) The RNA gel blot results (quantified as described above) show light-mediated induction of CAB1 and RBCS gene expression in wild-type (Col) and OE1 transgenic seedlings grown in dark (0) for 5 d and then transferred to WL (30 μmol/m2/s) for various time points.
(C) ChIP assays of CAB1 promoter from OE1, OE2, or OE3 transgenic seedlings using antibodies to c-Myc. The light-inducible NIA2 promoter fragment, which does not contain any Z- or G-box, was used as a control. Results of real-time quantitative PCR are presented as the ratio of the amount of DNA immunoprecipitated from overexpresser transgenic seedlings to nontransgenic control plants.
(D) Expression of CAB1-GUS or CAB1m-GUS reporter gene (reflected by GUS activities) relative to the 35S-GUS internal control in Arabidopsis protoplasts made from wild-type or CAM7 overexpresser (OE1) plants. Error bars indicate se (n = 5). The experiment was repeated three times.
(E) Normalized graph of the data in (A) (quantified as described in [A]).
We performed chromatin immunoprecipitation (ChIP) experiments to determine whether CAM7 binds to CAB1 minimal promoter in vivo. The CAM7-c-Myc fusion protein in transgenic plants was immmunoprecipitated by antibody to c-Myc. The genomic DNA fragments that coimmunoprecipitated with CAM7-c-Myc were analyzed by real-time quantitative PCR. The analyses of these data revealed that the amount of DNA fragment of CAB1 promoter coimmunoprecipitated from the transgenic seedlings was >30-fold higher than that precipitated from the nontransgenic seedlings, and ∼10-fold higher than the NIA2 promoter, which is induced by light but does not contain any Z-/G-box LRE (Figure 3C). These results demonstrate that CAM7 binds to the CAB1 minimal promoter in vivo.
To determine whether CAM7 binding to the Z-box is required for the in vivo activation of CAB1 promoter, we used CAB1 minimal promoter containing either wild type or mutated Z-box fused to the β-glucuronidase reporter gene construct (CAB1 promoter-GUS or CAB1m promoter-GUS). We examined the activity of these promoters in transiently transformed protoplasts made from wild-type or CAM7 overexpresser transgenic plants (OE1). The activity of CAB1 promoter was increased by more than twofold in OE1 compared with wild-type background, confirming that CAM7 promotes CAB1-GUS expression. By contrast, overexpression of CAM7 was unable to activate the CAB1m promoter in vivo (Figure 3D). Together, the above results demonstrate that CAM7 acts as a transcriptional activator of CAB1 promoter in vivo and the Z-box is essential for such activation mediated by CAM7 protein.
Overexpression of CAM7-M2 Confers Possible Dominant-Negative Effects
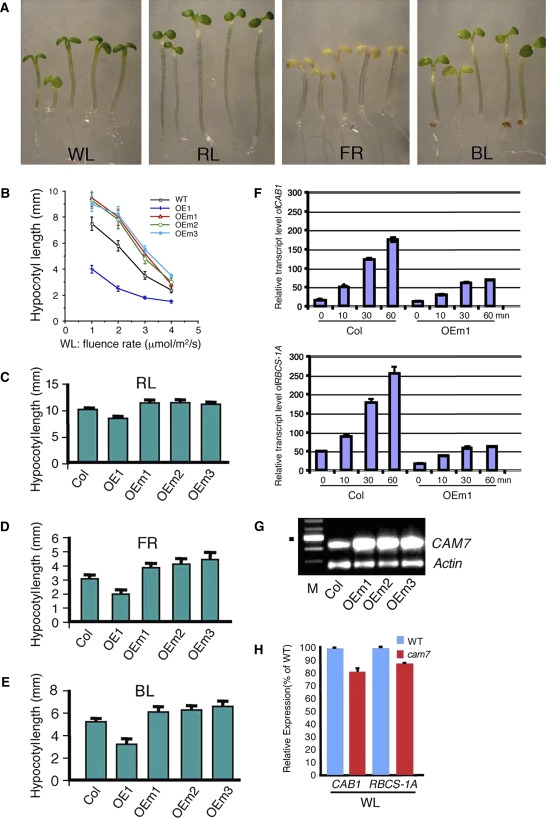
To investigate the physiological function of the mutated version of CAM7 protein, which lost DNA binding ability due to amino acid substitutions (Figure 1G), we constructed a series of 32 transgenic lines overexpressing CAM7-M2 (Figure 4G). Examination of 2- to 6-d-old transgenic seedlings did not show any altered morphology in the dark. However, 6-d-old WL-grown transgenic seedlings displayed a longer hypocotyl compared with the corresponding wild type (Figure 4A). Furthermore, the transgenic seedlings displayed elongated hypocotyls in all light conditions tested compared with the corresponding wild type (Figure 4A). The measurements of hypocotyl length revealed significant reduction (P value < 0.01; n = 3) in light-mediated inhibition of hypocotyl elongation in transgenic seedlings compared with the wild type in different light conditions (Figures 4B to 4E). The elongated hypocotyl phenotype conferred by the overexpression of CAM7-M2 could be attributable to dominant-negative interference of the light signaling pathways by CAM7-M2 protein. To determine whether overexpression of CAM7-M2 leads to similar effects on light-regulated gene expression, we performed quantitative real-time PCR experiments of CAB1 and RBCS-1A genes. The rate of light-mediated induction of CAB1 and RBCS-1A genes was significantly reduced in transgenic seedlings compared with wild-type background (Figure 4F).
Figure 4.
Overexpression of CAM7-M2 Results in Suppression of Photomorphogenic Growth Irrespective of Light Qualities.
(A) Visible phenotypes of 6-d-old wild-type (Col), and transgenic seedlings grown in various light conditions. In each panel, wild-type (Col), OE1, OEm1, OEm2, and OEm3 seedlings are shown from left to right, respectively.
(B) to (E) Quantification of hypocotyl length of 6-d-old seedlings grown in WL, RL (60 μmol/m2/s), FR (40 μmol/m2/s), or BL (20 μmol/m2/s). For each measurement of hypocotyl length, 25 to 30 seedlings were used. The error bars indicate sd. All the samples were significantly different from the wild type at each light condition (P < 0.01; n = 3).
(F) The abundance of CAB1 and RBCS-1A transcripts in total RNA from wild-type (Col) and CAM7-M2 overexpresser transgenic seedlings (OEm1) grown in dark (0) for 5 d and then transferred to white light (30 μmol/m2/s) for various time points was determined by quantitative real-time PCR, and the transcript levels were normalized to the level of ACTIN2 transcript abundance. Error bars represent sd (P < 0.01 between Col versus OEm1 at each time point exposed to light; n = 3).
(G) RT-PCR results (using CAM7-specific primers) show the level of expression of CAM7-M2 in overexpresser transgenic lines (OEm1, OEm2, and OEm3) or in the corresponding wild-type (Col) background. Actin band shows the loading control. M indicates molecular weight markers (100- bp ladder), and the dot shows a DNA fragment of 500 bp.
(H) Real-time quantitative PCR results show the relative expression of CAB1 and RBCS-1A in 6-d-old wild-type versus cam7 mutant seedlings (P < 0.03 between the wild type versus cam7 mutants; n = 4) grown in WL (30 μmol/m2/s).
Loss-of-Function Mutants of CAM7 Have Reduced Expression of Light-Inducible Genes
To determine whether loss of CAM7 function would lead to reduced photomorphogenesis, we searched for mutants in T-DNA knockout collections (Alonso et al., 2003). We identified two such T-DNA insertion knockout lines (cam7-1 and cam7-2) and performed PCR genotyping analyses to identify plants homozygous or heterozygous for a cam7 mutation (see Supplemental Figure 2 online). The segregation ratios of self-fertilized plants heterozygous for cam7, determined by the genotyping PCR on T2 progeny, suggested that a single T-DNA locus was present in each of the cam7 mutant lines. The junctions of T-DNA and CAM7 were amplified by PCR, and the DNA sequence analyses revealed that the T-DNA was inserted in nucleotide position 225 and 113 bp upstream to the ATG codon of CAM7 in cam7-1 and cam7-2 mutants, respectively (see Supplemental Figure 2A online). RT-PCR analyses were unable to detect any CAM7 mRNA in either of the cam7 mutant lines (see Supplemental Figure 2B online).
When the growth of cam7 mutant seedlings was examined in dark and in various light conditions, cam7 mutants grew normally in the dark and at various wavelengths of light tested, showing no sign of altered photomorphogenic growth (Figures 5A and 5C). These results indicate that CAM7-mediated inhibition of hypocotyl elongation is functionally redundant. However, the level of CAB1 and RBCS-1A expression was compromised in cam7 mutants (P value < 0.03; n = 4), thereby suggesting that CAM7 is required for the optimum expression of CAB1 and RBCS-1A genes (Figure 4H).
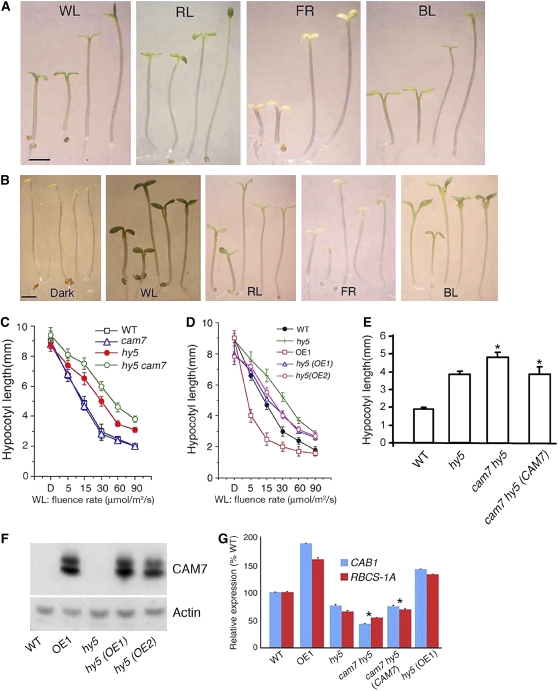
Figure 5.
The Elongated Hypocotyl Phenotype of hy5 Is Enhanced in cam7 hy5 Double Mutants.
(A) The visible phenotypes of 6-d-old cam7 hy5 double mutant seedlings grown in WL (30 μmol/m2/s), RL (60 μmol/m2/s), FR (40 μmol/m2/s), or BL (20 μmol/m2/s) are shown. In each panel, wild-type (segregated wild type in F2), cam7-1, hy5, and cam7-1 hy5 seedlings are shown from left to right. Bar = 1 mm.
(B) The visible phenotype of 6-d-old hy5 transgenic seedlings (hy5 [OE1] and hy5 [OE2]), containing 35S promoter-c-Myc-CAM7 transgene, grown in dark, WL (30 μmol/m2/s), RL (60 μmol/m2/s), FR (40 μmol/m2/s), or BL (20 μmol/m2/s) is shown. In each panel, wild-type, OE1, hy5, hy5 (OE1), and hy5 (OE2) seedlings are shown from left to right. Bar = 1 mm.
(C) and (D) Quantification of hypocotyl length in dark (D; on x-axis label) or various fluence rates of WL. Approximately 25 to 30 seedlings were used for the measurement of hypocotyl length. The error bars indicate sd.
(E) Quantification of hypocotyl length of 6-d-old seedlings grown in WL (60 μmol/m2/s). Approximately 25 to 30 seedlings were used for the measurement of hypocotyl length. The error bars indicate sd (P < 0.01, between cam7 hy5 versus cam7 hy5 [CAM7] as marked by asterisks; n = 3).
(F) Immunoblot (using c-Myc antibodies) of 20 μg of total protein prepared from wild-type, OE1, hy5, and hy5 (OE1), and hy5 (OE2) seedlings. The actin bands indicate approximate equal loading.
(G) Real-time quantitative PCR results (P < 0.01; between cam7 hy5 versus cam7 hy5 [CAM7] as marked by asterisks; n = 3) show the relative expression of CAB1 and RBCS-1A in 6-d-old seedlings grown in WL (30 μmol/m2/s).
CAM7 and HY5 Function in an Independent and Interdependent Manner to Promote Photomorphogenesis
HY5 is thus far the only known transcription factor in light signaling that promotes photomorphogenesis in RL, FR, and BL. Since higher-level accumulation of CAM7 also leads to hyperphotomorphogenic growth irrespective of light qualities, we asked whether HY5 and CAM7 are functionally interrelated. We constructed cam7 hy5 double mutants and examined the genetic interactions between cam7 and hy5. Similar to hy5 or cam7 single mutants, cam7 hy5 double mutants did not show any altered growth in the dark. However, the characteristic long hypocotyl phenotype of hy5 in WL irradiation was further enhanced in cam7 hy5 double mutants, exhibiting a super tall phenotype (Figure 5A). Furthermore, as shown in Figure 5A, cam7 hy5 double mutants also displayed reduced sensitivity in RL, FR, and BL compared with hy5 single mutants. Measurements revealed that the hypocotyl length of cam7 hy5 double mutants was strikingly increased compared with hy5 or cam7 alone, indicating a synergistic effect of cam7 and hy5 mutations on hypocotyl length irrespective of light qualities (Figure 5C; see Supplemental Figures 3A to 3C online). The expression of light-regulated genes has been shown to be downregulated in hy5 mutants (Ang et al., 1998; Chattopadhyay et al., 1998). When tested, the level of CAB1 and RBCS-1A gene expression was found to be further reduced in cam7 hy5 double mutants compared with the cam7 or hy5 mutant background (Figure 5G). A genomic fragment containing CAM7 and 1.5 kb of its upstream sequence was introduced into the cam7 hy5 double mutant plants for a complementation test. The transgenic seedlings did not display the super-tall phenotype, and the expression of light-regulated genes was also restored to hy5 mutant levels (Figures 5E and 5G). These results confirm that the observed super-tall phenotype of cam7 hy5 double mutants was caused by the additional loss of CAM7 function.
To further test this observation, we introduced the 35S-CAM7-c-Myc transgene from the overexpresser transgenic lines (OE1 and OE2) into hy5 mutant background by genetic crosses. The higher level of CAM7 protein in hy5 transgenic seedlings was indeed able to suppress the elongated hypocotyl phenotype of hy5 (Figures 5B, 5D, and 5F). When examined under various wavelengths of light, the hy5 phenotype was significantly suppressed in transgenic hy5 seedlings grown in RL, FR, and BL (Figure 5B; see Supplemental Figures 3D to 3F online). Furthermore, similar to OE1, hy5 transgenic seedlings also displayed partial photomorphogenic growth with slightly reduced hypocotyl length in the darkness. However, unlike OE1, the cotyledons remained closed with apical hooks in hy5 transgenic seedlings in the darkness (Figures 5B, dark, and 5D). The lower-level expression of light-inducible genes, such as CAB1 and RBCS-1A, in hy5 mutants was also restored in hy5 transgenic seedlings (Figure 5G). Taken together, these results suggest that CAM7 and HY5 function in an independent and interdependent manner to promote photomorphogenic growth and light-regulated gene expression.
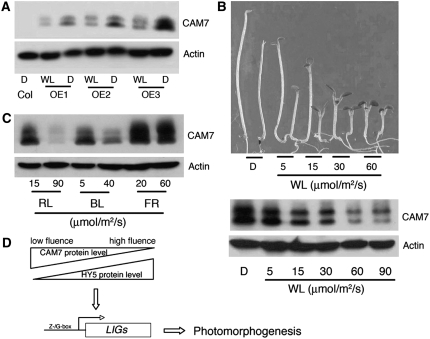
The Accumulation of CAM7 Protein Is Dependent on Light Intensity
Since abundance of HY5 protein has been correlated with the extent of photomorphogenic growth, we asked whether CAM7 protein also accumulated in a similar fashion correlating with photomorphogenic growth. To address this question, we first used 6-d-old constant dark- or WL-grown CAM7-c-Myc overexpresser transgenic seedlings for immunoblot analysis. The accumulation of CAM7 protein was significantly reduced in WL compared with dark-grown seedlings (Figure 6A), which is contrary to the accumulation pattern of HY5 protein under similar conditions.
Figure 6.
The Accumulation of CAM7 Protein Is Altered in Dark and Light.
(A) Six-day-old wild-type (Col) or OE1 transgenic seedlings (OE1, OE2, and OE3) grown in constant dark (D) or WL were used for immunoblot analyses of CAM7 protein (using c-Myc polyclonal antibodies). The actin bands (probed by anti-actin monoclonal antibodies) indicate approximate equal loading.
(B) Top panel: The visible phenotypes of seedlings grown in constant dark or various fluences of WL are shown. Six-day-old wild-type (Col) and CAM7 overexpresser transgenic seedlings (OE1) are shown alternatively from left to right. Bottom panel: The level of CAM7 protein in OE1 transgenic seedlings as described in the top panel. The actin bands indicate approximate equal loading.
(C) Six-day-old OE1 transgenic seedlings grown at various wavelengths of light and at different fluence rates were used for immunoblot detection of CAM7 protein. The actin bands indicate approximate equal loading.
(D) A schematic model for CAM7- and HY5-mediated regulatory pathways. The triangles indicate light intensity–dependent gradual decrease or increase of CAM7 or HY5 protein, respectively. LIGs, light-inducible genes.
We then examined whether the reduced accumulation of CAM7 protein in WL was dependent on light intensity. As shown in Figure 6B (bottom panel), the accumulation of CAM7 protein decreased with increase in fluence rates of WL in overexpresser transgenic lines (OE1). Whereas the level of accumulation of CAM7 protein was slightly reduced at 5 μmol/m2/s, it was further reduced at 15 or 30 μmol/m2/s and strikingly reduced at 60 μmol/m2/s or higher fluence rates of WL. The enhanced inhibition in hypocotyl elongation, caused by higher level of CAM7 protein, was also gradually reduced with higher fluence rates of WL, suggesting a likely correlation between the level of CAM7 protein and the extent of hyperphotomorphogenic growth of the transgenic seedlings. To determine whether WL-mediated reduction of CAM7 protein is wavelength specific, we examined the level of CAM7 protein in 6-d-old seedlings grown at low or relatively high intensities of RL, FR, and BL. As shown in Figure 6C, similar to WL, CAM7 protein was strikingly reduced at higher fluence rates of RL and BL, although the accumulation of CAM7 was largely maintained at higher fluence rates of FR.
DISCUSSION
The primary structures of CaMs are highly similar in plants and animals with respect to their Ca2+ binding loops and E and F helices. The topology of the EF-hand motif of CaM is similar to the helix-turn-helix DNA binding domain of various transcription factors that can recognize the major groove of DNA. Examination of amino acid sequences of all four subgroups of Arabidopsis CaM family reveals that all CAM proteins, except CAM7, have at least one amino acid substitution compared with CAM7 (McCormack et al., 2005). The binding of CAM7, but not CAM2/3/5, CAM7-M1, or CAM7-M2, to the Z-/G-box of light-regulated promoters supports the notion that although four Arabidopsis CaM isoforms have very similar amino acid sequences, substitution of amino acids in the EF-hand region may contribute to select target specificity. Consistent with this notion, the human DREAM, which has four consensus EF-hands, specifically binds to the DRE element (Carrion et al., 1999). It is worth mentioning here that the Z-box (ATACGTGT) and G-box (CACGTG) motifs recognized by CAM7 in this study have very similar (or identical in the case of the G-box) sequence to the recently identified Ca2+-responsive element (CACGTG[T/C/G]) (Kaplan et al., 2006). A detailed nuclear magnetic resonance study using various isoforms and mutated versions of CAM7 in the presence or absence of Z-/G-box would address the question of how amino acid substitution alters the target specificity of CAM7.
It has been shown that change in Ca2+ flux plays important regulatory functions in sensing dark–light transition of Arabidopsis seedlings (Sai and Johnson, 2002). Furthermore, the role of Ca2+/CaM in phytochrome signaling has been postulated, and the potential connection between light and Ca2+/CaM signaling has started emerging, especially with the identification and functional characterization of SUB1, a Ca2+ binding protein operative in both cryptochrome- and phytochrome-mediated light signaling (Guo et al., 2001). Recent studies have also shown the involvement of phototropins in blue light–mediated Ca2+ and H+ fluxes (Babourina et al., 2002). The data in this study collectively provide evidence that CAM7 acts as a transcriptional regulator and promotes photomorphogenic growth and light-regulated gene expression. However, the possible role of Ca2+ or other divalent cation in CAM7-mediated Arabidopsis seedling development remains to be elucidated. For example, recent studies have suggested that Mg2+ may structurally bridge the DREAM protein to DNA, whereas Ca2+-induced dimerization of DREAM disrupts DREAM–DNA interactions (Osawa et al., 2005).
It has been predicted that, similar to other proteins that have interacting protein partners, mutations in CaM might result in dominant-negative effects (Veitia, 2005). Recent protein microarray analysis data also support such prediction (Popescu et al., 2007). The elongated hypocotyl phenotype and reduced expression of light-regulated genes conferred by the overexpression of CAM7-M2 could be attributed to dominant-negative interference of the light signaling pathways by CAM7-M2 protein. The alternate possibility of cosuppression of the endogenous CAM7 gene expression caused by overexpression of CAM7-M2 seems to be less likely since the cam7 mutants do not display any altered photomorphogenic growth. Furthermore, cam7 cam3 double mutants also do not display any altered morphology. However, it could be possible that overexpression of CAM7-M2 cosuppresses endogenous CAM7 gene expression and one or more additional genes of the seven-member gene family of CAM (except CAM3) or the 50-member gene family of CML (CaM like) (McCormack et al., 2005). In either case, further study on identification and functional characterization of such genes is required to test the possibility.
HY5 is considered to be an important signal integration point of major branches downstream to all known photoreceptors (Jiao et al., 2007). The Z- and G-box have been shown to be functionally equivalent in the context of ZBF1/MYC2 and ZBF2/GBF1 transcription factors (Yadav et al., 2005; Mallappa et al., 2006). Recently, genome-wide promoter target studies using ChIP-chip analysis have revealed that the Z- and G-box sequences are enriched in the promoter region of HY5 target genes (Lee et al., 2007). Therefore, it is possible that CAM7 and HY5 regulate the expression of a common set of downstream genes in light signaling and have partially overlapping functions in light-dependent development. Although mutations in CAM7 do not cause any visible morphological defects, the expression of light-regulated genes is downregulated in cam7 mutants (Figure 4H). It could be envisioned that a coactivator (possibly one of the other CAMs or CAM-like proteins) may recognize both CAM7 and HY5 proteins, which are already bound to the respective promoter elements. In the absence of CAM7, the coactivator and HY5 interaction might be sufficient (omitting the requirement for DNA binding of CAM7) to promote photomorphogenic growth, thus making CAM7 protein functionally redundant. However, under this condition (in the absence of CAM7), the expression of the light-regulated genes is moderately downregulated. Alternatively, functional redundancy of CAM7 may be due to the overlapping functions of light and another signaling pathway working via CAM7 protein. For example, Ca2+/CaM-mediated signaling has been shown to be involved in brassinosteroid biosynthesis and auxin signaling pathways (Yang and Poovaiah, 2000; Du and Poovaiah, 2005). HY5 has also been shown to act as a regulatory protein in auxin signaling (Sibout et al., 2006). Recent studies have revealed that seedlings that are deficient in gibberellin synthesis or signaling exhibit photomorphogenic growth in the darkness. Furthermore, these studies have shown that HY5 is a point of crosstalk between light and jasmonic acid signaling pathways (Alabadi et al., 2004, 2008).
This study reveals that CAM7 protein accumulates at higher levels in dark or at lower intensity of WL, which directly correlates with its physiological functions under such conditions (Figures 2 and 6). Under certain growth conditions, two bands of CAM7 were detected in protein gel blot analyses, one of which might be a posttranslationally modified form of the protein. HY5 accumulates at a lower level at lower intensities of WL, and the level of HY5 protein increases with exposure to higher intensity of WL (Osterlund et al., 2000). The overexpression of full-length or truncated HY5 is unable to promote photomorphogenic growth or derepression of light-regulated genes in the darkness (Ang et al., 1998). On the other hand, CAM7 overexpresser transgenic seedlings not only display partial photomorphogenic growth in the dark, but the light-regulated genes are also expressed under the similar conditions. Overexpression of CAM7 in hy5 transgenic lines not only partially suppresses the hy5 phenotype in light, it also promotes photomorphogenic growth in the darkness (Figure 5B). Furthermore, accumulation of CAM7 in hy5 transgenic lines fully restored the expression of light-regulated genes (Figure 5G). Collectively, this study demonstrates that CAM7 acts as a positive regulator of photomorphogenic growth and light-regulated gene expression and highlights the concerted function of CAM7 and HY5 in Arabidopsis seedling development (Figure 6D).
METHODS
Transgenic Plants and Mutants
Plant growth and light conditions were as described by Yadav et al. (2005). The segregation ratios of self-fertilized plants heterozygous for cam7-1 or cam7-2 determined by the analyses of genotyping PCR (left border–specific primer LBP, 5′-GCGTGGACCGCTTGCTGCAACT-3′; and CAM7-specific primers LP15, 5′-GACCATCTCCTCTCCGTCTTTGTCGAA-3′, and RP15, 5′-CGAATGTGTTTCGTT TACAGTTCA-3′) in T2 progeny suggested that a single T-DNA locus was present in cam7-1 or cam7-2 mutant lines. The junctions of T-DNA and CAM7 were amplified by PCR, and the DNA sequence analyses revealed that the T-DNA was inserted at 225 and 113 bp upstream to ATG of CAM7 in cam7-1 (salk_074336) and cam7-2 (Flag_397A10), respectively. The hypocotyl length measurement data were analyzed by one-way analysis of variance using a post-hoc Dunnett test. All analyses were performed using the SPSS15.0 program.
For the generation of overexpresser transgenic lines, a 571-bp fragment of cDNA was PCR amplified using primers with c-Myc sequence: FP, 5′-CATGCCATGGCAATGAACATCTCAGAGTTCAAGGAGGCTT-3′; RP2A, 5′-GGAGATTAGCTTTTGTTCACCGTTCAAATCTTCTTCAGAAATCAACTTTTGTTCACCGTCGAGCTT AGCCATCATGACTTTGACAAACTC-3′; RP2B, 5′-GACTAGTACCGTCGAGTCCGTTCAAGTCTTCTTCTGAGATTAATTTTTGTTCACGTTCAAGTCTTCC TCGGAGATTAGCTTTTGTTCACCGTTCAAAT-3′. The PCR products were digested and cloned into the NcoI and SpeI sites of pCAMBIA1303, and the transgenic lines were generated as described by Mallappa et al. (2006).
The cam7 hy5 double mutants were constructed by genetic crosses using hy5-ks50 (Wassilewskija) and cam7-1 (Col) single mutants following similar methods described by Yadav et al. (2005). A segregated wild-type line of the T2 generation was used as a control to compare the phenotypic and molecular differences. The hy5 transgenic lines containing the 35S promoter-CAM7-c-Myc transgene were constructed by genetic crosses between hy5-KS50 and OE1 or OE2 transgenic plants containing the 35S promoter-CAM7-c-Myc transgene. For the complementation test, a 2.1-kb fragment containing CAM7 and its 1.5-kb upstream promoter region was cloned into the NcoI and SpeI sites of pCAMBIA1303, and the transgenic lines were generated as described by Mallappa et al. (2006).
Chlorophyll and Anthocyanin Measurements
Chlorophyll and anthocyanin levels were measured following protocols as described by Holm et al. (2002). Briefly, seedlings were collected into microcentrifuge tubes, weighed, and crushed by a pestle in 700 μL of chilled 80% acetone. Cellular debris was removed by centrifugation at 4°C, and the supernatant containing chlorophyll was collected into a fresh microcentrifuge tube, and volume was made up to 1 mL. Then the absorbance was measured at the wavelengths of 645 and 663 nm. The total chlorophyll content was calculated with the following formula: (20.2 × A645) – (8.02 × A663) = μg/g of fresh tissue weight.
Arabidopsis thaliana Protoplast Transfection Assays
Arabidopsis protoplasts were isolated and transfection assays were performed following the methods described by Wang et al. (2005). The mutated Z-box used in CAB1m promoter has been described by Yadav et al. (2002).
RNA Gel Blot Analysis
For RNA gel blots, total RNA was extracted using the RNeasy plant minikit (Qiagen) following the manufacturer's instructions. The DNA fragment of CAB1 or RBCS gene was used as probe as described (Yadav et al., 2005) using a random priming kit (Megaprime; Amersham). To quantify the RNA gel blot data, the intensity of each band was quantified by the Fluor-S-MultiImager (Bio-Rad) and ratios of the gene versus its corresponding rRNA band were determined and plotted (Fluor-S-MultiImager; Bio-Rad).
Protein Analysis
The ligand binding screen (DNA binding to filter-immobilized protein) was performed as described (Yadav et al., 2005). For EMSAs, CAM7 cDNA was cloned in pGEX4T-2 vector, and GST-CAM7 was induced using 1 mM isopropylthio-β-galactoside and overexpressed in Escherichia coli. The overexpressed GST-CAM7 was affinity purified following the manufacturer's protocol (GE). GST-CAM3, GST-CAM7-M1, or GST-CAM7-M2 proteins were also purified similarly. EMSAs were performed as described (Mallappa et al., 2006). Protein gel blot analysis was performed using the Super Signal West Pico chemiluminescent substrate kit (Pierce) following the instructions as described in the user's manual. Protein extracts were prepared from wild-type or transgenic seedlings. The seedlings (100 mg) were frozen in liquid nitrogen and ground in 300 μL of grinding buffer (400 mM sucrose, 50 mM Tris-Cl, pH 7.5, 10% glycerol, and 2.5 mM EDTA), and PMSF was added (0.5 μL for every 100 μL of grinding buffer). The protein extract was transferred to a fresh microcentrifuge tube and centrifuged at 5000 rpm for 5 min to pellet down the debris. The supernatant was transferred to a fresh tube, and an aliquot of 5 μL was taken out in a separate tube for the estimation of protein by Bradford assay. Proteins were separated by 10% SDS-PAGE. Prestained protein markers (GE) were used for molecular mass determination. The samples were then transferred to Hybond C-Extra (Fermentas) at 100 mA for 2 h in transfer buffer (7.56 g Tris, 47 g glycine, and 20% methanol in 2.5 liters) in a mini blot protein gel apparatus (GE). The membrane was blocked with 5% milk in PBS (10 mM Na2HPO4, 1.8 mM KH2PO4, 140 mM NaCl, and 2.7 mM KCl) and probed with c-Myc polyclonal antibodies or anti-Actin monoclonal antibodies (Sigma-Aldrich).
The ChIP assays were essentially performed as described (He et al., 2005). The sequence of primer pairs (resulting products of ∼500 bp) used were as follows: NIA2-PROMO-FP, 5′-CTATACATGTTTCCGAGACG-3′; NIA2-PROMO-RP, 5′-AGTATCGTGCCGAATCACACG-3′; CAB1-PROMO-FP, 5′-GGTTTACATTGATGCTCTCAGGATTTC-3′; CAB1-PROMO-RP, 5′-CGTGGTTAATGGCTCGCACTTCGC-3′.
Real-Time Quantitative PCR
Total RNA was isolated using the RNeasy plant minikit (Qiagen) extraction kit according to the manufacturer's protocol. cDNA was synthesized from 1 μg of the total RNA using RT-AMV reverse transcriptase (Roche). Real-time PCR was performed using Light Cycler faststart DNA Masterplus SYBR Green1 (Roche). Values were normalized with the amplification of Actin as a constitutively expressed internal control. Primers used were as follows: CAB1-FP, 5′-CCCATTTCTTGGCTTACAACAAC-3′; CAB1-RP, 5′-TCGGGGTCAGCTGAAAGTCCG-3′; RBCS-1A-FP, 5′-GAGTCACACAAAGAGTAAAGAAG-3′; RBCS-1A-RP, 5′-CTTAGCCAATTCGGAATCGGT-3′.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession numbers AM422556 (ZBF3/CAM7) and At1g37130 (NIA2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Comparison of Quality of Various Purified Proteins Used for EMSAs.
Supplemental Figure 2. Identification of cam7 Mutants.
Supplemental Figure 3. CAM7 and HY5 Promote Photomorphogenic Growth at Various Wavelengths of Light.
Supplementary Material
Acknowledgments
We thank Asis Datta, Sudhir K. Sopory, Sunil Mukherjee, and Ashis K. Nandi for critically reading and commenting on the manuscript. This work is supported by a research grant (Ramanna Fellowship) from the Department of Science and Technology, Government of India to S.C. R.K. and A.S. are recipients of fellowships from the University Grants Commission and the Council of Scientific and Industrial Research, respectively, of the Government of India.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Sudip Chattopadhyay (sudipchatto@yahoo.com).
Online version contains Web-only data.
References
- Alabadi, D., Gallego-Bartolome, J., Orlando, L., Garcia-Carcel, L., Rubio, V., Martinez, C., Frigerio, M., Iglesias-Pedraz, J.M., Espinosa, A., Deng, X.W., and Blázquez, M.A. (2008). Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J. 53 324–335. [DOI] [PubMed] [Google Scholar]
- Alabadi, D., Gil, J., Blazquez, M.A., and Garcia-Martinez, J.L. (2004). Gibberellins repress photomorphogenesis in darkness. Plant Physiol. 134 1050–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Ang, L.H., Chattopadhyay, S., Wei, N., Oyama, T., Okada, K., Batschauer, A., and Deng, X.W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1 213–222. [DOI] [PubMed] [Google Scholar]
- Babourina, O., Newman, I., and Shabala, S. (2002). Blue light-induced kinetics of H+ and Ca++ fluxes in etiolated wild type and phototropin-mutant Arabidopsis seedlings. Proc. Natl. Acad. Sci. USA 99 2433–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge, M.J., Lipp, P., and Bootman, M.D. (2000). The calcium entry pas de deux. Science 287 1604–1605. [DOI] [PubMed] [Google Scholar]
- Braam, J., and Davis, R.W. (1990). Rain, wind and touch induced expression of calmodulin and calmodulin related genes in Arabidopsis. Cell 60 357–364. [DOI] [PubMed] [Google Scholar]
- Bowler, C., Neuhaus, G., Yamagata, H., and Chua, N.H. (1994). Cyclic GMP and calcium mediate phytochrome phototransduction. Cell 77 73–81. [DOI] [PubMed] [Google Scholar]
- Carrion, A.M., Link, W.A., Ledo, F., Mellstorm, B., and Naranjo, J.R. (1999). DREAM is a Ca2+-regulated transcriptional repressor. Nature 398 80–84. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay, S., Ang, L.H., Puente, P., Deng, X.W., and Wei, N. (1998). Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M., Chory, J., and Fankhauser, C. (2004). Light signal transduction in higher plants. Annu. Rev. Genet. 38 87–117. [DOI] [PubMed] [Google Scholar]
- Corneliussen, B., Holm, M., Waltersson, Y., Onions, J., Hallberg, B., Thornell, A., and Grundstrom, T. (1994). Calcium/calmodulin inhibition of basic-helix-loop-helix transcription factors domains. Nature 368 760–764. [DOI] [PubMed] [Google Scholar]
- Craig, T.A., Benson, L.M., Venyaminov, S.Y., Klimtchuk, E.S., Bajzer, Z., Prendergast, F.G., Naylor, S., and Kumar, R. (2002). The metal-binding properties of DREAM: evidence for calcium-mediated changes in DREAM structure. J. Biol. Chem. 277 10955–10966. [DOI] [PubMed] [Google Scholar]
- Du, L., and Poovaiah, B.W. (2005). Ca2+/calmodulin is critical for brassinosteroid biosynthesis and plant growth. Nature 437 741–745. [DOI] [PubMed] [Google Scholar]
- Gilchrist, C.A., Holm, C.F., Hughes, M.A., Schaenman, J.M., Mann, B.J., Petri, W.A. (2001). Identification and characterization of an Entamoeba histolytica upstream regulatory element 3 sequence-specific DNA-binding protein containing EF-hand motifs. J. Biol. Chem. 276 11838–11843. [DOI] [PubMed] [Google Scholar]
- Guo, H., Mocker, T., Duong, H., and Lin, C. (2001). SUB1, an Arabidopsis Ca2+-binding protein involved in cryptochrome and phytochrome coaction. Science 291 487–490. [DOI] [PubMed] [Google Scholar]
- He, J.-X., Gendron, J.M., Sun, Y., Gampala, S.S.L., Gendron, N., Sun, C.Q., and Wang, Z.-Y. (2005). BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307 1634–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler, P.K. (2005). Calcium: A central regulator of plant growth and development. Plant Cell 17 2142–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq, E., and Quail, P.H. (2005). Phytochrome signaling. In Handbook of Photosensory Receptors, W.R. Briggs and J.L. Spudich, eds (Weinheim, Germany: Wiley), pp. 151–170.
- Jiao, Y., Lau, O.S., and Deng, X.W. (2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8 217–230. [DOI] [PubMed] [Google Scholar]
- Kaplan, B., Davydov, O., Galon, Y., Knight, M.R., Fluhr, R., and Fromm, H. (2006). Rapid transcriptome changes induced by cytosolic Ca2+ transients reveal ABRE-related sequences as Ca2+-responsive cis elements in Arabidopsis. Plant Cell 18 2733–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, M.R., Campbell, A.K., Smith, S.M., and Trewavas, A.J. (1991). Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature 352 524–526. [DOI] [PubMed] [Google Scholar]
- Lee, J., He, K., Stolc, V., Lee, H., Figueroa, P., Gao, Y., Tongprasit, W., Zao, H., Lee, I., and Deng, X.W. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19 731–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, J.C., and Jenkins, G.I. (1998). Involvement of plasma membrane redox activity and calcium homeostasis in the UV-B and UV-A/blue-light induction of gene expression in Arabidopsis. Plant Cell 10 2077–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallappa, C., Yadav, V., Negi, P., and Chattopadhyay, S. (2006). A bzip transcription factor, GBF1, regulates blue light mediated photomorphogenic growth in Arabidopsis. J. Biol. Chem. 281 22190–22199. [DOI] [PubMed] [Google Scholar]
- McCormack, E., Tsai, Y., and Braam, J. (2005). Handling calcium signalling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 10 383–389. [DOI] [PubMed] [Google Scholar]
- Miller, A.J., and Sanders, D. (1987). Depletion of cytosolic free calcium induced by photosynthesis. Nature 326 397–400. [Google Scholar]
- Nagy, F., and Schaefer, E. (2002). Phytochromes control photomorphogenesis by differentially regulated interacting signaling pathways in higher plant. Annu. Rev. Plant Biol. 53 329–355. [DOI] [PubMed] [Google Scholar]
- Neuhaus, G., Bowler, C., Hiratsuka, K., Yamagata, H., and Chua, N.H. (1997). Phytochrome-regulated repression of gene expression requires calcium and cGMP. EMBO J. 16 2554–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus, G., Bowler, C., Kern, R., and Chua, N.H. (1993). Calcium/CaM-dependent and independent phytochrome signal transduction pathways. Cell 73 937–952. [DOI] [PubMed] [Google Scholar]
- Osawa, M., Dace, A., Tong, K.I., Valiveti, A., Ikura, M., and Ames, J.B. (2005). Mg2+ and Ca2+ diffentially regulate DANN binding and dimerization of DREAM. J. Biol. Chem. 280 18008–18014. [DOI] [PubMed] [Google Scholar]
- Osterlund, M.T., Hardtke, C.S., Wei, N., and Deng, X.W. (2000). Targeted destabilization of HY5 during light regulated development of Arabidopsis. Nature 405 462–466. [DOI] [PubMed] [Google Scholar]
- Oyama, T., Shimura, Y., and Okada, K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11 2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu, S.C., Popescu, G.V., Bachan, S., Zhang, Z., Seay, M., Gerstein, M., Snyder, M., and Dinesh-Kumar, S.P. (2007). Differential binding of calmodulin related proteins to their targets revealed through high-density Arabidopsis protein microarrys. Proc. Natl. Acad. Sci. USA 104 4730–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sai, J., and Johnson, C.H. (2002). Dark-stimulated calcium ion fluxes in the chloroplast stroma and cytosol. Plant Cell 14 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibout, R., Sukumar, P., Hettiarachchi, C., Holm, M., Muday, G.K., and Hardtke, C. (2006). Opposite root growth phenotypes of hy5 versus hy5 hyh mutants correlate with increased constitutive auxin signalling. PLoS Genet. 2 1898–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski, D.B., Liao, B., and Zielinski, R.E. (1996). Calmodulin isoforms differentially enhance the binding of cauliflower nuclear proteins and recombinant TGA3 to a region derived from the Arabidopsis Cam-3 promoter. Plant Cell 8 1069–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitia, R.A. (2005). Paralogs in polyploids: One for all and all for one? Plant Cell 17 4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., Tiwari, S.B., Hagen, G., and Guilfoyle, T.J. (2005). AUXIN RESPONSE FACTOR7 restores the expression of auxin-responsive genes in mutant Arabidopsis leaf mesophyll protoplasts. Plant Cell 17 1979–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav, V., Kundu, S., Chattopadhyay, D., Negi, P., Wei, N., Deng, X.W., and Chattopadhyay, S. (2002). Light regulated modulation of Z-box containing promoters by photoreceptors and downstream regulatory components, COP1 and HY5, in Arabidopsis. Plant J. 31 741–753. [DOI] [PubMed] [Google Scholar]
- Yadav, V., Mallappa, C., Gangappa, N.S., Bhatia, S., and Chattopadhyay, S. (2005). A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell 17 1953–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, T., and Poovaiah, B.W. (2000). Molecular and biochemical evidences for the involvement of calcium/calmodulin in auxin action. J. Biol. Chem. 275 3137–3143. [DOI] [PubMed] [Google Scholar]
- Yang, T., and Poovaiah, B.W. (2002). A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signalling pathways in plants. J. Biol. Chem. 277 45049–45058. [DOI] [PubMed] [Google Scholar]
- Yoo, J.H., et al. (2004). Regulation of the dual specificity protein phosphatase, DsPTP1, through interactions with calmodulin. J. Biol. Chem. 279 848–858. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.