Abstract
A plant-specific family of WRKY transcription factors regulates plant responses to pathogens and abiotic stresses. Here, we identify two insect-responsive WRKY genes in the native tobacco Nicotiana attenuata: WRKY3, whose transcripts accumulate in response to wounding, and WRKY6, whose wound responses are significantly amplified when fatty acid–amino acid conjugates (FACs) in larval oral secretions are introduced into wounds during feeding. WRKY3 is required for WRKY6 elicitation, yet neither is elicited by treatment with the phytohormone wound signal jasmonic acid. Silencing either WRKY3 or WRKY6, or both, by stable transformation makes plants highly vulnerable to herbivores under glasshouse conditions and in their native habitat in the Great Basin Desert, Utah, as shown in three field seasons. This susceptibility is associated with impaired jasmonate (JA) accumulation and impairment of the direct (trypsin proteinase inhibitors) and indirect (volatiles) defenses that JA signaling mediates. The response to wounding and herbivore-specific signals (FACs) demonstrates that these WRKYs help plants to differentiate mechanical wounding from herbivore attack, mediating a plant's herbivore-specific defenses. Differences in responses to single and multiple elicitations indicate an important role of WRKY3 and WRKY6 in potentiating and/or sustaining active JA levels during continuous insect attack.
INTRODUCTION
In nature, plants battle continuously against pathogens and herbivores; these attack plants in various ways, possess diverse feeding methods, and evolve resistance to plant defenses (Agrawal, 2001). In return, plants have evolved structural and chemical barriers that, along with induced defenses, counter herbivore attacks. Induced defenses, which are deployed only when protection is needed, allow plants to forgo the costs of defense production (Heil and Baldwin, 2002) and optimize their ability to compete with other plants (Zavala et al., 2004) or to tolerate the damage caused by resistant herbivores (Stowe et al., 2000; Schwachtje et al., 2006). These defenses include the rapid accumulation of toxins, antidigestive proteins, and antifeedants expressed both locally at the feeding site and systemically in unattacked tissues. Direct defenses that diminish the suitability of plant tissues as food for herbivores can be simultaneously deployed with indirect defenses, such as the production of extra floral nectars (Heil et al., 2001) or volatile alarm calls that attract predators or parasitoids to eliminate feeding herbivores (Dicke and Sabelis, 1988; De Moraes et al., 1998; Kessler and Baldwin, 2002). Many of these defenses are coordinated by a jasmonate (JA)-dependent signaling cascade that is required to activate and rapidly disseminate the elicited responses throughout the plant (Halitschke and Baldwin, 2003; Kalde et al., 2003).
A complex suite of defenses is elicited when herbivore-specific elicitors are introduced into wounds during feeding; these rapidly activate large-scale transcriptional changes, phytohormone signaling, and posttranslational modifications of proteins (Halitschke et al., 2000; Halitschke and Baldwin, 2003; Kalde et al., 2003). Such responses are often found to be specific to the attacker (Walling, 2000; Kessler and Baldwin, 2002; Voelckel and Baldwin, 2004; Schmidt et al., 2005; Ralph et al., 2006; Wu et al., 2007). Large-scale and specific transcriptional responses, which are mediated by transcription factors that bind DNA in a sequence-specific manner, illustrate how adaptive phenotypic plasticity evolves. A number of the transcription factors that specifically mediate responses to pathogens, wounding, and abiotic stresses have been identified (Jakoby et al., 2002; Ulker and Somssich, 2004; Eulgem and Somssich, 2007).
The WRKY transcription factors, which make up one of the largest groups of transcription regulators, are unique to plants. These proteins typically contain one or two WRKY domains composed of ∼60 amino acids with the conserved amino acid sequence WRKYGQK, together with a novel zinc finger–like motif (Pfam database: http://pfam.sanger.ac.uk/family?acc=PF03106). WRKY proteins bind to specific W-boxes in the promoters of their transcriptional targets, either up- or downregulating the gene expression. WRKY genes often contain W-boxes in their own promoters, suggesting that they are self-regulated or that they are regulated by other WRKY transcription factors (Ulker and Somssich, 2004; Eulgem, 2006).
WRKY transcription factors regulate responses of plants to pathogen attack and to various abiotic stresses (see summary in Supplemental Figure 1 online). In rice (Oryza sativa), several recently described WRKY genes mediate defense responses to pathogens such as the rice blast fungus (Magnaporthe grisea) or bacterial blight (Xanthomonas oryzae) (Chuio et al., 2007; Liu et al., 2007; Qiu et al., 2007; Shimono et al., 2007; Wang et al., 2007a). Most of the WRKY genes characterized in Arabidopsis thaliana and other plant species also help defend plants against phytopathogens and abiotic stresses (Robatzek and Somssich, 2002; Mare et al., 2004; Park et al., 2006; Knoth et al., 2007; Marchive et al., 2007; Murray et al., 2007; Ulker et al., 2007; Xie et al., 2007; Zheng et al., 2007; Zou et al., 2007) and mediate the crosstalk between JA and salicylic acid (SA) signaling pathways (Li et al., 2004). While most WRKY genes identified so far seem to mediate antimicrobial defensive responses, in Arabidopsis, some WRKYs were found to regulate developmental processes, such as senescence and trichome development (Johnson et al., 2002; Miao et al., 2007).
To date, only a single WRKY gene has been directly associated with defense against herbivores: the ectopic overexpression of WRKY89 in rice enhanced resistance to the white-backed planthopper Sogatella furcifera, a rice herbivore. However, the resistance mechanisms regulated by WRKY89 remain unknown (Wang et al., 2007a). Here, we describe two novel WRKY transcription factors from the native tobacco Nicotiana attenuata, WRKY3 and WRKY6 (Figure 1A), which are strongly upregulated in plants attacked by Manduca sexta (Hui et al., 2003). Moreover, plants accumulate transcripts of these genes in response to attack by insects from other feeding guilds (Voelckel and Baldwin, 2004).
Figure 1.
WRKY3 Strongly Responds to Wounding, and WRKY6 Requires M. sexta's OS for Transcript Elicitation.
(A) Alignment of WRKY3 and WRKY6 proteins. Identities of both proteins are shaded with black, and conserved WRKY domains are enclosed in the black box.
(B) Mean (±se) WRKY3 transcript levels in elicited (filled symbols) and unelicited (systemic; open symbols) leaves in five replicate N. attenuata wild-type plants at each harvest time and treatment. Leaves were elicited by wounding and immediately treating the puncture wounds with 20 μL of water (W; dotted lines) or 1:1 diluted OS from M. sexta larvae (solid lines). Inset: induction of WRKY3 transcripts in an independent experiment using wounding and water (W; dotted line), 1:1 diluted OS (solid line), or a synthetic mixture of the two most abundant FACs in M. sexta OS, N-linolenoyl-l-glutamine and N-linolenoyl-l-glutamate (FAC; dashed line). Transcripts were analyzed by real-time PCR in arbitrary units after calibration with a 4× dilution series of cDNAs prepared from RNA samples containing WRKY3 or WRKY6 transcripts.
(C) WRKY6 transcript levels determined as above. The x axes in (B) and (C) are on a nonlinear scale to reveal the culmination of transcript levels between 0 and 60 min.
Realizing the potential importance of these genes in plant–herbivore interactions, we performed functional analysis with both genes in N. attenuata. While WRKY3 transcripts were already maximally induced in wounded leaves, we found that the herbivore-specific elicitors, fatty acid–amino acid conjugates (FACs) found in M. sexta oral secretions (OS), are required to amplify the moderately wound-induced accumulation of WRKY6 transcripts. Using gene silencing and an Agrobacterium tumefaciens–mediated transformation system (Kruegel et al., 2002), we show that WRKY3 and WRKY6 mediate both the direct and indirect defenses of N. attenuata by regulating JA-dependent signaling in herbivore-attacked plants. Moreover, when silenced plants are transplanted into their native habitat in the Great Basin Desert for three consecutive field seasons, they suffer extensive damage from the local herbivore community. This susceptibility is associated with impaired transcriptional responses, JA and JA-Ile/-Leu phytohormone accumulation, and the deployment of both direct (trypsin proteinase inhibitors) and indirect (volatile alarm calls) defenses in WRKY-silenced plants. Because exogenously applied JA restores all impaired direct and indirect defense responses in WRKY3- and WRKY6-silenced plants, we propose that WRKY3 and WRKY6 function upstream of JA biosynthesis to potentiate and/or sustain plants' defenses against herbivorous insects.
RESULTS
Specific Responses of WRKY3 and WRKY6 to Herbivory-Associated Signals
Previously, we conducted differential display RT-PCR (DDRT-PCR) analysis on M. sexta–attacked N. attenuata leaves and identified many early herbivory-induced genes that may help defend plants against insect herbivores (Hermsmeier et al., 2001; Hui et al., 2003). One of them, a gene fragment with sequence similarity to the WRKY gene family in plants, was particularly strongly upregulated in M. sexta–elicited leaves. Using a 270-bp DDRT-PCR fragment and N. attenuata cDNA library derived from M. sexta–attacked shoots, two full-length cDNA sequences belonging to group I of the WRKY protein family were isolated and named Na WRKY3 and Na WRKY6 (Figure 1A). At the protein level, Na WRKY3 was most similar to previously isolated Nicotiana tabacum WRKY2 (89% identity at protein level), while Na WRKY6 resembled Nt WRKY1 (96% identity at protein level) (Nishiuchi et al., 2004).
DDRT-PCR Results Suggested That at Least One of the Genes Should Be Regulated by Herbivory
Quantitative RT-PCR analysis revealed that both genes are differentially upregulated by herbivory-associated signals in gene-specific ways: WRKY3 transcript levels maximally increased in response to wounding (Figure 1B), those of WRKY6, in response to OS elicitation (Figure 1C). When synthetic FACs were applied to wounds at concentrations found in OS (Halitschke et al., 2001), the OS-elicited increase in WRKY6 transcripts was fully mimicked (inset in Figure 1C). Moreover, the expression of WRKY3 and WRKY6 transcription factors was shown to be independent of JA signaling, as comparable transcript accumulation was found in the wild type and in plants silenced in their expression of JA-biosynthetic enzyme lipoxygenase3 (LOX3) (see Supplemental Figure 2 online). OS- and FAC-elicited JA bursts are known to be impaired in these plants (Halitschke and Baldwin, 2003). Moreover, neither WRKY was elicited by direct treatment of the leaves with methyl jasmonate, an elicitor that reliably activates wound- and herbivore-specific defense responses in plants (Wu et al., 2008; see Supplemental Figure 2 online).
Overexpressing WRKY3 and WRKY6 Does Not Alter Resistance to M. sexta Larvae
To understand if WRKY3 or WRKY6 mediates defense responses to M. sexta attack, full-length cDNA fragments of both genes were overexpressed in N. attenuata. The ectopic overexpression of WRKY3 (ov-WRKY3) and WRKY6 (ov-WRKY6) with a constitutive 35S cauliflower mosaic virus promoter (see Supplemental Figures 3A and 3B online) substantially increased the basal and induced transcript levels of WRKY3 and of WRKY6 genes (see Supplemental Figure 4A online), without significantly affecting the plants' morphology. Subsequently, performance was assessed for M. sexta larvae that fed on two homozygous lines per construct, each harboring a single T-DNA insert (see Supplemental Figures 3C and 3D online), and compared with larval performance on wild-type plants. Ten days after larvae fed on either ov-WRKY3 or ov-WRKY6 lines, their masses did not differ from those of larvae that fed on wild-type plants (see Supplemental Figure 4B online). No metabolic changes or enhanced defense responses were observed in these transgenic plants: JA and JA-Ile/-Leu concentrations were similar in both overexpression lines and in wild-type plants after a single OS elicitation (data not shown) as well as after continuous caterpillar feeding (see Supplemental Figure 4C online). The absence of an overexpressing phenotype in plants transformed to ectopically express a transcription factor is not uncommon and likely reflects the functional requirements of being expressed in the correct molecular context: time, location, and/or the presence of specific interacting partners. It is also possible that WRKY genes may have to be posttranslationally modified during elicitation so they can act as regulators.
Individually Silencing WRKY3 and WRKY6 Shows Interaction between the Genes
Because ectopic overexpression of the WRKY genes in N. attenuata did not seem to enhance protection against herbivores, we next constructed plants with suppressed levels of WRKY transcripts and subjected them to extensive functional analysis. Fragments (1 kb) of WRKY3 or WRKY6 coding regions were introduced into an inverted-repeat silencing (ir) construct, and N. attenuata plants were transformed (Kruegel et al., 2002; see Supplemental Figure 5 online). Silencing of either WRKY3 or WRKY6 resulted in plants with normal morphology and development (see Supplemental Figure 6 online). WRKY3 basal transcript concentrations in WRKY3-silenced plants (ir-wrky3) were significantly reduced compared with those in wild-type plants, whereas transcript concentrations in WRKY6-silenced plants (ir-wrky6) did not differ from those in wild-type plants (Figure 2A). Surprisingly, the accumulation of WRKY6 transcripts was as reduced in WRKY3-silenced plants as in WRKY6-silenced plants (Figure 2B), suggesting that WRKY3 is required for efficient WRKY6 transcript accumulation.
Figure 2.
WRKY3 Is Required for WRKY6 Transcript Accumulation, and Both Are Required for Resistance to M. sexta Larvae.
(A) Mean (±se) WRKY3 basal transcript levels in five replicates of each wild-type, WRKY3-, and WRKY6-silenced plants. Transcripts were analyzed by real-time PCR. Different letters reflect significant differences at P < 0.05 from wild-type plants of the same treatment.
(B) Mean (±se) WRKY6 basal transcript levels in five replicates of each wild-type, WRKY3-, and WRKY6-silenced plants as above.
(C) Mean (±se) mass of M. sexta larvae that fed on wild-type (solid line), two lines of WRKY3-silenced (ir-wrky3; dotted lines), and two lines of WRKY6-silenced (ir-wrky6; dashed lines) N. attenuata plants in the glasshouse for 8 d (n = 12/genotype).
(D) Mean (±se) of JA and JA-Ile/-Leu concentrations and transcript levels of LOX3 after M. sexta larvae fed on the plants. FM, fresh mass.
(E) Mean (±se) of TPI activity measured 4 d after M. sexta larvae fed on the plants. Different letters reflect significant differences at P < 0.05 from wild-type plants of the same treatment.
When working with two WRKY genes that share a certain level of similarity, one must consider the possibility that the WRKY3 silencing construct might have silenced WRKY6 transcript accumulation. While the overall DNA sequence identity between WRKY3 and WRKY6 gene fragments used for silencing is only 55.9%, WRKY3 and WRKY6 do show local areas of higher identity, particularly in the conserved WRKY domains. However, in both silencing constructs, these DNA sequences were present, implying that if ir-wrky3 cosilenced WRKY6, reciprocal effects of ir-wrky6 on endogenous WRKY3 transcripts should also occur. Because this was not the case, we assume that the similarity between both genes was below the threshold required for cosilencing. In addition, we tested five other WRKY genes, each with conserved WRKY domains. In all cases, the transcripts of these genes were not significantly influenced by the silencing constructs (see Supplemental Figure 8 online).
Similar trends in WRKY3 and WRKY6 transcript levels were observed in plants elicited with M. sexta OS (data not shown), providing a strong demonstration of the functional silencing and the mutual relationship between WRKY3 and WRKY6 genes. These results indicate that WRKY6 is located downstream of WRKY3 in the signaling cascade, where it may tailor the WRKY3-mediated wound responses to the attack of a specific herbivore. This hypothesis is consistent with the observation that WRKY6 is mainly under the transcriptional control of FACs from insect OS and suggests that the herbivore signaling network consists of a wound-responsive component (WRKY3) and a nested component responsive to herbivore-specific elicitors such as FACs (WRKY6).
Silencing WRKY3 and WRKY6 Makes Plants Vulnerable to Herbivores
The performance of M. sexta larvae on WRKY3- and WRKY6-silenced plants was compared with the performance of these larvae on wild-type plants in the glasshouse: after 8 d of feeding on both WRKY3- and WRKY6-silenced lines, larvae weighed at least twice as much as those that fed on wild-type plants (Figure 2C; analysis of variance [ANOVA] Fisher's protected least significant difference [PLSD], all P values < 0.05). This enhanced caterpillar mass gain correlated with large reductions in the activity of trypsin proteinase inhibitors (TPIs), an important family of plant defense proteins that interfere with efficient digestion in the gut of herbivores (Figure 2E; ANOVA Fisher's PLSD, all P values < 0.05). By contrast, levels of another direct defense-related compound, nicotine, did not differ significantly between wild-type and WRKY-silenced lines (data not shown).
Silencing WRKY3 or WRKY6 Reduces M. sexta–Elicited JA and JA-Ile/-Leu Levels
JA signaling is known to mediate TPI activation (Halitschke and Baldwin, 2003); therefore, we analyzed JA and JA-Ile/-Leu levels in attacked leaves. We found significantly smaller concentrations of JA and JA-Ile/-Leu compared with wild-type plants measured 22 and 30 h after infestation with M. sexta larvae (Figure 2D), which provided an explanation for the low levels of TPI activity (Figure 2E). Moreover, decreased phytohormone levels in WRKY-silenced plants compared with wild-type plants correlated with lower levels of the gene transcripts that encode for the JA biosynthetic enzyme LOX3 (Figure 2D; Halitschke and Baldwin, 2003), suggesting that these WRKY transcription factors directly or indirectly mediate LOX transcriptional activation and, thus, JA accumulation.
Tobacco plants respond to simulated herbivory (puncture wounds created by rolling a pattern wheel the length of the leaf lamina followed by the immediate application of M. sexta OS) with a JA burst that usually attains maximum levels 60 min after elicitation. We examined the ability of WRKY-silenced plants to produce a JA burst in response to such standardized elicitation treatment. In contrast with previous results with herbivore-fed plants (Figure 2D), JA and JA-Ile/-Leu levels in WRKY3- and WRKY6-silenced plants were similar to those observed in the OS-elicited tissues of wild-type plants 60 min after elicitation (Figures 3A and 3C). To determine if the number of OS elicitations could account for the difference in phytohormone response between plants that were OS elicited just once and larvae-attacked plants (continuous elicitation), we elicited plants with OS every 30 min six times, with one row of puncture wounds each time being immediately treated with 10 μL OS. JA and JA-Ile/-Leu were measured 30 min after the final, or sixth, elicitation. Phytohormone levels were found to be significantly lower in multiply elicited WRKY-silenced plants than in multiply elicited wild-type plants: JA concentrations were significantly reduced in both ir-wrky3 and ir-wrky6 lines (Figure 3B; ANOVA Fisher's PLSD, all P values < 0.05); JA-Ile/-Leu concentrations, however, were significantly reduced only in ir-wrky6 lines (Figure 3D; ANOVA Fisher's PLSD, P < 0.05). Although JA-Ile/-Leu levels in ir-wrky3-silenced plants tended to be reduced, only one line provided a statistically significant reduction of JA-Ile/-Leu (Figure 3D; ANOVA Fisher's PLSD, P = 0.0421, P = 0.0563, respectively).
Figure 3.
Consecutively OS-Elicited WRKY-Silenced Plants Show Decreased JA and JA-Ile/-Leu Levels.
Mean (±se) of JA ([A] and [B]) and JA-Ile/-Leu ([C] and [D]) concentrations in OS-elicited leaves of separate plants of the wild type (black), two ir-wrky3 lines (gray), and two ir-wrky6 lines (white).
(A) and (C) Leaves harvested 60 min after a single OS elicitation, during which one leaf with six rows of holes was treated with 20 μL of OS (n = 5/genotype; * P < 0.05, as determined by Fisher's PLSD test from ANOVA).
(B) and (D) Leaves harvested after multiple OS elicitations, whereby one leaf was wounded with one row of holes and 10 μL of OS was applied to the wounds. Treatment was repeated six times every 30 min, and leaves were harvested 30 min after the last elicitation (n = 5/genotype; * P < 0.05, as determined by Fisher's PLSD test from ANOVA).
Performance of WRKY-Silenced Plants in the Natural Environment
Because only multiple OS elicitations, not single OS elicitations, induced differential responses in WRKY-silenced plants, we hypothesized that the endogenous function of WRKY genes may be to integrate multiple herbivory-associated signals. Such an ability could allow plants to respond differently to a continuously feeding herbivore and an herbivore that was only sampling. As a corollary to this hypothesis, we assumed that other environmental factors that change the phytohormone balance in plants may influence the behavior of WRKY-silenced plants and that the effects of WRKY silencing may be either enhanced or suppressed in nature. In a series of experiments over three successive field seasons in 2006, 2007, and 2008 in the natural habitat of N. attenuata in the Great Basin Desert of Utah, we assessed herbivore resistance in wild-type plants paired with either ir-wrky3 or ir-wrky6 lines. In addition, heterozygous plants that were silenced in both WRKY3 and WRKY6 were created by crossing ir-wrky3 with ir-wrky6 plants; these so-called ir-wrky3/6 plants were planted with size-matched wild-type pairs during the 2007 and 2008 field seasons. Although only the results of the 2007 season are presented, the results obtained in the 2006 and 2008 were very similar to these results (see Supplemental Figure 7 online).
Phytohormone Concentrations in Field-Grown Plants
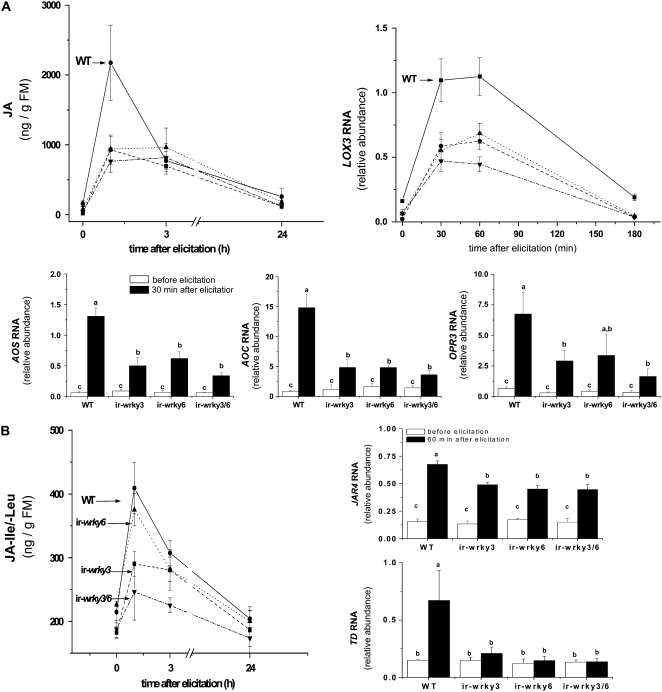
In the field, we first performed a simulated herbivory test on plants that were pre-elicited by exposure to the natural environment. When the plant pairs had grown in the field for 30 d, we elicited them with a single application of OS and harvested leaves both before and 1, 3, and 24 h after OS elicitation. Analysis of JA concentrations revealed that 1 h after OS elicitation ir-wrky3, ir-wrky6, and ir-wrky3/6 plants were significantly impaired in their ability to produce a JA burst (∼40% of the wild-type levels; ANOVA Fisher's PLSD, P = 0.0098, 0.0038, and 0.0074, respectively; Figure 4A). In addition, the constitutive JA levels measured just before OS elicitation revealed that plants silenced in either WRKY3 or WRKY6, or both, contained only 10 to 20% of the levels of JA in wild-type plants (ANOVA Fisher's PLSD, P = 0.0002, 0.0008, and 0.0024, respectively). Like the phytohormone analysis, real-time quantitative RT-PCR analysis revealed that the relative abundance of transcripts of LOX3, which encode the key enzyme in JA biosynthesis, was significantly reduced in ir-wrky3, ir-wrky6, and ir-wrky3/6 plants (Figure 4A; ANOVA Fisher's PLSD, P = 0.0039, 0.0023, and 0.0005, respectively). Similar results were obtained for AOS, AOC, and OPR3 transcript levels, which were also reduced in WRKY-silenced lines (Figure 4A).
Figure 4.
In Field-Grown Plants, Silencing WRKY3, WRKY6, or Both Reduces Constitutive and OS-Elicited Levels of JA.
(A) Mean (±se) concentrations of JA in leaves of wild-type (solid line), ir-wrky3 (dashed line), ir-wrky6 (dotted line), and ir-wrky3/6 (dashed/dotted line) plants grown in the Great Basin Desert in Utah after a single elicitation with OS. Mean (±se) LOX3 transcript levels of wild-type (solid line), ir-wrky3 (dashed line), ir-wrky6 (dotted line), ir-wrky3/6 (dashed/dotted line) plants (n = 6/harvest and genotype). Mean (± se) of AOS, AOC, and OPR3 transcript levels before (white) and 30 min after (black) OS elicitation (n = 6/harvest and genotype). Different letters reflect significant differences at P < 0.05 among plants that underwent the same treatment. The transcripts were analyzed by real-time PCR.
(B) Mean (±se) concentrations of JA-Ile/-Leu measured as above. Mean (±se) of Na TD and Na JAR4 transcript levels before (white) and 60 min after (black) OS elicitation (n = 6/harvest and genotype).
Until now, no specific differences between WRKY3- and WRKY6-silenced plants had been found. However, when the JA-Ile/-Leu pools from OS-elicited field-grown plants were analyzed, plants silenced in WRKY3 or in both transcription factors were impaired in the accumulation of JA-Ile/-Leu (ANOVA Fisher's PLSD, P = 0.0071 and 0.0018; respectively; Figure 4B), whereas plants lacking WRKY6 produced close to normal levels of JA-Ile/-Leu (ANOVA Fisher's PLSD, P = 0.3562; Figure 4B) 1 h after OS elicitation. The constitutive levels of JA-Ile/-Leu in the field were also significantly lower in ir-wrky3 and ir-wrky3/6 plants; these contained ∼80% of the JA-Ile/-Leu (ANOVA Fisher's PLSD, P = 0.0190 and 0.0425, respectively) found in ir-wrky6 and wild-type plants (ANOVA Fisher's PLSD, P = 0.9186). The reduced JA-Ile/-Leu accumulation in ir-wrky3 and ir-wrky3/6 plants was not due to the regulation of transcript levels of JA–amino acid synthetase JAR4 (Figure 4B) or Ile biosynthesis-involved gene transcripts, Thr deaminase (TD; Figure 4B); these transcripts were both similarly reduced in all WRKY-silenced lines, including ir-wrky6, which had wild-type levels of JA-Ile/-Leu.
Silencing WRKY3 and WRKY6 Impairs Both Direct and Indirect Defenses and Increases Plants' Susceptibility to Herbivores in Nature
In glasshouse experiments, ir-wrky3 and ir-wrky6 plants were found to be more susceptible to M. sexta larvae than were wild-type plants. To determine whether WRKY3 and WRKY6 transcription factors mediate resistance to other herbivores as well, we monitored herbivore damage on plants from WRKY-silenced lines and on wild-type plants in the natural environment. Although the growth of ir-wrky6, ir-wrky3, and ir-wrky3/6 plants was indistinguishable from that of pairwise planted wild-type plants, all transformed plants were heavily attacked by Trimerotropis spp grasshoppers (Figure 5A; ANOVA Fisher's PLSD, P = 0.0194, 0.0040, and 0.0465; respectively) and Tupiocoris notatus mirids (Figure 5A; ANOVA Fisher's PLSD, P = 0.0157, 0.0039, and 0.051, respectively), the two most abundant herbivores on N. attenuata at the time.
Figure 5.
WRKY-Silenced Plants Have Impaired Direct and Indirect Defenses and Are Highly Susceptible to N. attenuata's Native Herbivore Community When Transplanted into Their Native Habitat.
(A) Mean (±se) percentage canopy area damaged by grasshoppers (Trimerotropis ssp) observed on pairs of wild-type plants (white) paired with ir-wrky3, ir-wrky6, or ir-wrky3/6 (black) plants. Mean (±se) canopy damage caused by mirids (T. notatus). Mean (±se) percentage of M. sexta eggs predated per wild-type plant (solid line), ir-wrky3 (dashed line), ir-wrky6 (dotted line), or ir-wrky3/6 (dashed/dotted line) plants grown in Utah. Predation rates were monitored before and after elicitation with OS. Five eggs were glued on the second stem leaf of each plant (inset), and predation rates from G. pallens were monitored for 24 h before and for 36 h after OS elicitation.
(B) Mean (±se) of TPI activity levels in leaves from wild-type plants (black) or from lines silenced in WRKY3 (dark gray), WRKY6 (light gray), or both (open) 3 d after plants growing in the field were elicited with OS (n = 6). Different letters reflect significant differences at P < 0.05 from wild-type plants that were treated similarly. Mean (±se) DTG levels. Mean (±se) emissions of cis-α-bergamotene 24 h after OS elicitation trapped over an 8-h period. Mean (±se) emission of cis-3-hexenol. C6-compounds were trapped over an 8-h period from plants treated with wounding and OS.
In the glasshouse, the plants exposed to M. sexta herbivory showed lower levels of direct defense-related TPI transcripts and less TPI protein activity compared with wild-type plants (Figure 2E). In the field, the TPI protein activity of ir-wrky3, ir-wrky6, and ir-wrky3/6 OS-elicited plants was 45 to 55% lower than that of wild-type plants (Figure 5B; ANOVA Fisher's PLSD, P < 0.05). In addition, levels of diterpene glycosides (DTGs), which have been shown to reduce larval growth in tobacco budworm and to reduce consumption of M. sexta larvae (Jassbi et al., 2006, 2008), were also significantly lower in WRKY-silenced plants compared with wild-type plants (Figure 5B; ANOVA Fisher's PLSD, P < 0.05).
The release of volatile organic compounds (VOCs), which function as effective indirect defenses in nature by attracting Geocoris pallens predators to herbivore-attacked plants (Kessler and Baldwin, 2001), is activated by a JA burst (Halitschke and Baldwin, 2003). Because the JA burst was not as strong in the field-grown WRKY-silenced lines, we examined whether the OS-elicited VOC bouquet of wild-type plants differs from that of WRKY-silenced plants. The most active OS-elicited N. attenuata sesquiterpene known to attract predators is cis-α-bergamotene (Kessler and Baldwin, 2001). We used a whole-plant trapping system to determine the total amount of cis-α-bergamotene and C6-volatiles (green leaf volatiles) released from the induced plants. VOCs released from wild-type and WRKY-silenced plants 24 h after OS elicitation were collected over an 8-h period and measured by gas chromatography–mass spectrometry. After plants that had been exposed to natural herbivores were elicited once with OS, the emission of cis-α-bergamotene was lower in all three transformed lines, ir-wrky3, ir-wrky6, and ir-wrky3/6, than in wild-type plants (Figure 5B; ANOVA Fisher's PLSD, P < 0.0001, < 0.0001, and < 0.0001, respectively). In contrast with levels of the sesquiterpene, bergamotene, levels of green leaf volatile cis-3-hexenol emitted by WRKY plants upon OS elicitation were not significantly different from those emitted by wild-type plants (Figure 5B).
We assumed that the abnormal herbivore loads and damage suffered by WRKY-silenced plants resulted from both impaired direct defenses (as demonstrated above) and the inability of these plants to recruit predatory insects by releasing cis-α-bergamotene after being attacked. To estimate the role of indirect defenses in herbivore susceptibility, we glued M. sexta eggs to wild-type and WRKY-silenced plants and measured the rate at which Manduca eggs were predated (Figure 5A). Predation rates of eggs placed on ir-wrky3, ir-wrky6, and ir-wrky3/6 plants were determined in the habitats used previously to monitor herbivore-induced damage. Predation of M. sexta eggs began immediately after OS elicitation (and cis-α-bergamotene emission); 36 h later, the predation rates on ir-wrky3, ir-wrky6, and ir-wrky3/6 plants were significantly lower than those on wild-type plants (Figure 5A; ANOVA Fisher's PLSD, P < 0.0001, < 0.0001, and < 0.0001, respectively). All predated eggs carried the characteristic signature of attack from G. pallens, namely, a small puncture hole through which the contents had been emptied.
Exogenous Application of JA Complements WRKY Phenotypes
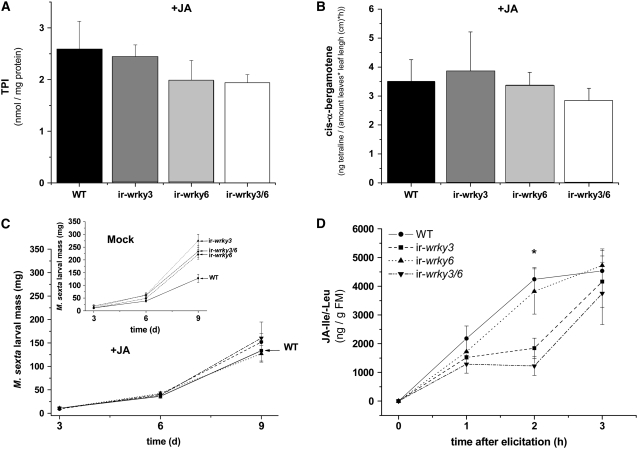
Overall, WRKY3 and WRKY6 seem to contribute to mounting increased levels of herbivore resistance by regulating JA levels and/or JA-dependent direct and indirect defenses. To determine whether impaired resistance to herbivores and compromised defenses in WRKY-silenced plants could be primarily due to lower JA levels (Figures 2D, 3B, and 4A), we treated plants with JA in the field and in the glasshouse. First, we treated leaves of field-grown plants with OS that had been supplemented with synthetic JA. This direct JA complementation restored TPI activity levels in ir-wrky3, ir-wrky6, and ir-wrky3/6 plants to the levels observed in wild-type plants (Figure 6A). In addition, the levels of cis-α-bergamotene released from OS-elicited ir-wrky3, ir-wrky6, and ir-wrky3/6 plants were restored by JA treatment to the levels released from wild-type plants (Figure 6B), implying that lower JA levels in WRKY-silenced plants were responsible for lower OS-elicited direct and indirect defenses.
Figure 6.
Impaired Direct and Indirect Defenses Are Restored by Applying JA, and Plants Silenced in WRKY3 Are Delayed in JA-Ile Conjugation.
(A) Mean (±se) of TPI activity measured after plants were elicited with JA-supplemented OS (10 μg/μL, n = 6) and harvested 3 d later.
(B) Mean (±se) emissions of cis-α-bergamotene from wild-type plants (black), lines silenced in WRKY3 (dark gray), WRKY6 (light gray), or both (open) 24 h after plants were treated with JA-supplemented OS (10 μg/μL).
(C) Larval mass of M. sexta that fed on wild-type plants (solid line), WRKY3 (dashed line), WRKY6 (dotted line), and WRKY3/6-silenced (dashed/dotted line) glasshouse-grown plants sprayed with 1 mM JA solution (diluted in 30% ethanol) (+JA; n = 12) or sprayed with 30% ethanol as control (inset; mock, n = 12)
(D) Mean (±se) of JA-Ile conjugation after adding both exogenous JA (0.625 μmol) and Ile (0.625 μmol) to the OS in the greenhouse (n = 4, * P < 0.05, as determined by Fisher's PLSD test from ANOVA).
In the field experiments, ir-wrky3 and ir-wrky3/6 plants were impaired in JA-Ile/-Leu accumulation. In similar glasshouse JA complementation experiments, both ir-wrky3 and ir-wrky3/6 lines were found to have delayed JA-Ile bursts, suggesting that these plants indeed possess lower conjugation capacity compared with wild-type or ir-wrky6 plants (Figure 6D). These results suggest that WRKY3 may play role in activating the JAR4 enzyme or regulating the transcription of an unknown JA conjugating enzyme in N. attenuata (Figure 7).
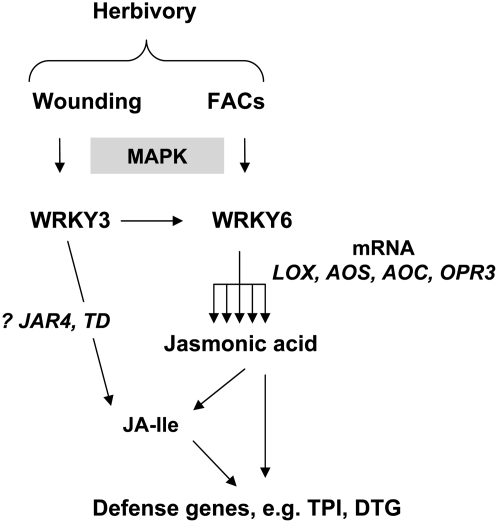
Figure 7.
Proposed Function and Inferred Relative Position of the Two N. attenuata WRKY Transcription Factors in the Defense Signaling Cascade against Herbivores.
Two N. attenuata WRKY transcription factors regulate transcript accumulation of JA biosynthesis genes. The JA derivate JA-Ile regulates direct and indirect defenses in tobacco plants. WRKY3 transcription factor is required for transcript accumulation of the WRKY6 gene, and both genes are under the control of MAPK signaling cascade, herbivory-specific signals, wounding, and herbivore elicitors (FACs).
Next, we sprayed wild-type, ir-wrky3, ir-wrky6, and ir-wrky3/6 rosette-stage plants in the glasshouse with a 1 mM JA solution (dissolved in 30% ethanol) and examined how M. sexta performed on them. In a control experiment, M. sexta larvae that fed on plants sprayed with an equivalent amount of 30% ethanol (used to suspend JA) retained their accelerated growth rates on ir-wrky3, ir-wrky6, and ir-wrky3/6 compared with wild-type plants (inset in Figure 6C), essentially as observed before in unsprayed plants (Figure 2C). However, the caterpillars that fed on WRKY-deficient plants sprayed with JA had the same low growth rates as caterpillars that fed on wild-type plants (Figure 6C), demonstrating that the levels of direct defenses in WRKY-deficient plants were restored to normal levels.
DISCUSSION
We describe two WRKY transcription factors from N. attenuata that are differentially regulated during herbivore attack; both factors play essential roles in the resistance of tobacco plants to herbivores. Plants silenced in the expression of their WRKY3 and WRKY6 genes are significantly more vulnerable to herbivores in the glasshouse and in nature. We show that direct and indirect defense strategies are strongly compromised in plants in which WRKY3- and WRKY6-dependent transcriptional control of gene expression has been suppressed.
Transcription of N. attenuata WRKY Genes
WRKY transcription factors that are involved in regulating various plant-specific responses, including pathogen defense, senescence, trichome development, and the biosynthesis of secondary metabolites, are often functionally redundant, complicating analysis of these genes (Ulker and Somssich, 2004; Zhang and Wang, 2005). Two WRKY genes isolated from N. attenuata contain highly conserved WRKY domains in their protein sequence; however, other parts of the proteins show significantly less similarity, suggesting that the functions of these proteins do not overlap. Indeed, the phenotypic similarity we found between ir-wrky3 and ir-wrky6 plants is due to the function of WRKY3, which is required to activate WRKY6 transcription. In other words, plants deficient in WRKY3 are also deficient in WRKY6 accumulation, resulting in a common phenotype that is determined by the downstream gene product of WRKY6 (Figure 7).
Two previously reported defense-related genes from N. tabacum that have strong similarity to N. attenuata WRKY3 and WRKY6 genes, Nt WRKY2 and Nt WRKY1, rapidly respond to wounding and treatment with fungal elicitors (Nishiuchi et al., 2004; Yamamoto et al., 2004). Molecular analysis of the recombinant proteins showed that these proteins bind specifically to the W-box sequence found in the promoter of tobacco class I basic chitinase gene CHN48; this promoter was also transactivated in transient expression assays with N. tabacum WRKY1, WRKY2, and WRKY4 proteins. In addition, the efficiency of the transactivation process was further enhanced in elicitor-treated cells, suggesting that the proteins were posttranslationally modified in response to elicitor-derived signals.
Positioning WRKY Genes in the Defense Cascade
We found that neither of the N. attenuata WRKY genes is transcriptionally induced by JAs. Thus, these genes may function in herbivore defense downstream of the hypothetical FAC receptor in plants and upstream of the JA signal (Figure 7). Recently, two wound- and FAC-elicited mitogen-activated protein kinases (MAPKs), salicylic acid–induced protein kinase (SIPK) and wound-induced protein kinase (WIPK), were characterized in N. attenuata (Wu et al., 2007). Plants whose SIPK or WIPK was silenced by virus-induced gene silencing had diminished JA and JA-Ile/-Leu bursts and lower levels of JA biosynthetic gene transcripts as well as lower WRKY6 transcript levels (Wu et al., 2007). Given the similarity in phenotypes and the dependence of WRKY6 gene transcription on SIPK and WIPK, we propose that WRKY3 and WRKY6 transcriptionally activate JA biosynthesis and downstream defenses in a MAPK-dependent manner. The obvious targets of N. attenuata WRKY3 and WRKY6 are the JA-biosynthetic genes, including LOX3, AOS, AOC, and OPR3, which are essential for JA accumulation after wounding and herbivore attack (Halitschke and Baldwin, 2003; Figure 4A). Accumulation of these transcripts was lower in field-grown plants silenced in WRKY expression; however, it is still unknown whether WRKY proteins directly regulate the transcription of these genes or if they use another transcription factor(s) to interact with their promoters.
Are WRKY Genes Targets of MAPKs?
The activation of several WRKY transcription factors is known to be regulated posttranslationally by phosphorylation (Ulker and Somssich, 2004). For example, N. tabacum WRKY1 (accession number AAD16138), which is structurally different from the tobacco WRKY1 gene described by Nishiuchi et al. (2004) (accession number BAA82107), is phosphorylated by SIPK; this process in turn enhances WRKY1 DNA binding activity to its cognate binding site, a W-box sequence from the tobacco chitinase gene CHN50 (Menke et al., 2005). In addition, the closest Arabidopsis relative to N. attenuata WRKY3 and WRKY6 genes, pathogen-, SA- and ethylene-responsive WRKY33 (Rushton et al., 2002), is phosphorylated by MAP kinase 4 (MPK4). Because WRKY33 also interacts with the MKS1 protein, MKS1 is thought to help couple the kinase to the WRKY33 transcription factor during defense activation (Andreasson et al., 2005). Interestingly, MPK4 functions to regulate hormone crosstalk and systemic acquired resistance in Arabidopsis plants: it is required both to repress SA-dependent resistance and to activate JA-dependent defense gene expression (Petersen et al., 2000). The involvement of MAPKs in regulating JA and herbivore defense (Wu et al., 2007) and the dependence of pathogen resistance on the same MAPK cascade (Seo et al., 2007) may facilitate crosstalk between plant defenses against pathogens and herbivores and two main phytohormone signals, SA and JA (Li et al., 2004, 2006; Qiu et al., 2007). Divergence of signaling pathways after the MAPK level probably involves specific transcription factors, including members of the WRKY family.
Overexpressing WRKY Genes Does Not Produce a Defense Phenotype
The ectopic overexpression of WRKY3 and WRKY6 transcription factors in our experiments did not result in the expected constitutive defense phenotype. Specific posttranslational modification(s) of WRKY proteins, including phosphorylation by MAPKs (Ulker and Somssich, 2004), during defense elicitation may be required to mediate downstream responses. The presence of transcriptional complexes consisting of several WRKY proteins and other scaffold proteins (such as MKS1) may also be required for WRKY3 and WRKY6 to function in planta. Several examples of such interactions have been reported in the literature: WRKY71 and WRKY51 proteins interact when the α-amylase gene is repressed in rice aleurone cells (Xie et al., 2006), and pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors interact in pathogen defense (Xu et al., 2006). Understanding the posttranslational modifications and interactions of N. attenuata WRKY proteins will make it easier to determine the molecular function of these proteins in herbivore defense.
Alternatively, WRKY transcripts in wild-type plants accumulate within 30 min after their first encounter with herbivores (Figures 1B and 1C). This could mean that the plants ectopically overexpressing WRKY genes gain only very limited time advantage from having the transcripts, while efficient herbivore defense responses take at least several hours to launch. The short time advantage of WRKY-overexpressing lines may be too short to be translated into a stronger defense response.
Using Gene Silencing to Understand the Role of WRKY3 and WRKY6 in Defense
To understand the ecological role of WRKY regulatory proteins, we used gene silencing in combination with bioassays and field releases; these techniques allowed us to monitor a large range of responses and interactions between various stress factors, including pathogens, herbivores, and abiotic stress, in a single experimental design (Baldwin, 2001). We performed parallel experiments with WRKY-silenced plants in the glasshouse and at our experimental station in the Great Basin Desert in Utah using M. sexta and natural populations of herbivores (including Manduca quinquemaculata), respectively. In the field, ir-wrky3 and ir-wrky6 plants were morphologically indistinguishable from wild-type plants; however, silencing WRKY3 and WRKY6 genes impaired the deployment of both direct and indirect defenses, thus making these plants more susceptible to herbivores in nature. As the field data showed, several novel phenotypic differences appeared when plants were grown in their natural habitat and highlight the value of studying highly pleiotropic responses, such as resistance traits, in natural environments.
JA Metabolism in Field-Grown WRKY-Silenced Plants
JA and JA-Ile/-Leu bursts in field-grown plants were different among ir-wrky3, ir-wrky6, and wild-type plants. In the controlled glasshouse environment, the phytohormone burst in inexperienced ir-wrky plants was identical to the burst in wild-type plants after a single OS elicitation (Figures 3A and 3C). However, in the field-grown plants that were silenced in WRKY3 or WRKY6 expression, we observed smaller JA bursts after OS elicitation than in wild-type plants (Figure 4A), which was similar to multiply elicited glasshouse-grown plants (Figures 3B and 3D). The additional differences in JA-Ile/-Leu levels observed in ir-wrky3 plants suggest that WRKY3 and WRKY6 genes, despite their common phenotype, affect JA metabolism differently. We confirmed the differences found in JA-Ile/-Leu concentrations in the field after OS elicitation in a separate laboratory experiment using glasshouse plants that were simultaneously elicited with OS and exogenous JA (the latter to mimic the plant's prior history of attack in the field). Such treatment again resulted in lower levels of JA-Ile/-Leu in ir-wrky3 plants compared with wild-type or ir-wrky6 plants. At present, we are analyzing the role of both WRKY genes in JA-Ile/-Leu formation and in the WRKY-mediated responses; in particular, we ask why ir-wrky6 plants that accumulate wild-type levels of JA-Ile/-Leu still show impaired defense responses. JA-Ile/-Leu is believed to be the form of JA that mediates the degradation of JAZ repressors and activates most defense genes at the transcriptional level (Chini et al., 2007; Thines et al., 2007). We are exploring two working hypotheses: that WRKY3 controls the expression of JA-conjugating enzymes and that WRKY6 interacts with the signaling pathway downstream of JA-Ile. Interestingly, N. attenuata plants silenced in their expression of the COI1 gene show normal or even increased levels of JA-Ile/-Leu, despite their low levels of elicited JA (Paschold et al., 2008). It suggests that WRKY6 may interact with JA signaling at the level of the COI1 protein; however, in contrast with true COI1 mutants, this deficiency can be restored by exogenously applied JA and supra-optimal levels of JA-Ile (Figures 6A to 6D).
The Priming Effect of N. attenuata WRKY3 and WRKY6 Genes
The fact that treatment with JA restored the expression of defenses that were impaired in both WRKY-silenced lines suggests that impaired accumulation of JA and its metabolites in these plants was primarily responsible. However, the JA burst in glasshouse-grown plants after a single OS elicitation did not differ significantly between ir-wrky and wild-type plants. The differences in JA content and subsequently compromised defenses appeared only when plants were exposed to (1) natural stresses, (2) continuous feeding by M. sexta larvae, or (3) repeated OS elicitations that closely mimicked herbivore feeding. These findings suggest that the main function of WRKY3 and WRKY6 genes might not be to elicit the primary JA burst but, rather, to integrate and maintain JA levels and defense responses during prolonged herbivore attack. According to our observations, the JA burst in N. attenuata elicited by a single OS elicitation lasts on average 3 h, after which levels of JA and JA-Ile/-Leu return to their pre-elicitation levels (Kang et al., 2006). How repeated elicitations, whose timing and duration vary due to variable insect feeding behavior (Van Dam et al., 2001b), are integrated to elicit cost-optimized defense responses in plants remains largely unknown. We propose that N. attenuata WRKY genes, being elicited by wounding and FACs, help to coordinate these defense signals generated from individual encounters with herbivores, potentiating and/or sustaining optimal defense output. Consistent with this hypothesis, both constitutive JA levels and the OS-elicited JA burst of field-grown WRKY-silenced plants were found to be significantly smaller than these levels or this burst in wild-type plants. Such a discrepancy could be viewed as the failure of WRKY-deficient plants to integrate environmental signals into an optimized defense response. Further experiments are now being conducted that are aimed at finding the direct transcriptional targets of WRKY3 and WRKY6 genes in native tobacco, which will help to further establish the role of these genes in mediating defense responses to herbivores.
METHODS
Isolation of WRKY3/6 cDNAs
WRKY3 and WRKY6 were isolated from cDNA libraries (λZAPll vector; Stratagene) prepared from the leaves of Nicotiana attenuata plants fed on by Manduca sexta for 24 h (Ziegler et al., 2001). We screened the library using a WRKY fragment isolated from a DDRT-PCR (Hui et al., 2003) according to the manufacturer's instructions. Sequencing was conducted using an ABI PRISM 377 automated sequencer (Applied Biosystems). Clones were sequenced in both sense and antisense directions. Using ClustalW (Multiple Sequence Alignment, http://align.genome.jp/), we aligned protein sequences from WRKYs with at least some published biological functions and constructed a neighbor-joining tree using MEGA 3.1 software (see Supplemental Figure 1A online).
Plant Culture in the Glasshouse and in the Field
Seeds of wild-type N. attenuata Torr. Ex Watts lines (synonymous with Nicotiana torreyana Nelson and Macbr.; Solanaceae), originally collected from a native population in southwestern Utah in 1988, were transformed (see below) and used for all experiments. The same inbred generation of seeds was used for the wild type and transformations.
For glasshouse experiments, seeds were germinated on sterile Gamborg B5 media (Sigma-Aldrich) and grown as described by Kruegel et al. (2002) and Qu et al. (2004). For field experiments in 2006, plants were transferred to 50 mm of peat pellets (Jiffy Products) after germinating in Petri dishes on agar medium. Seedlings were gradually exposed to the environmental conditions of the Great Basin Desert (high sun exposure and low relative humidity) over 2 weeks and afterwards situated in two different habitats: adapted seedlings of the same size were transplanted to a field plot at the Lytle Ranch Preserve research station (Santa Clara, UT) and into a natural population of N. attenuata growing in an area near Santa Clara, UT, that had been burned by a wildfire in 2005. Seedlings were watered every other day for 2 weeks until roots were established in the native soil. Releases of the transformed plants were conducted under APHIS notification number 06-003-08n.
In 2007, seeds were germinated directly in Jiffy 703 pots. These were watered every other day over 2 weeks and fertilized with iron solution (stock solution 2.78 g FeSO4·7 H2O and 3.93 g Triplex in 1 liter H2O, 100-fold diluted). After 1 month, plants were transplanted into natural habitats of N. attenuata in the Great Basin Desert. Wild-type and transformed plants were placed in a watered field plot at the Lytle Ranch Preserve research station in pairs. Releases of the transformed plants were conducted under APHIS notification number 062424-03r. Experiments in 2008 were conducted under APHIS notification numbers 06-242-02r and 06-242-03r.
Nucleic Acid Analysis
The isolation of DNA and RNA and DNA gel blot analysis were performed as described by Winz and Baldwin (2001). Blots were hybridized with radioactively labeled WRKY3 or WRKY6 full-length cDNA or a PCR fragment of the hygromycin phosphotransferase II gene specific for the selective marker located on the T-DNA.
A quantitative real-time PCR assay was used to analyze the transcript accumulation of WRKY3/6. Using TaqMan gene expression assay, specific amplicons for WRKY3 (WRKY3_F1, 5′-CAGGATATGCAAATTCAGAGGATTC-3′; WRKY3_R1, 5′-ATTCAATTCAGCAGAGCAATGTG-3′; and WRKY3_P1, 5′-AGCAAAGGACGAGCCTCGAGGTGACA-3′) and WRKY6 (WRKY6_F1, 5′-ACAAAACAAAGATGAAGTTCCAAAG-3′; WRKY6_R1, 5′-GGAGAAGCTGGTGATGAAGATG-3′; and WRKY6_P1, 5′-AAGTCATTTCCACCTTGTTCTTTGCCA-3′) detecting only the endogenous transcript were designed. Two additional set of primers were later used with the SYBR Green detection system for WRKY3 (WRKY3_5prime_FWD, 5′-CCAATTTGGACAATCTTTTGCATC-3′; WRKY3_5prime_RVS, 5′-TGCATAGTGTTGAATTCAGTTCTTCC-3′) and WRKY6 (WRKY6_3prime_FWD, 5′-TCGATGTAATTTATGATAGTTTTTGC-3′; WRKY6_3prime_RVS, 5′-CAAATATCTTAATGGGGAATTACAGC-3′) detection of endogenous transcripts. As both detection methods provided consistently similar results, only TaqMan data are presented in the figures. LOX3, AOS, AOC, OPR3, JAR4, and JAR6 transcript levels were detected with primers as described by Paschold et al. (2008).
Total RNA was isolated from four to five replicated wild-type plants, and each of the transgenic lines from each treatment and time point and 66.6 ng of total RNA of each samples was used for cDNA synthesis with the SuperScript first-strand synthesis system (Invitrogen). Real-time PCR was performed on a SDS7700 system (Applied Biosystems) using the qPCR core reagent kit for TaqMan and qPCR core kit for SYBR green analysis, respectively (Eurogentec). N. attenuata sulfite reductase (ECI) gene was used as an internal standard for normalizing cDNA concentration variations (Wang et al., 2007b).
Generation and Characterization of Transgenic Plants
Plants overexpressing WRKY3 and WRKY6 were generated by cloning full-length PCR-amplified cDNA fragments of WRKY3 (ov-WRKY3) and WRKY6 (ov-WRKY6) in a pRESC2 transformation vector after a constitutive 35S cauliflower mosaic virus promoter (see Supplemental Figures 3A and 3B online). Plants were transformed and screened for single T-DNA inserts by DNA gel blotting (see Supplemental Figures 3C and 3D online).
To generate stably transformed plants silenced in their expression of WRKY3, a 1-kb cDNA fragment of WRKY3 was PCR amplified using the primer sets of WRKY5-33 (5′-GCGGCGCCATGGCTCGAGGCTCGTCCTTTGCTC-3′), WRKY6-33 (5′-GCGGCGCTGCAGCTCAGAGCAATGGTGGGAACC-3′), and WRKY7-30 (5′-GCGGCGGAGCTCAGAGCAATGGTGGGAACC-3′) and was cloned in an inverted-repeat orientation (ir-wrky3) in a pSOL3 transformation vector (see Supplemental Figure 5A online) as described by Bubner et al. (2006) to silence the gene. In the case of WRKY6, a 1-kb cDNA fragment of WRKY6 was amplified with the primers WRKY8-34 (5′-GCGGCGCCATGGCGAATACAATGAATTGAGATGG-3′), WRKY10-34 (5′-GCGGCGCTCGAGCGAATACAATGAATTGAGATGG-3′), and WRKY9-21 (5′-GGAAATATGGGCAGAAGCAAG-3′), and PCR products were also cloned in an inverted-repeat orientation (ir-wrky6) in a pSOL3 transformation vector (see Supplemental Figure 5B online) using respective restriction sites or blunt-ended DNA fragments (Bubner et al., 2006).
Plant transformation using Agrobacterium tumefaciens was performed as described by Kruegel et al. (2002). Progeny of homozygous lines harboring a single T-DNA insertion were selected by hygromycin resistance screening, confirmed by DNA gel blotting (see Supplemental Figures 5C and 5D online) and determined to be diploid by flow cytometry (Bubner et al., 2006). T2 and T3 homozygous plants from two homozygous lines, which accumulated low basal and OS-elicited levels of WRKY3 and WRKY6 transcripts (Figures 2A and 2B), were used for subsequent analysis and creation of the double-silenced heterozygous plant. The impact of silencing WRKY3 and WRKY6 on other independent WRKY genes was assessed by measuring the transcript levels of five other independent WRKY genes in ir-wrky3, ir-wrky6, and ir-wrky3/6 plants. The transcripts of these genes were not influenced, suggesting that silencing constructs were specific for WRKY3 and WRKY6 genes (see Supplemental Figure 8 online).
Plant Treatments in the Glasshouse
For the kinetic analysis of WRKY expression in wild-type plants, five individual plants (biological replicates) were used at each harvest time for each treatment. Rosette-stage plants were randomly assigned to treatments. Leaves at nodes +1 and +2 (first and second fully expanded) were elicited. A fabric pattern wheel was used to create six rows of puncture wounds parallel to the midrib. Twenty microliters of deionized water or M. sexta OS (diluted in 1:1 deionized water) from fourth- or fifth-instar larvae (for the treatment referred to as OS-elicited) or an OS equivalent concentration of a synthetic mixture of the two most abundant FACs in M. sexta OS, N-linolenoyl-l-glutamine and N-linolenoyl-l-glutamate (0.012 mM N-linolenoyl-l-glutamine and 0.034 mM N-linolenoyl-l-glutamate, synthesized in-house), were immediately added to the puncture wounds. Nonwounded plants were treated with 150 μg methyl jasmonate in 20 μL of lanolin paste, which was applied to lamina of the leaves. The treated leaves or those at the next youngest positions (leaves at nodes 0 and −1) were harvested 0, 15, 30, 60, 120, 240, and 480 min after treatment and frozen in liquid nitrogen. Samples from the plants treated with FACs were harvested only until 60 min after elicitation.
To examine the effects of silencing WRKY3 or WRKY6 on the accumulation of JA and JA-Ile/-Leu at the site of attack from M. sexta larvae, neonates were placed on each of six replicate plants from each genotype and harvest, two each at leaf nodes +2, +1, 0, and −1, and enclosed in clip cages. Leaf tissue from within 2 cm of the feeding edge was collected 22 and 30 h after plants were infested with larvae. For the standardized elicitation of phytohormone accumulation, leaves at node +2 from plants at the rosette stage were OS elicited. The accumulation of JA and JA-Ile/-Leu was monitored 60 min after a single elicitation (one leaf elicited with six rows of holes and immediately treated with 20 μL of OS) and a multiple elicitation (six consecutive OS elicitations involving one leaf wounded with one row of holes to which 10 μL of OS was applied every 30 min for six times and harvested 30 min after the last elicitation). Identical nonelicited leaves were also harvested as controls.
Analysis of Herbivore Susceptibility in the Glasshouse and in the Field
In the glasshouse, M. sexta larvae were monitored on the stably transformed lines. M. sexta eggs were purchased from North Carolina State University. To examine how M. sexta performed on WRKY-overexpressed lines and WRKY-silenced lines and compare how they performed on wild-type plants in the glasshouse, we enclosed neonates on node +2 leaves and allowed the caterpillars to feed. Each caterpillar was weighed on days 4, 6, and 8 (and 10).
In the field, we observed the abundance of naturally occurring herbivores and the particular damage they caused to 23 pairs of wild-type and ir-wrky6 lines and to eight pairs of wild-type and ir-wrky3 lines in 2006 as well as to six to eight pairs of ir-wrky3, ir-wrky6, and ir-wrky3/6 lines in 2007, respectively. Plants were monitored starting 21 d after they were transplanted into the field in 2006 and 50 d after being transplanted in 2007, depending on environmental conditions. To examine the effect of silencing WRKYs on herbivore-induced indirect defense, we conducted an egg predation assay specifically designed to measure the ability of N. attenuata's herbivore-induced VOCs to attract the dominant predator of N. attenuata herbivores, Geocoris pallens, as previously described (Kessler and Baldwin, 2001). Five M. sexta eggs were glued on the second stem leaf using a natural glue that is known to have no effect on predation rates or on plants' VOC emissions (Kessler and Baldwin, 2001). Seven replicate wild-type and ir-wrky3 plant pairs and seven wild-type and ir-wrky6 plant pairs were used in this experiment in 2006 and six pairs, a wild-type and a transformed plant, in 2007. The number of M. sexta eggs predated was measured before elicitation and after the first stem leaf was OS elicited.
Analysis of Direct and Indirect Defense Traits
To analyze secondary metabolites and TPI activity, all of which are known to be regulated by M. sexta attack, elicited leaf tissue was harvested 72 h after continuous feeding by M. sexta or after OS elicitation; the accumulation of secondary metabolites (nicotine and DTGs) was analyzed by high-performance liquid chromatography as described by Keinanen et al. (2001). TPI activity was analyzed by the radial diffusion assay described by Van Dam et al. (2001a).
In 2006, terpenoid VOCs were trapped from individual leaves of field-grown plants 24 h after OS elicitation. In the next season, whole plants were used to collect sesquiterpenes and C6-volatiles (green leaf volatiles). The VOCs released from treated leaves or whole plants were trapped for 8 h using charcoal traps (OrboM32; Supelco). The traps were spiked with 80 ng of tetraline as an internal standard and eluted with 500 μL dichloromethane. Samples were analyzed by gas chromatography–mass spectrometry as described by Halitschke et al. (2000).
Analysis of Phytohormone Accumulation
Approximately 300 mg of tissues from each sample was homogenized on a FastPrep homogenizer (Thermo Electron) with 1 mL of ethyl acetate spiked with 200 ng of 1,2-13C-JA, paracoumaric acid and D4-SA as internal standards in the FastPrep tubes. After centrifugation at maximum speed for 10 min at 4°C, the supernatants were transferred to fresh 2-mL Eppendorf tubes, and the pellet was re-extracted with 1 mL of ethyl acetate and centrifuged. The supernatants were combined and evaporated completely on a vacuum concentrator. The residue was resuspended in 0.5 mL of 70% methanol (v/v) and centrifuged at maximum speed for 1 min. The supernatants were then used to analyze JA and JA-Ile/-Leu with a 1200L Varian liquid chromatography–triple-quadrupole-mass spectrometry system (Varian). Fifteen microliters of each sample was injected into a Pursuit C8 column (3 μm, 150 × 2 mm; Varian) at a flow rate of 0.1 mL/min. A gradient of mobile phase composed of solvent A (0.05% formic acid) and solvent B (0. 05% formic acid in methanol) was used for separation, and a negative electrospray ionization mode was used for detection. Ions with mass-to-charge ratios at 209, 211, 322, and 163 generated from endogenous JA, internal JA standard, JA-Ile/-Leu conjugates, and paracoumaric acid in the ion source were collected and fragmented with a collision energy of 12 V. Since JA-Ile and JA-Leu have the same molecular mass and retention time, our liquid chromatography–triple-quadrupole-tandem mass spectrometry method was not able to distinguish the two, as described by Wang et al. (2007b).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession numbers AY456271 (Na WRKY3) and AY456272 (Na WRKY6) (see Supplemental Figure 1 online).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Plant WRKY Gene Family: Phylogenetic Relationships, Biological Function, and Elicitors of Selected WRKY Genes.
Supplemental Figure 2. JA Signaling Does Not Influence WRKY Transcript Accumulation.
Supplemental Figure 3. Plant Transformation Vector pRESC2 and DNA Gel Blot Analysis of Overexpressing ov-WRKY3 and ov-WRKY6 Lines.
Supplemental Figure 4. Ectopic Overexpression of WRKY3 or WRKY6 Does Not Change M. sexta Performance or Levels of Jasmonic Acid and JA-Ile/-Leu.
Supplemental Figure 5. Plant Transformation Vector pSOL3 and DNA Gel Blot Analysis of Inverted-Repeat WRKY3 (ir-wrky3) and Inverted-Repeat WRKY6 (ir-wrky6) Lines.
Supplemental Figure 6. Silencing WRKY3, WRKY6, or Both Does Not Alter Plants' Physiology.
Supplemental Figure 7. WRKY-Silenced Plants Are Highly Susceptible to N. attenuata's Native Herbivore Community When Transplanted into a Native Habitat in 2006 and 2008.
Supplemental Figure 8. Expression Pattern of Five Other WRKY Genes Is Not Altered in ir-wrky3– and ir-wrky6–Silenced Plants.
Supplemental Data Set 1. Alignment Used to Generate Supplemental Figure 1.
Supplementary Material
Acknowledgments
This work was funded by the Max-Planck-Gesellschaft. We thank K. Gase, T. Hahn, S. Kutschbach, T. Krügel, and the glasshouse team for invaluable assistance with the experiments as well as E. Rothe, M. Schöttner, and N. Heinzel for technical assistance. For the help with the field work and volatile experiments, we thank D. Kessler, C. Diezel, E. Körner, M. Schuman, and G. Rayapuram as well as Brigham Young University for use of the Lytle Ranch Preserve field station and USDA-APHIS for constructive regulatory oversight of the release of genetically modified organisms.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Ian T. Baldwin (baldwin@ice.mpg.de).
Online version contains Web-only data.
References
- Agrawal, A.A. (2001). Ecology - Phenotypic plasticity in the interactions and evolution of species. Science 294 321–326. [DOI] [PubMed] [Google Scholar]
- Andreasson, E., (2005). The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 24 2579–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin, I.T. (2001). An ecologically motivated analysis of plant-herbivore interactions in native tobacco. Plant Physiol. 127 1449–1458. [PMC free article] [PubMed] [Google Scholar]
- Bubner, B., Gase, K., Berger, B., Link, D., and Baldwin, I.T. (2006). Occurrence of tetraploidy in Nicotiana attenuata plants after Agrobacterium-mediated transformation is genotype specific but independent of polysomaty of explant tissue. Plant Cell Rep. 25 668–675. [DOI] [PubMed] [Google Scholar]
- Chini, A., Fonseca, S., Fernandez, G., Adie, B., Chico, J.M., Lorenzo, O., Garcia-Casado, G., Lopez-Vidriero, I., Lozano, F.M., Ponce, M.R., Micol, J.L., and Solano, R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448 666–671. [DOI] [PubMed] [Google Scholar]
- Chuio, T., et al. (2007). Involvement of the elicitor-induced gene OsWRKY53 in the expression of defense-related genes in rice. Biochim. Biophys. Acta 1769 497–505. [DOI] [PubMed] [Google Scholar]
- De Moraes, C.M., Lewis, W.J., Pare, P.W., Alborn, H.T., and Tumlinson, J.H. (1998). Herbivore-infested plants selectively attract parasitoids. Nature 393 570–573. [Google Scholar]
- Dicke, M., and Sabelis, M.W. (1988). How plants obtain predatory mites as bodyguards. Neth. J. Zool. 38 148–165. [Google Scholar]
- Eulgem, T. (2006). Dissecting the WRKY web of plant defense regulators. PLoS Pathog. 2 1028–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem, T., and Somssich, I.E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10 366–371. [DOI] [PubMed] [Google Scholar]
- Halitschke, R., and Baldwin, I.T. (2003). Antisense LOX expression increases herbivore performance by decreasing defense responses and inhibiting growth-related transcriptional reorganization in Nicotiana attenuata. Plant J. 36 794–807. [DOI] [PubMed] [Google Scholar]
- Halitschke, R., Kessler, A., Kahl, J., Lorenz, A., and Baldwin, I.T. (2000). Ecophysiological comparison of direct and indirect defenses in Nicotiana attenuata. Oecologia 124 408–417. [DOI] [PubMed] [Google Scholar]
- Halitschke, R., Schittko, U., Pohnert, G., Boland, W., and Baldwin, I.T. (2001). Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. III. Fatty acid-amino acid conjugates in herbivore oral secretions are necessary and sufficient for herbivore-specific plant responses. Plant Physiol. 125 711–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil, M., and Baldwin, I.T. (2002). Fitness costs of induced resistance: Emerging experimental support for a slippery concept. Trends Plant Sci. 7 61–67. [DOI] [PubMed] [Google Scholar]
- Heil, M., Koch, T., Hilpert, A., Fiala, B., Boland, W., and Linsenmair, K.E. (2001). Extrafloral nectar production of the ant-associated plant, Macaranga tanarius, is an induced, indirect, defensive response elicited by jasmonic acid. Proc. Natl. Acad. Sci. USA 98 1083–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermsmeier, D., Schittko, U., and Baldwin, I.T. (2001). Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. I. Large-scale changes in the accumulation of growth- and defense-related plant mRNAs. Plant Physiol. 125 683–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui, D.Q., Iqbal, J., Lehmann, K., Gase, K., Saluz, H.P., and Baldwin, I.T. (2003). Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. V. Microarray analysis and further characterization of large-scale changes in herbivore-induced mRNAs. Plant Physiol. 131 1877–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby, M., Weisshaar, B., Droge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., and Parcy, F. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7 106–111. [DOI] [PubMed] [Google Scholar]
- Jassbi, A.R., Gase, K., Hettenhausen, C., Schmidt, A., and Baldwin, I.T. (2008). Silencing geranylgeranyl diphosphate synthase in Nicotiana attenuata dramatically impairs resistance to Manduca sexta. Plant Physiol. 146 974–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassbi, A.R., Zamanizadehnajari, S., Kessler, D., and Baldwin, I.T. (2006). A new acyclic diterpene glycoside from Nicotiana attenuata with a mild deterrent effect on feeding Manduca sexta larvae. Zeitschrift Fur Naturforschung Section B A J. Chem. Sci. 61 1138–1142. [Google Scholar]
- Johnson, C.S., Kolevski, B., and Smyth, D.R. (2002). TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14 1359–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalde, M., Barth, M., Somssich, I.E., and Lippok, B. (2003). Members of the Arabidopsis WRKY group III transcription factors are part of different plant defense signaling pathways. Mol. Plant Microbe Interact. 16 295–305. [DOI] [PubMed] [Google Scholar]
- Kang, J.H., Wang, L., Giri, A., and Baldwin, I.T. (2006). Silencing threonine deaminase and JAR4 in Nicotiana attenuata impairs jasmonic acid-isoleucine-mediated defenses against Manduca sexta. Plant Cell 18 3303–3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinanen, M., Oldham, N.J., and Baldwin, I.T. (2001). Rapid HPLC screening of jasmonate-induced increases in tobacco alkaloids, phenolics, and diterpene glycosides in Nicotiana attenuata. J. Agric. Food Chem. 49 3553–3558. [DOI] [PubMed] [Google Scholar]
- Kessler, A., and Baldwin, I.T. (2001). Defensive function of herbivore-induced plant volatile emissions in nature. Science 291 2141–2144. [DOI] [PubMed] [Google Scholar]
- Kessler, A., and Baldwin, I.T. (2002). Plant responses to insect herbivory: The emerging molecular analysis. Annu. Rev. Plant Biol. 53 299–328. [DOI] [PubMed] [Google Scholar]
- Knoth, C., Ringler, J., Dangl, J.L., and Eulgem, T. (2007). Arabidopsis WRKY70 is required for full RPP4-mediated disease resistance and basal defense against Hyaloperonospora parasitica. Mol. Plant Microbe Interact. 20 120–128. [DOI] [PubMed] [Google Scholar]
- Kruegel, T., Lim, M., Gase, K., Halitschke, R., and Baldwin, I.T. (2002). Agrobacterium-mediated transformation of Nicotiana attenuata, a model ecological expression system. Chemoecology 12 177–183. [Google Scholar]
- Li, J., Brader, G., Kariola, T., and Palva, E.T. (2006). WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 46 477–491. [DOI] [PubMed] [Google Scholar]
- Li, J., Brader, G., and Palva, E.T. (2004). The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X.Q., Bai, X.Q., Wang, X.J., and Chu, C.C. (2007). OsWRKY71, a rice transcription factor, is involved in rice defense response. J. Plant Physiol. 164 969–979. [DOI] [PubMed] [Google Scholar]
- Marchive, C., Mzid, R., Deluc, L., Barrieu, F., Pirrello, J., Gauthier, A., Corio-Costet, M.F., Regad, F., Cailleteau, B., Hamdi, S., and Lauvergeat, V. (2007). Isolation and characterization of a Vitis vinifera transcription factor, VvWRKY1, and its effect on responses to fungal pathogens in transgenic tobacco plants. J. Exp. Bot. 58 1999–2010. [DOI] [PubMed] [Google Scholar]
- Mare, C., Mazzucotelli, E., Crosatti, C., Francia, E., Stanca, A.M., and Cattivelli, L. (2004). Hv-WRKY38: A new transcription factor involved in cold- and drought-response in barley. Plant Mol. Biol. 55 399–416. [DOI] [PubMed] [Google Scholar]
- Menke, F.L.H., Kang, H.G., Chen, Z.X., Park, J.M., Kumar, D., and Klessig, D.F. (2005). Tobacco transcription factor WRKY1 is phosphorylated by the MAP kinase SIPK and mediates HR-like cell death in tobacco. Mol. Plant Microbe Interact. 18 1027–1034. [DOI] [PubMed] [Google Scholar]
- Miao, Y., Laun, T.M., Smykowski, A., and Zentgraf, U. (2007). Arabidopsis MEKK1 can take a short cut: It can directly interact with senescence-related WRKY53 transcription factor on the protein level and can bind to its promoter. Plant Mol. Biol. 65 63–76. [DOI] [PubMed] [Google Scholar]
- Murray, S.L., Ingle, R.A., Petersen, L.N., and Denby, K.J. (2007). Basal resistance against Pseudomonas syringae in Arabidopsis involves WRKY53 and a protein with homology to a nematode resistance protein. Mol. Plant Microbe Interact. 20 1431–1438. [DOI] [PubMed] [Google Scholar]
- Nishiuchi, T., Shinshi, H., and Suzuki, K. (2004). Rapid and transient activation of transcription of the ERF3 gene by wounding in tobacco leaves - Possible involvement of NtWRKYs and autorepression. J. Biol. Chem. 279 55355–55361. [DOI] [PubMed] [Google Scholar]
- Park, C.J., Shin, Y.C., Lee, B.J., Kim, K.J., Kim, J.K., and Paek, K.H. (2006). A hot pepper gene encoding WRKY transcription factor is induced during hypersensitive response to Tobacco mosaic virus and Xanthomonas campestris. Planta 223 168–179. [DOI] [PubMed] [Google Scholar]
- Paschold, A., Bonaventure, G., Kant, M.R., and Baldwin, I.T. (June 17, 2008). Jasmonate perception regulates jasmonate biosynthesis and JA-Ile metabolism: The case of COI1 in Nicotiana attenuata. Plant Cell Physiol. http://dx.doi.org/10.1093/pcp/pcn091. [DOI] [PubMed]
- Petersen, M., et al. (2000). Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 103 1111–1120. [DOI] [PubMed] [Google Scholar]
- Qiu, D.Y., Xiao, J., Ding, X.H., Xiong, M., Cai, M., Cao, C.L., Li, X.H., Xu, C.G., and Wang, S.P. (2007). OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol. Plant Microbe Interact. 20 492–499. [DOI] [PubMed] [Google Scholar]
- Qu, N., Schittko, U., and Baldwin, I.T. (2004). Consistency of Nicotiana attenuata's herbivore- and jasmonate-induced transcriptional responses in the allotetraploid species Nicotiana quadrivalvis and Nicotiana clevelandii. Plant Physiol. 135 539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph, S.G., et al. (2006). Conifer defence against insects: Microarray gene expression profiling of Sitka spruce (Picea sitchensis) induced by mechanical wounding or feeding by spruce budworms (Choristoneura occidentalis) or white pine weevils (Pissodes strobi) reveals large-scale changes of the host transcriptome. Plant Cell Environ. 29 1545–1570. [DOI] [PubMed] [Google Scholar]
- Robatzek, S., and Somssich, I.E. (2002). Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 16 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton, P.J., Reinstadler, A., Lipka, V., Lippok, B., and Somssich, I.E. (2002). Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen- and wound-induced signaling. Plant Cell 14 749–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, D.D., Voelckel, C., Hartl, M., Schmidt, S., and Baldwin, I.T. (2005). Specificity in ecological interactions. Attack from the same lepidopteran herbivore results in species-specific transcriptional responses in two Solanaceous host plants. Plant Physiol. 138 1763–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwachtje, J., Minchin, P.E.H., Jahnke, S., van Dongen, J.T., Schittko, U., and Baldwin, I.T. (2006). SNF1-related kinases allow plants to tolerate herbivory by allocating carbon to roots. Proc. Natl. Acad. Sci. USA 103 12935–12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, S., Katou, S., Seto, H., Gomi, K., and Ohashi, Y. (2007). The mitogen-activated protein kinases WIPK and SIPK regulate the levels of jasmonic and salicylic acids in wounded tobacco plants. Plant J. 49 899–909. [DOI] [PubMed] [Google Scholar]
- Shimono, M., Sugano, S., Nakayama, A., Jiang, C.J., Ono, K., Toki, S., and Takatsuji, H. (2007). Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 19 2064–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowe, K.A., Marquis, R.J., Hochwender, C.G., and Simms, E.L. (2000). The evolutionary ecology of tolerance to consumer damage. Annu. Rev. Ecol. Syst. 31 565–595. [Google Scholar]
- Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G.H., Nomura, K., He, S.Y., Howe, G.A., and Browse, J. (2007). JAZ repressor proteins are targets of the SCFCO11 complex during jasmonate signalling. Nature 448 661–662. [DOI] [PubMed] [Google Scholar]
- Ulker, B., Mukhtar, M.S., and Somssich, I.E. (2007). The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta 226 125–137. [DOI] [PubMed] [Google Scholar]
- Ulker, B., and Somssich, I.E. (2004). WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 7 491–498. [DOI] [PubMed] [Google Scholar]
- Van Dam, N.M., Hermenau, U., and Baldwin, I.T. (2001. a). Instar-specific sensitivity of specialist Manduca sexta larvae to induced defences in their host plant Nicotiana attenuata. Ecol. Entomol. 26 578–586. [Google Scholar]
- Van Dam, N.M., Horn, M., Mares, M., and Baldwin, I.T. (2001. b). Ontogeny constrains systemic protease inhibitor response in Nicotiana attenuata. J. Chem. Ecol. 27 547–568. [DOI] [PubMed] [Google Scholar]
- Voelckel, C., and Baldwin, I.T. (2004). Herbivore-induced plant vaccination. Part II. Array-studies reveal the transience of herbivore-specific transcriptional imprints and a distinct imprint from stress combinations. Plant J. 38 650–663. [DOI] [PubMed] [Google Scholar]
- Walling, L.L. (2000). The myriad plant responses to herbivores. J. Plant Growth Regul. 19 195–216. [DOI] [PubMed] [Google Scholar]
- Wang, H.H., Hao, J.J., Chen, X.J., Hao, Z.N., Wang, X., Lou, Y.G., Peng, Y.L., and Guo, Z.J. (2007. a). Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol. Biol. 65 799–815. [DOI] [PubMed] [Google Scholar]
- Wang, L., Halitschke, R., Kang, J.H., Berg, A., Harnisch, F., and Baldwin, I.T. (2007. b). Independently silencing two JAR family members impairs levels of trypsin proteinase inhibitors but not nicotine. Planta 226 159–167. [DOI] [PubMed] [Google Scholar]
- Winz, R.A., and Baldwin, I.T. (2001). Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. IV. Insect-induced ethylene reduces jasmonate-induced nicotine accumulation by regulating putrescine N-methyltransferase transcripts. Plant Physiol. 125 2189–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., Wang, L., and Baldwin, I.T. (2008). Methyl jasmonate-elicited herbivore resistance: Does MeJA function as a signal without being hydrolyzed to JA? Planta 227 1161–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J.Q., Hettenhausen, C., Meldau, S., and Baldwin, I.T. (2007). Herbivory rapidly activates MAPK signaling in attacked and unattacked leaf regions but not between leaves of Nicotiana attenuata. Plant Cell 19 1096–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Zhang, Z.L., Hanzlik, S., Cook, E., and Shen, Q.X.J. (2007). Salicylic acid inhibits gibberellin-induced alpha-amylase expression and seed germination via a pathway involving an abscisic-acid-inducible WRKY gene. Plant Mol. Biol. 64 293–303. [DOI] [PubMed] [Google Scholar]
- Xie, Z., Zhang, Z.L., Zou, X.L., Yang, G.X., Komatsu, S., and Shen, Q.X.J. (2006). Interactions of two abscisic-acid induced WRKY genes in repressing gibberellin signaling in aleurone cells. Plant J. 46 231–242. [DOI] [PubMed] [Google Scholar]
- Xu, X.P., Chen, C.H., Fan, B.F., and Chen, Z.X. (2006). Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18 1310–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, S., Nakano, T., Suzuki, K., and Shinshi, H. (2004). Elicitor-induced activation of transcription via W box-related cis-acting elements from a basic chitinase gene by WRKY transcription factors in tobacco. Biochim. Biophys. Acta 1679 279–287. [DOI] [PubMed] [Google Scholar]
- Zavala, J.A., Patankar, A.G., Gase, K., and Baldwin, I.T. (2004). Constitutive and inducible trypsin proteinase inhibitor production incurs large fitness costs in Nicotiana attenuata. Proc. Natl. Acad. Sci. USA 101 16071612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y.J., and Wang, L.J. (2005). The WRKY transcription factor superfamily: Its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 5 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Z.Y., Mosher, S.L., Fan, B.F., Klessig, D.F., and Chen, Z.X. (2007). Functional analysis of Arabidopsis WRKY25 transcription factor in plant defense against Pseudomonas syringae. BMC Plant Biol. 7 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler, J., Keinanen, M., and Baldwin, I.T. (2001). Herbivore-induced allene oxide synthase transcripts and jasmonic acid in Nicotiana attenuata. Phytochemistry 58 729–738. [DOI] [PubMed] [Google Scholar]
- Zou, X.L., Shen, Q.J.X., and Neuman, D. (2007). An ABA inducible WRKY gene integrates responses of creosote bush (Larrea tridentata) to elevated CO2 and abiotic stresses. Plant Sci. 172 997–1004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.