Abstract
The GTPase dynamin has been clearly implicated in clathrin-mediated endocytosis of synaptic vesicle membranes at the presynaptic nerve terminal. Here we describe a novel 52-kDa protein in rat brain that binds the proline-rich C terminus of dynamin. Syndapin I (synaptic, dynamin-associated protein I) is highly enriched in brain where it exists in a high molecular weight complex. Syndapin I can be involved in multiple protein–protein interactions via a src homology 3 (SH3) domain at the C terminus and two predicted coiled-coil stretches. Coprecipitation studies and blot overlay analyses revealed that syndapin I binds the brain-specific proteins dynamin I, synaptojanin, and synapsin I via an SH3 domain-specific interaction. Coimmunoprecipitation of dynamin I with antibodies recognizing syndapin I and colocalization of syndapin I with dynamin I at vesicular structures in primary neurons indicate that syndapin I associates with dynamin I in vivo and may play a role in synaptic vesicle endocytosis. Furthermore, syndapin I associates with the neural Wiskott-Aldrich syndrome protein, an actin-depolymerizing protein that regulates cytoskeletal rearrangement. These characteristics of syndapin I suggest a molecular link between cytoskeletal dynamics and synaptic vesicle recycling in the nerve terminal.
INTRODUCTION
Neurotransmitter release requires that synaptic vesicles fuse with the plasma membrane when intraterminal calcium rises, after which the synaptic vesicle membrane is rapidly recycled and refilled with neurotransmitter. Recovery of plasma membrane after stimulated exocytosis is commonly referred to as compensatory endocytosis. Compensatory endocytosis of synaptic vesicle membrane proteins from the plasma membrane was originally attributed to only two cytoplasmic proteins, clathrin and the heterotetrameric adaptor complex, adaptor protein 2 (AP2)1 (for review, see Schmid, 1997). A more complex model of endocytosis became necessary when the Drosophila shibire mutant that could not recycle synaptic vesicle membranes was shown to be defective in a GTPase, dynamin (Kosaka and Ikeda, 1983; Koenig et al., 1989; Koenig and Ikeda, 1989). Dynamin is now known to form tightly wound helical structures that participate in pinching off the constricted neck of a clathrin-coated pit (Hinshaw and Schmid, 1995; Takei et al., 1995, 1998; Sweitzer and Hinshaw, 1998). It is likely that dynamin is normally part of a much larger molecular machine that is responsible for compensatory endocytosis after exocytosis. Dynamin binds with high affinity via a proline-rich domain (PRD) to four brain-specific proteins: amphiphysin I (David et al., 1996), amphiphysin II (Leprince et al., 1997; Ramjaun et al., 1997; Wigge et al., 1997a), endophilin (Micheva et al., 1997; Ringstad et al., 1997), and an undescribed protein of ∼52 kDa (Roos and Kelly, 1998). Amphiphysin I, amphiphysin II, and endophilin bind to dynamin and other PRD-containing proteins such as synapsin I (De Camilli et al., 1983) and synaptojanin (McPherson et al., 1996) via their src homology 3 (SH3) domains.
Interaction of dynamin via its PRD with SH3 domain-containing proteins is essential to dynamin function. SH3 domains stimulate the GTPase activity of dynamin I in vitro by binding its C-terminal PRD (Gout et al., 1993; Herskovits et al., 1993). Recruitment of dynamin to clathrin-coated pits is critically dependent on the amino acid sequence of a potential SH3 domain recognition site in its PRD (Shpetner et al., 1996), suggesting the involvement of SH3 domain-containing proteins in dynamin targeting. PRD–SH3 domain interactions have been implicated in endocytotic function. When injected into the presynaptic compartment of lamprey neurons, the SH3 domain of amphiphysin or a dynamin peptide containing the SH3-binding site impaired synaptic vesicle recycling and caused the accumulation of invaginated clathrin-coated pits (Shupliakov et al., 1997). A similar dynamin peptide inhibited synaptic vesicle endocytosis in synaptosomes (Marks and McMahon, 1998). COS-7 cells transfected with the amphiphysin SH3 domain were blocked in receptor-mediated endocytosis; normal endocytosis was rescued when dynamin was cotransfected (Wigge et al., 1997b). The PRD of Drosophila dynamin impaired the in vitro formation of synaptic vesicles in pheochromocytoma (PC12) cells (Shi et al., 1998). Furthermore, from immunoprecipitation studies, PRD- and SH3 domain-containing proteins in the nerve terminal are known to interact with each other to form large, stable protein complexes (David et al., 1996; Micheva et al., 1997; Wigge et al., 1997a). An appealing conjecture, therefore, is that these molecules are linked together via their PRDs and their SH3 domains into protein machines that help form synaptic vesicles at endocytosis “hot spots” (Estes et al., 1996; González-Gaitán and Jäckle, 1997; Roos and Kelly, 1998) on the nerve terminal plasma membrane.
To understand how a dynamin-based molecular machine might participate in synaptic vesicle recycling, the first step is to identify the major components of the machine. While testing the binding specificity of the PRD of Drosophila dynamin, we noted strong binding in rat brain extracts to amphiphysin I, amphiphysin II, endophilin, and a 52-kDa protein (Roos and Kelly, 1998). Since the protein of 52 kDa had not yet been identified, we used the binding assay to purify it and obtained the sequence. We find that the 52-kDa protein is a dynamin-binding protein with an SH3 domain and sequence homology to a chicken focal adhesion protein 52 (FAP52) (Meriläinen et al., 1997). Furthermore, the 52-kDa protein interacts with another major nerve terminal enriched protein, neuronal Wiskott-Aldrich syndrome protein (N-WASP) (Miki et al., 1996). There is strong evidence that N-WASP plays a major role in regulating the actin cytoskeleton via its Cdc42-binding domain and a verprolin/cofilin homology domain (Miki et al., 1996; Miki and Takenawa, 1998). Thus the 52-kDa, synapse-specific protein we have characterized might serve to link the dynamin-mediated processes of endocytosis to actin cytoskeleton rearrangements within the terminal.
We have called the 52-kDa protein syndapin I because it is the first member of a family of synaptic, dynamin-associated proteins.
MATERIALS AND METHODS
Blot Overlay
Blot overlays with recombinant fusion proteins were performed as described previously (Roos and Kelly, 1998). Briefly, protein fractions were resolved on 4–15% SDS-PAGE gels and transferred to nitrocellulose. Membranes were blocked in 5% nonfat dry milk powder in PBS and 0.05% Tween 20 for 1 h and incubated overnight at 4°C with the GST fusion protein. Bound fusion protein was subsequently detected with affinity-purified antibodies directed against GST or dynamin, goat anti-rabbit HRP-conjugated secondary antibody (Cappel/ICN, Aurora, OH), and the ECL detection system (Amersham, Buckinghamshire, United Kingdom).
Purification of Syndapin I
Adult rat brains (Pel-Freez Biologicals, Rogers, AR) were homogenized in 1:3 (wt/vol) HEPES buffer (10 mM HEPES, pH 7.4, 1 mM EDTA, 0.1 mM MgCl2), and extracts were centrifuged at 235,000 × g (maximum) for 75 min. Saturated ammonium sulfate solution was added to the supernatant to achieve 25% saturation. The sample was incubated on ice for 30 min and centrifuged at 17,000 × g for 15 min. The resulting supernatant was then adjusted to 40% saturation with ammonium sulfate, and the precipitation procedure was repeated. Lastly, saturated ammonium sulfate was added to bring the final concentration to 60% saturation. The resulting pellet was resuspended and dialyzed overnight against HEPES buffer. The dialysate was then applied to a MonoQ anion exchange fast-performance liquid chromatography column (Pharmacia, Piscataway, NJ), and bound proteins were eluted with a linear gradient (buffer 1: HEPES buffer; buffer 2: HEPES buffer and 1 M NaCl). Syndapin I-positive fractions were detected by the overlay assay, pooled, and dialyzed overnight against 0.25 M bis-Tris, pH 7.1. The dialysate was applied to a MonoP fast-performance liquid chromatography column (Pharmacia). After running a linear polybuffer (Pharmacia), pH 7–4 gradient, syndapin I was eluted with a linear gradient (buffer 1: HEPES buffer; buffer 2: HEPES buffer and 1 M NaCl). Syndapin I-positive fractions were pooled and resolved by SDS-PAGE. Syndapin I was excised, and protein microsequencing was performed by the Protein/DNA Technology Center of the Rockefeller University (New York, NY) (Fernandez et al., 1994.). Protein concentrations were determined using the bicinchoninic acid assay (Pierce, Rockford, IL).
Cloning of Syndapin I
PCR cloning was performed with a forward primer (5′-CGCGGATCCATGTCTGGCTCCTACGATG-3′) and a reverse primer (5′-CGGAATTCKATMGCCTCMACRTARTTKGC-3′) matching with the nucleotide sequence of the mouse h74 clone. Ten nanograms of cDNA prepared from a rat brain MATCHMAKER cDNA library in pGAD10 (Clontech, Palo Alto, CA) were used per reaction. The PCR product (∼1300 bp) was digested with BamHI and EcoRI and subcloned into the pGEX-2T vector (Pharmacia). The predicted amino acid sequence of the 1323-bp product included the peptide sequences obtained from the protein microsequencing project.
Extension of the nucleotide sequence to the 5′- and 3′-ends was achieved by PCR using primers complementary to the syndapin I sequence (at the 5′-end: 5′-CGTTGCACAAGCGGTGC-3′; at the 3′-end: 5′-CCGATGAGAGCGGAAACC-3′) and primers complementary to the vector sequences flanking the inserts, followed by a second round of PCR using nested primers complementary to the syndapin I sequence (at the 5′-end: 5′-CGTCCGCTTGTAGTTCCC-3′; at the 3′-end: 5′-TCACCAAGCTCGGAGAGG-3′) and nested primers complementary to the vector sequences. PCR products from the second round were ligated into the pT-Adv vector (Clontech).
All PCRs were performed with the Expand High Fidelity PCR System (Boehringer Mannheim, Indianapolis, IN). DNA of all PCR products was sequenced in the University of California, San Francisco/Hormone Research Institute sequencing facility with an ABI model 373A sequencer.
Recombinant Proteins
Rat brain cDNA was used as a template to generate the following constructs for GST fusion proteins: full-length syndapin I (SdpI) (amino acid residues 1–441), SdpI-SH3 (residues 376–441), and the N-terminal part of syndapin I (SdpI-N; residues 1–382). Full-length SdpI was generated with primer BQ023 (5′-CGCGGATCCATGTCTGGCCCCTACGATG-3′) and primer BQ026 (5′-CGGAATTCCTATATAGCCTCAACGTAG-3′), SdpI-SH3 was generated with primer BQ041 (5′-CGCGGATCCAACCCCTTCGAGGACGATGC) and primer BQ026, and SdpI-N was generated with primer BQ023 and primer BQ027 (5′-CGGAATTCCTAGGCATCGTCCTCGAA-GGG-3′). The PCR products were digested with BamHI and EcoRI and cloned in frame into the BamHI-EcoRI sites of pGEX-2T.
Constructs to express maltose-binding protein (MBP) fusion proteins of SdpI-SH3 and SdpI-N for affinity purification of anti-syndapin I antibodies were obtained as follows. PCR using the GST-SdpI plasmid as a template generated SdpI-SH3 and SdpI-N with an EcoRI site at the 5′-end and a BamHI site at the 3′-end of the gene products. The resulting DNA fragments were cloned into the EcoRI-BamHI sites of the pMAL-c2 vector (New England Biolabs, Beverly, MA). MBP fusion proteins were expressed and purified over an amylose column following the recommendations of the manufacturers (New England Biolabs).
Plasmids encoding for full-length SdpI and SdpI-SH3 harboring a point mutation in the SH3 domain (SdpIm and SdpI-SH3m) were obtained by PCR using wild-type syndapin I cDNA as a template with a primer partially matching the 3′-end (5′-CGGAATTCCTATATAGCCTCAACGTAGTTGGCAAGATAGAG-3′), which included a one-base exchange resulting in the substitution of leucine for proline at amino acid residue 434. Full-length SdpIm was generated with the forward primer BQ023; SdpI-SH3m was generated with the forward primer BQ041. PCR products were cloned into the BamHI-EcoRI sites of pGEX-2T, and mutagenesis was verified by DNA sequencing.
Generation of a plasmid encoding a GST fusion protein of the PRD of Drosophila dynamin (GST-Ddyn[PRD]) was described previously (Roos and Kelly, 1998). An analogous construct encoding the rat dynamin I PRD (amino acid 746–851) was obtained as follows. PCR was performed on rat brain cDNA using the forward primer 5′-CGGAATTCAACACGACCACCGTCAGCA-3′ and the reverse primer 5′-ACGCGTCGACTCAGGGGTCACTGATAGTG-3′. The ∼300-bp product was cloned into the EcoRI-SalI sites of pGEX-5X (Pharmacia).
GST fusion proteins were expressed in Escherichia coli BL21 cells according to standard methods and were purified from cell lysates on glutathione-agarose (Sigma, St. Louis, MO) columns. Fusion proteins were eluted with 20 mM glutathione in 120 mM NaCl and 50 mM Tris, pH 8.0, concentrated, and dialyzed against PBS. GST for control experiments was expressed from the plasmid pGEX-2T.
Antibodies
Polyclonal antibodies against syndapin I were raised in rabbits by Alpha Diagnostic (San Antonio, TX). GST-SdpI-SH3 fusion protein as an antigen generated antiserum 2521; GST-SdpI-N generated antiserum 2704. Antibodies were affinity purified (Smith and Fisher, 1984) on MBP fusion proteins of SdpI-SH3 and SdpI-N. Fusion proteins were resolved on preparative gels by SDS-PAGE and transferred to nitrocellulose. Pieces of nitrocellulose carrying the fusion protein were blocked with 1% BSA in PBS and 0.05% Tween 20, washed with 0.1% BSA in PBS and 0.05% Tween 20, and incubated with serum for 3 h at room temperature. After washing the nitrocellulose with 0.1% BSA in PBS and 0.05% Tween 20, antibodies were sequentially eluted with 5 mM glycine, pH 2.3, 0.5 M NaCl, 0.5% Tween 20, and 0.01% BSA. Eluates were neutralized immediately with 1 M Na2HPO4. Rabbit antisera 2521 and 2704 also served as the source for affinity-purified anti-GST antibodies.
Polyclonal rabbit antibodies against dynamin (2072) were raised and purified as described previously for anti-dynamin antibody 2073 (Estes et al., 1996). Anti-dynamin monoclonal antibody hudy-1 was the generous gift of Dr. Sandra Schmid (Scripps Research Institute, La Jolla, CA), anti-synaptojanin and anti-endophilin antibodies were kindly provided by Dr. Peter McPherson (McGill University, Montreal, Quebec, Canada), and rabbit polyclonal antibody raised against N-WASP was kindly provided by Dr. H. Miki (University of Tokyo, Tokyo, Japan). Monoclonal antibodies against synaptophysin, synapsin I, and synapsin IIa were purchased from Boehringer Mannheim, Stressgen Biotechnologies (Victoria, British Columbia, Canada), and Transduction Laboratories (Lexington, KY), respectively.
Preparation of PC12 Cell Extracts and Rat Tissue Homogenates and Subcellular Fractionation
PC12 cells were lysed with 0.1% Triton X-100 in buffer A (10 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM EGTA, 0.1 mM MgCl2), supplemented with protease inhibitors (10 μg/ml aprotinin, 5 μg/ml leupeptin, 2 μg/ml antipain, 10 μg/ml benzamidine, 1 μg/ml chymostatin, 5 μg/ml pepstatin, 1 mM PMSF [Sigma, Boehringer Mannheim]), for 30 min, and the lysates were then centrifuged for 10 min at 16,000 × g to obtain cellular extracts.
Kidney, liver, lung, spleen, heart, and skeletal muscle were dissected from female rats, diced, and homogenized (1:3, wt/vol) in buffer A supplemented with protease inhibitors via 20 passes in a glass-Teflon Dounce homogenizer. Rat brains (Pel-Freez Biologicals) were treated the same way. To obtain postnuclear supernatants (S1), the homogenates were centrifuged at 1000 × g for 20 min, yielding S1 and P1. Postnuclear supernatants were further fractionated by differential centrifugation. The S1 fraction was spun for 30 min at 25,000 × g, generating fractions P2 and S2. The S2 fraction was centrifuged for an additional 60 min at 200,000 × g, resulting in fractions P3 and S3. All pellets were resuspended in buffer A + protease inhibitors.
All procedures were performed on ice or at 4°C. Protein fractions were resolved by SDS-PAGE on 10% continuous or 4–15% gradient gels.
Size exclusion chromatography of rat brain cytosol was performed at the analytical scale as follows. Five milligrams of rat brain S3 were charged on a Superose 6 column (Pharmacia) and eluted with buffer A; 50 μl of the 0.5 ml fractions were resolved by SDS-PAGE on 6–15% gradient gels and electroblotted to nitrocellulose membranes. After Ponceau-S (Sigma) staining, the protein patterns were analyzed by overlay analysis or antibody staining as described above.
Coprecipitation Assays (Affinity Chromatography)
Recombinant GST fusion proteins or the GST protein were immobilized on glutathione Sepharose 4B beads (Pharmacia) in PBS. Twenty-five microliters of saturated beads were incubated with rat brain cytosol (1 mg of protein in 250 μl of buffer A, prepared as described above) overnight at 4°C with end-over-end rotation. Beads were washed extensively with buffer A and eluted with 20 mM glutathione in 50 mM Tris, pH 8.0, and 120 mM NaCl for 30 min at room temperature. Proteins were then separated on 4–15% SDS-PAGE and assayed by Coomassie staining, by Western blotting with various antibodies, and by blot overlay.
Immunoprecipitation
Dissected rat brains were homogenized 1:3 (wt/vol) in 10 mM HEPES, 1 mM EGTA, and 0.1 mM MgCl2, pH 7.4, supplemented with protease inhibitors (see above) via 20 passes in a glass-Teflon Dounce homogenizer and centrifuged at 130,000 × g for 45 min. Triton X-100 (1% final) and NaCl (10 mM final) were added to the supernatant. Three micrograms of affinity-purified anti-syndapin antibodies or unrelated rabbit immunoglobulin G (IgG) were immobilized onto protein G Sepharose (Pharmacia) in the presence of 5% BSA and, after several washes with immunoprecipitation (IP) buffer (10 mM HEPES, pH 7.4, 1 mM EGTA, 0.1 mM MgCl2, 10 mM NaCl, 1% Triton X-100), were incubated with 1 mg of rat brain high-speed supernatant overnight at 4°C. Beads were washed four times with IP buffer and eluted with SDS sample buffer containing 4 M urea. Eluates were separated on 8% SDS-PAGE and analyzed after immunoblotting by antibody staining.
Primary Culture of Neurons
Neuronal cultures were prepared from whole forebrains dissected from newborn rats. The cells were dissociated by trypsin treatment (0.25% trypsin, 30 min, 37°C) followed by trituration in a fire-polished Pasteur pipette. Approximately 60 neurons per cm2 were plated onto poly-l-lysine– and collagen-coated coverslips. Neurons were grown at 37°C in 5% CO2 in Neurobasal medium containing B27 serum-free supplement, 0.5 mM glutamine, and antibiotics (Life Technologies, Gaithersburg, MD) for 10–14 d.
Immunofluorescence
Neuronal cultures were processed for double immunofluorescence as follows. Cells were washed three times with PBS (supplemented with 0.3 mM CaCl2 and 0.3 mM MgCl2) before fixation in 4% freshly depolymerized paraformaldehyde in PBS for 20 min. The remaining aldehyde groups were quenched by two washes in 25 mM glycine in PBS, followed by an additional PBS wash. Neurons were blocked and permeabilized for 1 h in 2% BSA, 1% fish skin gelatin, and 0.02% saponin in PBS (block solution). The coverslips were incubated with primary antibodies (anti-syndapin I 2704, anti-synaptophysin, and anti-dynamin hudy-1 in block solution) for 90 min. After three washes in block solution (each 15 min), a mixture of the secondary antibodies, FITC-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch, West Grove, PA) and Texas Red-conjugated rabbit anti-mouse IgG (Jackson ImmunoResearch), both 1:200 in block solution, was applied for 1 h. After several washes, neurons were mounted in 0.1% p-phenylenediamine in 90% glycerol.
Incubations were viewed under a Leica TCS NT laser confocal microscope using the Leica TCS software package (Leica, Wetzlar, Germany).
RESULTS
Isolation of a 52-kDa Dynamin-binding Protein from Rat Brain Extracts
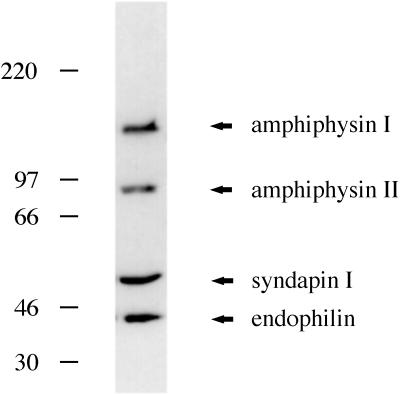
A protein of ∼52 kDa was initially identified in a screen for dynamin (PRD)-interacting proteins present in rat brain cytosol (Roos and Kelly, 1998). We have now sequenced the protein and named it syndapin I. Syndapin I was one of four putative SH3 domain-containing proteins that were abundant in rat brain cytosol and identifiable in fusion protein overlay assays (Figure 1). We have reported earlier that MonoQ anion exchange chromatography was able to separate amphiphysin I and II from syndapin I and endophilin (Roos and Kelly, 1998). Here, a significant purification of syndapin I was achieved by differential ammonium sulfate precipitation. Whereas endophilin precipitates at 25–40% ammonium sulfate, syndapin I was precipitated at 40–60% ammonium sulfate. Syndapin I was subsequently purified by MonoQ anion exchange chromatography and then by MonoP chromatography as described in Materials and Methods.
Figure 1.
Dynamin (PRD) overlays identify amphiphysin I, amphiphysin II, syndapin I, and endophilin. Overlay analysis of rat brain cytosol fractions with GST-Ddyn(PRD) reveals a 52-kDa protein, in addition to bands comigrating with amphiphysin I, amphiphysin II, and endophilin. Molecular weight markers on the left are in kilodaltons.
Primary Structure of Syndapin I
Protein sequence was obtained for two peptide fragments of syndapin I. The amino acid sequences were compared with protein databases and were found to align exactly to two internal sequences of the predicted gene product of the h74 clone (GenBank accession number 728604), obtained from a mouse hippocampal cDNA library. Oligonucleotides for PCR amplification of the syndapin I gene were designed according to the h74 sequence. PCR using a rat brain cDNA library as template yielded a 1323-bp product whose deduced amino acid sequence includes a perfect match of the peptide sequences generated from the 52-kDa band. Nucleotide sequences at the 5′- and 3′-ends of the gene were obtained by nested PCR. The primers for the nested PCR were synthesized to hybridize with the gene sequence and with vector sequences flanking the cDNA inserts.
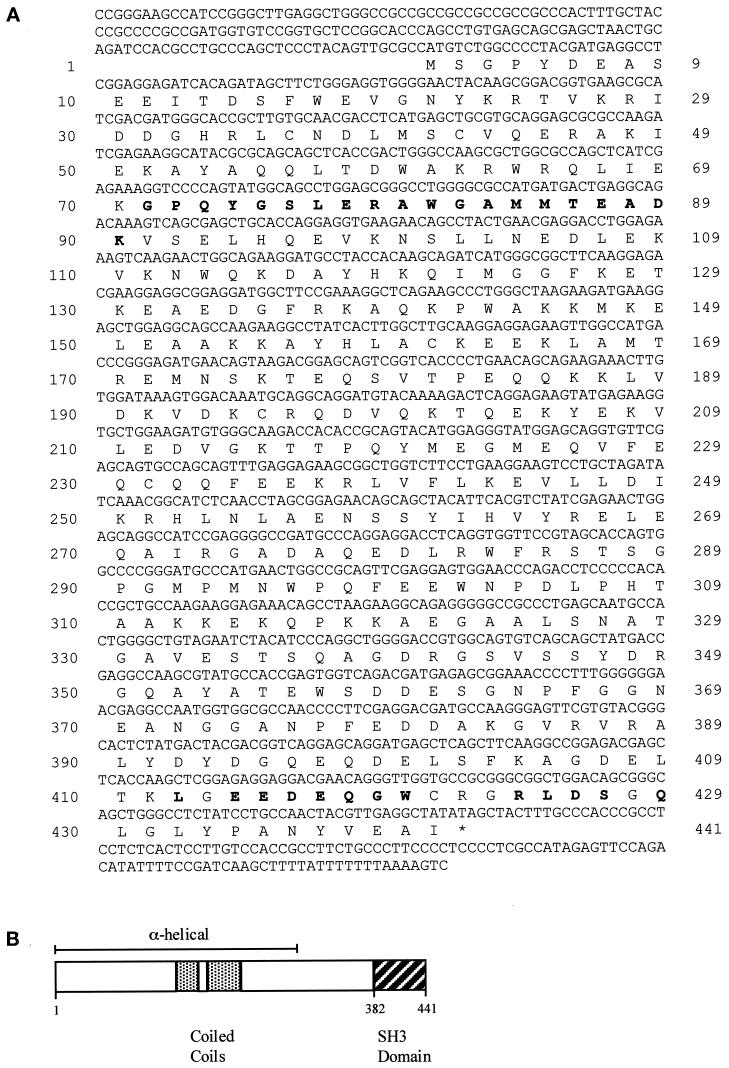
Figure 2A shows the obtained nucleotide and deduced amino acid sequence of syndapin I (GenBank accession number AF104402). The coding sequence begins at a methionine codon at nucleotides 156–158 and is followed by an open reading frame extending to a TAG stop codon (Figure 2A, *) at positions 1479–1481. The open reading frame encodes a putative protein of 441 amino acids with a predicted molecular mass of 50.4 kDa and an isoelectric point of 4.93. Within the coding region, no obvious signal sequences or putative transmembrane domains could be observed. A high degree of sequence identity was observed between the rat cDNA coding for syndapin I and the murine h74 gene (96%). The N-terminal two-thirds of syndapin I are predicted to be α-helical and to include a region with the heptad periodicity characteristic of coiled-coil domains. The C-terminal end of syndapin I encodes an SH3 domain, encompassing residues 382–441 (Figure 2B).
Figure 2.
.Molecular structure of syndapin I. (A) Nucleotide (upper) and deduced amino acid (lower) sequence of syndapin I (GenBank accession number AF104402). Amino acid residues in bold letters represent peptide sequences (peptide 1: GPQYGSLERAWGAMMTEADK; peptide 2: LXEEDEQGWXXXRL/EDSXQ) obtained from the 52-kDa protein purified from rat brain. (B) Schematic diagram revealing predictions for the secondary structure and domain structure of syndapin I. The secondary structure prediction was performed by PHD (Rost et al., 1994); domain predictions were run with the programs Coils (Lupas, 1996), Paircoil (Berger et al., 1995), and Pfam HMM Search (Sonnhammer et al., 1997).
The deduced amino acid sequence of syndapin I shares ∼65% identity with the chicken protein FAP52, an SH3 domain-containing protein localized to focal adhesions in cultured fibroblasts (Meriläinen et al., 1997). Unlike syndapin I, which is highly enriched in brain tissue, FAP52 was present in all chicken tissues tested. Other SH3 domain-containing proteins such as Grb2, paxillin, cortactin, yes, and the spectrin α-chain show >80% identity between mammalian and chicken forms, suggesting that syndapin I and FAP52 are in the same family of proteins but are not homologues. We have recently identified an additional brain-specific isoform of syndapin I, which we call syndapin II, that is more homologous to FAP52 in sequence (our unpublished results). Syndapin II has at least two alternatively spliced forms. In having an extended family, syndapin is reminiscent of another brain-specific, SH3 domain-containing protein, endophilin, also called SH3P4 (Sparks et al., 1996; deHeuvel et al., 1997). SH3P4 or endophilin was detected in brain only, whereas mRNA encoding the related proteins SH3P8 and SH3P13 was found predominantly in testis (SH3P13) or in all tissues examined (SH3P8) (Ringstad et al., 1997). This tissue distribution closely resembles the expression pattern of the three mammalian dynamin isoforms (for review, see Urrutia et al., 1997).
Syndapin I Is a Highly Brain-enriched Protein
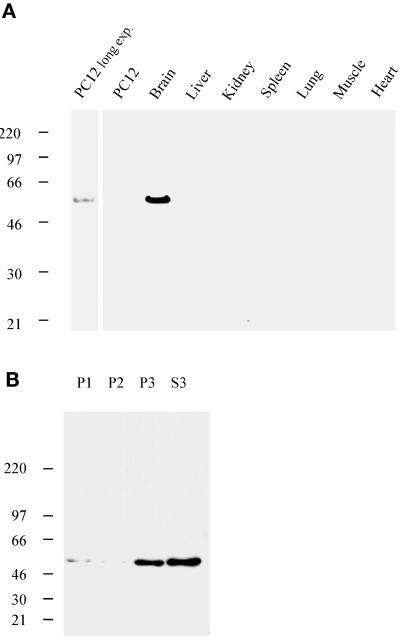
To characterize syndapin I further, polyclonal antisera were raised against recombinant GST fusion proteins of the SH3 domain (residues 376–441) and the N-terminal part (residues 1–382) of syndapin I. After affinity purification, antibodies from serum 2521 (anti-SdpI-SH3) and serum 2704 (anti-SdpI-N) recognized a single band of 52 kDa in rat brain postnuclear supernatants (Figure 3A). The electrophoretic mobility corresponded to the band recognized by GST fusion proteins of the PRD of Drosophila (Figure 1) and rat dynamin (our unpublished results) in blot overlays. In screens of postnuclear supernatants from various rat tissues, syndapin I immunoreactivity was only detected in brain. The protein was not expressed at a detectable level in liver, kidney, spleen, lung, heart, or skeletal muscle (Figure 3A), not even after further enrichment by subcellular fractionation (our unpublished results). At a low expression level, syndapin I was present in detergent extracts from undifferentiated PC12 cells. Thus, syndapin I appears to be a brain-enriched protein.
Figure 3.
Tissue and subcellular distribution of syndapin I. (A) Proteins from postnuclear supernatants of various rat tissues or from detergent extracts of PC12 cells (100 μg per lane) were resolved by SDS-PAGE, transferred to nitrocellulose, and processed for Western blots with affinity-purified antibodies against SdpI-N. Protein expression was exclusively detected in brain and, at low levels, in PC12 cells after a long exposure (exp.). (B) Proteins from subcellular fractions obtained by differential centrifugation of rat brains homogenized in buffer A were resolved by SDS-PAGE (20 μg per lane), transferred to nitrocellulose, and processed for Western blots with affinity-purified anti-SdpI-N–specific antibodies. Syndapin I was present both in soluble and sedimentable fractions (predominantly S3 and P3, corresponding to the 200,000 × g supernatant and pellet, respectively).
Western blot and overlay analyses on subcellular fractions of rat brain homogenates demonstrated that syndapin I was present in both soluble (S3) as well as sedimentable fractions (mainly P3), independent of the ionic strength of the extraction buffer (Figure 3B). At physiological ionic strengths (buffer A), the overall distribution of syndapin I during subcellular fractionation was similar to that of dynamin I and synaptojanin.
Syndapin I Interacts via Its SH3 Domain with a Subset of Rat Brain Proteins Including Dynamin I, Synaptojanin, and N-WASP
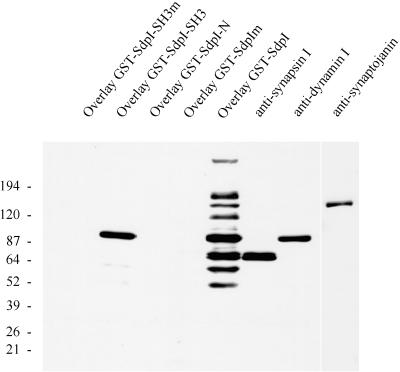
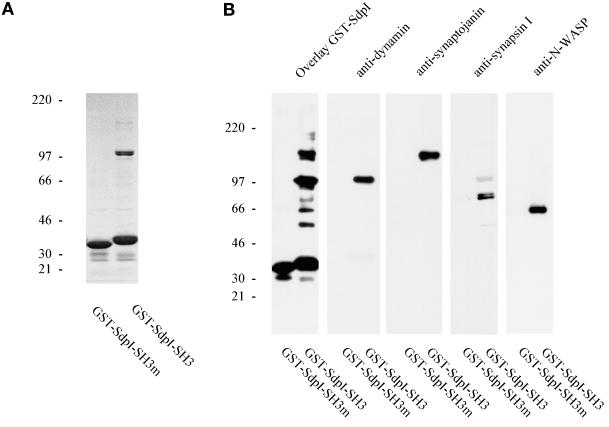
The blot overlay (far Western) technique was used to confirm the interaction between syndapin I and dynamin and to screen for the major syndapin I-binding partners in brain. Full-length syndapin I (SdpI), the SH3 domain alone (SdpI-SH3), and the N-terminal part of the protein lacking the SH3 domain (SdpI-N) were expressed as GST fusion proteins. Rat brain subcellular fractions, resolved on SDS-PAGE and transferred to nitrocellulose membranes, were overlaid with the purified GST fusion proteins. The full-length protein recognized eight major bands in rat brain cytosol with electrophoretic mobilities of ∼170, 145, 120, 100, 80, 75, 65, and 55 kDa (Figure 4); none of these were observed in liver, kidney, and heart cytosol (our unpublished results). Dynamin in rat brain cytosol comigrated with the 100-kDa protein, and synaptojanin comigrated with the 145-kDa protein recognized by syndapin I. A protein doublet of 80 and 75 kDa, whose appearance in the soluble phase was salt dependent, was recognized by antibodies against synapsin Ia and Ib and by the syndapin I overlay assay. A GST fusion protein containing the SH3 domain of syndapin I bound preferentially to the 100-kDa band identified as dynamin and to a lesser extent to the other seven proteins. A recombinant fusion protein encompassing the entire non-SH3 (residues 1–382) region did not exhibit any binding to proteins in rat brain subcellular fractions.
Figure 4.
Overlay analysis of rat brain cytosol with various GST-syndapin I constructs. Rat brain cytosol (20 μg per lane) was separated on 4–15% SDS-PAGE gels, transferred to nitrocellulose membranes, and either overlaid with 1 μM GST fusion proteins of syndapin I (wild-type and P434L mutant of full-length syndapin I and SH3 domain only and SdpI-N, lacking the SH3 domain) or incubated with antibodies against synapsin I, dynamin (2072), and synaptojanin. Binding of GST fusion proteins was detected with affinity-purified anti-GST antibodies. Full-length syndapin I fusion protein (GST-SdpI) reveals major bands at 170 kDa, 145 kDa (comigrating with synaptojanin), 120 kDa, 100 kDa (comigrating with dynamin), 80 and 75 kDa (doublet comigrating with synapsin Ia and Ib), 65 kDa, and 55 kDa, whereas the SH3 domain showed a high preference for the 100-kDa band corresponding to dynamin (GST-SdpI-SH3).
To address whether the observed interactions of syndapin I are critically dependent on the classical SH3 domain-binding interface, we mutated amino acid residue 434 from proline to leucine. Such a mutation in the SH3 domain of the Grb2 homologue sem-5 caused a lethal phenotype in Caenorhabditis elegans (Clark et al., 1992). The P434L point mutation completely abolished the binding capacity of both full-length GST-SdpI and GST-SdpI-SH3 in overlays on fractionated rat brain homogenate (Figure 4). These results indicate that the interactions are specific and highly dependent on the SH3 domain consensus sequence.
To characterize the interactions between syndapin I and its binding partners further, GST fusion proteins comprising either the wild-type or the mutant syndapin I SH3 domain, immobilized on glutathione-Sepharose beads were incubated, with high-speed supernatant fractions of rat brains homogenized in buffer A. Bead-bound material was eluted with glutathione, separated by SDS-PAGE, and assayed by protein staining, immunoblot, and blot overlay. As shown in Figure 5, dynamin I and synaptojanin were specifically bound by the wild-type SH3 domain of syndapin I but not by the mutant form. Coomassie blue staining of bands of 145- and 100-kDa electrophoretic mobility (Figure 5A) further indicated that these two proteins were major binding partners of syndapin I in rat brain cytosol. In addition, overlay analysis of the bead-bound material revealed the synapsin I doublet and bands at 65 and 55 kDa (Figure 5B). Antibodies against synapsin I demonstrated that the affinity-purified bands of 80 and 75 kDa correspond to synapsin Ia and Ib.
Figure 5.
Identification of major binding partners of syndapin I as dynamin, synaptojanin, synapsin I, and N-WASP. Soluble proteins (S3) from rat brains after homogenization in buffer A were affinity purified onto either wild-type or mutant GST-SdpI-SH3 immobilized on glutathione-Sepharose. Material specifically bound to the beads was eluted with glutathione-containing buffer and analyzed on 4–15% SDS-PAGE. (A) Coomassie protein staining revealed a protein doublet at 100 kDa and a minor band at 145 kDa specifically coprecipitated with wild-type SdpI-SH3 but not with SdpI-SH3m harboring the P434L point mutation. (B) Precipitated material was analyzed by overlay with GST-SdpI and by Western blot analyses. Antibodies against dynamin, synaptojanin, and synapsin I demonstrate that the affinity-purified bands of 100, 145, and 80 and 75 kDa correspond to dynamin, synaptojanin, and synapsin Ia and Ib, respectively. N-WASP–specific antibodies revealed that syndapin I coprecipitated N-WASP because of an SH3 domain-dependent interaction.
Western blot analysis identified the 65-kDa protein that coprecipitated with the SH3 domain of syndapin I as N-WASP (Miki et al., 1996; Fukuoka et al., 1997), a highly brain-enriched multidomain protein sharing ∼50% homology with the Wiskott-Aldrich syndrome protein WASP (Derry et al., 1994). N-WASP was identified via its interaction with the SH3 domains of Grb2, mediated via proline-rich stretches in the N-WASP sequence (Miki et al., 1996).
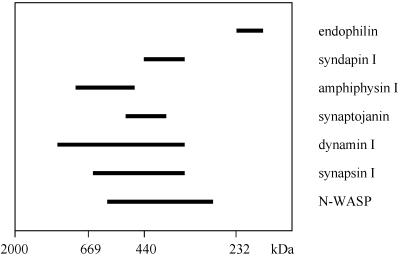
Analysis of Syndapin I-containing Complexes in Rat Brain
In high-speed supernatants of rat brains homogenized in medium ionic strength (buffer A), syndapin I existed as a high molecular weight complex that elutes from gel filtration on a Superose 6 column with an apparent molecular mass of >350 kDa (Figure 6). The elution profile of syndapin I overlapped but did not exactly match with those of synaptojanin, dynamin, synapsin I, and N-WASP, the four PRD-containing proteins recognized by syndapin I in overlay analysis. Dynamin also comigrated with amphiphysin I but not with endophilin under these conditions (Figure 6). At low ionic strength (10 mM HEPES), the elution profiles were shifted to even higher molecular weights (our unpublished results).
Figure 6.
Syndapin I exists in a high molecular weight complex. Rat brain high-speed supernatant was analyzed by gel filtration over Superose 6. Aliquots of each fraction were resolved on 6–15% SDS-PAGE and blotted to nitrocellulose. Proteins were detected by antibody reaction and overlay analyses with GST-Ddyn(PRD) and GST-SdpI, and the staining intensities of the fractions were plotted. Some proteins eluted in sharp peaks, whereas others were more broadly distributed. The horizontal bars end at the position at which the concentration of the protein in a fraction was half-maximal and give an estimate of the distribution. Half-maximum widths are shown for endophilin, syndapin I, amphiphysin I, synaptojanin, dynamin I, synapsin I, and N-WASP. Standards for column calibration correspond to dextran 2000 (2000 kDa), thyroglobulin (669 kDa), ferritin (440 kDa), and catalase (232 kDa).
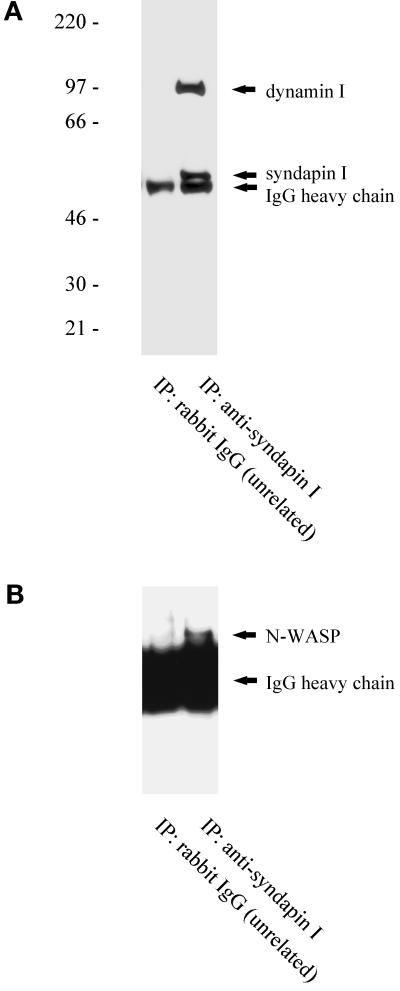
To investigate further the nature of syndapin I-containing complexes in the neuronal cytoplasm, we analyzed whether immunoprecipitation of syndapin I from rat brain high-speed supernatants resulted in the coprecipitation of dynamin I and N-WASP (Figure 7). Western blot analysis demonstrated that both dynamin I (Figure 7A) and N-WASP (Figure 7B) were specifically coimmunoprecipitated with affinity-purifiedanti-syndapin I antibodies. Thus, dynamin I and N-WASP are associated with syndapin I in rat brain.
Figure 7.
Coimmunoprecipitation of dynamin I and N-WASP with syndapin I. Rat brain high-speed supernatant was immunoprecipitated with affinity-purified antibodies against syndapin I or with unrelated rabbit IgG. Precipitated material was separated on SDS-PAGE, transferred to nitrocellulose, and analyzed with antibodies directed against syndapin I and dynamin I (A) or directed against N-WASP (B).
Colocalization of Syndapin I with Dynamin I and Synaptic Vesicle Markers in Cultured Neurons
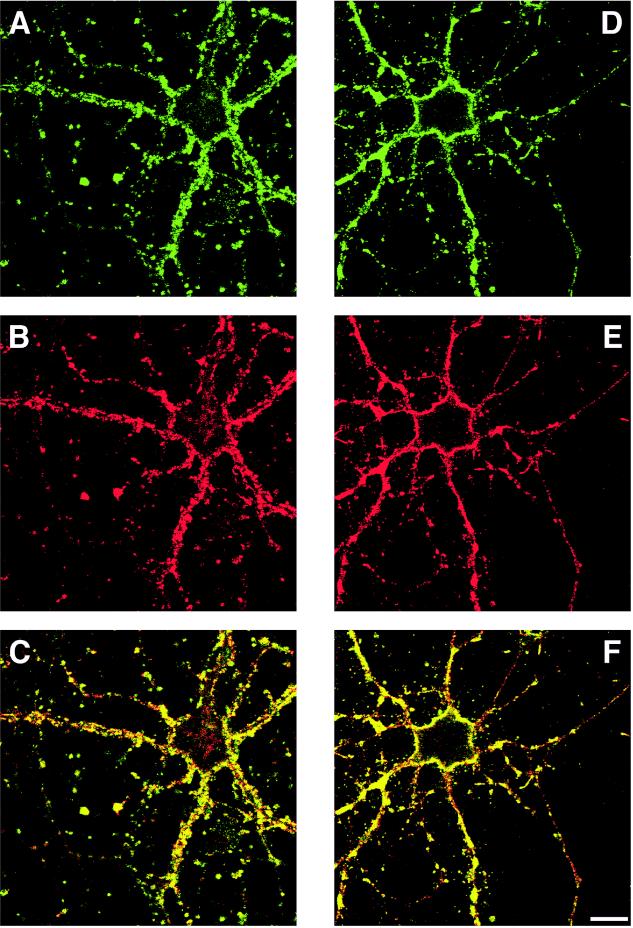
To determine whether the subcellular distribution of syndapin I was consistent with an in situ interaction with dynamin I and a possible role in synaptic vesicle recycling, double immunostaining in primary neuronal cultures was performed using antibodies against syndapin I, synaptophysin, and dynamin. Syndapin I immunoreactivity revealed an intense punctate staining in the processes and surrounding the cell bodies of the neurons; nuclei were immunonegative (Figure 8, A and D). Colocalization with synaptophysin at these sites indicates that syndapin I is present in presynaptic nerve terminals at a high level (Figure 8, A–C). Consistent with data from the subcellular fractionation, an additional diffuse cytoplasmic staining for syndapin I over the perikarya was observed. In another set of incubations, syndapin I and dynamin I immunoreactivities were compared by double-label immunofluorescence using affinity-purified antibodies against the N-terminal part of syndapin I and the monoclonal antibody hudy-1 against dynamin I. Syndapin I immunoreactivity overlapped with the dynamin I distribution (Figure 8, D–F). In addition to a diffuse cytoplasmic fluorescence (different focus plain), dynamin I and syndapin I colocalized at vesicular structures in the cell body and neurites that were also stained by antibodies specific for nerve terminal marker proteins such as synaptophysin.
Figure 8.
Colocalization of syndapin I with dynamin I and the synaptic vesicle marker synaptophysin in cultured neurons. Ten- to fourteen-day-old cultures of neurons from the forebrains of newborn rats were fixed and double labeled with affinity-purified antibodies against syndapin I (A and D) and anti-synaptophysin antibodies (B) or anti-dynamin I antibodies (E). Syndapin I immunoreactivity colocalizes with both synaptophysin and dynamin I (C and F, respectively) in presynaptic nerve terminals visible as punctate staining surrounding the perikarya and in processes of the neurons. Scale bar, 15 μm.
DISCUSSION
The protein we describe was identified by its ability to bind the PRD of both rat and Drosophila dynamin. It coimmunoprecipitates with dynamin and is enriched in synapses. We therefore propose to call it syndapin I for synaptic dynamin-associated protein I.
Syndapin I exhibited an SH3 domain-mediated, direct binding to three major nerve terminal proteins implicated in the trafficking of synaptic vesicles: synaptojanin, dynamin I, and synapsin I. SH3 domains appear to show preferences for different PRDs. In the experiments described here, the SH3 domain of syndapin I clearly preferred dynamin. The amphiphysin I SH3 domain has also been shown to prefer dynamin to synaptojanin (David et al., 1996). In contrast, proteins of the endophilin family recognized predominantly synaptojanin in rat brain extracts (Ringstad et al., 1997). The interactions between the major SH3-containing proteins of the nerve terminal (amphiphysin I and II, syndapin I, and endophilin [SH3P4]) and the PRD-containing proteins mentioned above are sufficiently strong to allow detection by the relatively stringent overlay assay. A high affinity may let the proteins form a large protein complex that is either a protein machine that executes or regulates endocytosis, a protein scaffold that holds the machinery close to the site at which it will be needed, or both. The existence of large multiprotein complexes at sites of synaptic vesicle endocytosis is suggested by the presence of hot spots in Drosophila nerve terminals that are enriched in dynamin, AP2, and the dynamin-scaffolding protein DAP160 (Estes et al., 1996; González-Gaitán and Jäckle, 1997; Roos and Kelly, 1998). These hot spots are present both in resting terminals and after complete exocytosis of the vesicle content of a terminal. Formation of protein matrices requires more than two binding sites per protein, which is true for several members of the nerve terminal PRD- and SH3 domain-containing proteins. Amphiphysin I and II, dynamin, and endophilin have predicted coiled-coil domains that could mediate homo- or heterotypic interactions. Syndapin I has two predicted coiled-coil domains at amino acids 145–170 and 183–220. It is also possible that one PRD can bind more than one SH3 domain-containing protein because endophilin and the amphiphysins did not compete for binding to the same PRD (Micheva et al., 1997). A plausible hypothesis is that the rapid speed of the endocytosis process in nerve terminals results from several factors, including the 10- to 50-fold higher concentration of clathrin, dynamin, and AP complexes in neuronal compared with non-neuronal cells (Morris and Schmid, 1995), the association of these complexes into hot spots, and the neuron-specific isoforms or splice variants of proteins such as dynamin and adaptor proteins.
Syndapin is similar to the recently described chicken protein FAP52, a ubiquitously expressed SH3 domain-containing protein (Meriläinen et al., 1997). Whereas syndapin I colocalizes with dynamin I to presynaptic nerve terminals, FAP52 was localized to focal adhesions in cultured chicken fibroblasts. The sequence homology between these two proteins suggests some relationship between the endocytotic machinery at the synapse and the machinery of cellular adhesion. Both focal adhesions and synapses are regions at which intracellular machinery is anchored to specialized regions of plasma membrane.
We have also shown that syndapin I binds via its SH3 domain to the N-WASP protein. N-WASP, which shows ∼50% homology to the Wiskott-Aldrich syndrome protein WASP (Derry et al., 1994), is found predominantly in brain nerve terminals (Fukuoka et al., 1997) and possesses several functional motifs. A pleckstrin homology domain near the N terminus that binds phosphatidylinositol 4,5-bisphosphate is important in localizing N-WASP to the cortical cytoskeleton (Miki et al., 1996). A GTPase-binding domain is recognized by Cdc42, and the interaction between N-WASP and Cdc42 has been shown to regulate the formation of actin microspikes (Miki et al., 1998). In addition to the PRD that is presumably binding syndapin I, N-WASP has verpolin-homology and cofilin-homology domains that bind and sever actin filaments (Miki et al., 1996; Miki and Takenawa, 1998; Suzuki et al., 1998). Thus, syndapin I binds a nerve terminal protein, N-WASP, that associates with and regulates the dynamic of the actin cytoskeleton.
Endocytosis has been linked to the actin cytoskeleton by both genetic and pharmacological experiments. In yeast, several of the endocytosis mutations also affect the organization of the actin cytoskeleton (for review, see Riezman et al., 1996). Association between the actin cytoskeleton and the endocytotic machinery has also been observed in mammalian cells (Geli and Riezman, 1998). Both the actin-stabilizing drug jasplakinolide and actin monomer-sequestering reagents such as latrunculin A and thymosin β4 were shown to affect endocytotic processes (Lamaze et al., 1997; Shurety et al., 1998). The link between endocytosis and the actin cytoskeleton will be further strengthened if N-WASP is found to affect synaptic vesicle formation in the synapse. Strikingly, the dynamin-associated protein syndapin I is capable of interacting with three proteins implicated in cytoskeletal reorganization: N-WASP exhibiting actin-depolymerizing activity, the actin-bundling synaptic vesicle protein synapsin I (Bähler and Greengard, 1987; Petrucci et al., 1988), and synaptojanin, which is thought to regulate actin dynamics via hydrolysis of phosphatidylinositol 4,5-bisphosphate bound to actin-regulatory proteins (Sakisaka et al., 1997).
Proteins associated with membrane traffic or with the actin cytoskeleton often have PRDs or SH3 domains. Although such domains might mediate a direct interaction between the actin cytoskeleton and the endocytotic machinery, it is also possible that the domains allow two separate processes to be controlled by the same regulatory proteins. Establishing that direct linkage occurs in nerve terminals will need careful and complete analysis of the steps of synaptic vesicle formation.
Note.
While this manuscript was in review, the sequence of a mouse homologue of syndapin I was reported and called PACSIN by Plomann et al. (1998). In this study, PACSIN was discovered as a hippocampal protein whose mRNA level decreased dramatically after entorhinal-cortex lesion. Since we find that syndapin I is a nerve terminal protein likely to be involved in synaptic vesicle recycling, we can infer that the lesions cause changes in synaptic number or function.
ACKNOWLEDGMENTS
We thank Drs. V. Faúndez, M.M. Kessels, and Y. Lichtenstein for critically reading the manuscript. We thank J. Zamanian for help with the neuronal cultures. We are grateful to Dr. H. Miki for anti-N-WASP antibody, Dr. P. McPherson for anti-synaptojanin and anti-endophilin antibodies, and Dr. S. L. Schmid for anti-dynamin antibody hudy-1. We thank L. Spector for help in preparing the manuscript. This work was supported by National Institutes of Health grants NS-09878, NS-15927, and DA-10154 to R.B.K and by postdoctoral fellowships from the Deutsche Forschungsgemeinschaft to B.Q. and from the American Cancer Society to J.R.
Abbreviations used:
- AP
adaptor protein
- Ddyn
Drosophila dynamin
- FAP52
focal adhesion protein 52
- IgG
immunoglobulin G
- IP
immunoprecipitation
- MBP
maltose-binding protein
- N-WASP
neuronal Wiskott-Aldrich syndrome protein
- PC12
pheochromocytoma
- PRD
proline-rich domain
- SdpI
syndapin I (synaptic, dynamin-associated protein I)
- SdpIm
mutant form of SdpI
- SdpI-N
N-terminal part of syndapin I
- SH3
src homology 3
- SH3m
mutant form of SH3
REFERENCES
- Bähler M, Greengard P. Synapsin I bundles F-actin in a phosphorylation-dependent manner. Nature. 1987;326:704–707. doi: 10.1038/326704a0. [DOI] [PubMed] [Google Scholar]
- Berger B, Wilson DB, Wolf E, Tonchev T, Milla M, Kim PS. Predicting coiled coils by use of pairwise residue correlations. Proc Natl Acad Sci USA. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SG, Stern MJ, Horvitz HR. C. elegans cell-signalling gene sem-5 encodes a protein with SH2 and SH3 domains. Nature. 1992;356:340–344. doi: 10.1038/356340a0. [DOI] [PubMed] [Google Scholar]
- David C, McPherson PS, Mundigl O, De Camilli P. A role of amphiphysin in synaptic vesicle endocytosis suggested by its binding to dynamin in nerve terminals. Proc Natl Acad Sci USA. 1996;93:331–335. doi: 10.1073/pnas.93.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Camilli P, Cameron R, Greengard P. Synapsin I (protein I), a nerve terminal-specific phosphoprotein. I. Its general distribution in synapses of the central and peripheral nervous system demonstrated by immunofluorescence in frozen and plastic sections. J Cell Biol. 1983;96:1337–1354. doi: 10.1083/jcb.96.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deHeuvel E, Bell AW, Ramjaun AR, Wong K, Sossin WS, McPherson PS. Identification of the major synaptojanin-binding proteins in brain. J Biol Chem. 1997;272:8710–8716. doi: 10.1074/jbc.272.13.8710. [DOI] [PubMed] [Google Scholar]
- Derry JMJ, Ochs HD, Francke U. Isolation of a novel gene mutated in Wiskott-Aldrich syndrome. Cell. 1994;78:635–644. [PubMed] [Google Scholar]
- Estes PS, Roos J, van der Bliek A, Kelly RB, Krishnan KS, Ramaswami M. Traffic of dynamin within individual Drosophila synaptic boutons relative to compartment-specific markers. J Neurosci. 1996;16:5443–5456. doi: 10.1523/JNEUROSCI.16-17-05443.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez J, Andrews L, Mische SM. An improved procedure for enzymatic digestion of polyvinylidene difluoride-bound proteins for internal sequence analysis. Anal Biochem. 1994;218:112–117. doi: 10.1006/abio.1994.1148. [DOI] [PubMed] [Google Scholar]
- Fukuoka M, Miki H, Takenawa T. Identification of N-WASP homologs in human and rat brain. Gene. 1997;196:43–48. doi: 10.1016/s0378-1119(97)00184-4. [DOI] [PubMed] [Google Scholar]
- Geli MI, Riezman H. Endocytic internalization in yeast and animal cells: similar and different. J Cell Sci. 1998;111:1031–1037. doi: 10.1242/jcs.111.8.1031. [DOI] [PubMed] [Google Scholar]
- González-Gaitán M, Jäckle H. Role of Drosophila α-adaptin in presynaptic vesicle recycling. Cell. 1997;88:767–776. doi: 10.1016/s0092-8674(00)81923-6. [DOI] [PubMed] [Google Scholar]
- Gout I, et al. The GTPase dynamin binds to and is activated by a subset of SH3 domains. Cell. 1993;75:25–36. [PubMed] [Google Scholar]
- Herskovits JS, Shpetner HS, Burgess CC, Vallee RB. Microtubules and Src homology 3 domains stimulate the dynamin GTPase via its C-terminal domain. Proc Natl Acad Sci USA. 1993;90:11468–11472. doi: 10.1073/pnas.90.24.11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw JE, Schmid SL. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature. 1995;374:190–192. doi: 10.1038/374190a0. [DOI] [PubMed] [Google Scholar]
- Koenig JH, Ikeda K. Disappearance and reformation of synaptic vesicle membrane upon transmitter release observed under reversible blockage of membrane retrieval. J Neurosci. 1989;9:3844–3860. doi: 10.1523/JNEUROSCI.09-11-03844.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig JH, Kosaka T, Ikeda K. The relationship between the number of synaptic vesicles and the amount of transmitter released. J Neurosci. 1989;9:1937–1942. doi: 10.1523/JNEUROSCI.09-06-01937.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka T, Ikeda K. Possible temperature-dependent blockage of synaptic vesicle recycling induced by a single gene mutation in Drosophila. J Neurobiol. 1983;14:207–225. doi: 10.1002/neu.480140305. [DOI] [PubMed] [Google Scholar]
- Lamaze C, Fujimoto LM, Yin HL, Schmid SL. The actin cytoskeleton is required for receptor-mediated endocytosis in mammalian cells. J Biol Chem. 1997;272:20332–20335. doi: 10.1074/jbc.272.33.20332. [DOI] [PubMed] [Google Scholar]
- Leprince C, Romero F, Cussac D, Vayssiere B, Berger R, Tavitian A, Camonis JH. A new member of the amphiphysin family connecting endocytosis and signal transduction pathways. J Biol Chem. 1997;272:15101–15105. doi: 10.1074/jbc.272.24.15101. [DOI] [PubMed] [Google Scholar]
- Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- Marks B, McMahon HT. Calcium triggers calcineurin-dependent synaptic vesicle recycling in mammalian nerve terminals. Curr Biol. 1998;8:740–749. doi: 10.1016/s0960-9822(98)70297-0. [DOI] [PubMed] [Google Scholar]
- McPherson PS, Garcia EP, Slepnev VI, David C, Zhang X, Grabs D, Sossin WS, Bauerfeind R, Nemoto Y, De Camilli P. A presynaptic inositol-5-phosphatase. Nature. 1996;379:353–357. doi: 10.1038/379353a0. [DOI] [PubMed] [Google Scholar]
- Meriläinen J, Lehto V-P, Wasenius V-M. FAP52, a novel, SH3 domain-containing focal adhesion protein. J Biol Chem. 1997;272:23278–23284. doi: 10.1074/jbc.272.37.23278. [DOI] [PubMed] [Google Scholar]
- Micheva KD, Kay BK, McPherson PS. Synaptojanin forms two separate complexes in the nerve terminal. Interactions with endophilin and amphiphysin. J Biol Chem. 1997;272:27239–27245. doi: 10.1074/jbc.272.43.27239. [DOI] [PubMed] [Google Scholar]
- Miki H, Miura K, Takenawa T. N-WASP, a novel actin-depolymerizing protein, regulates the cortical cytoskeletal rearrangement in a PIP2-dependent manner downstream of tyrosine kinases. EMBO J. 1996;15:5326–5335. [PMC free article] [PubMed] [Google Scholar]
- Miki H, Sasaki T, Takai Y, Takenawa T. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP. Nature. 1998;391:93–96. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- Miki H, Takenawa T. Direct binding of the verprolin-homology domain in N-WASP to actin is essential for cytoskeletal reorganization. Biochem Biophys Res Commun. 1998;243:73–78. doi: 10.1006/bbrc.1997.8064. [DOI] [PubMed] [Google Scholar]
- Morris SA, Schmid SL. Synaptic vesicle recycling. The Ferrari of endocytosis? Curr Biol. 1995;5:113–115. doi: 10.1016/s0960-9822(95)00028-5. [DOI] [PubMed] [Google Scholar]
- Petrucci TC, Mooseker MS, Morrow JS. A domain of synapsin I involved with actin bundling shares immunologic cross-reactivity with villin. J Cell Biochem. 1988;36:25–35. doi: 10.1002/jcb.240360104. [DOI] [PubMed] [Google Scholar]
- Plomann, et al. PACSIN, a brain protein that is upregulated upon differentiation into neuronal cells. Eur J Biochem. 1998;256:201–211. doi: 10.1046/j.1432-1327.1998.2560201.x. [DOI] [PubMed] [Google Scholar]
- Ramjaun AR, Micheva KD, Bouchelet I, McPherson PS. Identification and characterization of a nerve terminal-enriched amphiphysin isoform. J Biol Chem. 1997;272:16700–16706. doi: 10.1074/jbc.272.26.16700. [DOI] [PubMed] [Google Scholar]
- Riezman H, Munn A, Geli MI, Hicke L. Actin-, myosin- and ubiquitin-dependent endocytosis. Experientia. 1996;52:1033–1041. doi: 10.1007/BF01952099. [DOI] [PubMed] [Google Scholar]
- Ringstad N, Nemoto Y, De Camilli P. The SH3p4/Sh3p8/SH3p13 protein family: binding partners for synaptojanin and dynamin via a Grb2-like Src homology 3 domain. Proc Natl Acad Sci USA. 1997;94:8569–8574. doi: 10.1073/pnas.94.16.8569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos J, Kelly RB. Dap160, a neural-specific eps15 homology and multiple SH3 domain-containing protein that interacts with Drosophila dynamin. J Biol Chem. 1998;273:19108–19119. doi: 10.1074/jbc.273.30.19108. [DOI] [PubMed] [Google Scholar]
- Rost B, Sander C, Schneider R. PHD—an automatic mail server for protein secondary structure prediction. Comput Appl Biosci. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- Sakisaka T, Itoh T, Miura K, Takenawa T. Phosphatidylinositol 4,5-bisphosphate phosphatase regulates the rearrangement of actin filaments. Mol Cell Biol. 1997;17:3841–3849. doi: 10.1128/mcb.17.7.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid SL. Clathrin-coated vesicle formation and protein sorting: an integrated process. Annu Rev Biochem. 1997;66:511–548. doi: 10.1146/annurev.biochem.66.1.511. [DOI] [PubMed] [Google Scholar]
- Shi G, Faúndez V, Roos J, Dell’Angelica EC, Kelly RB. Neuroendocrine synaptic vesicles are formed in vitro by both clathrin-dependent and -independent pathways. J Cell Biol. 1998;143:947–956. doi: 10.1083/jcb.143.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpetner HS, Herskovits JS, Vallee RB. A binding site for SH3 domains targets dynamin to coated pits. J Biol Chem. 1996;271:13–16. doi: 10.1074/jbc.271.1.13. [DOI] [PubMed] [Google Scholar]
- Shupliakov O, Löw P, Grabs D, Gad H, Chen H, David C, Takei K, De Camilli P, Brodin L. Synaptic vesicle endocytosis impaired by disruption of dynamin-SH3 domain interactions. Science. 1997;276:259–263. doi: 10.1126/science.276.5310.259. [DOI] [PubMed] [Google Scholar]
- Shurety W, Stewart NL, Stow JL. Fluid-phase markers in the basolateral endocytic pathway accumulate in response to the actin assembly-promoting drug Jasplakinolide. Mol Biol Cell. 1998;9:957–975. doi: 10.1091/mbc.9.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DE, Fisher PA. Identification, developmental regulation, and response to heat shock of two antigenically related forms of a major nuclear envelope protein in Drosophila embryos: application of an improved method for affinity purification of antibodies using polypeptides immobilized on nitrocellulose blots. J Cell Biol. 1984;99:20–28. doi: 10.1083/jcb.99.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer ELL, Eddy SR, Durbin R. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins. 1997;28:405–420. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Sparks AB, Hoffman NG, McConnell SJ, Fowlkes DM, Kay BK. Cloning of ligand targets: systematic isolation of SH3 domain-containing proteins. Nat Biotechnol. 1996;14:741–744. doi: 10.1038/nbt0696-741. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Miki H, Takenawa T, Sasakawa C. Neural Wiskott-Aldrich syndrome protein is implicated in the actin-based motility of Shigella flexneri. EMBO J. 1998;17:2767–2776. doi: 10.1093/emboj/17.10.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweitzer SM, Hinshaw JE. Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell. 1998;93:1021–1029. doi: 10.1016/s0092-8674(00)81207-6. [DOI] [PubMed] [Google Scholar]
- Takei K, Haucke V, Slepnev V, Farsad K, Salazar M, Chen H, De Camilli P. Generation of coated intermediates of clathrin-mediated endocytosis on protein-free liposomes. Cell. 1998;94:131–141. doi: 10.1016/s0092-8674(00)81228-3. [DOI] [PubMed] [Google Scholar]
- Takei K, McPherson PS, Schmid SL, De Camilli P. Tubular membrane invaginations coated by dynamin rings are induced by GTP-γS in nerve terminals. Nature. 1995;374:186–190. doi: 10.1038/374186a0. [DOI] [PubMed] [Google Scholar]
- Urrutia R, Henley JR, Cook T, McNiven MA. The dynamins: redundant or distinct functions for an expanding family of related GTPases? Proc Natl Acad Sci USA. 1997;94:377–384. doi: 10.1073/pnas.94.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge P, Köhler K, Vallis Y, Doyle CA, Owen D, Hunt SP, McMahon HT. Amphiphysin heterodimers: potential role in clathrin-mediated endocytosis. Mol Biol Cell. 1997a;8:2003–2015. doi: 10.1091/mbc.8.10.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge P, Vallis Y, McMahon HT. Inhibition of receptor-mediated endocytosis by the amphiphysin SH3 domain. Curr Biol. 1997b;7:554–560. doi: 10.1016/s0960-9822(06)00254-5. [DOI] [PubMed] [Google Scholar]