Abstract
The replication initiator protein, π, plays an essential role in the initiation of replication of plasmid R6K. Both monomers and dimers of π bind to iterons in the γ origin of plasmid R6K, yet monomers facilitate open complex formation while dimers, the predominant form in the cell, do not. Consequently, π monomers activate replication while π dimers inhibit replication. Recently, it was shown that the monomeric form of π binds multiple tandem iterons in a strongly cooperative fashion, which might explain how monomers out-compete dimers for replication initiation when plasmid copy number and π supply are low. Here, we examine cooperative binding of π dimers and explore the role these interactions may have in the inactivation of γ origin. To examine π dimer/iteron interactions in the absence of competing π monomer/iteron interactions using wild-type π, constructs were made with key base changes to each iteron that eliminate π monomer binding yet have no impact on π dimer binding. Our results indicate that in the absence of π monomers, π dimers bind with greater cooperativity to alternate iterons than adjacent iterons, thus preferentially leaving intervening iterons unbound and the origin unsaturated. We discuss new insights into plasmid replication control by π dimers.
It is believed that all naturally occurring plasmids employ efficient copy-control mechanisms to ensure their maintenance at a reasonably constant copy number from cell to cell. The antibiotic resistance plasmid, R6K, is maintained at a steady 15–20 copies per chromosome1 in a wide variety of bacterial hosts.2 For this to occur, regulatory controls at the step of replication initiation work to increase low plasmid copy numbers and reduce elevated ones.3
Controlled replication of plasmid R6K requires two plasmid-encoded elements: the iterons in the γ origin of replication (γ ori) and the pir gene that encodes the replication (Rep) protein, π (Figure 1)4–8 γ ori activation requires the binding of monomers of π protein to the seven 22 base pair (bp) iterons within γ ori that are adjacent to an A+T-rich region of the plasmid.9–11 The binding of π monomers to iterons causes an apparent bending of the origin DNA, allowing the nearby A+T-rich region to melt, the replication complex to bind, and replication to start uni-directionally from a specific site within the A+T-rich region.11–13
Figure 1.
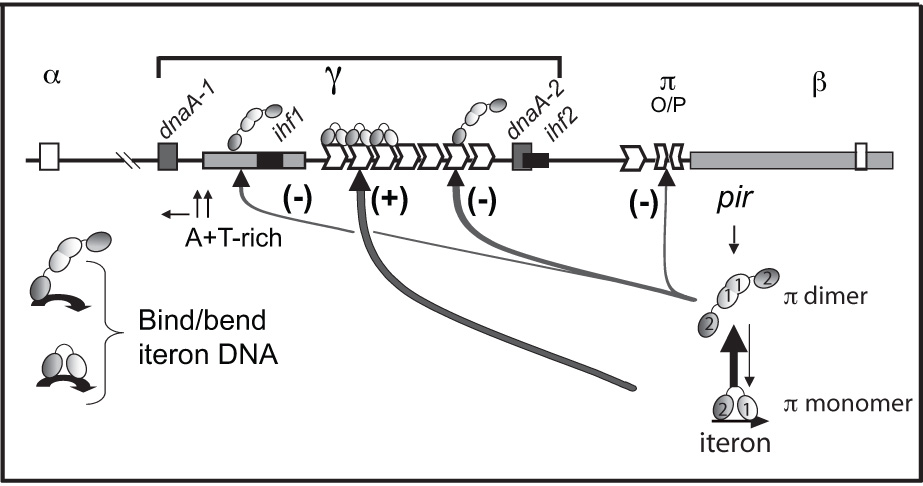
Plasmid-encoded elements involved in the regulation of replication of γ ori plasmids. A physical map of the replicon of R6K showing the relative locations of the seven tandem iterons of γ ori, the single iteron and the half-iteron of α ori and β ori, respectively, the inverted half-repeats in the operator site adjacent to the pir gene, two DnaA sites, the integration host factor (IHF) site, the A+T-rich region of γ ori, and different paths of ori activation and inhibition by π monomers and dimers, respectively. Symbols representing monomers and dimers are labeled and the WH1 and WH2 recognition helices of both monomers and dimers are represented by ‘1’ and ‘2’, respectively. Shading indicates that two monomer subunits of a dimer make head-to-head contacts while two monomers bound to two tandem iterons are proposed to make head-to-tail contacts. A monomer contacts the iteron with both WH1 and WH224 while a dimer most likely contacts the iteron only with WH2 of one of the two subunits.
While monomers of π activate replication, dimers of π appear to inhibit replication through several different mechanisms (Figure 1).9,14–17 π dimers bind a non-iteron site within the A+T-rich region in proximity to the start sites for leading strand synthesis.18 It has been hypothesized that π dimers negatively modulate the priming step of the replication process by binding to this site.18–20 π dimers also inactivate γ ori by binding iterons. π was the first of its family of Rep proteins that was shown to be capable of binding iterons as a dimer.9 Since then, two more Reps were also shown or inferred to bind iterons as dimers, thus establishing a new trend in Rep/iteron control.21–23 Although a π dimer can bend iteron DNA to the same extent as a π monomer,12 π dimers cannot induce DNA strand separation,16 yet the binding of π dimers to iterons has several consequences for γ ori replication control.9,15–17
A π monomer contacts both the 5’ half and the 3’ half of the iteron through the C- terminal winged-helix (WH2) and the N-terminal winged helix (WH1), respectively, but a π dimer only contacts the 5’ half of each iteron, including a highly conserved TGAGnG motif, with the WH2 of one of its subunits.24–27 This potentially allows the WH2 motif of the second subunit of the dimer to contact another iteron-bearing sequence,9,26 for instance, either of the other two functional oris of R6K, the α iteron or the β half-iteron, which are the active oris in vivo.28,29 Such ‘looping’ is believed to transmit the replication signal from the internal iteron cluster at γ ori to the distant oris.30 The second subunit of the dimer may also participate in handcuffing, which is the coupling of two oris by Rep-Rep interactions, blocking the initiation of replication for each plasmid involved.31 Finally, dimers may inhibit over-replication by simply prohibiting monomers from binding to a sufficient number of iterons for activation (this number might be as little as 5),32 preventing the ori from achieving the bent conformation required for open complex formation.
Because π is not limiting in the cell and both π monomers and dimers bind to iterons, there is likely to be a continuing competition for π binding sites. Therefore, a mechanism must be employed by which monomers out-compete dimers for iteron binding and ori activation when plasmid copy number is low while dimers out-compete monomers to prevent over-replication when plasmid copy number is high.
A partial explanation was provided recently when it was shown that monomers of π bind cooperatively to two adjacent iterons.33 It was also shown that a dimer-biased variant of π, π•M36A^M38A binds independently to two adjacent iterons.33 However, these studies did not reveal how dimers bind to more than two iterons, or if trends are the same for adjacent and alternate iterons. Also, it remains unclear whether the binding properties of π•M36A^M38A are similar to π•wt. This work attempts to answer these questions and demonstrates that dimers of π•wt bind poorly to nearest neighbor iterons but exhibit strong cooperative binding to alternate iterons. These findings cast a new light on how replication control of this complex replicon can be achieved.
Mutations in the “right” side of the iteron prevent π monomer binding but not π dimer binding
To accurately quantify the binding of dimers of π•wt, it was necessary to first eliminate binding of the monomeric form. Since both monomers and dimers bind to iterons, and preparations of π•wt contain both monomers and dimers, in order to use π•wt in these experiments, iterons had to be mutated in a way that prevented monomers from binding yet had no impact on dimer binding. Our lab previously demonstrated through contact probing with a single iteron that monomers of π contact bases throughout the 22 bp iteron while dimers only contact the “left-half” of the iteron.26 Furthermore, it was previously reported that collective mutations in the 14th, 17th, and 18th bp of a single iteron prevented monomers from binding both in vitro and in vivo.26 However, these mutations were tested in one isolated iteron and were not tested with two or more iterons in tandem. Thus, EMSA was carried out with π•wt and labeled DNA probes containing either three tandem wt iterons or three tandem iterons with mutations in the 14th, 17th, and 18th bp (Figure 2). To distinguish complexes containing monomers from complexes containing dimers, π•M36A^M38A was used as a size control because this variant binds iterons predominantly as a dimer.16,25
Figure 2.
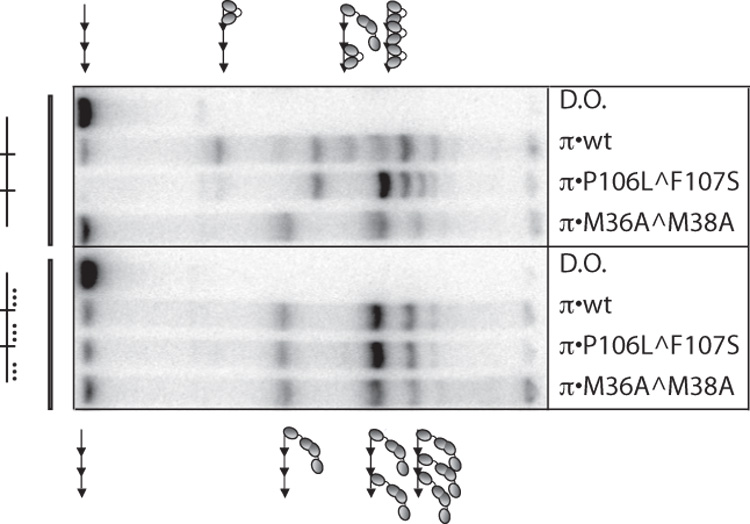
In vitro binding patterns of π•wt to three tandem iterons with and without mutations in the “monomer-only” side of each iteron. Binding assays were performed with a probe containing three wt iterons in the left four lanes and three monomer-deficient iterons (mutations in the 14th, 17th, and 18th, bp) in the right four lanes. These iteron mutations are depicted by (…). π variants are labeled. DNA probe preparation and gel shift titrations were carried out exactly as previously described33 except that: 110 pg labeled iteron-containing probe was used in the binding reactions and Promega (Madison, WI) 6X loading dye was added prior to loading the gel. Probe sequences and construction are described in Table 1 of the Supplementary Materials. 200 ng of π was added to each binding reaction. His π•WT, His-π•P106L^F107S, and His-π•M36A^M38A were purified as described.38,39 D.O. is DNA only. Symbols representing monomers and dimers of π are the same as in Figure 1.
Results in Figure 2 show that mutations in the 14th, 17th, and 18th bp of each iteron, collectively, eliminate π monomer binding to three tandem iterons in vitro. This is true for π•wt and a monomer-biased variant, π•P106L^F107S. Conversely, πdimers bind equally well to wt iterons and iterons with mutations in the 14th, 17th, and 18th positions (the “monomer-only” side) of the iteron in vitro.
Quantification of cooperative π dimer binding to adjacent and alternate iterons
The above monomer-deficient iterons with mutations in the 14th, 17th, and 18th bp were used to quantify the cooperativity of π dimer binding to two adjacent iterons and two iterons separated by 22 bp or 44 bp (one or two iterons, respectively). Because changes in the sequence of the iterons might change the intrinsic architecture of the DNA, which could affect binding of π, a minimal number of base changes were made to the intervening iterons to prevent dimer binding. It was shown previously that mutating the 7th and 9th bp of the iteron greatly reduces π dimer binding (and π monomer binding as well).26,34 Iterons with mutations at the 14th, 17th, and 18th bp (the monomer-only side) that are still proficient for dimer binding, are represented as (_…). Additionally, iterons with mutations in the 7th and 9th bp as well as the 14th, 17th, and 18th bp are incapable of binding dimers and monomers and are represented here as (X…). Thus, three constructs of equal length were created with two dimer-binding-proficient iterons adjacent to each other (_…|_…|X…|X…), separated by 22 bp (_…|X…|_…|X…) and separated by 44 bp (_…|X…|X…|_…).
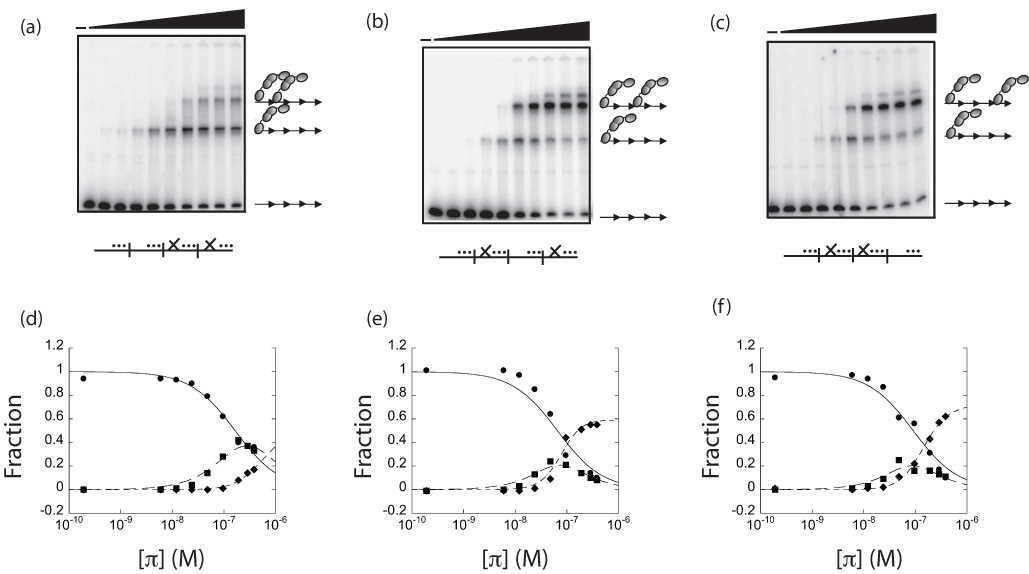
Cooperativity is expressed as the value k12, a constant obtained from binding equations derived by the statistical mechanical approach,35 and fit to data from titrations of protein with DNA. Values of k12>1 represent positive cooperativity. Figure 3(a–c) shows a representative gel shift titration of π • wt with each of the above probes. The fraction of total DNA that was free, bound to one dimer, and bound to two dimers was quantified and plotted as a function of π concentration (Figure 3(d–f)). To calculate the cooperativity coefficient, k12, data in Figure 3(d–f) were subjected to a least-squares linear regression analysis using equation (1c). This analysis provided the macroscopic binding constants, K1 and K2, which were used to calculate k12 using equation (2), as described previously.33 The values for K1 and K2 are displayed in Supplementary Table 2.
Figure 3.
| (Eq. 1a) |
| (Eq. 1b) |
| (Eq. 1c) |
| (Eq. 2) |
It was found that dimers of π•□ □ bind poorly to nearest-neighbor iterons (_…| …|X…|X…) (k12 = 11.5±9.6) but in contrast, they bind with significant cooperativity to alternate iterons (_…|X…| …|X…) (k12 = 92.2±7.3). Furthermore, it was found that π dimers bind with similar cooperativity to iterons separated by 22 and 44 bp (_…|X…| …|X…) and (_…|X…|X…| …) (k12=97.7±3.3). Thus, these data demonstrate that the cooperative binding mode of π dimers is dependent on iteron spacing.
Discussion
A major complication in the study of mechanisms that regulate γ ori activation is the intriguing quality of π that monomers and dimers bind to overlapping sequences of DNA.26 This property of π has made it difficult to examine the binding of one form in the absence of the other. Many π variants that are biased toward monomer or dimer binding have been used to circumvent this complication.16,17,25,36 However, an accurate assessment of the interaction of iteron DNA with strictly monomers or dimers of π•wt as been difficult to achieve because it has not been possible to eliminate πdimers from binding iterons without also eliminating π monomer binding. However, this work shows that it is possible to do the reverse and use key iteron mutations in multiple tandem iterons to completely eliminate π monomer binding while leaving π dimer binding unaffected. This has allowed us to calculate the binding cooperativity of π dimers without monomer interference.
The current study was based on our previous observation that monomers of π•wt bind iterons cooperatively in vivo and monomers of the monomer-biased variant π•P106L^F107S bind cooperatively in vitro.33 With this study, we have demonstrated that dimers of π•wt bind with greater cooperativity to alternate iterons than adjacent iterons. The magnitude of this cooperativity (k12≈100) is less than that of two π monomers binding to tandem iterons (k12≈210).33 Thus, the hierarchy of π/iteron cooperativity in vitro appears to be as follows: monomers bind with the highest cooperativity to adjacent iterons, then dimers bind with moderate cooperativity to alternate iterons and dimers bind with very low cooperativity to adjacent iterons.
These results fit into a working model for the regulation of replication initiation by the disparate cooperative binding properties of monomers and dimers of π(Figure (6)). In this model, at low concentrations of intracellular π•□□, the monomeric form of π binds with strong cooperativity to a sufficient number of iterons to activate γ ori and initiate replication.33 Likewise, at slightly elevated levels of π, host factors such as Integration Host Factor (IHF) and DnaA bind to neighboring sites in γ ori and may work to inhibit binding of dimers to iterons,13,37 thus allowing monomers with stronger cooperative binding to continue to bind and activate γ ori. This fits with previous data showing that IHF and DnaA are only needed at moderate and high levels of π protein in vivo.13,37 However, as the concentration of π increases, dimers become the predominant form of π in the cell, outnumbering monomers, and perhaps override the relief of inhibition provided by IHF and other host factors. At this level of π, dimers bind preferentially to alternate iterons of γ ori, resulting in incomplete saturation of the ori. Hypothetically, incomplete saturation would explain why dimers do not induce DNA strand separation even though they can bind to an iteron and bend DNA to a similar degree as do monomers. When dimers bind to alternate iterons, they are insufficient to facilitate the localized bending of γ ori iteron DNA necessary to induce strand separation of the nearby A+T rich segment of DNA. They may also initiate looping to shuttle the replication complex to the α and/or β origins of the same plasmid.30 At the highest concentrations of π, which occur at the highest plasmid copy number, dimers may engage in origin pairing (handcuffing) with other origin-bound π dimers, thus negatively regulating replication initiation by yet another mechanism.31 Thus, R6K has evolved multiple mechanisms to control its replication initiation, and the hierarchical employment of these mechanisms may be based on the disparate binding cooperativities of the monomer/activator and dimer/inhibitor forms of its Rep protein, π.
Supplementary Material
Table 1. Plasmids and oligonucleotides. Oligonucleotides were purchased from IDT (Coralville, IA). To create plasmids pFL530 through pFL536, each of the corresponding double-strand oligonucleotides (above) were inserted blunt-end into the HpaI site of pBend5.41 The oligonucleotides have a SnaBI sequence followed by three ‘iteron-like’ sequences. To create plasmids pFL537, pFL538, and pFL539, plasmids pFL532, pFL535, and pFL536 (respectively) were cleaved with SnaBI and ligated with the double-strand oligonucleotide LB37+LB38, resulting in four tandem ‘iteron-like’ sequences. Standard cloning techniques were followed42 and new clones were verified by sequencing.(__) represents a wt iteron while (_…) represents an iteron with mutations in the 14th, 17th, and 18th bp. (X) represents mutations in the 7th and 9th bp of the iteron. These mutations are highlighted in the oligonucleotide sequences by bold italics. The top strand is shown in the 5’-3’ direction.
Table 2. πdimers bind with greater cooperativity to two alternate iterons than two adjacent iterons in vitro. K1 and K2 were obtained from the least squares linear regression analysis and k12 was derived from K1 and K2 by Equation (2), as described previously.33 The data represent the average of three independent experiments.
Figure 4.
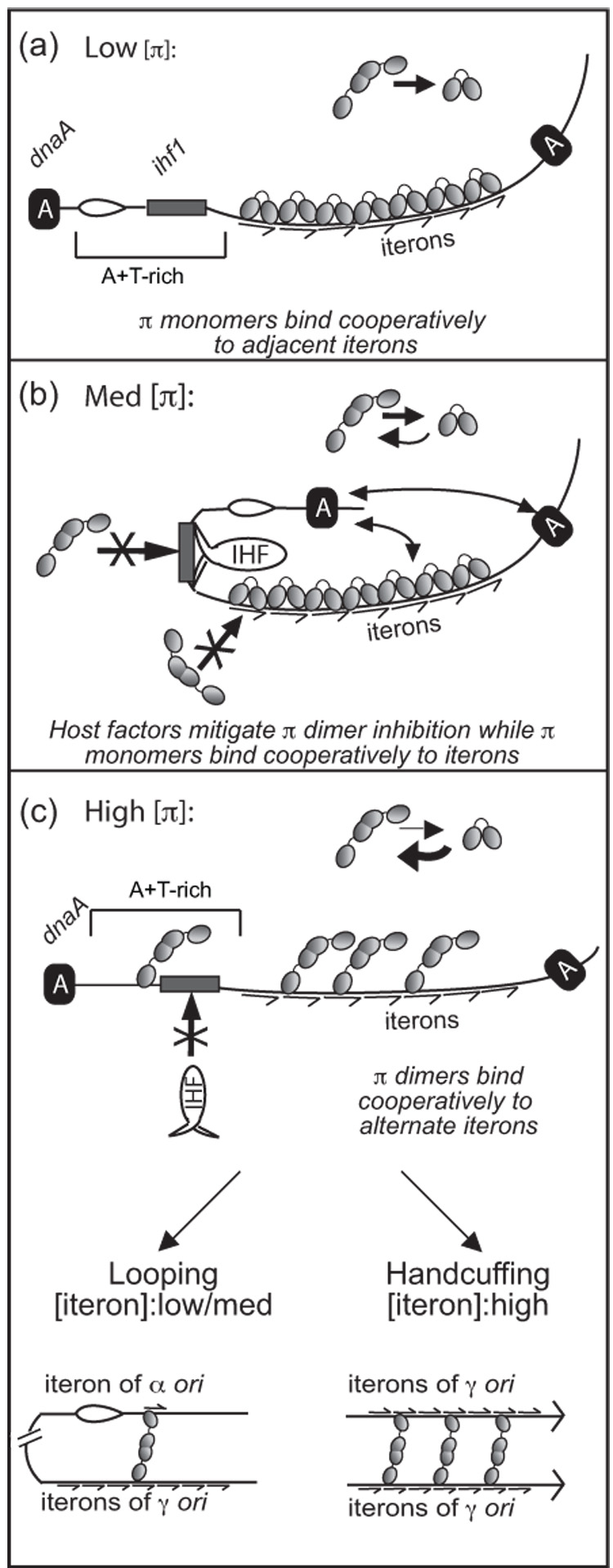
Proposed role of the disparate cooperative binding properties of π monomers and dimers in the regulation of γ ori replication. Symbols representing monomers and dimers of π are the same as in Figure 1. Iterons are depicted by half-arrows. DnaA is labeled as ‘A’. Full arrows represent contacts between different molecules of DnaA and between DnaA and π. The open complex is shown as a bubble. (a), (b), and (c) represent a model for the interactions that occur between π, iteron DNA, and host factors at low, medium, and high levels of intracellular π, respectively. In (a), the host factors IHF and DnaA are not needed at low concentrations of π and monomers of π bind cooperatively to all seven iterons, facilitating γ ori activation. In (b), as π concentration increases, IHF and DnaA help to mitigate the inhibitory effect of πdimers, allowing monomers of π to bind cooperatively to all seven iterons and facilitate γ ori activation. In (c), at high π concentrations, the dimer out-competes IHF for the ihf1 site and occupies alternate iterons of γ ori. The result may be intramolecular looping or intermolecular handcuffing, depending on the concentration of iteron DNA. This model is explained in more depth in the Discussion section.
Acknowledgments
We are grateful to Sheryl Rakowski and Selvi Kunnimalaiyaan for their support, advice, and many helpful discussions. This work was supported by the National Institutes of Health grant GM40314 to M.F.
Abbreviations
- ori
origin of replication
- Rep
replication initiator
- bp
base pair
- wt
wild type
- WH
winged-helix
- IHF
integration host factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kontomichalou P, Mitani M, Clowes RC. Circular R-factor molecules controlling penicillinase synthesis, replicating in Escherichia coli under relaxed or stringent control. J Bacteriol. 1970;104:34–44. doi: 10.1128/jb.104.1.34-44.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wild J, Czyz A, Rakowski SA, Filutowicz M. γ origin plasmids of R6K lineage replicate in diverse genera of Gram-negative bacteria. Annals Microbiol. 2004;54:471–480. [Google Scholar]
- 3.Pritchard RH, Barth PT, Collins J. Control of DNA synthesis in bacteria. In: Meadow P, Pirt SJ, editors. Microbial growth: nineteenth symposium of the Society for General Microbiology held at University College, London, April 1969 19th: 1969. Vol. 19. Cambridge U.P., London: University College edit.; 1969. pp. 263–297. [Google Scholar]
- 4.Stalker DM, Kolter R, Helinski DR. Nucleotide sequence of the region of an origin of replication of the antibiotic resistance plasmid R6K. Proc Natl Acad Sci U S A. 1979;76:1150–1154. doi: 10.1073/pnas.76.3.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacAllister TW, Kelley WL, Miron A, Stenzel TT, Bastia D. Replication of plasmid R6K origin γ in vitro. Dependence on dual initiator proteins and inhibition by transcription. J Biol Chem. 1991;266:16056–16062. [PubMed] [Google Scholar]
- 6.York D, Ivanov V, Gan J, Filutowicz M. Translational options for the pir gene of plasmid R6K: multiple forms of the replication initiator protein π. Gene. 1992;116:7–12. doi: 10.1016/0378-1119(92)90622-v. [DOI] [PubMed] [Google Scholar]
- 7.Levchenko I, Inman RB, Filutowicz M. Replication of the R6K γ origin in vitro: dependence on wt π and hyperactive πS87N protein variant. Gene. 1997;193:97–103. doi: 10.1016/s0378-1119(97)00092-9. [DOI] [PubMed] [Google Scholar]
- 8.Chen D, Feng J, Krüger R, Urh M, Inman RB, Filutowicz M. Replication of R6K γ origin in vitro: discrete start sites for DNA synthesis dependent on π and its copy-up variants. J Mol Biol. 1998;282:775–787. doi: 10.1006/jmbi.1998.2055. [DOI] [PubMed] [Google Scholar]
- 9.Urh M, Wu J, Forest K, Inman RB, Filutowicz M. Assemblies of replication initiator protein on symmetric and asymmetric DNA sequences depend on multiple protein oligomerization surfaces. J Mol Biol. 1998;283:619–631. doi: 10.1006/jmbi.1998.2120. [DOI] [PubMed] [Google Scholar]
- 10.Urh M, York D, Filutowicz M. Buffer composition mediates a switch between cooperative and independent binding of an initiator protein to DNA. Gene. 1995;164:1–7. doi: 10.1016/0378-1119(95)00493-p. [DOI] [PubMed] [Google Scholar]
- 11.Abhyankar MM, Zzaman S, Bastia D. Reconstitution of R6K DNA replication in vitro using 22 purified proteins. J Biol Chem. 2003;278:45476–45484. doi: 10.1074/jbc.M308516200. [DOI] [PubMed] [Google Scholar]
- 12.Krüger R, Rakowski SA, Filutowicz M. Isomerization and apparent DNA bending by π, the replication protein of plasmid R6K. Biochem Biophys Res Commun. 2004;313:834–840. doi: 10.1016/j.bbrc.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 13.Dellis S, Feng J, Filutowicz M. Replication of plasmid R6K γ origin in vivo and in vitro: dependence on IHF binding to the ihf1 site. J Mol Biol. 1996;257:550–560. doi: 10.1006/jmbi.1996.0184. [DOI] [PubMed] [Google Scholar]
- 14.Filutowicz M, McEachern MJ, Helinski DR. Positive and negative roles of an initiator protein at an origin of replication. Proc Natl Acad Sci U S A. 1986;83:9645–9649. doi: 10.1073/pnas.83.24.9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu J, Sektas M, Chen D, Filutowicz M. Two forms of replication initiator protein: positive and negative controls. Proc Natl Acad Sci U S A. 1997;94:13967–13972. doi: 10.1073/pnas.94.25.13967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krüger R, Konieczny I, Filutowicz M. Monomer/dimer ratios of replication protein modulate the DNA strand- opening in a replication origin. J Mol Biol. 2001;306:945–955. doi: 10.1006/jmbi.2000.4426. [DOI] [PubMed] [Google Scholar]
- 17.Abhyankar MM, Reddy JM, Sharma R, Bullesbach E, Bastia D. Biochemical investigations of control of replication initiation of plasmid R6K. J Biol Chem. 2004;279:6711–6719. doi: 10.1074/jbc.M312052200. [DOI] [PubMed] [Google Scholar]
- 18.Levchenko I, Filutowicz M. Initiator protein π can bind independently to two domains of the γ origin core of plasmid R6K: the direct repeats and the A+T-ich segment. Nucleic Acids Res. 1996;24:1936–1942. doi: 10.1093/nar/24.10.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Filutowicz M, Uhlenhopp E, Helinski DR. Binding of purified wild-type and mutant π initiation proteins to a replication origin region of plasmid R6K. J Mol Biol. 1986;187:225–239. doi: 10.1016/0022-2836(86)90230-5. [DOI] [PubMed] [Google Scholar]
- 20.Krüger R, Filutowicz M. Dimers of π protein bind the A+T-rich region of the R6K γ origin near the leading-strand synthesis start sites: regulatory implications. J Bacteriol. 2000;182:2461–2467. doi: 10.1128/jb.182.9.2461-2467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das N, Chattoraj DK. Origin pairing ('handcuffing') and unpairing in the control of P1 plasmid replication. Mol Microbiol. 2004;54:836–849. doi: 10.1111/j.1365-2958.2004.04322.x. [DOI] [PubMed] [Google Scholar]
- 22.Giraldo R, Andreu JM, Díaz-Orejas R. Protein domains and conformational changes in the activation of RepA, a DNA replication initiator. Embo J. 1998;17:4511–4526. doi: 10.1093/emboj/17.15.4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diaz-Lopez T, Davila-Fajardo C, Blaesing F, Lillo MP, Giraldo R. Early events in the binding of the pPS10 replication protein RepA to single iteron and operator DNA sequences. J Mol Biol. 2006;364:909–920. doi: 10.1016/j.jmb.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 24.Swan MK, Bastia D, Davies C. Crystal structure of π initiator protein-iteron complex of plasmid R6K: implications for initiation of plasmid DNA replication. Proc Natl Acad Sci U S A. 2006;103:18481–18486. doi: 10.1073/pnas.0609046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunnimalaiyaan S, Inman RB, Rakowski SA, Filutowicz M. Role of π dimers in coupling ("handcuffing") of plasmid R6K's γ ori iterons. J Bacteriol. 2005;187:3779–3785. doi: 10.1128/JB.187.11.3779-3785.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunnimalaiyaan S, Krüger R, Ross W, Rakowski SA, Filutowicz M. Binding modes of the initiator and inhibitor forms of the replication protein π to the γ ori iteron of plasmid R6K. J Biol Chem. 2004;279:41058–41066. doi: 10.1074/jbc.M403151200. [DOI] [PubMed] [Google Scholar]
- 27.Kunnimalaiyaan S, Rakowski SA, Filutowicz M. Structure-based functional analysis of the replication protein of plasmid R6K: key amino acids at the π/DNA interface. J Bacteriol. 2007;189:4953–4956. doi: 10.1128/JB.00109-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelley WL, Patel I, Bastia D. Structural and functional analysis of a replication enhancer: separation of the enhancer activity from origin function by mutational dissection of the replication origin γ of plasmid R6K. Proc Natl Acad Sci U S A. 1992;89:5078–5082. doi: 10.1073/pnas.89.11.5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukherjee S, Erickson H, Bastia D. Enhancer-origin interaction in plasmid R6K involves a DNA loop mediated by initiator protein. Cell. 1988;52:375–383. doi: 10.1016/s0092-8674(88)80030-8. [DOI] [PubMed] [Google Scholar]
- 30.Mukherjee S, Erickson H, Bastia D. Detection of DNA looping due to simultaneous interaction of a DNA- binding protein with two spatially separated binding sites on DNA. Proc Natl Acad Sci U S A. 1988;85:6287–6291. doi: 10.1073/pnas.85.17.6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McEachern MJ, Bott MA, Tooker PA, Helinski DR. Negative control of plasmid R6K replication: possible role of intermolecular coupling of replication origins. Proc Natl Acad Sci U S A. 1989;86:7942–7946. doi: 10.1073/pnas.86.20.7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolter R. Replication properties of plasmid R6K. Plasmid. 1981;5:2–9. doi: 10.1016/0147-619x(81)90073-1. [DOI] [PubMed] [Google Scholar]
- 33.Bowers LM, Krüger R, Filutowicz M. Mechanism of origin activation by monomers of R6K-encoded π protein. J Mol Biol. 2007;368:928–938. doi: 10.1016/j.jmb.2007.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McEachern MJ, Filutowicz M, Helinski DR. Mutations in direct repeat sequences and in a conserved sequence adjacent to the repeats result in a defective replication origin in plasmid R6K. Proc Natl Acad Sci U S A. 1985;82:1480–1484. doi: 10.1073/pnas.82.5.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brenowitz M, Senear DF, Shea MA, Ackers GK. "Footprint" titrations yield valid thermodynamic isotherms. Proc Natl Acad Sci U S A. 1986;83:8462–8466. doi: 10.1073/pnas.83.22.8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Filutowicz M, Dellis S, Levchenko I, Urh M, Wu F, York D. Regulation of replication of an iteron-containing DNA molecule. In: Cohn W, Moldave K, editors. Progress in Nucleic Acid Research and Molecular Biology. Vol. 48. San Diego: Academic Press; 1994. pp. 239–273. [DOI] [PubMed] [Google Scholar]
- 37.Wu F, Levchenko I, Filutowicz M. Binding of DnaA protein to a replication enhancer counteracts the inhibition of plasmid R6K γ origin replication mediated by elevated levels of R6K π protein. J Bacteriol. 1994;176:6795–6801. doi: 10.1128/jb.176.22.6795-6801.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu J, Filutowicz M. Hexahistidine (His6)-tag dependent protein dimerization: A cautionary tale. Acta Biochimica Polonica. 1999;46:591–599. [PubMed] [Google Scholar]
- 39.Krüger R, Filutowicz M. Characterization of His-tagged, R6K-encoded π protein variants. Plasmid. 2003;50:80–85. doi: 10.1016/s0147-619x(03)00043-x. [DOI] [PubMed] [Google Scholar]
- 40.Senear DF, Brenowitz M. Determination of binding constants for cooperative site-specific protein-DNA interactions using the gel mobility-shift assay. J Biol Chem. 1991;266:13661–13671. [PubMed] [Google Scholar]
- 41.Kim J, Zwieb C, Wu C, Adhya S. Bending of DNA by gene-regulatory proteins: construction and use of a DNA bending vector. Gene. 1989;85:15–23. doi: 10.1016/0378-1119(89)90459-9. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3rd edit. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1. Plasmids and oligonucleotides. Oligonucleotides were purchased from IDT (Coralville, IA). To create plasmids pFL530 through pFL536, each of the corresponding double-strand oligonucleotides (above) were inserted blunt-end into the HpaI site of pBend5.41 The oligonucleotides have a SnaBI sequence followed by three ‘iteron-like’ sequences. To create plasmids pFL537, pFL538, and pFL539, plasmids pFL532, pFL535, and pFL536 (respectively) were cleaved with SnaBI and ligated with the double-strand oligonucleotide LB37+LB38, resulting in four tandem ‘iteron-like’ sequences. Standard cloning techniques were followed42 and new clones were verified by sequencing.(__) represents a wt iteron while (_…) represents an iteron with mutations in the 14th, 17th, and 18th bp. (X) represents mutations in the 7th and 9th bp of the iteron. These mutations are highlighted in the oligonucleotide sequences by bold italics. The top strand is shown in the 5’-3’ direction.
Table 2. πdimers bind with greater cooperativity to two alternate iterons than two adjacent iterons in vitro. K1 and K2 were obtained from the least squares linear regression analysis and k12 was derived from K1 and K2 by Equation (2), as described previously.33 The data represent the average of three independent experiments.


