Abstract
In many organisms nonsense mutations decrease the level of mRNA. In the case of mammalian cells, it is still controversial whether translation is required for this nonsense-mediated RNA decrease (NMD). Although previous analyzes have shown that conditions that impede translation termination at nonsense codons also prevent NMD, the residual level of termination was unknown in these experiments. Moreover, the conditions used to impede termination might also have interfered with NMD in other ways. Because of these uncertainties, we have tested the effects of limiting translation of a nonsense codon in a different way, using two mutations in the immunoglobulin μ heavy chain gene. For this purpose we exploited an exceptional nonsense mutation at codon 3, which efficiently terminates translation but nonetheless maintains a high level of μ mRNA. We have shown 1) that translation of Ter462 in the double mutant occurs at only ∼4% the normal frequency, and 2) that Ter462 in cis with Ter3 can induce NMD. That is, translation of Ter462 at this low (4%) frequency is sufficient to induce NMD.
INTRODUCTION
Many and diverse organisms, bacteria, yeast, and metazoa, have been shown to have a decreased level of mRNA when that RNA bears a nonsense or frame shift mutation (Maquat, 1995; Jacobson and Peltz, 1996; Li and Wilkinson, 1998). The mechanisms that contribute to this nonsense-mediated RNA decrease (NMD) are still unclear, although some important structural features and trans-acting factors have been defined. Analysis in yeast has detected a specific, albeit commonly occurring, nucleotide segment, which is required 3′ of the nonsense codon for NMD to occur (Ruiz-Echevarria and Peltz, 1996; Ruiz-Echevarria et al., 1998). This element thus defines 5′ and 3′ domains in the RNA, such that NMD is induced by more 5′ but not by more 3′ termination codons. In the case of mammalian RNAs, NMD requires that there be at least one intron 3′ of the nonsense codon (Urlaub et al., 1989; Maquat, 1995; Carter et al., 1996; Zhang et al., 1998a); the minimum interval between the nonsense codon and the more 3′ intron has been variously estimated to be 8–10 nuclotides for a T cell receptor β RNA and ∼50 nucleotides for triosephosphate isomerase and β globin (Carter et al., 1996; Zhang et al., 1998a,b). The requirement for this intron initially suggested that nonsense codons in the more 5′ exons might be recognized before the 3′-most intron has been excised, and two models, the translational translocation and nuclear scanning models, were proposed on this basis (Urlaub et al., 1989). According to the translational translocation model, NMD operates on RNA molecules that are in transit from the nucleus to the cytoplasm, such that translation of the 5′ segment in the cytoplasm drives splicing of the more 3′ segment in the nucleus, with the result that nonsense mutations interrupt splicing, which in turn leads to RNA degradation. This model is contradicted by two lines of evidence. First, this model predicts that the 3′-most intron must be the last intron to be excised, contrary to observations involving both the APRT and DHFR genes (Kessler et al., 1993). Second, as noted above, analysis of a T cell receptor gene indicated that as few as 8–10 nucleotides are required between the splice donor and the most 3′ NMD-effective mutation, an interval that is seemingly too short to span the nuclear membrane (Carter et al., 1996). The nuclear scanning model postulated that some mechanism in the nucleus scans RNA for the occurrence of nonsense mutations. This mechanism was not defined, except that it was presumed not to rely on translation to detect the nonsense mutations. Evidence of a nontranslational mechanism of recognizing nonsense mutations has also been inferred from the finding that some nonsense mutations affect the level of alternatively spliced forms of RNA (Naeger et al., 1992; Dietz et al., 1993; Dietz and Kendzior, 1994; Lozano et al., 1994; Aoufouchi et al., 1996). As well, the level of nuclear-associated mutant RNA is reduced by nonsense codons under conditions that were considered to exclude cytoplasmic contamination (Cheng and Maquat, 1993; Belgrader and Maquat, 1994; Li et al., 1997), a finding that is consistent with the hypothesis that nonsense codons can be recognized in the nucleus or in a closely associated compartment, which copurifies with the nucleus. However, the conceptual difficulties associated with a translation-independent nuclear scanning mechanism are daunting. Moreover, multiple studies argue that NMD occurs after translation has initiated and that translation of the nonsense codon is required for NMD. Thus, NMD is abrogated by reinitiation of translation (Zhang and Maquat, 1997), and NMD is lessened by conditions that prevent termination at the nonsense codon, namely provision of suppressor tRNA, introduction of a 5′ hairpin into the mRNA, mutation of initiator ATG codons, and treatment with general inhibitors of protein synthesis (Belgrader et al., 1993; Carter et al., 1995; Li et al., 1997). To accommodate this panoply of observations, it has been proposed that splicing introduces a “mark” on the RNA near the splice site (Cheng et al., 1994; Carter et al., 1996; Jacobson and Peltz, 1996; Maquat, 1996). The mark might be permanent, e.g., methylation of a specific nucleotide, with the property that termination 5′ of the mark causes degradation of that RNA. Alternatively, the mark might be erased by translation through the splice junction; e.g., the mark might correspond to a protein that is bound to the exon junction and is displaced by the ribosomes in the course of translation.
Although showing that translation of the nonsense codon is required for NMD, the foregoing experiments did not permit an estimate of the amount of translation that is necessary or sufficient for NMD. Moreover, these different treatments, although lessening translation termination, might have also have depressed NMD by other mechanisms. For example, a 5′ hairpin might itself impede RNA degradation. Similarly, provision of suppressor tRNA typically yields incomplete suppression with the result that the nonsense codon is translated sometimes as “sense” and sometimes as “nonsense.” Effects of the residual nonsense translation might be masked if sense translation erases a splice-induced mark that is required for NMD. In an effort to avoid these ambiguities we have used another method to limit translation of a nonsense codon in the immunoglobulin (Ig) μ heavy chain gene. In this case we have made use of an exceptional nonsense mutation, Ter3, which efficiently terminates translation at codon 3 but which maintains a much higher level of μ mRNA than is the case for more 3′ nonsense mutations viz., Ter462 (Connor et al., 1994). We have generated a μ gene bearing Ter3 in cis with Ter462 and thus used termination at codon 3 to limit translation of Ter462. Our results indicate that Ter462 induces NMD even when in cis with Ter3. To estimate the extent of translation, we have exploited the expectation that in-frame translation of Ter462 should yield a truncated μ chain. The only detectable in-frame protein product of the Ter3 mutant μ gene corresponded to initiation at codon 100 (Met100) and occurred at an efficiency of ∼4% the normal rate. These results indicate that that infrequent (∼4% normal) translation of this premature termination codon is sufficient for NMD.
MATERIALS AND METHODS
Cell Lines and Vectors
The cell lines have been described previously and have been renamed for simplicity as follows. The parental hybridoma, Sp6/HL (Baumann et al., 1985), is designated WT. The igm482 mutation, which has a one-nucleotide deletion in the Cμ3 exon causing a UGA termination at codon 462 (Baumann et al., 1985), is denoted Ter462*. The mutants N89 and N114 have similarly been renamed Ter3 and Ter73, respectively, and the mutant X10, which lacks the entire μ gene, is denoted Δμ (Connor et al., 1994). The targeting vector, pTer462*, previously denoted pIΔCμ482 (Buzina and Shulman, 1996), was used to introduce the igm482 frame shift mutation into the endogenous μ gene.
Analysis of RNA
Cells were grown to 3 × 105 cells/ml and harvested by centrifugation. Total RNA was isolated using the single-step guanidinium thiocyanate-phenol-chloroform extraction (Chomczynski and Sacchi, 1987). Ten micrograms of RNA were electrophoresed in 1.4% agarose-formaldehyde gels and transferred to a nylon membrane (Boehringer Mannheim, Indianapolis, IN) in 10× SSC. Membranes were probed with DNA fragments, labeled with 32P by random priming. The level of hybridization was quantified using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA) or visualized by autoradiography.
Analysis of DNA Structure
PCR amplifications were performed using Taq polymerase (Boehringer Manheim) according to the following protocol for 30 cycles: denaturation, 1 min at 94°C; reannealing, 2 min at 65°C; and extension, 3 min at 72°C, which was increased by 3 s/cycle. Oligonucleotide primers were 1, 5′-TTCCTCAGCAAGTCCGCTAACCTGAC-3′; 2, 5′-TTGGGGCAAGAGTTGCCCTCTCTGAA-3′; 3, 5′-CGAT-ACGGTGATTGGCTACCG-3′; 4, 5′-GGACACCCAGCCACATGA-GG-3′; 5, 5′-TTACCTGGGTCTATGGCAGT-3′; and 6, 5′-GTCACTGTAAATGCTTCGGG-3′.
Analysis of μ Heavy Chains
To analyze intracellular μ chains, cells were grown to 4 × 105 cells/ml, harvested, washed twice with PBS, and placed in 1 ml of methionine-free medium for 30 min at a density of 2 × 107 cells/ml. Four hundred microcuries of [35S]methionine were then added for the times indicated in the figure legends ranging from 4 to 30 min. Cells were washed in cold PBS and suspended in 400 μl of lysis buffer (PBS supplemented with 1% NP40, 1 mM PMSF, 1 mM iodoacetic acid, and 20 μg/ml leupeptin, pepstatin A, aprotinin, antipain, and chymostatin). After 15 min at 4°C, lysates were cleared by centrifugation, diluted twice with precipitation buffer (PBS containing the same protease inhibitors), and incubated overnight at 4°C with 30 μg of rabbit antibody specific for mouse IgM. To prepare agarose G beads for immunoadsorptions, the beads were washed with PBS and preincubated for 2 h at 4°C with lysate prepared from the (μ-deleted) X10 cell line to reduce nonspecific binding of proteins. The beads were then washed in PBS supplemented with 0.5% NP40, 1 mM PMSF, 1 mM iodoacetic acid, and 1 mM EDTA. For immunoadsorptions, 20 μl of beads were added to each lysate. After incubation for 4 h at 4°C, beads were recovered by centrifugation, washed, and resuspended in 2× sample buffer (0.125 M Tris, pH 7.4, 20% glycerol, and 4% SDS containing bromphenol blue) in the presence of 5% β-mercaptoethanol. This material was then incubated at 100°C for 3 min, cleared by centrifugation, and analyzed by SDS-PAGE using 12% acrylamide (Laemmli, 1970). Gels were dried and analyzed by autoradiography or by PhosphorImager to quantitate radioactivity.
To estimate the stability of the intracellular μ-related material from the continuous radiolabeling experiments, we used the differential equation dR/dt = Nα − Rβ, where R is the incorporation of radioactivity into a particular protein by N cells in a time interval, t, α is the rate constant for synthesis of the protein, and β is the rate constant for decay of that protein. The solution to this equation is R = Nα/β (1 − e−βt), and R reaches half its maximum value in an interval t½ = (1/β)ln2.
For in vitro translation, total RNA was isolated as described above and used with the rabbit reticulocyte lysate system (Amersham, Arlington Heights, IL) according to the manufacture’s instructions. Thus, RNA from the indicated cell lines was denatured at 65°C, and 10 or 20 μg of RNA were added to make up a 50-μl mixture containing amino acids (including [35S]methionine), 100 mM potassium acetate, 1 mM magnesium acetate, 33 U RNAguard (Amersham), and reticulocyte lysate. This mixture was incubated for 60 min at 30°C and then placed on ice. The IgM-related material was then immunoprecipitated and analyzed by SDS-PAGE, as described above.
RESULTS
The effect of nonsense mutations on the Ig μ heavy chain mRNA has been studied in mutant mouse hybridoma cells (Baumann et al., 1985; Jack et al., 1989; Connor et al., 1994). As described in MATERIALS AND METHODS, the nonsense and frame shift mutations used in this study, N89, N114, and igm482, have been renamed simply to indicate the site of termination, Ter3, Ter73, and Ter462*, respectively. The asterisk on Ter462* designates that the site of termination is different from the site of mutation, which in this case is a more 5′, single-nucleotide deletion.
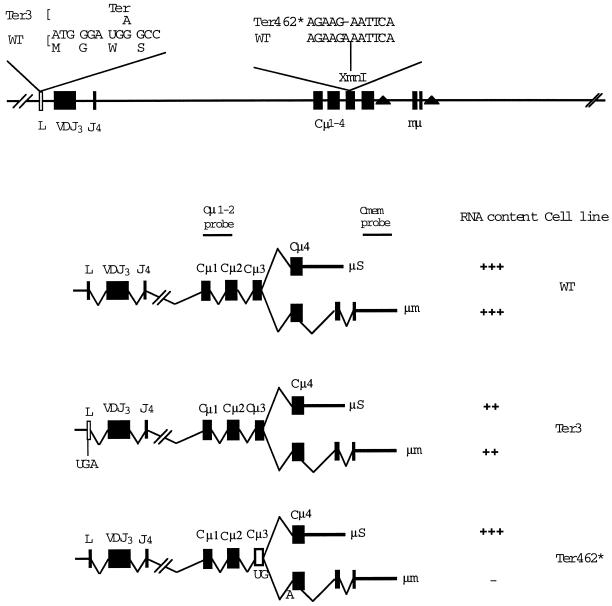
As illustrated in Figure 1, the μ gene yields two forms of μ mRNA, μs and μm, encoding the secreted and membrane forms of the μ heavy chain, respectively. Mutant hybridoma cells in which premature termination occurs in Cμ4 have approximately normal levels of μs mRNA, whereas mutants terminating in more 5′ exons have strongly decreased levels, ranging from 1 to 10% of the normal μs level (Baumann et al., 1985; Jack et al., 1989; Connor et al., 1994). The mutant Ter3 is atypical in that it has ∼50% of the normal level of μ mRNA, thus much more than mutants such as Ter73, which lies in the VDJ exon and has only ∼2% of the normal level of μ mRNA (Connor et al., 1994). The Ter3 mutant was originally isolated in a screening for mutant hybridoma cells that produced no detectable IgM, using both an ELISA and Western blot, which could detect IgM production at 0.1 and 1%, respectively, of the normal level. Considering that Ter3 has ∼50% of the normal level of μ mRNA, the absence of detectable IgM in the ELISA indicated that termination at Ter3 is >99.8% efficient. Moreover, analysis of cytoplasmic extracts of Ter3 implied that in-frame initiation 3′ of Ter 3 is infrequent, at most (Connor et al., 1994). These results suggested that the Ter3 mutation could be used in cis with a more 3′ nonsense codon to test whether translation at the normal, or near normal, frequency is required for that 3′ nonsense codon to induce NMD.
Figure 1.
Production of μ mRNA. The diagram indicates the structure of the μ gene and its relationship to the μs and μm forms of mRNA. The filled triangles indicate the poly(A) addition sites, which function in these two forms of μ mRNA. The position of the nonsense codons in the gene and their effects on μs and μm mRNA are shown. This diagram also shows the origin of the probes used in measuring μm and μs RNA. The Cμ1-2 probe corresponds to the 0.9-kb XbaI–BamHI fragment. The μm-specific probe corresponds to the 0.9-kb XbaI–KpnI fragment.
μ mRNA Content of Double-nonsense Mutant
To test whether a more 3′ nonsense codon in cis with the Ter3 mutation would induce NMD, we used the frame shift mutation, Ter462*, which has the effect that codon 462 formed at the junction of the Cμ3 and Cμ4 exons is changed to UGA (Figure 1) (Baumann et al., 1985). In keeping with the fact that the Cμ4 is the most 3′ exon of μs but is a relatively 5′ exon in μm mRNA, Ter462* depresses the level of μm RNA but has little or no effect on the level of μs mRNA (Connor et al., 1994). We have used this effect of Ter462* on μm to test whether Ter462* in cis with Ter3 causes NMD.
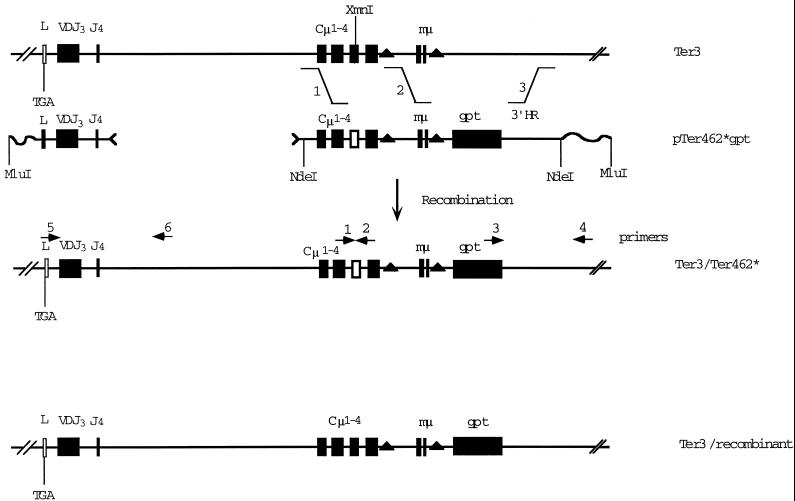
Expression of (randomly inserted) μ transgenes in hybridoma cell lines can vary over a wide (1000-fold) range (Davis et al., 1989; Wiersma and Shulman, 1995), making it very difficult to discern effects on NMD by comparing independently generated transfectants. To reduce this variability, we have analyzed the effects of targeted mutations in the endogenous μ gene. To construct the targeted recombinants, we used the vector pTer462*, which bears the selectable marker, the gpt gene, flanked by 5′ and 3′ homology regions, as illustrated in Figure 2. Our previous work with this system has shown that ∼10% of the gpt+ (MHXR) transfectants are properly targeted (Oancea and Shulman, 1994; Buzina and Shulman, 1996). As described in Figure 2, we transfected Ter3 cells with the linearized (MluI) vector pTer462* or the indicated NdeI fragment. MHXR transfectants were selected at limiting dilution, and individual colonies were screened as follows. To detect recombinants of Ter3 that acquired the Ter462* mutation, we took advantage of the fact that the Ter462* mutation destroys an XmnI site, which was assayed as described in the legend to Figure 2. Those colonies in which the Ter462* mutation had replaced the wild-type sequence were then tested for the expected 5′ and 3′ junction fragments by PCR using primers unique to the endogenous and transfected DNA (Figure 2). The Ter3 recombinant control cell line bearing the adjoining gpt gene but not the Ter462* mutation was obtained in a similar way; i.e., gpt+ transfectants were tested by PCR for the proper 3′ and 5′ junctions and the absence of Ter462*. We then examined how these mutations affected the level of μm mRNA.
Figure 2.
Construction of recombinant IgH loci bearing Ter3 and Ter462* mutations. Illustrated is the system used to construct the IgH recombinants. In the case of constructing the Ter3/Ter462* double mutant, the pTer462* vector was cut at the indicated NdeI sites, and the Cμ-gpt–containing fragment was purified by electrophoresis. In the case of constructing the Ter3 recombinant, the pTer462* vector was simply linearized at the MluI site. The DNA was introduced into the Ter3 cell line by electroporation and plated in MHX medium at limiting dilution, as described (Oancea and Shulman, 1994; Buzina and Shulman, 1996). MHXR (gpt+) transformants were tested for the replacement of the Cμ region by amplifying the indicated segment by PCR and testing its sensitivity to XmnI. We then tested for proper targeting by amplifying the 3′ junction segment by PCR, as illustrated.
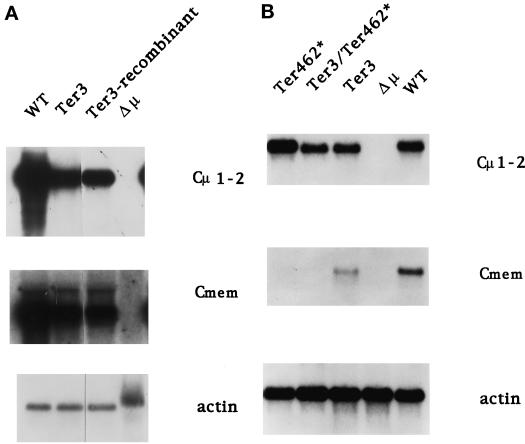
It was first necessary to test whether the recombinant structure, e.g., insertion of the gpt gene, did not alter the transcription of the μ gene. We isolated total RNA from the recombinants depicted in Figure 2 and analyzed μ RNA with a Cμ1-2 probe to detect total μ RNA and a Cmem probe to detect only μm RNA (probes defined in Figure 2). In each case we also measured the amount of actin mRNA as a loading control. Because μs RNA constitutes 90% of the total μ RNA in the wild-type (Connor et al., 1994) and mutant cell lines (see below), the values obtained with the Cμ1-2 probe will be referred to as measurements of μs. As shown in Figure 3A and Table 1, our analysis of multiple independent RNA preparations indicated that the Ter3 mutant produced both μs and μm transcripts at substantial levels, which ranged from ∼20 to 80% of the value obtained for the wild-type parental cell line. The Ter3 recombinant cell line yielded μs and μm mRNA levels that were comparable to the original Ter3 mutant cells; i.e., the gpt gene did not affect the level of μ mRNA production, as noted in an earlier analysis (Oancea and Shulman, 1994).
Figure 3.
Analysis of μs and μm RNA. RNA was isolated from the indicated cell lines and analyzed by Northern blot using probes for Cμ1-2, Cmem, (Figure 1), and actin. (A) Comparison of the original Ter3 mutant with the Ter3 recombinant (Figure 2). (B) Typical results of experiments to measure the effects of Ter462* on μ mRNA. The WT, Δμ, Ter3, and Ter462* lanes correspond to the original hybridoma and mutant cell lines (see MATERIALS AND METHODS). The Ter3/Ter462* lane corresponds to the recombinants illustrated in Figure 2. The radioactivity in the bands was quantified by PhosphorImager analysis (see Table 1). In related experiments the frame shift mutation U26, which is a 1-bp deletion 39 nucleotides 5′ of the igm482 mutation and creates the same terminator at codon 462, behaved like igm482 in depressing the level of μm but not of μs. The actin band from Δμ is diffuse and more slowly migrating because of DNA present in this preparation.
Table 1.
Effect of mutations on the level of μs and μm RNA
| Cell line | μs/actin
|
μm/actin
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Exp
|
Mean ± SD | Exp
|
Mean ± SD | ||||||
| 1 | 2 | 3 | 4 | 2 | 3 | 4 | |||
| Original | |||||||||
| WT | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Ter3 | 55 | 22 | 80 | 75 | 58 ± 23 | 39 | 26 | 28 | 31 ± 6 |
| Ter462* | 103 | 90 | 138 | 100 | 108 ± 18 | <0.1 | <0.1 | ||
| Recombinant | |||||||||
| Ter3 | 18 | 18 | 37 | 37 | |||||
| Ter3/Ter462* | 71 | 83 | 77 ± 6 | <0.1 | <0.1 | <0.1 | |||
For each independent experiment (exp) total RNA was isolated from the indicated cell lines and analyzed by Northern blots using the probes defined in Figure 1 to detect μs, μm, and actin RNA, as shown in Figure 3. The radioactivity in each band was quantified by PhosphorImager, and the values obtained for μs and μm were divided by the value obtained for actin. The quotient obtained for each cell line is expressed as a percentage of the WT quotient. Experiments 2 and 3 correspond to the blots shown in Figure 3, A and B, respectively.
To test whether the Ter462* mutation in cis with Ter3 could induce NMD, we compared the levels of μs and μm by Northern blot analyzes of μ RNA in the normal, single, and double mutant cell lines (Figure 3B and Table 1). As reported previously (Connor et al., 1994), the Ter462* mutation depressed the level of μm by >100 fold, whereas it had virtually no effect on μs mRNA. Interestingly, the Ter462* mutation in cis with Ter3 also decreased the level of μm RNA and was without effect on the level of μs. We conclude from these results that the termination codon Ter462* is recognized and then results in NMD even in cis with Ter3.
Detection of Truncated μ Chains
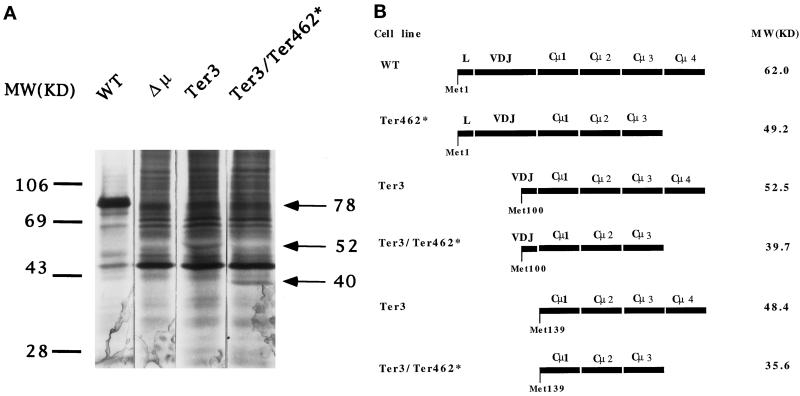
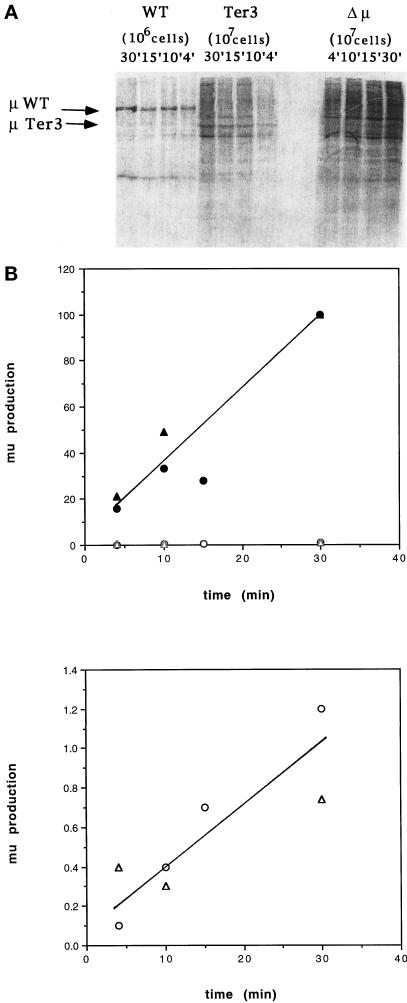
As noted above, the finding that the Ter3 mutant secretes <0.1% of the normal level of IgM indicates that the Ter3 mutation is not misread as sense (Connor et al., 1994). However, it was possible that in-frame translation initiated (or reinitiated) 3′ of Ter3, for example, at Met100 in V or at Met139 in Cμ1, and thus allowed translation of Ter462*. Truncated μ chains of this type would lack the leader segment and are therefore expected not to be glycosylated or secreted. We have previously estimated from Western blot analysis of Ter3 that such truncated proteins constitute <1% of the normal level of intracellular μ-related protein. However, our estimate of sensitivity presupposed that the mutant protein has normal stability and bears all the normal μ epitopes. To avoid these uncertainties, we retested for an abnormal translation product by examining intracellular material for biosynthetically labeled μ chains, thus testing for μ-specific bands obtained from Ter3 and the double mutant Ter3/Ter462*, which were absent in the Δμ cell line lacking the μ gene. For this purpose 2 × 107 cells were incubated for 30 min in the presence of [35S]methionine, and total cell lysates were immunoprecipitated with rabbit IgM-specific antibodies. These precipitates (2 × 106 cell equivalents for wild-type cells and 2 × 107 cell equivalents for the mutants) were then analyzed by SDS-PAGE. As illustrated in Figure 4A, we found bands at ∼52 and ∼40 kDa, which were specific to Ter3 and Ter3/Ter462*, respectively; we detected no other specific bands. We have confirmed that the Ter3 protein is not glycosylated; i.e., the Ter3-specific material has the same mobility when produced in the presence of tunicamycin, an inhibitor of N-linked glycosylation (our unpublished results).
Figure 4.
Production of truncated μ chains by Ter3 and Ter3/Ter462* mutants. (A) The indicated cell lines were grown in [35S]methionine-containing medium for 30 min at 2 × 107 cells/ml. The μ chain from 2 × 107 cell equivalents was precipitated with anti-IgM and protein G-agarose. The immunoprecipitated material was dissolved in SDS and reduced with mercaptoethanol. The full amount was analyzed by SDS-PAGE for all cell lines except the wild type, for which only 2 × 106 cell equivalents were analyzed. Immunoprecipitation was sufficiently complete, in that <25% of the total μ-specific activity could be recovered in secondary precipitations. The results were then visualized by autoradiography. (B) Predicted sizes of μ chains which result from initiation at the indicated internal AUG codons. As noted in the text, the truncated μ chains that result from initiation at Met100 or Met139 are expected to be unglycosylated, and therefore only the amino acid component has been calculated. The size listed for μ WT also represents only the polypeptide chain (62 kDa) and does not include its ∼26-kDa oligosaccharide component.
As noted above and illustrated in Figure 4B, the μ gene presents two ATG codons, which might account for these bands. Initiation at Met100 would yield μ proteins with molecular masses of 52.5 and 39.7 kDa for termination at the normal and Ter462 sites, respectively; initiation at Met139 would yield 48.4- and 35.6-kDa proteins. The observed sizes corresponded more closely to the expectations for initiation at Met100 results, and this designation is therefore used to describe the truncated proteins.
We have estimated the frequency (efficiency) at which initiation occurred at Met100 by measuring the rate at which the Ter3 protein was produced. To measure this rate we incubated the mutant and normal cells with [35S]methionine for various time intervals. As described above, the cell lysates were immunoprecipitated with IgM-specific antibodies, and the radiolabeled material was analyzed by SDS-PAGE (Figure 5A). Table 2 presents the PhosphorImager analysis of both this particular gel and another; the values thus calculated for μ biosynthesis are plotted in Figure 5B. 35S incorporation into both the normal and truncated μ chains increased steadily over the 30-min course of these experiments. We obtained comparable results when protein was produced in the presence of the protease inhibitor N-acety leucyl leucyl norleucine. As presented in MATERIALS AND METHODS, the radiolabeling, R, is expected to follow the equation R = Nα/β(1 − e−βt). The imprecision of the data allows for somewhat different estimates of the rate of synthesis. On the one hand, the results are consistent with a constant incorporation rate, i.e., no significant degradation of the normal and truncated μ chains during the 30-min labeling, thus indicating that the Ter-3 protein was produced at ∼1% of the rate of normal μ chain. On the other hand, the data are also consistent with limited decay; e.g., the incorporation at 30 min might be only 1.5 times the incorporation at 15 min, indicating a value of β = 1/22 min, i.e., a half-life of 15 min. In this case we estimate that the rate of synthesis of the Met100-initiated μ chain to be ∼1.8% of the rate for the normal μ. Considering that Ter3 contains ∼50% of the normal level of μs mRNA, the frequency of initiation at Met100 is therefore ∼3% (2–3.6%) of the rate of initiation at the normal AUG initiator codon.
Figure 5.
Kinetics of μ production. (A) The indicated cell lines (2 × 107 cells) were incubated in [35S]methionine-containing medium for various intervals. μ chains were analyzed by SDS-PAGE and then visualized by PhosphorImager. As described in Figure 4, 107 cell equivalents were analyzed in the case of mutant cell lines, and 106 cell equivalents were analyzed for the wild type. (B) PhosphorImager results presented in Table 2. Upper panel, results for both wild type (filled symbols) and Ter3 (open symbols); lower panel, Ter3 results on an expanded scale to show the nearly linear incorporation. Two independent experiments are plotted: the circles and triangles correspond to experiments 1 and 2 (Table 2), respectively.
Table 2.
Quantitative analysis of μ synthesis by wild-type and Ter3 cell lines
| Incubation (min) | Arbitrary units
|
Normalized μ production
|
|||
|---|---|---|---|---|---|
| μ band activity | BG activity | μ-specific activity | 107 cells | 106 cells | |
| Activity for 106 wild-type cells | |||||
| 4 | |||||
| Exp 1 | 226 | 97 | 129 | 16 | |
| Exp 2 | 362 | 127 | 235 | 21 | |
| 10 | |||||
| Exp 1 | 400 | 138 | 263 | 33 | |
| Exp 2 | 879 | 343 | 536 | 49 | |
| 15 | |||||
| Exp 1 | 364 | 142 | 222 | 28 | |
| 30 | |||||
| Exp 1 | 1128 | 331 | 797 | 100 | |
| Exp 2 | 1889 | 793 | 1096 | 100 | |
| Activity for 107 Ter3 cells | |||||
| 4 | |||||
| Exp 1 | 128 | 119 | 9 | 1 | 0.1 |
| Exp 2 | 161 | 117 | 44 | 4 | 0.4 |
| 10 | |||||
| Exp 1 | 197 | 164 | 33 | 4 | 0.4 |
| Exp 2 | 195 | 161 | 34 | 3 | 0.3 |
| 15 | |||||
| Exp 1 | 220 | 161 | 59 | 7 | 0.7 |
| 30 | |||||
| Exp 1 | 410 | 312 | 98 | 12 | 1.2 |
| Exp 2 | 504 | 423 | 81 | 7 | 0.7 |
Production of normal and truncated μ chains was assessed by SDS-PAGE analysis of immunoprecipitated, radiolabeled material sysnthesized during the indicated time period by the wild-type and Ter3 cell lines. Experiment 1 (Exp 1) corresponds to the blot shown in Figure 5A. The blot was exposed to a PhosphorImager plate, and the activity in each band of interest was measured in arbitrary units (column 2). The background (BG) contribution was estimated by the activity in the adjacent area of each lane (column 3). The difference between columns 2 and 3 is listed as the μ-specific activity in column 4. These values were then normalized to the 30-min value obtained for μ production by the wild-type cells. Because the SDS-PAGE analysis was applied to 107 mutant cells but only 106 normal cells, the values for Ter3 were further reduced by 10-fold to compare the rates of μ synthesis by an equal number of cells. Experiment 2 (Exp 2) is an analysis of a similar blot of independently labeled material.
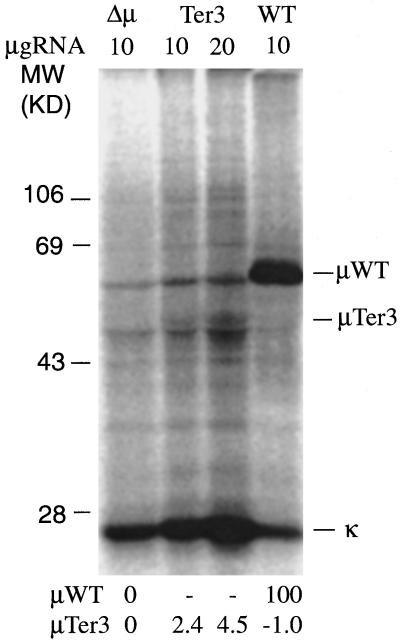
Out of concern that there might be in-frame initiation, which yielded μ-related proteins that were degraded so rapidly that they could not be detected even with the 4-min labeling, we examined the products of in vitro translation, in which such degradation is not expected to occur. As described in Figure 6, we immunoprecipitated μ-related material that was produced using rabbit reticulotcyte lysate to translate RNA from the wild-type, Ter3, and Δμ cell lines. We detected only one Ter3-specific band. This band had a mobility corresponding to ∼48 kDa, thus comparable to the Ter-specific material detected from intracellular lysates. As illustrated in Figure 6, quantitation of the μ bands by PhosphorImager analysis indicated that the Ter3 truncated μ chain was synthesized at ∼2% of the rate of μWT, again similar to the rate of synthesis inferred from the intracellular labeling experiments. Considering that the Ter3 μ mRNA is present at half the level of the wild-type mRNA, these results imply that initiation 3′ of Ter3 occurs at ∼4% of the frequency of the normal initiation rate,
Figure 6.
In vitro translation of μ RNA. As described in MATERIALS AND METHODS, total RNA (10 or 20 μg) from the indicated cell lines was translated in vitro using rabbit reticulocyte lysate and [35S]methionine. The IgM-related material was then immunoprecipitated with rabbit anti-IgM serum and analyzed by SDS-PAGE. The intensity of the indicated μWT and μTer3 bands was quantified by PhosphorImager. The background for the Δμ cell line was subtracted, and the resulting values were then normalized to the value obtained for the μWT band. These normalized values are listed below each lane.
DISCUSSION
Although numerous experiments have adduced evidence that NMD depends on translation of nonsense codons, it has not been possible previously to estimate how much translation is required. Our analysis of the truncated μ chains produced in the Ter3 mutant suggests that initiation at Met100/Met139 occurred ∼4% as often as initiation normally occurs at Met1. Therefore, translation of Ter462* in the Ter3 mutant occurred ∼4% as often as termination normally occurs for the wild-type μ chain. Our results thus imply that termination at no more than ∼4% of the normal rate was sufficient to induce NMD. This conclusion rests on the assumption that immunoprecipitation and SDS-PAGE analysis detected most of the in-frame, μ-related material, which results from initiation (or reinitiation) 3′ of Ter3. This assumption is supported by our consistent detection of only one μ-specific band using multiple independent batches of polyclonal μ-specific antibodies. Nevertheless, our results do not rule out low-level initiation from other 3′ in-frame initiation codons or initiation from sites that generate μ-related proteins, which, although in frame, are not recognized by the polyclonal anti-IgM sera.
As summarized in INTRODUCTION, the role of translation in NMD is controversial. On the one hand, our observation that very little if any translation is required for NMD is consistent with proposals that there is a nontranslational mechanism of recognizing nonsense mutations. On the other hand, our results indicate that Ter462* is translated at a detectable level, and perhaps even this very low-level translation is sufficient for a translation-dependent mechanism of NMD. Other treatments, viz., introduction of a 5′ hairpin and provision of suppressor tRNA, also reduced the frequency at which translation terminates at nonsense codons. However, in contrast to the effect of Ter3, these other treatments prevented NMD (Belgrader et al., 1993; Li et al., 1997). Assuming that 4% of the normal level of translation is generally sufficient for NMD, the comparison of these various treatments has interesting implications for models in which nonsense codons are recognized via a translation-based mechanism. For example, if the 5′ hairpin allowed >4% normal translation, then the RNA-sparing effect of the hairpin might reflect a second inhibitory effect of the hairpin on RNA degradation. Also, the 4% limit on translation of Ter462* is much lower than the level at which termination typically occurs in the presence of suppressor tRNA, which has been variously estimated to be 60–97% (Young et al., 1983; Laski et al., 1984). The RNA-sparing effect of suppressor tRNA might therefore occur because translation of the codon as sense plays an active role in preventing NMD. These comparisons suggest an interesting interpretation in the context of the models, which invoke a splice-associated mark to distinguish normal and abnormal terminators. As noted in INTRODUCTION, both permanent and erasable marks can be envisaged. If, in fact, substantial termination still occurred in the presence of suppressor tRNA, the sparing of mutant RNA by suppressor tRNA then argues that translation of the nonsense codon as sense erases the mark.
ACKNOWLEDGMENTS
This work was supported by a grant from the Medical Research Council of Canada.
REFERENCES
- Aoufouchi S, Yelamos J, Milstein C. Nonsense mutations inhibit RNA splicing in a cell-free system: recognition of mutant codon is independent of protein synthesis. Cell. 1996;85:415–422. doi: 10.1016/s0092-8674(00)81119-8. [DOI] [PubMed] [Google Scholar]
- Baumann B, Potash MJ, Kohler G. Consequences of frameshift mutations at the Ig heavy chain locus of the mouse. EMBO J. 1985;4:351–359. doi: 10.1002/j.1460-2075.1985.tb03636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgrader P, Cheng J, Maquat LE. Evidence to implicate translation by ribosomes in the mechanism by which nonsense codons reduce the nuclear level of human triosephosphate isomerase mRNA. Proc Natl Acad Sci USA. 1993;90:482–486. doi: 10.1073/pnas.90.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgrader P, Maquat LE. Nonsense but not missense mutations can decrease the abundance of nuclear mRNA for the mouse major urinary protein, while both types of mutations facilitate exon skipping. Mol Cell Biol. 1994;14:6326–6336. doi: 10.1128/mcb.14.9.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzina A, Shulman MJ. An element in the endogenous IgH locus stimulates gene targeting in hybridoma cells. Nucleic Acids Res. 1996;24:1525–1530. doi: 10.1093/nar/24.8.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter MS, Doskow J, Morris P, Li S, Nhim RP, Sandstedt S, Wilkinson MF. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem. 1995;270:28995–29003. doi: 10.1074/jbc.270.48.28995. [DOI] [PubMed] [Google Scholar]
- Carter MS, Li S, Wilkinson MF. A splicing-dependent regulatory mechanism that detects translation signals. EMBO J. 1996;15:5965–5975. [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Belgrader P, Zhou X, Maquat LE. Introns are cis effectors of the nonsense-codon-mediated reduction in nuclear mRNA abundance. Mol Cell Biol. 1994;14:6317–6325. doi: 10.1128/mcb.14.9.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Maquat LE. Nonsense codons can reduce the abundance of nuclear mRNA without affecting the abundance of premRNA or the half-life of cytoplasmic mRNA. Mol Cell Biol. 1993;13:1892–1902. doi: 10.1128/mcb.13.3.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Connor A, Wiersma E, Shulman MJ. On the linkage between RNA processing and RNA translatability. J Biol Chem. 1994;269:25178–25184. [PubMed] [Google Scholar]
- Davis AC, Roux KH, Pursey J, Shulman MJ. Intermolecular disulfide bonding in IgM: effects of replacing cysteine residues in the μ heavy chain. EMBO J. 1989;8:2519–2526. doi: 10.1002/j.1460-2075.1989.tb08389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz HC, Kendzior RJ. Maintenance of an open reading frame as an additional level of scrutiny during splice site selection. Nat Genet. 1994;8:183–188. doi: 10.1038/ng1094-183. [DOI] [PubMed] [Google Scholar]
- Dietz HC, Valle D, Francomano CA, Kendzior RJ, Pyeritz RE, Cutting GR. The skipping of constitutive exons in vivo induced by nonsense mutations. Science. 1993;259:680–683. doi: 10.1126/science.8430317. [DOI] [PubMed] [Google Scholar]
- Jack H, Berg J, Wabl M. Translation affects Ig mRNA stability. Eur J Immunol. 1989;1989:843–847. doi: 10.1002/eji.1830190510. [DOI] [PubMed] [Google Scholar]
- Jacobson A, Peltz SW. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- Kessler O, Jiang Y, Chasin LA. Order of intron removal during splicing of endogenous adenine phophoribosyltransferase and dihydrofolate reductase premRNA. Mol Cell Biol. 1993;13:6211–6222. doi: 10.1128/mcb.13.10.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laski FA, Belagaje R, Hudziak RM, Capecchi MR, Norton GP, Palese P, RajBhandary UL, Sharp PA. Synthesis of an ochre suppressor tRNA gene and expression in mammalian cells. EMBO J. 1984;3:2445–2452. doi: 10.1002/j.1460-2075.1984.tb02154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Leonard D, Wilkinson MF. T cell receptor (TCR) mini-gene mRNA expression regulated by nonsense codons: a nuclear-associated translation-like mechanism. J Exp Med. 1997;185:985–992. doi: 10.1084/jem.185.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wilkinson MF. Nonsense surveillance in lymphocytes? Immunity. 1998;8:135–141. doi: 10.1016/s1074-7613(00)80466-5. [DOI] [PubMed] [Google Scholar]
- Lozano F, Maertzdorf B, Pannell R, Milstein C. Low cytoplasmic mRNA levels of Ig κ light chain genes containing nonsense codons correlate with inefficient splicing. EMBO J. 1994;13:4617–4622. doi: 10.1002/j.1460-2075.1994.tb06783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE. Defects in RNA splicing and the consequences of shortened translational reading frames. Am J Hum Genet. 1996;59:279–286. [PMC free article] [PubMed] [Google Scholar]
- Maquat LE. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- Naeger LK, Schoberg RV, Zhao Q, Tullis GE, Pintel DJ. Nonsense mutations inhibit splicing of MVM RNA in cis when they interrupt the reading frame of either exon of the final spliced product. Genes & Dev. 1992;6:1107–1119. doi: 10.1101/gad.6.6.1107. [DOI] [PubMed] [Google Scholar]
- Oancea AE, Shulman MJ. An improved system of somatic cell molecular genetics for analyzing the requirements of Ig synthesis and function. Int Immunol. 1994;6:1161–1168. doi: 10.1093/intimm/6.8.1161. [DOI] [PubMed] [Google Scholar]
- Ruiz-Echevarria MJ, Gonzalez CI, Peltz SW. Identifying the right stop: determining how the surveillance complex recognizes and degrades an aberrant RNA. EMBO J. 1998;17:575–589. doi: 10.1093/emboj/17.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Echevarria MJ, Peltz SW. Utilizing the GCN4 leader region to investigate the role of the sequence determinants in nonsense-mediated mRNA decay. EMBO J. 1996;15:2810–2819. [PMC free article] [PubMed] [Google Scholar]
- Urlaub G, Mitchell PJ, Ciudad CJ, Chasin LA. Nonsense mutations in the dihydrofolate reductase gene affect RNA processing. Mol Cell Biol. 1989;9:2868–2880. doi: 10.1128/mcb.9.7.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersma EJ, Shulman MJ. Assembly of IgM: role of disulfide bonding and noncovalent interactions. J Immunol. 1995;154:5265–5272. [PubMed] [Google Scholar]
- Young JF, Capecchi M, Laski FA, RajBhandary UL, Sharp PA, Palese P. Measurement of suppressor tRNA activity. Science. 1983;221:873–875. doi: 10.1126/science.6308765. [DOI] [PubMed] [Google Scholar]
- Zhang J, Maquat LE. Evidence that translation reinitiation abrogates nonsense-mediated mRNA decay in mammalian cells. EMBO J. 1997;16:826–833. doi: 10.1093/emboj/16.4.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Sun X, Qian Y, LaDuca JP, Maquat LE. At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol Cell Biol. 1998a;18:5272–5283. doi: 10.1128/mcb.18.9.5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Sun X, Qian Y, Maquat LE. Intron function in the nonsense-mediated decay of β-globin mRNA: indications that premRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA. 1998b;4:801–815. doi: 10.1017/s1355838298971849. [DOI] [PMC free article] [PubMed] [Google Scholar]