Abstract
The sodium phosphate co-transporters Npt2a and Npt2c play important roles in the regulation of phosphate homeostasis. Slc34a1, the gene encoding Npt2a, resides downstream of the gene encoding coagulation factor XII (f12) and was inadvertently modified while generating f12−/− mice. In this report, the renal consequences of this modification are described. The combined single allelic mutant Slc34a1m contains two point mutations in exon 13: A499V is located in intracellular loop 5, and V528M is located in transmembrane domain 11. In addition to the expected coagulopathy of the f12−/− phenotype, mice homozygous for the double allelic modification (f12−/−/slc34a1m/m) displayed hypophosphatemia, hypercalcemia, elevated levels of alkaline phosphatase, urolithiasis, and hydronephrosis. Strategic cross-breedings demonstrated that the kidney-related pathology was associated only with autosomal recessive transmission of the slc34a1m gene and was not influenced by the simultaneous inactivation of f12. Npt2a[V528M] could be properly expressed in opossum kidney cells, but Npt2a[A499V] could not. These results suggest that a single amino acid substitution in Npt2a can lead to improper translocation of the protein to the cell membrane, disturbance of phosphate homeostasis, and renal calcification. Whether point mutations in the SLC34A1 gene can lead to hypophosphatemia and nephrolithiasis in humans remains unknown.
Three classes of sodium phosphate (Na/Pi) transporters have been identified. The most important of these for regulation of inorganic Pi in mouse kidney are members of the type 2 family, viz, Npt2a (gene slc34a1) and Npt2c (gene slc34a3). These transport proteins are specifically expressed in the brush border membranes of renal proximal tubules, where the bulk of filtered Pi is reabsorbed, and mediate Na gradient–dependent transport of Pi from primary urine to proximal tubular cells.1 Regulation of Npt2a protein abundance in the apical membrane by factors such as dietary phosphate, parathyroid hormone,2–4 and phosphatonins5 occurs via the action of scaffolding proteins and protein kinases.6,7
Inactivation of murine slc34a1 in mice led to a decrease in renal Pi reabsorption arising from a defect in Na/Pi co-transport.8 The accompanying hypophosphatemia in slc34a1−/− mice was not fully compensated by the consequent 2.8-fold increase in Npt2c protein abundance in kidney proximal tubules.9 These biochemical abnormalities, as well as hypercalcemia and hypercalciuria in slc34a1−/− mice, were associated with low body weight, mild skeletal abnormalities,8 and urolithiasis.10
The phenotype of slc34a1−/− mice resembles that of patients with hereditary hypophosphatemic rickets with hypercalciuria (HHRH), an inherited disorder of Pi homeostasis,11 with the exception that the mice have a milder skeletal phenotype.12,13 On this basis, it was hypothesized that mutations in the human orthologue SLC34A1 were responsible for HHRH. However, genome-based studies on kindreds with HHRH indicated that mutations of NPT2a were not responsible for this disease,14 and subsequent studies identified mutations in SLC34A3 as possibly causative in these and other HHRH patient populations.15,16 Whether point mutations in the SLC34A1 gene can lead to hypophosphatemia and nephrolithiasis in humans is an unresolved issue. An approach to addressing this question can be made through the generation of mice harboring known mutations in slc34a1. However, to date, no point mutations in Npt2a have been mechanistically linked to renal pathologies in controlled mouse studies.
The gene arrangement in the vicinity of slc34a1 is, consecutively, 5′-[coagulation factor 12 (f12)/profilin-3 (pfn3)/slc34a1]-3′, with this latter gene arranged in reverse orientation. To generate mice with a total inactivation of the coagulation f12 gene, we first designed a targeting vector (TV) with homologous recombination 5′ and 3′ sites external to the f12 gene at its untranslated regions (UTR). Although we confirmed that the resulting mice lacked f12 mRNA or FXII protein, we also found unexpected kidney-related phenotypes in these mice (unpublished observations). Since then, other lines of f12−/− mice have been generated by us and by others17,18 through exon-deletion targeting strategies, with homologous recombination sites in the TV designed entirely within the f12 gene sequence. In these mice (f12−/−-L2), f12 message and FXII protein were entirely eliminated, but kidney-related pathologies were not found. Furthermore, renal disease has not been linked to FXII deficiencies in humans.
The unexpected phenotypes in the f12−/−-L1 mice with total exon deletions were reminiscent of those in slc34a1−/− mice,8,10 rather than the phenotypes of the f12−/−-L2 mice.17,18 Thus, in this study we examined the integrity of the slc34a1 gene in f12−/−-L1 mice as a candidate mechanism for the renal pathologies that were observed. These in-depth studies allowed novel mechanistic revelations to be made in regard to significant kidney disease in a mouse line containing minimal point mutations in slc34a1.
RESULTS
Targeted Inactivation of the f12 Gene to Generate f12-L1 Mice
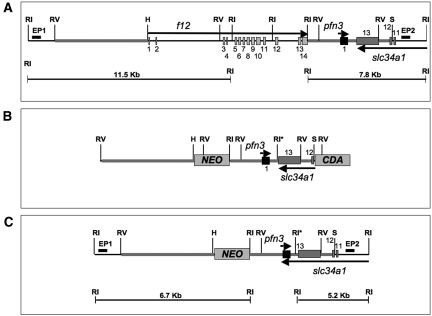
On the basis of the mouse genomic arrangement of Figure 1A, a targeting vector TV (Figure 1B) was constructed and electroporated in mouse embryonic stem cells (ESC). After screening the ESC with positive (G418)/negative (5′-fluorocytosine) selection, approximately 400 properly targeted cells were identified. DNA samples from these cells were digested with EcoRI, after which the DNA fragments were subjected to Southern blotting using the external probes EP1 and EP2 (Figure 1C). From these analyses, two independent and correctly targeted homologous recombinants were identified (Figure 2A).
Figure 1.
Generation of f12−/−-L1 mice. (A) The top line represents the f12 gene with its 14 exons (light gray boxes), the pfn3 gene with its one exon (black box), and exons 11 through 13 of the slc34a1 gene (dark gray boxes). Some key restriction enzyme sites are indicated (RI, EcoRI; RV, EcoRV; H, HindIII; S, SalI). (B) Construction of the TV for f12−/−-L1 mice. An RI-H digestion region of the 5′-UTR (A) and an RI-S digestion fragment of the 3′-UTR (A) of the f12 gene were used to flank the NEO cDNA, which was used for positive selection after homologous recombination in mouse embryonic stem cells. A cytidine deaminase (CDA) cassette was inserted downstream of the RI-S fragment and used for negative selection of recombinants. This process resulted in replacement of the entire f12 coding sequence with NEO. *One RI site was newly generated for purposes of screening. EP1 and EP2 in A represent external probes used for the 5′ and 3′ flanks of the TV, respectively. (C) Expected mutated allele.
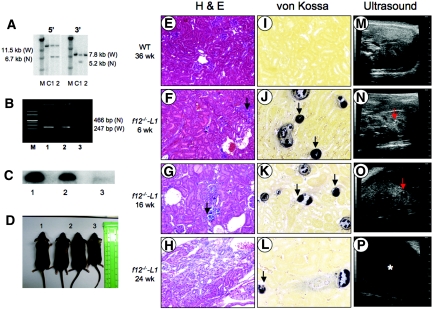
Figure 2.
Screening and characterization of f12−/−-L1 mice. (A) Southern blot analysis with the 5′- and 3′-external probes of digested DNA from ESC transfected with f12. The 5′ external probe (EP1 in Figure 1A) distinguished the WT-f12 (11.5 kb) and null (6.7 kb) alleles with RI-digested DNA, and the 3′ external probe (EP2 in Figure 1A) distinguished the WT-f12 (W; 7.8 kb) and null (N; 5.2 kb) alleles with RI-digested DNA. Molecular weight standards (M) are shown in lane 1; control 129/SvJ DNA (C) is shown in lane 2, and individual targeted ESC samples are enumerated in the remaining lanes. (B) PCR-based genotyping of typical WT (lane 2), f12+/−-L1 (lane 3) and f12−/−-L2 (lane 4) mouse tail-tip DNA. (C) Western blot analysis of typical WT (lane 1), f12+/−-L1 (lane 2), and f12−/−-L1 (lane 3) mouse plasmas, using a polyclonal antibody to human FXII, showing the absence of FXII antigen in f12−/−-L1 mouse plasma. (D) Body sizes of WT, f12+/−-L1, and f12−/−-L1 mice. (E) Hematoxylin II and eosin Y (H & E) stains for WT mouse kidney at 36 wk of age. (F through H) H & E stains of f12−/−-L1 mouse kidney at 6 wk of age (F), 16 wk of age (G), and 24 wk of age (H). The arrows in F and G indicate areas of kidney with abnormal cellularity, with widespread kidney architecture disruption in G. (I) von Kossa stains for WT mouse kidney at 36 wk of age and of f12−/−-L1 mouse kidneys at 6 wk of age (J), 16 wk of age (K), and 24 wk of age (L). The arrows point to examples of calcifications in J through L. Transabdominal ultrasound images for WT mouse kidney at 36 wk of age (M), f12−/−-L1 mouse kidney at 6 wk of age (N), 16 wk of age (O), and 24 wk of age (P). Red arrows in (N) and (O) indicate the high echogenic parts, which are typical indications of calcification. The arrow in N shows a calcification in a low echogenic cystic region of the kidney. An asterisk in (P) indicates a large area of low echogenicity, which is a typical signature of renal hydronephritis. Magnification, ×200.
Generation of f12−/−-L1 Mice
Blastocyst injections of the ESC yielded five male chimeric mice, which were mated with C57Bl/6J female mice. Germline transmission was observed with at least three pairs, and f12+/− mice (F1, 129SvJ/C57Bl6J; 50/50) were generated. Male F1 f12+/−-L1 mice were intermated with female f12+/−-L1 mice, thus providing f12−/−-L1 mice (Figure 2B).
Characterization of f12−/−-L1 mice showed the complete absence of f12 mRNA in liver (data not shown) and of FXII antigen in plasma (Figure 2C). Coagulation assays of f12−/−-L1 plasma also demonstrated greatly elevated activated partial thromboplastin times (aPTT; data not shown), similar to the activated partial thromboplastin times found in our f12−/−-L2 mice.18
Low Body Weight, Urolithiasis, and Hydronephrosis in the f12−/−-L1 Mice
The body sizes of the f12−/−-L1 mice were consistently smaller than those of wild-type (WT) and of f12+/−-L1 mice at 6 wk of age (Figure 2D). Massive calcifications were observed in f12−/−-L1 kidneys (Figure 2, J through L), even as young as 6 wk of age (data not shown). In 16-wk-old f12−/−-L1 mice, kidney cysts developed (Figure 2, F and G), along with severe hydronephrosis at 24 wk (Figure 2H). In contrast, neither WT mice (Figure 2, E and I) nor f12−/−-L2 mice18 presented with these kidney abnormalities, even to >1 yr of age, suggesting the absence of functional mutations in the slc34a1 gene in this latter mouse line. In agreement with these observations, plasma Pi levels were 9.3 ± 0.31 (n = 9), 6.4 ± 0.1 (n = 13), and 9.6 ± 0.48 (n = 6) in WT, f12−/−-L1, and f12−/−-L2 mice, respectively, showing that hypophosphatemia is found only in the f12−/−-L1 mice, consistent with the kidney abnormalities in this same line of mice.
These results were supported by transabdominal ultrasound analysis, which revealed high echogenic regions in f12−/−-L1 kidneys at 6 wk of age. These data are entirely consistent with renal calcification (Figure 2N). Similar regions were abundantly found in the entire kidneys and were associated with low echogenic cystic regions in 16-wk-old f12−/−-L1 mice (Figure 2O). Hydronephrotic echo patterns were obtained from 24-wk-old mice (Figure 2P), demonstrating significant kidney destruction. In contrast, no abnormalities in were found WT mice, even at 36 wk of age (Figure 2M).
Sequence Analysis of pfn3 and slc34a1 in f12−/−-L1 Mice
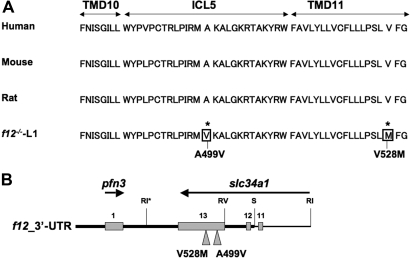
Because the genes pfn3 and slc34a1 are present in the 3′-UTR homologous recombination region of the TV used to construct f12−/−-L1 mice, we believed that the hypophosphatemia and kidney abnormalities seen in the f12−/−-L1 mice were the result of mutations in slc34a1. Thus, the sequences of both pfn3 and slc34a1 were determined from f12−/−-L1 mouse genomic DNA. The portion of the slc34a1 3′ sequence that was used for homologous recombination in f12−/−-L1 mice is shown in Figure 3. Two point mutations, viz., A499V and V528M, were discovered in exon 13 of the slc34a1 gene in the f12−/−-L1 mouse genome (Figure 3), which were not detected in WT and f12−/−-L2 mice or in the human or rat counterpart slc34a1 genes (Figure 3); therefore, we redefined f12−/−-L1 mice as f12−/−/slc34a1m/m to avoid any confusion with f12−/−-L2 mice, the genotype of which has been experimentally determined through genomic sequence analysis to be f12−/−/slc34a1+/+. Last, the pfn3 gene (Figure 3) was fully sequenced and found not to be altered in the f12−/−-L1 genome.
Figure 3.
Sequence analysis of the 3′-flanking region of f12−/−-L1. (A) Amino acid sequence comparisons of a region of the slc34a1 gene in humans and rodents with f12−/−-L1 mice, showing the two mutations (boxed) in the latter. TMD, transmembrane domain; ICL, intracellular loop. (B) The two point mutations (*) in exon 13 resulting from the recombination event in the 3′ genomic region of f12−/−-L1 mice are indicated and represented in slc34a1 in the bottom panel. This gene is oriented in the opposite direction of the f12 and pfn3 genes, and only exons 11, 12, and 13 (gray boxes) are shown because the 3′ flank of the TV does not extend beyond this region. The relative location of the pfn3 gene is also shown with its single exon (gray box).
Additional Breedings of Mice
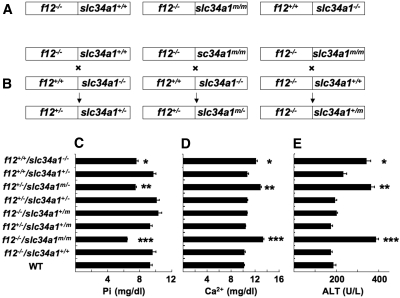
Because the f12−/− and slc34a1m/m genes were linked in f12−/−/slc34a1m/m mice, it was not possible to investigate the individual contributions of each genotype in vivo with this mouse line. Thus, we created new mouse lines by multiple cross-breedings to resolve this issue. We cross-bred f12−/−-L2 mice18 (i.e., f12−/−/slc34a1+/+) with f12+/+/slc34a1−/−8 mice to generate f12+/−/slc34a1+/− mice. We also cross-bred f12−/−/slc34a1m/m mice with f12+/+/slc34a1−/− mice to generate f12+/−/slc34a1m/− mice and f12−/−/slc34a1m/m mice with f12−/−/slc34a1+/+ mice to provide f12−/−/slc34a1+/m mice, each after multiple cross-breeding (Figure 4, A and B). Neither the coagulopathy nor renal abnormalities that were exhibited by singly deficient f12−/− or slc34a1−/− mice, respectively, or by the double-deficient f12−/−/slc34a1m/m mice were found in f12+/−/slc34a1+/−, fXII+/−/slc34a1+/−, or f12+/−/slc34a1+/m mice. Thus, the genetic traits were transmitted in an autosomal recessive manner for each of the f12 and slc34a1 genes, and the cross-breeding strategies provided us with the mice necessary to separate the phenotypes of the linked defects in the f12 and slc34a1 genes in the f12−/−/slc34a1m/m mice.
Figure 4.
A scheme of genotype variations and plasma chemistries of f12 and slc34a1 gene altered mice. (A) Genotypes of three different lines of f12/slc34a1 mice that were the starting points of the cross-breedings. (B) Genotypes of mice that were produced from the cross-breedings. (C through E) Serum Pi levels (C), serum Ca2+ levels (D), and serum ALP levels (E) of the various genotypic combinations of mice obtained from the cross-breedings. *The levels in f12+/+/slc34a1−/− sera were significantly different from those of WT, f12−/−/slc34a1+/+, f12+/−/slc34a1+/m, f12−/−/slc34a1+/m, f12+/−/slc34a1+/−, and f12+/+/slc34a1+/− sera but not significantly different from those of f12−/−/slc34a1m/m and f12+/−/slc34a1m/− sera. #The levels in f12+/−/slc34a1m/− were significantly different from those of WT, f12−/−/slc34a1+/+, f12+/−/slc34a1+/m, f12−/−/slc34a1+/m, f12+/−/slc34a1+/−, and f12+/+/slc34a1+/− sera, but not significantly different from those in f12+/+/slc34a1−/− and f12−/−/slc34a1m/m sera. †The levels in f12−/−/slc34a1m/m serum were significantly different from those in WT, f12−/−/slc34a1+/+, f12+/−/slc34a1+/m, f12−/−/slc34a1+/m, f12+/−/slc34a1+/−, and f12+/+/slc34a1+/− sera but not significantly different from those in f12+/+/slc34a1−/− and f12+/−/slc34a1m/− sera.
Serum Pi, Ca2+, and ALP Levels in Mice
Serum Pi levels in f12−/−/slc34a1m/m mice were significantly lower than those in WT and f12−/−/slc34a1+/+ mice (Figure 4C). In contrast, serum Ca2+ (Figure 4C) and ALP (Figure 4D) levels in f12−/−/slc34a1m/m mice were significantly higher than those in WT and f12−/−/slc34a1+/+ mice. To rule out the possibility that lack of FXII affected Npt2a functions, f12−/−/slc34a1+/+, f12−/−/slc34a11m/m, and f12+/+/slc34a1−/− mice were intercrossed to generate several double heterozygote combinations depicted in Figure 4, A and B. In this series, only f12+/−/slc34a1m/− mice displayed similar profiles of Pi (Figure 4C), Ca2+ (Figure 4D), and ALP (Figure 4E) as f12−/−/slc34a1m/m and f12+/+/slc34a1−/− mice. These results clearly indicate that the FXII deletion did not affect Npt2a function and that one intact copy of slc34a1 with one mutated slc34a1 copy was sufficient to maintain normal serum concentrations of Pi, Ca2+, and ALP. In addition, these data ruled out dominant negative effects of the slc34a1m gene on WT slc34a1.
Transcript Levels of slc34a1 in Kidneys
Kidney levels of slc34a1 transcript were similar in WT mice (1.9 ± 0.4 × 108 copies/100 ng total RNA [n = 6]), and f12−/−/slc34a1m/m mice (1.9 ± 0.5 × 108 copies/100 ng total RNA [n = 6]). Testicular transcript levels of pfn3 in WT and in f12−/−/slc34a1m/m mice were 6.1 ± 1.2 × 106 copies/100 ng total RNA (n = 6) and 11.0 ± 2.3 × 106 100 ng total RNA (n = 6), respectively. These data show that transcription of the two immediate downstream genes is not affected by the mutation in the f12 gene.
Immunohistochemistry of Npt2a in Kidney
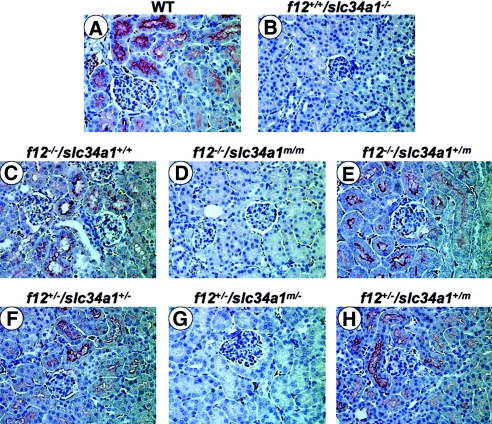
Anti-Npt2a immunostainings of kidney sections from the mice of Figure 4B were positive in the epithelia of proximal tubules from WT mice (Figure 5A). As expected, Npt2a immunostaining was negative in f12+/+/slc34a1−/− mice (Figure 5B). Whereas f12−/−/slc34a1+/+ mice also expressed Npt2a (Figure 5C), immunostaining for Npt2a was not evident in f12−/−/slc34a1m/m mice (Figure 5D), despite that its transcript levels were similar to those in WT mice. Furthermore, f12−/−/slc34a1+/m mice expressed Npt2a (Figure 5E). For further probing of the reason for interference in Npt2a expression in f12−/−/slc34a1m/m mice, f12+/−/slc34a1+/−, f12+/−/slc34a1m/−, and f12+/−/slc34a1+/m mice were generated. As clearly illustrated, f12+/−/slc34a1+/− (Figure 5F) and f12+/−/slc34a1m/+ (Figure 5H) mice stained positively for Npt2a. Conversely, f12+/−/slc34a1m/− mice did not express Npt2a antigen (Figure 5G). Thus, there was no interference in Npt2a expression from the absence of FXII, indicating that the double-mutated alleles in the slc34a1 gene were the cause of the abnormal renal phenotype in f12−/−/slc34a1m/m mice.
Figure 5.
(A through H) Immunostains of Npt2a in WT (A), f12+/+/slc34a1−/− (B), f12−/−/slc34a1+/+ (C), f12−/−/slc34a1m/m (D), f12−/−/slc34a1+/m (E), f12+/−/slc34a1+/− (F), f12+/−/slc34a1m/−, (G) and f12+/−/slc34a1+/m (H) kidneys. Magnification, ×400.
Urinary Analyses for Ca2+ and Pi Excretion
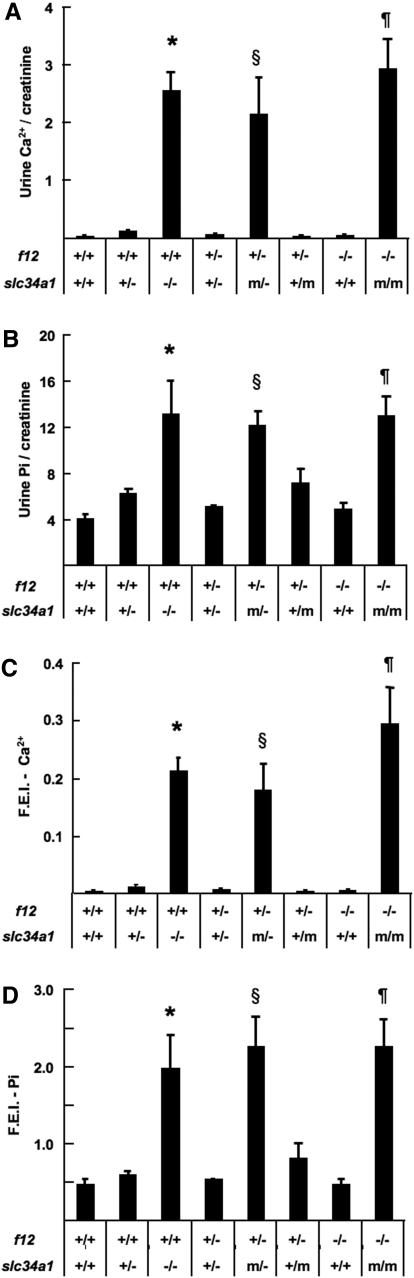
Urinary analyses clearly indicated that creatinine-normalized urine levels of Pi and Ca2+ were increased in f12+/+/slc34a1+/− and further elevated in f12+/+/slc34a1−/− mice (Figure 6, A and B). These alterations also corresponded to the fractional excretion of them (Figure 6, C and D). These values in f12−/−/slc34a1+/+ were similar to those in WT mice. These values in f12+/−/slc34a1+/m and f12+/−/slc34a1+/− were close to those in f12+/+/slc34a1+/− mice, and those in f12+/−/slc34a1m/− and f12−/−/slc34a1m/m were close to those in f12+/+/slc34a1−/− mice (Figure 6).
Figure 6.
Urinary analysis for Pi and Ca2+ excretion. (A) Urine Ca2+/creatinine. (B) Urine Pi/creatinine. (C) fractional excretion indexes (FEI) of Ca2+. (D) FEI of Pi. *The levels in f12+/+/slc34a1−/− were significantly different from those of WT, f12−/−/slc34a1+/+, f12+/−/slc34a1+/m, f12+/−/slc34a1+/−, and f12+/+/slc34a1+/− but not significantly different from those of f12−/−/slc34a1m/m and f12+/−/slc34a1m/−. §The levels in f12+/−/slc34a1m/− were significantly different from those of WT, f12−/−/slc34a1+/+, f12+/−/slc34a1+/m, f12+/−/slc34a1+/−, and f12+/+/slc34a1+/− but not significantly different from those in f12+/+/slc34a1−/− and f12−/−/slc34a1m/m. ¶The levels in f12−/−/slc34a1m/m were significantly different from those in WT, f12−/−/slc34a1+/+, f12+/−/slc34a1+/m, f12+/−/slc34a1+/−, and f12+/+/slc34a1+/−, but not significantly different from those in f12+/+/slc34a1−/− and f12+/−/slc34a1m/−.
Fluorescence Microscopy of Transfected Opossum Kidney Cells
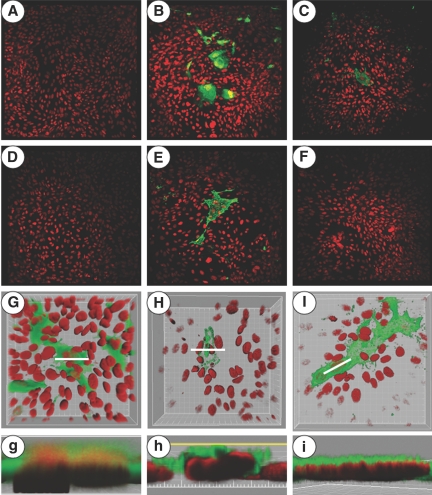
Fluorescence microscopic analysis showed that there were no endogenous positive recombinant green fluorescence protein (GFP) signals in opossum kidney (OK) cells transfected with pCS2-empty plasmid (Figure 7A) or in OK cells transfected with pCS2-containing cDNA that express WT-Npt2a, Npt2a[A499V], Npt2a[V528M], or Npt2a[A499V/V528M] (data not shown). Conversely, as expected, strongly positive GFP signals were found in OK cells transfected with pCS2-GFP (Figure 7B). Alexa Fluora 488–positive stainings were found in OK cells expressing recombinant WT-Npt2a or Npt2a[V528M] (Figure 7, C and E, respectively), showing expression of Npt2a. In contrast, Alexa Fluora 488–positive stainings were not found in OK cells expressing heterologous Npt2a[A499V] or Npt2a[A499V/V528M] (Figure 7, D and F, respectively). These results show that there was no expression of Npt2a[A499V] or Npt2a[A499V/V528M] and/or that the mutant proteins were conformationally altered and not recognized by the antibody to WT-Npt2a. Five separate transient transfections confirmed that neither Npt2a[A499V] nor Npt2a[A499V/V528M] antigen was observed in the cells. Higher magnified and cross-sectional images demonstrated that the positive GFP signals were diffusely observed in the cytoplasm of OK cells expressing GFP (Figure 7, G and g). Apical anti-Npt2a stains were seen in OK cells expressing recombinant WT-Npt2a (Figure 7, H and h) or Npt2a[V528M] (Figure 7, I and i).
Figure 7.
Fluorescence and laser scanning confocal microscopy for transfected OK cells. (A through F) Immunostains of Npt2a in OK cells transfected with pCS2-empty plasmid (A), pCS2-GFP (B), pCS2-slc34a1 (C), pCS2-slc34a1[A499V] (D), pCS2-slc34a1[V528M] (E), and pCS2-slc34a1[A499/V528M] (F). (G through I) Immunostains of Npt2a in OK cells transfected with pCS2-GFP (G), pCS2-slc34a1 (H), and pCS2-slc34a1[V528M] (I) using an oil immersion objective. (g through i) Cross-sections along the planes from G through I represented by white lines, respectively. Clipping planes were placed at the regions of interest, which are indicated as white lines in G through I, and used to expose the GFP-positive (g) or NPT2 Alexa Fluor 488–positive (h and i) labeling and its cellular localization. The images were rendered to 120° perspective projection and captured at a zoom factor of 0.857 pixel/voxel. XY and XZ orthogonal images were captured. Magnifications: ×20 in A through F; ×60 in G through I.
DISCUSSION
The kidney is primarily responsible for phosphate homeostasis and, in general, is a determinant for blood phosphate levels through its regulation of phosphate reabsorption. The movement of Pi in and out of kidney cells depends on two Na+/Pi2− transport protein ion channels, Npt2a and Npt2c, both of which are expressed in brush border membranes of proximal tubular cells, where the filtered Pi is reabsorbed. The trafficking of Npt2a and Npt2c to the brush border membrane of kidney tubular cells, a feature that governs their abilities to uptake Pi into cells, is regulated by several processes, such as the integrity of the binding of Npt2a with PDZ-adapter proteins (e.g., NHERF-1) which is influenced by parathyroid hormone.7 Apical membrane abundance of Npt2a is also affected by dietary Pi and by the gene products of phex and the fgf23, the latter of which is a vitamin D3–dependent hormone produced in bone.
Hypophosphatemia and hyperphosphaturia result from decreased renal Pi reabsorption in slc34a1−/− mice, thus providing a critical link between properly targeted Npt2a abundance in brush border membranes and Pi homeostasis. Although its is known that a total Npt2a deficiency leads to hypophosphatemia and hyperphosphaturia, there are no known mechanistic links between mutations in Npt2a and animal pathophysiology. The one study that demonstrated a relationship between two putative dominant negative point mutations in Npt2a in humans was responsible for their presentation of hypophosphatemia19 was not reproduced by another group.20 In addition, in this study, we were not able to confirm dominant negative effects of the Npt2a point mutants on WT Npt2a, and we found that genetic transmission of our mutated slc34a1 gene is autosomal recessive.
Xenopus oocytes have been used extensively for the expression of slc34a1.21 OK cells have also been used for study of the trafficking of Npt2a.22 Although both systems are seemingly appropriate for the purposes intended, a concern emerges regarding the relationships of Npt2a expressions between these cells and the physiologic epithelial cells in proximal tubules in kidney. Although Xenopus oocytes and OK cells can express both WT and mutated forms of Npt2a, no assurances can be made that epithelial cells in a physiologic environment are able to target mutated forms of Npt2a to apical renal tubule membranes properly, a property necessary for its activity.
A novel contribution of our work concerns the biology of the slc34a1 gene, with mechanistic correlations both in vivo and in vitro, despite that the positions of the chance mutations in the slc34a1 gene were not identical to the mutations reported in patients with HHRF. In our case, two point mutations in Npt2a, viz., A499V in intracellular loop-5 and V528M in transmembrane domain-11 of Npt2a, were identified in our f12−/−/slc34a1m/m mice. These mutations obviously affected the levels of Pi, Ca2+, and ALP in vivo, which led to hypophosphatemia in these mice. Only mice homozygous for the double mutation in Npt2a displayed renal pathologies, consistent with altered Na+/Pi2− transport. Furthermore, the presence of this protein in kidney was diminished in both f12−/−/slc34a1m/m and f12+/−/slc34a1m/− mice. This diminution of protein expression might be a result of mutation-based misfolding, because we show nearly normal levels of slc34a1 mRNA in kidneys of f12−/−/slc34a1m/m mice.
In vitro expression experiments demonstrated that renal proximal tubular OK cells that were transfected with slc34a1 or with the mutated slc34a1 gene that led to Npt2a[V528M] correctly expressed Npt2a; however, OK cells transfected with genes mutated to produce Npt2a[A499V] or Npt2a[A499V/V528M] did not express Npt2a. The mechanisms required for successful expression of Npt2a on apical membranes of polarized cells, in vitro, depend on the cell types used and on amino acid residues in the C-terminal intracellular tails of this protein.23 Our results suggested that Npt2a protein folding is important for its correct localized expression and is likely mediated by specific chaperones and by interactions with scaffolding proteins, which are present in the brush border membrane of renal proximal tubules; therefore, Npt2a[A499V] protein likely is not recognized by these chaperones and/or scaffolds, and this could result in its premature degradation in the cells.
Additional examples of point mutations in transporter proteins that lead to severe pathologies are known (e.g., cystic fibrosis [CF], a disease caused by mutations in the gene that encodes the CF transmembrane conductance regulator [CFTR]). Many reports of point mutations in this protein that result in its malfunction are known, and the most common of these CF-causing mutations (δF508) reaches the plasma membrane in reduced amounts as a result of protein misfolding.24 Other single amino acid substitutions in the CFTR gene also result in protein misfolding and lead to reduced expression on the cell surface.25,26 Thus, it seems that point mutations in the Npt2a co-transporter may have a similar impact on protein processing. In conclusion, this work has shown that abnormal kidney pathologies can mechanistically result from simple point mutations in Npt2a.
CONCISE METHODS
Screening of λ-Phage Libraries
Oligonucleotide primer set X1UP (5′-GGTCTCTGCTGATGAGTCTG, cDNA position 43 to 62) and X2RP (5′-GTGCGTCCTTAAATTTCTTG, cDNA position 98 to 117), or X13UP (5′-GGCTGAAGAATACTCCACCT, cDNA position 1499 to 1518) and X14RP (5′-CACTAGCTTCACTGTGACCC, cDNA position 1880 to 1899), based on the murine f12 cDNA sequence in the NCBI server (NM 021489), was used to obtain the fragment of the f12 gene that contained partial exon 1/intron A/partial exon 2 (F1) or partial exon 13/intron M/partial exon 14 (F2), respectively, from tail genomic DNA of 129/SvJ mice (Jackson Laboratories, Bar Harbor, ME) using PCR. PCR products resulting from amplifications with these 418-bp F1 or 486-bp F2 PCR probes were used to isolate genomic clones containing the f12 gene from a 129/SvJ λ-FIXII genomic library (Stratagene, La Jolla, CA), and several appropriate DNA clones were obtained. The resulting genomic clones spanned approximately 9 kb of the 5′-UTR, followed by the entire f12 gene (14 exons and 13 introns) and approximately 1 kb of the 3′-UTR. Despite many rescreening attempts to obtain a longer 3′-UTR for more effective genomic recombination, no additional useful clones were obtained. Previous results27 suggested that use of λ-libraries containing genomic sequences led to rearrangements of sequences between the 3′-UTR of the f12 and slc34a1 genes, and correct sequences were not obtained using this approach. The strategy for screening was then changed to vectorette and suppression long and accurate PCR (VS-LA-PCR), and 6 kb of the 3′-UTR was obtained. This allowed a TV to be constructed with ample flanking sequences of the 5′- and 3′-UTR regions of mouse f12.
Extended 3′-UTR Sequences of the f12 Gene
Genome walking with VS-LA-PCR was performed as described previously.28 Genomic DNA (50 mg) from 129/SvJ mice, prepared as in the previous section, was digested by the restriction endonucleases DraI, EcoICR1, EcoRV, PvuII, ScaI, SspI, and StuI (Promega, Madison, WI), individually. After the digestion, each fragment was ligated to an adaptor consisting of two oligonucleotides, 5′-pACCAGCCC-NH2 and 5′-GTAATACGACTCACTATAGGGCACGCGTGGTCGACGGCCCGGGCTGGT. VS-LA-PCR was performed several times with a gene-specific primer and the adaptor-specific primer to obtain sufficient sequence lengths.
The TV was constructed similar to those that we have previously described29 from a basic plasmid containing the neomycin-resistant gene (NEO) for positive selection along with 5′- and 3′-flanking sequences from the murine f12 gene. For the latter, a 5.2-kb EcoRV/HindIII fragment and a 4.8-kb EcoRI/SalI fragment of the f12 gene were used as the 5′ and 3′ flanks of f12, respectively, of the NEO gene in the TV. The cytosine deaminase (CDA) cassette was inserted downstream of the 3′-flanking region of the TV for f12 for negative selection against random chromosomal integrants.
Homologous Recombination in ESC
The TV was used to replace, by homologous recombination, a 9.0-kb fragment of the f12 gene with the NEO gene. This process eliminated the entire f12 coding sequence from the mouse genome. The generation of targeted ESC and subsequent blastocyst injections were performed.
Southern blot analyses with external probes on ESC DNA, after digestion with EcoRI, were carried out to assess the integrity of the homologous recombination at both the 5′ and 3′ terminal regions. The 5′ external probe (EP1; 421 bp, spanning bp 5846 to 5446 upstream of the ATG initiation codon of the f12 gene) was obtained by PCR using primers 5′-CTCTCAGGAGAAAAAGTATACC and 5′-CAGACCACATTCTCTTCATCC. The 3′ external probe (EP2; 471 bp, spanning bp 5453 to 5923 from the TAA stop codon of the f12 gene) was also obtained by PCR using the primers 5′-GTCTACATAGTTCTAACCAG and 5′-GGGTCCATGTGTTACCTTGTC.
Genotyping
Genomic DNA was obtained by ear-punch biopsy. PCR was performed with oligonucleotide primer sets: A common forward primer (5′-TCTTCAGTCCGCTACTCCCAC, spanning bp 200 to 180 upstream of the ATG initiation codon of the f12 gene), and a reverse primer in exon 1 of the f12 gene (5′-AGATCCAGACTCATCAGCAG, spanning bp 48 to 67 of the f12 cDNA) to detect the WT allele (247 bp), or the aforementioned common forward primer along with a reverse primer residing within NEO (5′-ACAAGCAAAACCAAATTAAGGGCCA) to detect the null allele (466 bp).
Histochemistry and Immunohistochemistry
Kidneys were obtained from mice at 6 wk of age, fixed with periodate-lysine-paraformaldehyde, embedded in paraffin, and sectioned at a thickness of 4 μm. The slides were stained with hematoxylin II and eosin Y (Richard Allen Scientific, Kalamazoo, MI) for morphologic analysis and von Kossa stain to detect calcification.
For immunostaining, tissue sections were deparaffinized and placed in avidin and biotin blocking solutions (Zymed Laboratories, South San Francisco, CA), followed by Peroxo-block (Zymed) to inhibit endogenous peroxidase activity. Anti-Npt2a immunostains were conducted using the ImmPress Kit (Vector Laboratories, Burlingame, CA). The sections were incubated with rabbit anti-human primary antibodies (Alpha Diagnostics, San Antonio, TX), followed by horseradish peroxidase–conjugated goat anti-rabbit IgG. The slides were developed with 3-amino-9-ethylcarbazole and counterstained.
Transabdominal Ultrasound Analysis
Mice were sedated by inhalation of 1.5% isoflurane sufficient to maintain their heart rates at approximately 500 bpm. The abdomens of the mice were cleaned with a chemical hair remover (Nair) to minimize ultrasound attenuation. Transabdominal ultrasound analysis was accomplished with a Vevo 770 system (VisualSonics, Toronto, ON, Canada).
Sequence Analysis of slc34a1 cDNA
Total RNA was obtained from kidneys using Trizol (Invitrogen, Carlsbad, CA). First-strand cDNA was synthesized using Superscript-III reverse transcriptase (Invitrogen). Reverse transcriptase–PCR was performed with the primers in Table 1 to obtain the designated fragments for sequence analysis. After purification of PCR products, they were directly sequenced using the same primers at the DNA Core Sequencing Facility at the University of Illinois at Urbana-Champaign.
Table 1.
Primers and probes used for sequence analysis
| Gene | Type | Sequence | Position | Size (bp) |
|---|---|---|---|---|
| pfn3 | Forward | 5′-CTAATGCAAGCACAAGTTGC | 20 to 39 | 506 |
| Reverse | 5′-GTTCACGGTTTATTCTGGTC | 506 to 525 | ||
| slc34a1 | Forward | 5′-GAGCTGAGCCACAGTCAAGG | 14 to 33 | 864 |
| Fragment-1 | Reverse | 5′-GTGAAGGGCTCTGTGATGAC | 858 to 877 | |
| slc34a1 | Forward | 5′-CTTTTGCAGGGGCGACCGTG | 700 to 719 | 735 |
| Fragment-2 | Reverse | 5′-GGCTGGCAACGGCTGCCAGG | 1415 to 1434 | |
| slc34a1 | Forward | 5′-GTGTGATCAGCATTGAGCGG | 1345 to 1364 | 780 |
| Fragment-3 | Reverse | 5′-CTTAATATGCACAGGTACC | 2106 to 2124 |
Other Mice
Slc34a1−/− mice were obtained from Dr. Beate Lanske (Harvard School of Medicine, Boston, MA). The generation and characterization of other mice used in this study, viz., f12+/+/slc34a1−/−8 and f12−/−/slc34a1+/+,18 have been described.
Measurements of Serum and Urine Levels of Pi, Ca2+, Creatinine, and ALP
Serum and urine levels of Pi, Ca2+, creatinine, and ALP were measured using the VetTest chemistry analyzer (IDEXX Laboratories, Westbrook, ME). The fractional excretion indexes Pi or Ca2+ were calculated as follows: Urine Pi or Ca2+/(urine creatinine × serum Pi or Ca2+).
Npt2a Constructs
An expression vector backbone, pCS2, that contains the cytomegalovirus immediate early promoter to drive downstream inserted genes, was obtained from Dr. Paul Huber (University of Notre Dame, Notre Dame, Indiana) and first digested with BamHI and XbaI. A fragment of recombinant GFP (Stratagene, La Jolla, CA) was amplified with the forward primer hrGFP.F (sequences provided next), which contained a BglII site (italic) and Kozak sequence (underlined), followed by the initiation codon (bold), along with the reverse primer phrGFP.R, which contained XbaI site (italic) just downstream of the termination codon (bold) of phrGFP.R. All amplifications were performed using PrimeStar DNA polymerase (TakaraBio, Madison, WI). The PCR amplicon was digested by BglII and XbaI. The BglII- and XbaI-digested fragment was ligated to pCS2 that was digested with BamHI and XbaI. The resulting plasmid was designated pCS2-hrGFP. A fragment of murine slc34a1 was amplified with the forward primer Slc34a1.F, which contained a BglII site (italics) and the Kozak sequence (underlined), followed by the initiation codon (bold), and the reverse primer Sc34a1.R, which contained a XbaI site (italics), just downstream of the termination codon (bold) using an NIH Mammalian Gene Collection (MGC) clone (clone 4223484; Invitrogen) as the template. The PCR amplicon was digested by BglII and XbaI, and the fragment obtained was ligated to pCS2 and digested by BamHI and XbaI. The plasmid was designated pCS2-slc34a1.
hrGFP.F: 5′-GAAGATCTGCCGCCACCATGGTGAGCAAGCAGATCCT
hrGFP.R: 5′-GTTCTAGATTACACCCACTCGTGCAGGC
Slc34a1.F: 5′-GAAGATCTGCCGCCACCATGATGTCCTACAGCGAGAGATTG
Slc34a1.R: 5′-GTTCTAGACTAGAGACGGGTGGCATTGTG
Site-Directed Mutagenesis
The single Npt2a mutations, viz., A499V and V528M, and the double mutation, Npt2a[A499V/V528M], were introduced into pCS2-slc34a1 by site-directed mutagenesis to construct a vector to express Npt2a[A499V], Npt2a[V528M], and Npt2a[A499V/V528M]. For this, an inverse PCR reaction was performed with forward primer A499V.F, which contained the underlined mutation (next), along with reverse primer, A499V.R, using pCS2-slc34a1 as the template to construct an expression plasmid for Npt2a[A499V]. Also, two inverse PCR reactions were performed with forward primer, V528M.F, which contains the underlined mutation, in combination with reverse primer, V528M.R, using pCS2-slc34a1 as the template to construct a plasmid expressing Npt2a[V528M]. This same strategy, with pCS2-slc34a1[A499V] as the template, was used to construct the plasmid expressing Npt2a[A499V/V528M]. After the PCR reactions, these amplicons were treated by TaKaRa BKL Kit (TakaraBio) for self-ligation.
A499V.F: 5′-TCAAGGCCCTGGGCAAACGCAC
A499V.R: 5′-CCATGCGTATGGGCAGGCGTG
V528M.F: 5′-ATGTTTGGCATTTCCATGGCAGGCTGG
V528M.R: 5′-CAGCGAGGGCAAGAGCAGGAAGC
Cell Culture, Transfection, and Fluorescence Microscopy
OK cells were purchased from ATCC (Manassas, VA) and maintained in DMEM/Ham's F-12 medium (1:1), containing 10% FBS and 1× antibiotic solution (Mediatech, Herndon, VA). Cells were seeded into each chamber of a four-chamber culture slide (BD Biosciences, Bedford, MA), and 50% confluent cultures were transiently transfected with pCS2-empty plasmid, pCS2-GFP, and pCS2 plasmids containing cDNA expressing WT-Npt2a, Npt2a[A499V], Npt2a[V528M], and Npt2a[A499V/V528M]. Transfections were performed using 1 μg of plasmid DNA and 3 μl of FuGENE6 (Roche Applied Science, Indianapolis, IN) per well. After the transfections, the cells were fixed with periodate-lysine-paraformaldehyde and stained with rabbit anti-human Npt2a, followed by an Alexa Fluora 488–labeled goat-anti-rabbit IgG (Invitrogen). These cells were further stained with DAPI (Invitrogen). Laser Scanning Confocal Microscopy was conducted with a Nikon Eclipse C1si confocal system on a Nikon TE2000-E microscope with Spectral Imaging. An EZ-C1 imaging software (Nikon, Melville, NY) was used for image acquisition. For gaining three-dimensional visualization control of the regions of interest, images were processed with Imaris (Bitplane AG, Zürich, Switzerland). The DAPI-labeled nuclei were pseudocolored (red) for enhanced contrast against the GFP-transfected cells or the Alexa Fluor 488 NPT2 positively labeled cells (green). The Surpass mode was used to create a volume reconstruction of the data set (Z-stack). The 20× data sets’ volume properties were rendered to a maximum intensity projection (MIP) mode of all layers along the viewing direction (black background). A Shadow Projection mode was used for the 60× data sets with a transparency adjustment of the blend opacity to 33.6% for the green channel. The nuclei's (red) blend opacity was maintained at 100%.
DISCLOSURES
None.
Acknowledgments
This work was supported in part by grant HL073750 (to F.J.C.).
We thank Diana Cruz-Topete for assistance of histologic analysis, Kyle Kisiel at Nikon Imaging Center at Northwestern University for fluorescence and confocal imaging for transfected OK cells, and the veterinary staff of the Freimann Life Sciences Facility for providing husbandry of the mice.
Published online ahead of print. Publication date available at www.jasn.org.
See related editorial, “Of Mice and Men: Who Is in Control of Renal Phosphate Reabsorption?” on pages 1625–1626.
REFERENCES
- 1.Forster IC, Kohler K, Biber J, Murer H: Forging the link between structure and function of electrogenic cotransporters: The renal type IIa Na+/Pi cotransporter as a case study. Prog Biophys Mol Biol 80: 69–108, 2002 [DOI] [PubMed] [Google Scholar]
- 2.Murer H, Forster I, Biber J: The sodium phosphate cotransporter family SLC34. Pflugers Arch 447: 763–767, 2004 [DOI] [PubMed] [Google Scholar]
- 3.Forster IC, Hernando N, Biber J, Murer H: Proximal tubular handling of phosphate: A molecular perspective. Kidney Int 70: 1548–1559, 2006 [DOI] [PubMed] [Google Scholar]
- 4.Tenenhouse HS: Phosphate transport: Molecular basis, regulation and pathophysiology. J Steroid Biochem Mol Biol 103: 572–577, 2007 [DOI] [PubMed] [Google Scholar]
- 5.Shimada T, Hasegawa H, Yamazaki Y, Muto T, Hino R, Takeuchi Y, Fujita T, Nakahara K, Fukumoto S, Yamashita T: FGF-23 is a potent regulator of vitamin D metabolism and phosphate homeostasis. J Bone Miner Res 19: 429–435, 2004 [DOI] [PubMed] [Google Scholar]
- 6.Khadeer MA, Tang Z, Tenenhouse HS, Eiden MV, Murer H, Hernando N, Weinman EJ, Chellaiah MA, Gupta A: Na+-dependent phosphate transporters in the murine osteoclast: Cellular distribution and protein interactions. Am J Physiol Cell Physiol 284: C1633–C1644, 2003 [DOI] [PubMed] [Google Scholar]
- 7.Weinman EJ, Biswas RS, Peng Q, Shen L, Turner CL, Steplock D, Shenolikar S, Cunningham R: Parathyroid hormone inhibits renal phosphate transport by phosphorylation of serine 77 of sodium-hydrogen exchanger regulatory factor-1. J Clin Invest 117: 3412–3420, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Beck L, Karaplis AC, Amizuka N, Hewson AS, Ozawa H, Tenenhouse HS: Targeted inactivation of Npt2 in mice leads to severe renal phosphate wasting, hypercalciuria, and skeletal abnormalities. Proc Natl Acad Sci U S A 95: 5372–5377, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tenenhouse HS, Martel J, Gauthier C, Segawa H, Miyamoto K: Differential effects of Npt2a gene ablation and X-linked Hyp mutation on renal expression of Npt2c. Am J Physiol Renal Physiol 285: F1281–F1278, 2003 [DOI] [PubMed] [Google Scholar]
- 10.Chau H, El-Maadawy S, McKee MD, Tenenhouse HS: Renal calcification in mice homozygous for the disrupted type IIa Na/Pi cotransporter gene Npt2. J Bone Miner Res 18: 6446–6457, 2003 [DOI] [PubMed] [Google Scholar]
- 11.Tenenhouse HS, Sabbagh Y: Novel phosphate-regulating genes in the pathogenesis of renal phosphate wasting disorders. Pflugers Arch 444: 317–326, 2002 [DOI] [PubMed] [Google Scholar]
- 12.Tieder M, Modai D, Samuel R, Arie R, Halabe A, Bab I, Gabizon D, Liberman UA: Hereditary hypophosphatemic rickets with hypercalciuria. N Engl J Med 312: 611–617, 1985 [DOI] [PubMed] [Google Scholar]
- 13.Tieder M, Modai D, Shaked U, Samuel R, Arie R, Halabe A, Maor J, Weissgarten J, Averbukh Z, Cohen N: “Idiopathic” hypercalciuria and hereditary hypophosphatemic rickets: Two phenotypical expressions of a common genetic defect. N Engl J Med 316: 125–129, 1987 [DOI] [PubMed] [Google Scholar]
- 14.Jones A, Tzenova J, Frappier D, Crumley M, Roslin N, Kos C, Tieder M, Langman C, Proesmans W, Carpenter T, Rice A, Anderson D, Morgan K, Fujiwara T, Tenenhouse H: Hereditary hypophosphatemic rickets with hypercalciuria is not caused by mutations in the Na/Pi cotransporter NPT2 gene. J Am Soc Nephrol 12: 507–514, 2001 [DOI] [PubMed] [Google Scholar]
- 15.Bergwitz C, Roslin NM, Tieder M, Loredo-Osti JC, Bastepe M, Abu-Zahra H, Frappier D, Burkett K, Carpenter TO, Anderson D, Garabedian M, Sermet I, Fujiwara TM, Morgan K, Tenenhouse HS, Juppner H: SLC34A3 mutations in patients with hereditary hypophosphatemic rickets with hypercalciuria predict a key role for the sodium-phosphate cotransporter NaPi-IIc in maintaining phosphate homeostasis. Am J Hum Genet 78: 179–192, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorenz-Depiereux B, Benet-Pages A, Eckstein G, Tenenbaum-Rakover Y, Wagenstaller J, Tiosano D, Gershoni-Baruch R, Albers N, Lichtner P, Schnabel D, Hochberg Z, Strom TM: Hereditary hypophosphatemic rickets with hypercalciuria is caused by mutations in the sodium-phosphate cotransporter gene SLC34A3. Am J Hum Genet 78: 193–201, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pauer HU, Burfeind P, Kostering H, Emons G, Hinney B: Factor XII deficiency is strongly associated with primary recurrent abortions. Fertil Steril 80: 590–594, 2003 [DOI] [PubMed] [Google Scholar]
- 18.Iwaki T, Castellino FJ: Plasma levels of bradykinin are suppressed in factor XII-deficient mice. Thromb Haemost 95: 1003–1010, 2006 [DOI] [PubMed] [Google Scholar]
- 19.Prie D, Huart V, Bakouh N, Planelles G, Dellis O, Gerard B, Hulin P, Benque-Blanchet F, Silve C, Grandchamp B, Friedlander G: Nephrolithiasis and osteoporosis associated with hypophosphatemia caused by mutations in the type 2a sodium-phosphate cotransporter. N Engl J Med 347: 983–991, 2002 [DOI] [PubMed] [Google Scholar]
- 20.Virkki LV, Forster IC, Hernando N, Biber J, Murer H: Functional characterization of two naturally occurring mutations in the human sodium-phosphate cotransporter type IIa. J Bone Miner Res 18: 2135–2141, 2003 [DOI] [PubMed] [Google Scholar]
- 21.Werner A, Biber J, Forgo J, Palacin M, Murer H: Expression of renal transport systems for inorganic phosphate and sulfate in Xenopus laevis oocytes. J Biol Chem 265: 12331–12336, 1990 [PubMed] [Google Scholar]
- 22.Sorribas V, Markovich D, Hayes G, Stange G, Forgo J, Biber J, Murer H: Cloning of a Na/Pi cotransporter from opossum kidney cells. J Biol Chem 269: 6615–6621, 1994 [PubMed] [Google Scholar]
- 23.Hernando N, Sheikh S, Karim-Jimenez Z, Galliker H, Forgo J, Biber J, Murer H: Asymmetrical targeting of type II Na-P(i) cotransporters in renal and intestinal epithelial cell lines. Am J Physiol Renal Physiol 278: F361–F368, 2000 [DOI] [PubMed] [Google Scholar]
- 24.Thomas PJ, Ko YH, Pedersen PL: Altered protein folding may be the molecular basis of most cases of cystic fibrosis. FEBS Lett 312: 7–9, 1992 [DOI] [PubMed] [Google Scholar]
- 25.Ostedgaard LS, Zeiher B, Welsh MJ: Processing of CFTR bearing the P574H mutation differs from wild-type and deltaF508-CFTR. J Cell Sci 112: 2091–2098, 1999 [DOI] [PubMed] [Google Scholar]
- 26.Mendes F, Roxo-Rosa M, Dragomir A, Farinha CM, Roomans GM, Amaral MD, Penque D: Unusually common cystic fibrosis mutation in Portugal encodes a misprocessed protein. Biochem Biophys Res Commun 311: 665–671, 2003 [DOI] [PubMed] [Google Scholar]
- 27.Braun A, Aszodi A, Hellebrand H, Berna A, Fassler R, Brandau O: Genomic organization of profilin-III and evidence for a transcript expressed exclusively in testis. Gene 283: 219–225, 2002 [DOI] [PubMed] [Google Scholar]
- 28.Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA: An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res 23: 1087–1088, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosen E, Chan JC, Idusogie E, Clotman F, Vlasuk G, Luther T, Jalbert L, Albrecht S, Zhong L, Lissens A, Schoonjans L, Moons L, Collen D, Castellino FJ, Carmeliet P: Mice lacking Factor VII develop normally but suffer fatal perinatal bleeding. Nature 390: 290–294, 1997 [DOI] [PubMed] [Google Scholar]