Abstract
The RNA interference pathway functions as an antiviral defense in invertebrates. In order to generate a phenotypic marker which “senses” the status of the RNAi pathway in Aedes aegypti, transgenic strains were developed to express EGFP and DsRED marker genes in the eye, as well as double-stranded RNA homologous to a portion of the EGFP gene. Transgenic “sensor” mosquitoes exhibited robust eye-specific DsRED expression with little EGFP, indicating RNAi-based silencing. Cloning and high-throughput sequencing of small RNAs confirmed that the inverted-repeat transgene was successfully processed into short-interfering RNAs by the mosquito RNAi pathway. When the Ae. aegypti homologues of the genes DCR-2 or AGO-2 were knocked-down, a clear increase in EGFP fluorescence was observed in the mosquito eyes. Knockdown of DCR-2 was also associated with an increase in EGFP mRNA levels, as determined by Northern blot and real-time PCR. Knockdown of AGO-3, a gene involved in the germline-specific piRNA pathway, did not restore EGFP expression at either the mRNA or protein level. This transgenic sensor strain can now be used to identify other components of the mosquito RNAi pathway and has the potential to be used in the identification of arboviral suppressors of RNAi.
Keywords: Aedes aegypti, RNAi, transgenic mosquito, dicer, argonaute
1. Introduction
Mosquito-borne viruses constitute a worldwide public health problem. Particularly important examples of arboviruses of concern include members of the family Flaviviridae such as the dengue (DEN; genus Flavivirus) viruses, serotypes 1–4, and yellow fever virus (YFV; genus Flavivirus). An estimated 50–100 million dengue fever cases, and several hundred thousand cases of the more severe dengue hemorrhagic fever and dengue shock syndrome, occur annually with more than 2.5 billion people at risk (Halstead, 2007). YFV also causes a severe viral hemorrhagic fever. Despite the existence of a highly effective vaccine, YFV still affects as many as 200,000 persons a year (Monath, 2001). Members of the family Togaviridae also contribute to the worldwide problem of arboviral disease. Chikungunya virus (CHIKV, genus Alphavirus) has re-emerged in Southeast Asia and India (Kamath et al., 2006) and has recently been imported to Europe (Rezza et al., 2007). Aedes aegypti is an important epidemic vector of dengue viruses, yellow fever virus and chikungunya virus. Although several studies have recently suggested an antiviral role for the RNA interference (RNAi) pathway in mosquitoes (Adelman et al., 2002b; Keene et al., 2004; Li et al., 2004; Sanchez-Vargas et al., 2004), it remains unclear if RNAi affects the ability of mosquitoes to serve as disease vectors.
Induction of the RNAi pathway results in specific inhibition of gene expression. RNAi is triggered by double stranded RNA (dsRNA). dsRNA is processed by Dicer (DCR) into short RNA sequences ~21–23 nucleotides in length (Bernstein et al., 2001). Short interfering RNA (siRNA) guide sequences derived from the small RNA duplexes are assembled in the RNA-induced silencing complex (RISC). In addition to the guide sequences, Argonaute (AGO) proteins are essential components of the RISC, with Ago-2 proteins required for RISC activity in both Drosophila and mammalian cells (Liu et al., 2004; Rand et al., 2004). Using the guide strand, active RISC degrades single stranded RNA in a homology dependent manner. siRNAs corresponding to a strain of Sindbis virus (SINV, genus Alphavirus) have been detected in mosquito cells and tissues following virus infection (Sanchez-Vargas et al., 2004). SINV expression systems also have been shown to generate siRNAs corresponding to inserted host gene sequences following infection of mosquitoes (Tamang et al., 2004). These siRNAs are functional, as specific silencing of host genes has been observed following infection of mosquitoes with recombinant SINV expression systems (Attardo et al., 2003; Shiao et al., 2001; Tamang et al., 2004). These results demonstrate that double-stranded RNA generated during alphavirus replication is recognized by the RNAi pathway of the mosquito. The biological significance of this, however, remains unknown.
The discovery that many genes essential for virulence in plant viruses are actually suppressors of RNA silencing supports an antiviral function for the RNAi pathway (Li and Ding, 2006). Suppressors of RNAi have also been identified from several animal virus families [reviewed in (Li and Ding, 2006)], though not yet from any arboviruses. However, it is clear that arboviruses are targeted by the RNAi pathway (Adelman et al., 2002b; Keene et al., 2004; Sanchez-Vargas et al., 2004). It seems possible then, that the methods these viruses use to evade, escape or suppress RNAi might only be effective in their natural vectors. This might help explain why most arboviruses are only transmitted efficiently by a handful of vectors.
The Mos1 mariner transposon has been used routinely to insert genetic material into the Ae. aegypti genome (Coates et al., 1998). Our report describes the generation and successful validation of two novel Mos1-generated transgenic strains of Ae. aegypti which express EGFP on a conditional basis. That is, EGFP is only expressed when the RNAi pathway has been compromised by knock-down of presumed RNAi components. Using these transgenic “sensor” strains, we confirm that Ae. aegypti dicer-2 (AaDCR-2) and argonaute-2 (AaAGO-2) are critical for the initiation and maintenance of RNA silencing in this mosquito. These transgenic strains can now be used to identify novel mosquito genes which contribute to, or regulate the RNAi response, as well as to identify potential arboviral suppressors of RNAi.
2. Materials and methods
2.1. Cloning of 3xP3-sensor construct
The transformation plasmid pMos3xP3-DsRED-3xP3-EGFP-3xP3-EGFPir (pMos/3xP3-Sensor) was assembled using standard techniques. The EGFP inverted repeat was generated by placing the first 505 nt of the EGFP ORF in sense, followed by antisense orientation. A 68 bp intron from the Ae. aegypti sialokinin gene was used as a spacer as in previous experiments (Franz et al., 2006). The EGFP inverted repeat was placed downstream of the previously described eye-specific 3xP3 artificial promoter (Sheng et al., 1997), and was followed by the 83 bp HDV ribozyme (Perrotta and Been, 1991) and SV40 3′UTR. This cassette was transferred into a vector containing a full length 3xP3-EGFP cassette (Berghammer et al., 1999). The full length 3xP3-EGFP and 3xP3-EGFPir cassettes were transferred together into the Asc I site of the previously described modified pMos3xP3-DsRED transformation vector (Adelman et al., 2007).
2.2. Mosquito rearing and generation of transgenic strains
Aedes aegypti (khw strain and Liverpool strain) were reared at 28°C and 60–70% humidity with a photoperiod of 15 h light/9 h dark. Mosquito colonies were maintained entirely on defibrinated sheep blood (Colorado Serum Company, Denver, CO) offered through a parafilm membrane feeder. Embryo microinjections were performed as described (Coates et al., 1998) using a micromanipulator (Leica Microsystems, Bannockburn, IL) and Fempto-Jet microinjector (Eppendorf, Hamburg, Germany). Progeny of injected embryos were screened for DsRED expression using a Leica MZ-16FL stereofluorescence microscope.
2.3. DNA extraction and Southern analysis
Genomic DNA was isolated from Liverpool, khw or transgenic strains as described previously (Adelman et al., 2007; Adelman et al., 2004). For Southern analysis, DNA from 6 females or 10 males was digested overnight with each restriction enzyme in the buffer recommended by the manufacturer. Blots were hybridized to a 32P-labeled random-primed probe (GE Healthcare, Piscataway, NJ) corresponding to a 1.2 kb Hind III fragment isolated from pMos3xP3-DsRED.
2.4. Cloning of RNAi genes and generation of recombinant Sindbis viruses
A pair of oligonucleotides encoding Asc I, Pac I, Not I, Sph I and Mlu I recognition sites (5′-ctagaggcgcgccttaattaagcggccgcatgcgtttaaacgcgtc-3′ + 5′-ctaggacgcgtttaaacgcatgcggccgcttaattaaggcgcgcct-3′) were annealed and ligated into the Xba I site of pTE/3′2J (Hahn et al., 1992), generating pTE/3′2J/mcs. Aedes aegypti gene fragments were amplified by ONE-STEP RT-PCR (Qiagen, Valencia, CA) using total RNA isolated from Liverpool strain mosquitoes and the following primers [VectorBase (Lawson et al., 2007) gene ID numbers or supercontig locations are indicated in parentheses]: AaDCR-2 (AAEL006794) 5′-tttgaattcTGGACTCTGGAAACGACTTTGCCG-3′ and 5′-ttttctagaGAGTCAATGGTTTCATGTAAACAG-3′, AaAGO-2 (supercont1.89 387915–388612) 5′-ttttgaattcGGAACTATCGTTGACCGTTACATC-3′ and 5′-ttttctagaGGTATAGCGGAATCACATTAAACG-3′, AaAGO-3 (AAEL007823) 5′-ttttgcggccgcgAGATAACATTCATTGTCGTCCAGAAGCG-3′ and 5′-ttttttaattaaTCATGTCGCTGATTAACAAGGCACTC-3′, where lower case letters indicate bases added to the native sequence to generate EcoR I, Xba I, Asc I or Pac I restriction enzyme sites. AaDCR-2 and AaAGO-2 amplification products were digested directly with EcoR I/Xba I and ligated into the EcoR I/Xba I sites of pLitmus28i (NEB, Ipswich, MA). Gene fragments in pLitmus28i were re-amplified using Pfx high-fidelity DNA polymerase (Invitrogen, Carlsbad, CA) with primers primers 5′-ttttttattaaATGCTTCCGGCTCGTATGTTGTGT-3′ and 5′-ttttggcgcgccCTGGCGAAAGGGGGATGTGCT-3′. Re-amplified AaDCR-2 and AaAGO-2 products, and directly amplified AaAGO-3 product were digested with Asc I/Pac I and ligated into the Asc I/Pac I sites of pTE/3′2J/mcs so that each insert was in the antisense orientation relative to the viral second subgenomic promoter. All clones were verified by sequencing. All pTE/3′2J/mcs clones were linearized with Xho I or PspX I prior to in vitro transcription with SP6 RNA polymerase (NEB). In vitro transcribed RNA was electroporated into BHK-21 cells as described previously (Myles et al., 2006). Infectious virus was harvested, titered in Vero cells by plaque assay, and stored at −80°C until use.
2.5. Intrathoracic inoculation of viruses and scoring of transgenic heads
One to three day old adult transgenic female mosquitoes were anesthetized and injected intrathoracically with approximately 0.4 μl of recombinant Sindbis virus suspension (~1 × 104 infectious particles/mosquito). Injected mosquitoes were kept under standard conditions for 17 days, at which time levels of EGFP or DsRED expression were determined using a Leica MZ-16FL stereofluorescence microscope. All fluorescent-light photos of mosquito heads were taken in a room with 800 lux of ambient lighting using a Canon Powershot IS3 digital camera at a constant shutter speed of 0.8 s at a total magnification of 40X prior to scoring.
2.6. RNA extractions, Northern analysis and Real-time PCR
Total RNA was extracted from heads or whole mosquitoes using the Trizol reagent (Invitrogen). For Northern analysis, 5 μg of total RNA was electrophoresed in a formaldehyde/agarose gel and blotted to a nylon membrane using standard procedures. Blots were hybridized to a 32P-labeled random-primed probe (GE Healthcare) corresponding to the 3′ 216 bp of the EGFP ORF, which is not present in the inverted repeat construct. For real-time PCR, total RNA was DNase treated, phenol-chloroform extracted and ethanol precipitated prior to first strand cDNA synthesis using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Power SYBR® Green PCR Master Mix (Applied Biosystems, UK) (supplied as 2X) was used for PCR reactions with final primer concentrations at 250nM and using 0.5 μl cDNA on a 7300 Real Time PCR System (Applied Biosystems) as follows: 95°C (10′), 40 cycles of 95°C (15 s), 54°C (30 s), 72°C (30 s); followed by a dissociation curve of 95°C (15 s), 60°C (60 s) with a slow ramp to 95°C taking constant fluorescence measurements (hold at 95 C for 15sec). Real-time PCR was performed in triplicate for each cDNA using primers for EGFP, a housekeeping gene (AaElav) or a putative RNAi gene (AaDcr-2, AaAGO-2, AaAGO-3). Primers pairs were as follows: EGFP: F 5′-AGCTGGACGGCGACGTAAA-3′ and R 5′-TCGCCGATGGGGGTGTTCTGC-3′; AaElav: F 5′-AAAGAAGCTGAACGTGCCATTG -3′ and R 5′-TCTCCTCCCATCGGTGAAAAG -3′; AaDcr-2: F 5′-AATCATTCCGCCCGAGTGCTAT -3′ and R 5′-GTCCCCCATGGTCTGCTGTGA -3′; AaAgo-2: F 5′-AGGTAATCAACGCCAAAACGAACG -3′ and R 5′-AAGCGGCCACTCCCACCACACT -3′; AaAgo-3: F 5′-TCCCGGAGATTTGCTACCTTACC -3′ and R 5′-AATTTCCCGGGCTTCCTTGTTT -3′. Data was analyzed using the ddCt method on the 7300 System SDS Software (Applied Biosystems) using the AaElav values as the endogenous control and the uninjected sample as the Calibrator. The Ct threshold was manually set to 0.2 and the baseline was set to cycles 3 to 15.
2.7. Small RNA cloning and identification of EGFP siRNAs
Total RNA was extracted from transgenic sensor strain heads and size fractionated by polyacrylamide gel electrophoresis (PAGE). Small RNAs (18–30nt) were isolated and ligated to 5′ and 3′ RNA adapters. A cDNA library was generated from the small RNAs with adapters by reverse transcription followed by polymerase chain reaction (RT-PCR). The cDNA library generated was sequenced on an Illumina (San Diego, CA) Genome Analyzer System. Sequences for the RNA adapters, RT-PCR primers, and the DNA sequencing primer can be provided upon request. In order to identify small RNAs corresponding to EGFP, the 720 nt EGFP ORF was used to Query a database (blastn program; e 100) containing all of the sequences obtained from high-throughput sequencing (~4 million individual reads).
3. Results
3.1. Generation of transgenic “sensor” strain
In order to generate a phenotypic marker which reflects, or “senses” the status of the RNA interference pathway in Ae. aegypti, a series of artificial gene cassettes was assembled in a single Mos1 transformation construct (Fig. 1A). The first two genes encoded for the red and green fluorescent proteins, DsRED and EGFP. The third cassette was an inverted repeat sequence designed to express dsRNA derived from a portion of the EGFP coding sequence. All three cassettes were controlled individually from the eye-specific artificial promoter 3xP3 (Sheng et al., 1997). To generate transgenic Ae. aegypti, embryos (n=1478) of the white-eyed khw strain were injected with a combination of pMos/3xP3-sensor (donor plasmid) and pKhsp82-MOS1 (source of transposase) at 0.5 and 0.3 μg/μl, respectively. From these, 142 G0 survivors (9.6% survival) were mated to khw males or females and combined into 8 male G0 families (#1–8) and 1 female G0 pool (#9). G1 progeny were screened as early larvae for DsRED expression in the eyes. Three DsRED+ G1 individuals survived to adulthood. Of these, two males from family #9 were mated separately to khw females to establish sublines #9.1 and #9.6. The final DsRED+ individual was a single female, derived from family #2. Progeny from lines #2, 9.1 and 9.6 were analyzed by Southern to determine the integration status of the Mos1 transposable element (Fig. 1B). Genomic DNA isolated from lines #2, 9.1 and 9.6 was digested with EcoR I, Sac I or Pst I. The relative locations of these restriction sites in the pMos/3xP3-sensor transformation construct are indicated in Figure 1A. Only a single hybridization signal was detected in EcoR I-digested genomic DNA. As this enzyme does not have a recognition site within the transformation construct, this result is consistent with the presence of a single insertion (Fig. 1B). Sac I-digestion was expected to produce two small internal fragments (not shown), as well as two fragments of variable size containing the left- and right-hand junction fragments, while Pst I-digestion was expected to produce one internal fragment (indicated by small arrowhead), as well as two unique junction fragments. As expected, two hybridization signals were detected in Sac I-digested genomic DNA, with three hybridization signals in the Pst I-digested DNA (Fig. 1B). The pattern of hybridization signals for transformed lines #9.1 and #9.6 were identical for all three digestions, indicating that the two founder males used to establish these lines were both descended from the same initial transformation event. Thus these two lines were collapsed into a single line, referred to as #9. These analyses indicate that two independent transformation events were recovered, yielding an estimated transformation frequency of ~2.8% (Adelman et al., 2002a).
Figure 1.
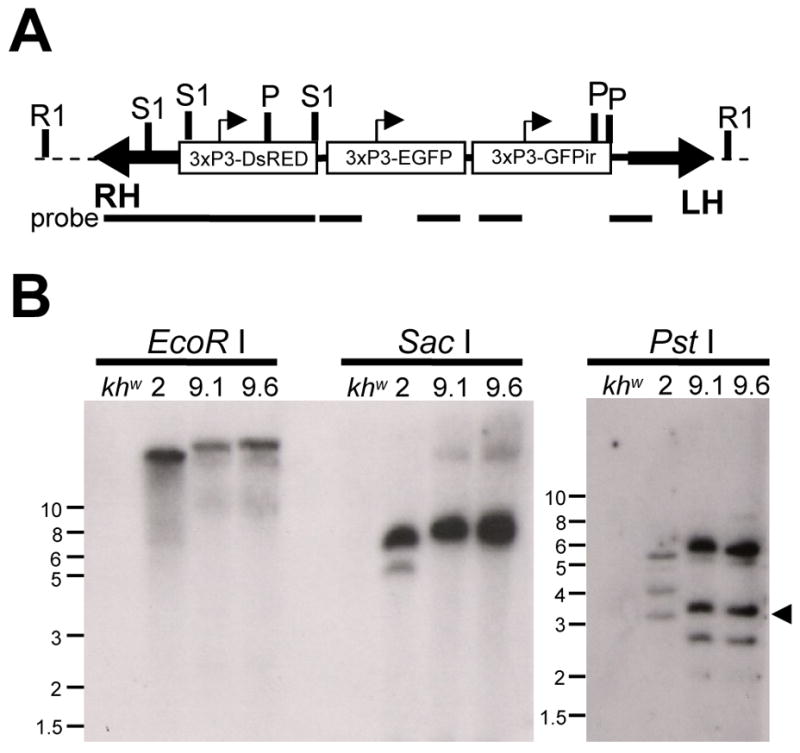
Transformation of Ae. aegypti with the pMos3xP3-sensor construct. (A) Schematic representation of a hypothetical insertion. Block arrows indicate the left (LH) and right (RH) arms of the Mos1 transposon, dotted line indicates mosquito chromosomal DNA. Small arrowheads indicate the approximate start of transcription sites for each 3xP3 promoter. Restriction enzyme recognition sites for EcoR I (R1), Sac I (S1) and Pst I (P) are indicated. Portions of the 3xP3-sensor construct recognized by the probe during Southern analysis are indicated with solid bars. (B) Southern analysis of genomic DNA isolated from lines #2, 9.1 and 9.6 or host khw, digested with restriction enzymes EcoR I, Sac I or Pst I. Approximate molecular size markers are shown at left (in kbps). Arrowhead indicates the expected size of common Pst I-generated hybridization fragment.
3.2. Sex-linkage of 3xP3-sensor transgene
While expanding each of these transgenic lines for the analyses described below, it was observed that the transgene insertion in line #2 appeared to be linked to the female-determining locus. Unlike other Dipterans, Aedes mosquitoes do not have separate sex chromosomes, but rather a sex-determining locus which maps to chromosome 1 (Craig, 1967). The male-determining allele (M) is dominant, with the female allele (m) recessive. To confirm the presumed linkage of the 3xP3-sensor transgene in line #2, three replicate crosses were performed. Each cross consisted of 75 3xP3-sensor #2 males (hypothetical genotype M/m-sensor) mated to 300 khw females (genotype m/m). The resulting progeny were scored as adults for both DsRED fluorescence and sex, with the results presented in Table 1. While the overall DsRED+:DsRED- and male:female ratios were essentially 1:1, as would be expected of the inheritance of a single gene, the 3xP3-sensor transgene was inherited by greater than 99% of female progeny (genotype m/m-sensor). Inversely, the estimated recombination distance between the 3xP3-sensor transgene and the sex locus (m) was ~0.4%. Thus with experiments involving transgenic line 3xP3-sensor #2 there is no need to sort individuals based on marker gene expression prior to experimentation, sorting by sex is sufficient. In Aedes, this is easily done in the pupal stage, where females are much larger than males. This makes transgenic line 3xP3-sensor #2 ideal for experiments involving high-throughput analysis. For this reason, transgenic line #2 is featured in the remaining experiments, which were all conducted using the heterozygous m/m-sensor genotype.
Table 1.
3xP3-sensor transgene in line #2 is tightly linked to the female determining locus.a
| ♂+ | ♂− | Total ♂ (%) | Total ♀ (%) | ♀+ | ♀;− | Total | % Lb | % Rc | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 399 | 400 (48.4) | 426 (51.6) | 426 | 0 | 826 | 99.88 | 0.12 |
| 2 | 3 | 765 | 768 (52.0) | 708 (48.0) | 707 | 1 | 1476 | 99.73 | 0.27 |
| 3 | 6 | 908 | 914 (53.1) | 806 (46.9) | 801 | 5 | 1720 | 99.30 | 0.64 |
| Total | 10 | 2072 | 2082 (51.8) | 1940 (48.2) | 1934 | 6 | 4022 | 99.58 | 0.40 |
each row represents the progeny scored from an independent genetic cross.
percentage of progeny where transgene remained linked to parental chromosome.
percentage of progeny containing a presumed recombination event.
3.3. Cloning and analysis of small RNAs from 3xP3-sensor mosquitoes
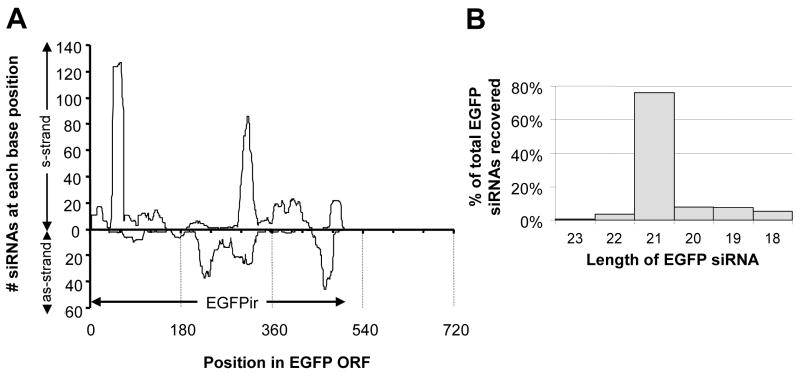
The 3xP3-sensor inverted repeat transgene is designed to produce dsRNA targeting a portion of the EGFP gene. Dicer-mediated cleavage of the EGFPir-derived dsRNA would be expected to result in the accumulation of 21 nt siRNAs corresponding to the double-stranded region. To determine if this was indeed the case, small RNAs were cloned from total RNA isolated from transgenic line #2 heads taken from adult females. High-throughput sequencing of these small RNAs revealed a population of siRNAs with homology to the entire EGFP inverted repeat (Fig. 2A), which were derived from both the sense and antisense strand. siRNAs corresponding to discreet regions of the inverted repeat sequence were present in higher copy numbers than those derived from other regions (Fig. 2A). No sequences corresponding to the EGFP gene region outside of the inverted repeat region were present in the small RNA library (Fig. 2A). The predominant length of EGFP siRNAs was 21 nt (76%), though longer and shorter EGFP siRNAs were also recovered (Fig. 2B). No small RNAs with more than 23 nt homology to EGFP were recovered, and sequences shorter than 18 nt were not included in our analysis to prevent the inclusion of host material with short stretches of homology to EGFP. No sequences (≥18 nt) matching EGFP were detected following high-throughput sequencing and analysis of small RNAs from non-transgenic Ae. aegypti (data not shown), confirming that the sequences we identified were indeed produced as a result of the insertion of the EGFPir transgene. A complete list of the EGFP-derived siRNAs recovered is presented in Table S1.
Figure 2.
Identification of EGFP siRNAs. (A) Short-interfering RNAs cloned from 3xP3-sensor heads (line #2) correspond to the EGFP inverted repeat sequence. Each base in the 720 nucleotides of the EGFP ORF was assigned a score based on the number of times it appeared in an siRNA. Separate scores were calculated for each strand, with siRNAs corresponding to the sense (s) strand plotted in the positive direction and those corresponding to the antisense (as) strand plotted in the negative direction. The region corresponding to the 505 bp inverted repeat (EGFPir) is noted. (B) Length of EGFP siRNA sequence as determined by blastn of siRNA library following removal of linker sequences, as described in Materials and Methods.
3.4. Conditional expression of EGFP in 3xP3-sensor transgenic mosquitoes
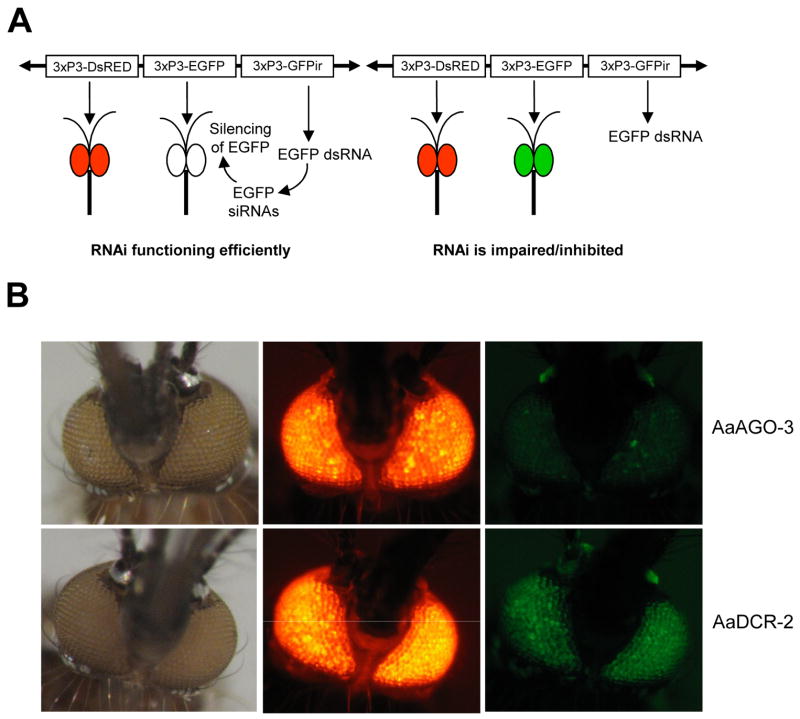
Transformed “sensor” mosquitoes from both line #2 and line #9 displayed robust DsRED expression in the eyes, while EGFP fluorescence was barely detectable. This is consistent with the presence of EGFP-derived siRNAs acting as part of the RNAi pathway to silence EGFP expression (Fig. 3A). In order to determine if EGFP expression could be restored, we injected sensor strain mosquitoes with recombinant double-subgenomic Sindbis viruses designed to express dsRNA derived from Ae. aegypti genes potentially involved in the RNAi pathway. The ability of Sindbis virus expression systems to generate knock-down phenotypes of endogenous mosquito genes has been well documented (Attardo et al., 2003; Shiao et al., 2001; Tamang et al., 2004). Following intrathoracic inoculation of recombinant double-subgenomic Sindbis viruses (dsSINV) targeting AaDCR-2, AaAGO-2 or AaAGO-3, 3xP3-sensor mosquitoes from transgenic line #2 were photographed under EGFP or DsRED filters (Fig. 3B). Similar to uninfected mosquitoes, 3xP3-sensor transgenic mosquitoes infected with dsSINV-AGO-3 displayed little to no EGFP fluorescence. In contrast, strong EGFP fluorescence was observed in dsSINV-AaDCR-2 (Fig. 3B) and dsSINV-AaAGO-2 (data not shown) infected 3xP3-sensor mosquitoes.
Figure 3.
Transgenic 3xP3-sensor mosquitoes express EGFP when RNAi is compromised. (A) Schematic representation of the conditional expression of EGFP in 3xP3-sensor mosquitoes. (B) Two representative 3xP3-sensor mosquitoes (line #2) photographed under white light (left), DsRED fluorescence (middle) and EGFP fluorescence (right) following knock-down of either AaAGO-3 or AaDCR-2.
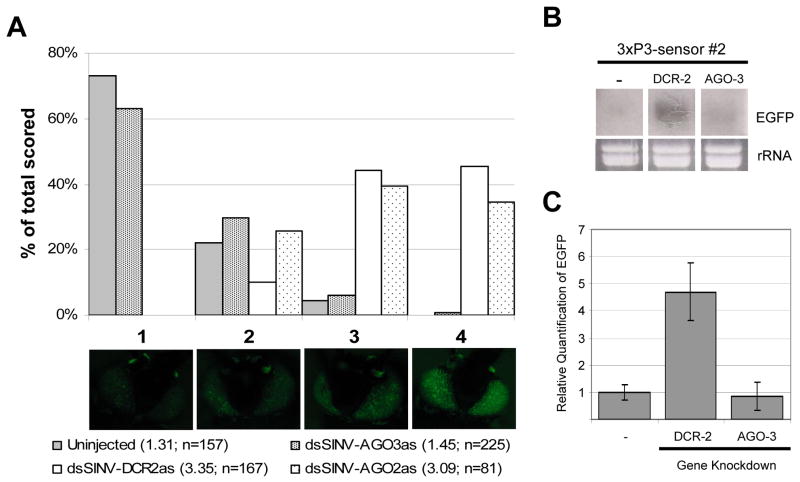
It was possible that the reason dsSINV-AGO-3-injected mosquitoes did not exhibit strong EGFP fluorescence was because the AGO-3 mRNA was not being silenced. To test this, we performed cDNA synthesis followed by real-time PCR on total RNA isolated from 3xP3-sensor heads following infection with the AaDCR-2 or AaAGO-3 recombinant viruses (Fig. 4). Our results show that both genes were knocked-down to the same extent. This suggests that both AaDCR-2 and AaAGO-2 are important for the continuous silencing of the EGFP transgene, while AaAGO-3 is not.
Figure 4.
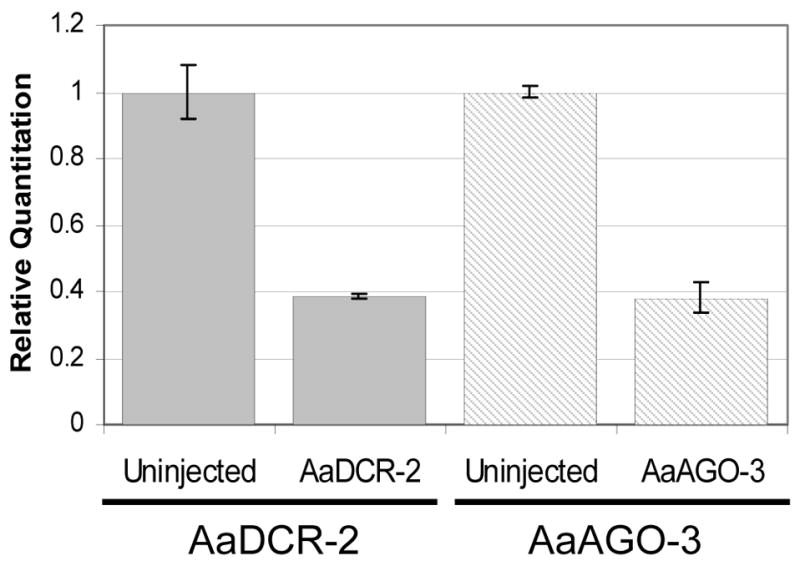
Recombinant dsSINVs induce effective gene knock-down. Real-time PCR was performed on cDNA derived from 3xP3-sensor transgenic mosquitoes. Levels of AaDCR-2 and AaAGO-3 transcript were normalized with AaElav, an endogenous gene expressed in mosquito head tissue (Adelman, unpublished). The first two bars represent relative levels of AaDCR-2 transcript in uninjected and dsSINV-AaDCR-2-injected 3xP3-sensor mosquito heads, while the second two bars represent relative levels of AaAGO-3 transcript in uninjected and dsSINV-AaAGO-3-injected 3xP3-sensor mosquito heads.
3xP3-sensor transgenic line #9 displayed a similar phenotype to line #2 after injection of recombinant dsSINVs, as dsSINV-AaAGO-3 injected individuals were indistinguishable from uninjected mosquitoes while dsSINV-AaDCR-2 injected individuals displayed strong EGFP expression (data not shown). Thus both 3xP3-sensor transgenic lines function as phenotypic sensors of the RNAi pathway.
While most of the 3xP3-sensor mosquitoes scored were clearly EGFP+ or EGFP-, there did appear to be some variation within each of the two 3xP3-sensor transgenic lines. To account for this, a qualitative scale was developed similar to that of Kim et al., (2005). 3xP3-sensor mosquitoes were scored based on the level of EGFP visible in the eyes, according to the scale shown in Figure 5A [scale of 1–4, where uniformly dim EGFP = 1, weakly mosaic = 2, strongly mosaic = 3, uniformly strong EGFP = 4]. The distribution of scores, the average score for each gene, as well as the total number of sensor mosquitoes screened is also presented in Figure 5A. Scoring of dsSINV-AaAGO-3 individuals followed the same pattern as uninjected 3xP3-sensor #2 mosquitoes. Both of these groups had small numbers of individuals (<10%) with strong EGFP expression (scores of 3 or 4), in contrast to the >75% seen in dsSINV-AaDCR-2 and dsSINV-AaAGO-2 injected individuals (Fig. 5A). Similar results were obtained with line #9. Greater than 80% of dsSINV-AaDCR-2-injected mosquitoes from line #9 were scored as 3 or 4 (65% were scored as a 4), as compared with 12% for dsSINV-AGO-3-injected mosquitoes (3% scored as a 4). The average scores were also similar, as averages of 3.50, 1.69 and 1.74 were obtained after scoring dsSINV-AaDCR-2 (n = 74), dsSINV-AGO-3 (n = 93), and uninjected mosquitoes (n = 42) from 3xP3-sensor transgenic line #9.
Figure 5.
Qualitative analysis of 3xP3-sensor mosquitoes following knock-down of putative RNAi genes. (A) 3xP3-sensor mosquitoes (line #2) were scored based on intensity of EGFP fluorescence. The percent of total mosquitoes scored at each value (1–4) is plotted, with the average for each sample and the total number of mosquitoes scored (n) given in parentheses. Results represent the sum of three replicate experiments performed during the G4-G8 generations for uninjected, dsSINV-DCR-2 and dsSINV-AGO-3 and two replicates for dsSINV-AGO-2. Sample photographs below the graph illustrate the approximate scoring guide. (B) Northern analysis of 3xP3-sensor mosquitoes following knock-down of putative RNAi genes. RNA from uninjected, dsSINV-AaDCR-2, or dsSINV-AaAGO-3 injected heads was probed for EGFP mRNA. Gel photograph of rRNA prior to transfer serves as loading control. (C) Real-time RT-PCR (two-step) to detect EGFP mRNA expression in 3xP3-sensor #2 heads following knockdown of AaDCR-2 or AaAGO-3. Samples were compared to EGFP expression in uninjected 3xP3-sensor #2 mosquito heads (−).
Knock-down of AaDCR-2 in both 3xP3-sensor transgenic strains results in an increase in EGFP protein expression. To confirm that this increase corresponded with an increase in EGFP mRNA levels, we conducted Northern analysis using total RNA isolated from the heads of 3xP3-sensor mosquitoes. Using a probe sequence based on the last 216 nt of the EGFP ORF (a region not present in the EGFPir), we observed an increase in EGFP mRNA expression in dsSINV-AaDCR-2-injected mosquitoes, but not in dsSINV-AaAGO-3 as compared with uninjected (Fig. 5B). This was further confirmed by using real-time PCR, which showed a 4–6 fold increase in EGFP mRNA expression when AaDCR-2 was knocked-down, while EGFP mRNA levels were unaffected by the knockdown of AaAGO-3 (Fig. 5C).
4. Discussion
This paper describes the development and validation of two transformed Ae. aegypti RNAi “sensor” strains. Southern analysis and subsequent inheritance data both suggest that each of these strains contains a single transgene insertion. The observation of DsRED and conditional EGFP fluorescence indicates that all three transgenes were successfully integrated with no perturbations to gene function. Short-interfering RNAs derived from the EGFP inverted repeat were successfully recovered from 3xP3-sensor mosquitoes. The overwhelming majority of EGFP siRNAs were found to be exactly 21 nt, confirming that the observed silencing phenotype was due to RNAi. While the distribution of siRNAs derived from the inverted repeat sequence did not appear to be random, to date we have been unable to identify any similarities between siRNAs that might facilitate future predictions of preferred target sequences. More information in this area, such as cloning of siRNAs from other inverted-repeat transgenes or from alternative EGFPir sequences might be useful and would aid in the design of future dsRNA constructs.
The transformation construct used in this study contains three tandem duplications of the 3xP3 artificial promoter, as well as a tandem duplication of EGFP sequence (in the translatable EGFP and inverted repeat cassettes). Given these duplications, there was an initial concern on our part about the potential for recombination which could result in a loss of one or more of the gene cassettes. However, this turned out not to be a significant danger, as both transgenic lines #2 and #9 have been reared out to the G8 and G6 generations, respectively, with no evidence as of yet for any loss of the DsRED, EGFP or EGFPir genes. Even the few outlier 3xP3-sensor mosquitoes which showed elevated levels of EGFP expression did not come close to rivaling that normally seen in mosquitoes transformed with only the 3xP3-EGFP cassette (Adelman et al., 2004; Horn et al., 2000), indicating that silencing was still occurring to some extent. In fact, it is worth noting that even AaDCR-2 or AaAGO-2 knockdown did not restore EGFP expression to levels normally observed with the 3xP3-EGFP cassette. This is not surprising, however, as we only achieved knock-down of these mRNAs (Fig. 4), not complete knockout. Also, we only knocked down one RNAi component at a time. Thus even with reduced AaDCR-2 levels, all the downstream steps (RISC) would be expected to function normally. The level of redundancy in the mosquito RNAi pathway, including the contributions of such genes as DCR-1 and AGO-1, is also unknown. The critical fact, however, is that the phenotypes of 3xP3-sensor mosquitoes when RNAi is functioning and when it is compromised are easily distinguished from each other.
While the majority of 3xP3-sensor mosquitoes displayed similar levels of EGFP activation or silencing, we consistently observed low numbers of outliers who either did not silence or activate EGFP effectively. The reasons for this variation are unknown, but may be due to unknown genetic differences within the colony. Further investigation into this variance might reveal other genetic loci which are important to the successful initiation or maintenance of RNAi.
Only heterozygous 3xP3-sensor mosquitoes (genotype m/m-sensor) were used in these experiments. At present, we do not know what the effect would be of having two copies of the 3xP3-sensor transgene, as both the source of dsRNA and the source of EGFP mRNA would be expected to be doubled. However, strong negative fitness effects have been observed with some homozygous transgenes due to the presence of deleterious recessive alleles linked to the transgene (Irvin et al., 2004), and we cannot predict what effect this might have on the RNAi pathway.
Initially, we attempted to knockdown putative RNAi genes by intrathoracic inoculation of dsRNA (Blandin et al., 2002). Using this method, we were unable to induce sufficient gene knockdown in the eye tissue to test the 3xP3-sensor transgene. Thinking that this may be related to the ability of the dsRNA to penetrate into the nervous system, specifically the eye tissue, we performed injections of dsRNA directly into the eye. Preliminary results were promising, as we did observe an increase in EGFP expression following intraocular injection of AaAGO-2 dsRNA. However, intraocular injections proved difficult, and were associated with low survival. Thus, to validate our transgenic strains, we utilized Sindbis virus-based expression systems, which are easily inoculated into the thorax and infect eye tissue (Higgs et al., 1996). Boussin et al (2006) found that ten times the standard amount of dsRNA is required to achieve gene knockdown in the salivary glands of An. gambiae due to low penetrance. Future investigations may explore the possibility of using non-standard amounts of dsRNA to achieve strong knockdown of genes in eye tissue of 3xP3-sensor mosquitoes, following intrathoracic injection.
Using our transgenic “sensor” strain, we have confirmed that AaDCR-2 and AaAGO-2 are both required for RNA silencing in the yellow fever mosquito, Aedes aegypti. This is not unexpected, as both the Drosophila and Anopheles homologues of these genes have been shown to be involved in RNAi (Hammond et al., 2001; Hoa et al., 2003). However, this validates our experimental approach, and demonstrates that our transgenic construct was indeed triggering a reversible silencing phenotype in the mosquito eye. Knockdown of AGO-3, a gene now known to be involved in the germline-specific piwi-RNA pathway in Drosophila (Brennecke et al., 2007; Gunawardane et al., 2007), did not restore EGFP expression. In flies, AGO-3 has been found to be associated with 23–26 nt small RNAs, with 24 nt being the most prominent (Gunawardane et al., 2007). In our sensor mosquitoes, EGFP-derived small RNAs were found to be 18–23 nt, with 21 nt being the most prominent. This again supports a role for DCR-2 in processing the EGFPir double-stranded RNA, and does not support a role for AGO-3. This indicates that the phenotypic change we observed from weak to strong expression of EGFP is specific for the knockdown of genes involved in RNAi (Fig. 3, 5). Previous work in Anopheles gambiae cell lines suggested that AgAGO-3 might play a role in RNAi (Hoa et al., 2003). Although results obtained in cultured cells do not always correlate with results obtained in whole organisms, studies involving o’nyong-nyong virus in Anopheles gambiae also suggest that AgAGO-3 plays a role in RNAi (Keene et al., 2004). It is possible that AGO-3 protein is highly stable, and thus even though we successfully reduced mRNA levels, protein levels might not have been affected to the point where a phenotype could be detected. Thus, additional investigations will be necessary to clarify the role of AGO-3 in the mosquito RNAi pathway. Nevertheless, our results are consistent with a role for AGO-3 recently described in other Dipterans (Brennecke et al., 2007; Gunawardane et al., 2007; Zamore, 2007).
The data presented here confirm that EGFP expression can be used to differentiate between mosquitoes possessing a fully functional or a compromised RNAi pathway. Several applications for this strain are obvious, with many more possible uses likely still waiting to be discovered. Similar to the studies presented here which tested for the involvement of AaDCR-2, AaAGO-2 and AaAGO-3, these sensor strains can be used to test the involvement of any Ae. aegypti gene product in the RNAi pathway. With the recent publication of the Ae. aegypti genome (Nene et al., 2007), ample targets for such a reverse genetics approach are available. This is especially relevant for genes already described to play a role in RNAi in non-vector model organisms. While possible homologues of such genes can be suggested through bioinformatics means, there is no substitute for functional confirmation in genome annotation. Therefore, as new components of the RNAi machinery are identified in model organisms, their counterparts in vector mosquitoes can quickly be tested for functional conservation. In addition, suspected mosquito genes with no clear homologues in model organisms can be tested for involvement in RNAi either individually, or through the creative design of forward genetic screens, such as mutagenesis followed by identification of mutants deficient in RNAi as described by Lee et al (2004). This should help paint a picture of what is conserved in the RNAi pathway, and what is unique to vector mosquitoes.
Investigations into arbovirus-vector interactions are naturally biased towards female subjects. This can present a logistical problem when scaling up experiments which utilize heterozygous transgenic mosquitoes, as the labor associated with manually separating transgenic from non-transgenic individuals prior to virus infection can be extensive. The sheer number of individuals reared also increases, as 75% of progeny are ultimately not used in the experiment (all males and non-transgenic females). While this can be overcome with automated sorting (Catteruccia et al., 2005), such technology is costly and not widely available. The fortuitous linkage of our 3xP3-sensor transgene to the female-determining locus makes this transgenic line ideal for downstream applications such as virus infection where only females are required, as 50% of progeny will be useful and no additional sorting based on marker gene expression is required.
The RNA interference pathway has been suggested to play a major role in anti-viral innate immunity (Galiana-Arnoux et al., 2006; Li et al., 2002; Wang et al., 2006). However, detailed studies of this pathway have only been performed in model organisms such as Drosophila and C. elegans. Studies designed to understand the role of RNAi in anti-viral defense in natural virus-vector systems have been lacking. In addition to identifying mosquito components of the RNAi pathway, the transgenic sensor strains described here can potentially be used to identify arboviral proteins or sequences which perturb or interfere with the functioning of RNAi. Ae. aegypti is a natural vector of both flaviviruses (dengue viruses, yellow fever virus) and alphaviruses (chikungunya virus), and viruses from both of these families show robust replication in the Ae. aegypti nervous system, including eye tissue (Pierro et al., 2003; Linthicum et al., 1996). Thus we are now in a position to use these transgenic sensor strains to investigate the interactions between these important human pathogens and the arthropod innate anti-viral RNAi pathway.
Supplementary Material
Table S1.
Acknowledgments
We thank Jessica M. Overcash, Mallory Brangan, Lisa Burley and Tiffany Gross as well as other members of the Myles/Adelman laboratories for technical assistance. This project was supported by NIAID grant 1R03-AI070198-01A1 and by Virginia Tech startup funds to K.M.M. and Z.N.A.
Footnotes
Other author affiliations: Kevin M. Myles, 306 Fralin Biotechnology Center, West Campus Dr., Virginia Polytechnic Institute and State University, Blacksburg, VA 24061
Michelle A. E. Anderson, 313 Fralin Biotechnology Center, West Campus Dr., Virginia Polytechnic Institute and State University, Blacksburg, VA 24061
Elaine M. Morazzani, 314 Fralin Biotechnology Center, West Campus Dr., Virginia Polytechnic Institute and State University, Blacksburg, VA 24061
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adelman ZN, Jasinskiene N, James AA. Development and applications of transgenesis in the yellow fever mosquito, Aedes aegypti. Mol Biochem Parasitol. 2002a;121:1–10. doi: 10.1016/s0166-6851(02)00028-2. [DOI] [PubMed] [Google Scholar]
- Adelman ZN, Sanchez-Vargas I, Travanty EA, Carlson JO, Beaty BJ, Blair CD, Olson KE. RNA silencing of dengue virus type 2 replication in transformed C6/36 mosquito cells transcribing an inverted-repeat RNA derived from the virus genome. J Virol. 2002b;76:12925–33. doi: 10.1128/JVI.76.24.12925-12933.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman ZN, Jasinskiene N, Onal S, Juhn J, Ashikyan A, Salampessy M, MacCauley T, James AA. nanos gene control DNA mediates developmentally regulated transposition in the yellow fever mosquito Aedes aegypti. Proc Natl Acad Sci U S A. 2007;104:9970–5. doi: 10.1073/pnas.0701515104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman ZN, Jasinskiene N, Vally KJ, Peek C, Travanty EA, Olson KE, Brown SE, Stephens JL, Knudson DL, Coates CJ, James AA. Formation and loss of large, unstable tandem arrays of the piggyBac transposable element in the yellow fever mosquito, Aedes aegypti. Transgenic Res. 2004;13:411–25. doi: 10.1007/s11248-004-6067-2. [DOI] [PubMed] [Google Scholar]
- Attardo GM, Higgs S, Klingler KA, Vanlandingham DL, Raikhel AS. RNA interference-mediated knockdown of a GATA factor reveals a link to anautogeny in the mosquito Aedes aegypti. Proc Natl Acad Sci U S A. 2003;100:13374–9. doi: 10.1073/pnas.2235649100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghammer AJ, Klingler M, Wimmer EA. A universal marker for transgenic insects. Nature. 1999;402:370–1. doi: 10.1038/46463. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–6. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Blandin S, Moita LF, Kocher T, Wilm M, Kafatos FC, Levashina EA. Reverse genetics in the mosquito Anopheles gambiae: targeted disruption of the Defensin gene. EMBO reports. 2002;3:852–6. doi: 10.1093/embo-reports/kvf180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson B, Jacques JC, Choumet V, Martin E, Xu J, Vernick K, Bourgouin C. Gene silencing in mosquito salivary glands by RNAi. FEBS Lett. 2006;580:1988–92. doi: 10.1016/j.febslet.2006.02.069. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Catteruccia F, Benton JP, Crisanti A. An Anopheles transgenic sexing strain for vector control. Nat Biotechnol. 2005;23:1414–7. doi: 10.1038/nbt1152. [DOI] [PubMed] [Google Scholar]
- Coates CJ, Jasinskiene N, Miyashiro L, James AA. Mariner transposition and transformation of the yellow fever mosquito, Aedes aegypti. Proc Natl Acad Sci U S A. 1998;95:3748–51. doi: 10.1073/pnas.95.7.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig GB. Genetics of Aedes aegypti. In: Wright JW, Pal R, editors. Genetics of Insect Vectors of Disease. Elsevier Publishing Co.; Amsterdam: 1967. pp. 67–132. [Google Scholar]
- Franz AW, Sanchez-Vargas I, Adelman ZN, Blair CD, Beaty BJ, James AA, Olson KE. Engineering RNA interference-based resistance to dengue virus type 2 in genetically modified Aedes aegypti. Proc Natl Acad Sci U S A. 2006;103:4198–203. doi: 10.1073/pnas.0600479103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat Immunol. 2006;7:590–7. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–90. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Hahn CS, Hahn YS, Braciale TJ, Rice CM. Infectious Sindbis virus transient expression vectors for studying antigen processing and presentation. Proc Natl Acad Sci U S A. 1992;89:2679–83. doi: 10.1073/pnas.89.7.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead SB. Dengue. Lancet. 2007;370:1644–52. doi: 10.1016/S0140-6736(07)61687-0. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–50. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Higgs S, Traul D, Davis BS, Kamrud KI, Wilcox CL, Beaty BJ. Green fluorescent protein expressed in living mosquitoes--without the requirement of transformation. Biotechniques. 1996;21:660–4. doi: 10.2144/96214st03. [DOI] [PubMed] [Google Scholar]
- Hoa NT, Keene KM, Olson KE, Zheng L. Characterization of RNA interference in an Anopheles gambiae cell line. Insect Biochem Mol Biol. 2003;33:949–57. doi: 10.1016/s0965-1748(03)00101-2. [DOI] [PubMed] [Google Scholar]
- Horn C, Jaunich B, Wimmer EA. Highly sensitive, fluorescent transformation marker for Drosophila transgenesis. Dev Genes Evol. 2000;210:623–9. doi: 10.1007/s004270000111. [DOI] [PubMed] [Google Scholar]
- Irvin N, Hoddle MS, O’Brochta DA, Carey B, Atkinson PW. Assessing fitness costs for transgenic Aedes aegypti expressing the GFP marker and transposase genes. Proc Natl Acad Sci U S A. 2004;101:891–6. doi: 10.1073/pnas.0305511101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath S, Das AK, Parikh FS. Chikungunya. The Journal of the Association of Physicians of India. 2006;54:725–6. [PubMed] [Google Scholar]
- Keene KM, Foy BD, Sanchez-Vargas I, Beaty BJ, Blair CD, Olson KE. RNA interference acts as a natural antiviral response to O’nyong-nyong virus (Alphavirus; Togaviridae) infection of Anopheles gambiae. Proc Natl Acad Sci U S A. 2004;101:17240–5. doi: 10.1073/pnas.0406983101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Gabel HW, Kamath RS, Tewari M, Pasquinelli A, Rual JF, Kennedy S, Dybbs M, Bertin N, Kaplan JM, Vidal M, Ruvkun G. Functional genomic analysis of RNA interference in C. elegans. Science. 2005;308:1164–7. doi: 10.1126/science.1109267. [DOI] [PubMed] [Google Scholar]
- Lawson D, Arensburger P, Atkinson P, Besansky NJ, Bruggner RV, Butler R, Campbell KS, Christophides GK, Christley S, Dialynas E, Emmert D, Hammond M, Hill CA, Kennedy RC, Lobo NF, MacCallum MR, Madey G, Megy K, Redmond S, Russo S, Severson DW, Stinson EO, Topalis P, Zdobnov EM, Birney E, Gelbart WM, Kafatos FC, Louis C, Collins FH. VectorBase: a home for invertebrate vectors of human pathogens. Nucleic Acids Res. 2007;35:D503–5. doi: 10.1093/nar/gkl960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Li F, Ding SW. Virus Counterdefense: Diverse Strategies for Evading the RNA-Silencing Immunity. Annual review of microbiology. 2006;60:503–31. doi: 10.1146/annurev.micro.60.080805.142205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–21. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- Li WX, Li H, Lu R, Li F, Dus M, Atkinson P, Brydon EW, Johnson KL, Garcia-Sastre A, Ball LA, Palese P, Ding SW. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci U S A. 2004;101:1350–5. doi: 10.1073/pnas.0308308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linthicum KJ, Platt K, Myint KS, Lerdthusnee K, Innis BL, Vaughn DW. Dengue 3 virus distribution in the mosquito Aedes aegypti: an immunocytochemical study. Med Vet Entomol. 1996;10:87–92. doi: 10.1111/j.1365-2915.1996.tb00086.x. [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–41. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Monath TP. Yellow fever: an update. Lancet Infect Dis. 2001;1:11–20. doi: 10.1016/S1473-3099(01)00016-0. [DOI] [PubMed] [Google Scholar]
- Myles KM, Kelly CL, Ledermann JP, Powers AM. Effects of an opal termination codon preceding the nsP4 gene sequence in the O’Nyong-Nyong virus genome on Anopheles gambiae infectivity. J Virol. 2006;80:4992–7. doi: 10.1128/JVI.80.10.4992-4997.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, Xi Z, Megy K, Grabherr M, Ren Q, Zdobnov EM, Lobo NF, Campbell KS, Brown SE, Bonaldo MF, Zhu J, Sinkins SP, Hogenkamp DG, Amedo P, Arsenburger P, Atkinson PW, Bidwell S, Biedler J, Birney E, Bruggner RV, Costas J, Coy MR, Crabtree J, Crawford M, Debruyn B, Decaprio D, Eiglmeier K, Eisenstadt E, El-Dorry H, Gelbart WM, Gomes SL, Hammond M, Hannick LI, Hogan JR, Holmes MH, Jaffe D, Johnston SJ, Kennedy RC, Koo H, Kravitz S, Kriventseva EV, Kulp D, Labutti K, Lee E, Li S, Lovin DD, Mao C, Mauceli E, Menck CF, Miller JR, Montgomery P, Mori A, Nascimento AL, Naveira HF, Nusbaum C, O’Leary BS, Orvis J, Pertea M, Quesneville H, Reidenbach KR, Rogers YH, Roth CW, Schneider JR, Schatz M, Shumway M, Stanke M, Stinson EO, Tubio JM, Vanzee JP, Verjovski-Almeida S, Werner D, White O, Wyder S, Zeng Q, Zhao Q, Zhao Y, Hill CA, Raikhel AS, Soares MB, Knudson DL, Lee NH, Galagan J, Salzberg SL, Paulsen IT, Dimopoulos G, Collins FH, Bruce B, Fraser-Liggett CM, Severson DW. Genome Sequence of Aedes aegypti, a Major Arbovirus Vector. Science. 2007 doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotta AT, Been MD. A pseudoknot-like structure required for efficient self-cleavage of hepatitis delta virus RNA. Nature. 1991;350:434–6. doi: 10.1038/350434a0. [DOI] [PubMed] [Google Scholar]
- Pierro DJ, Myles KM, Foy BD, Beaty BJ, Olson KE. Development of an orally infectious Sindbis virus transducing system that efficiently disseminates and expresses green fluorescent protein in Aedes aegypti. Insect Mol Biol. 2003;12:107–16. doi: 10.1046/j.1365-2583.2003.00392.x. [DOI] [PubMed] [Google Scholar]
- Rand TA, Ginalski K, Grishin NV, Wang X. Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity. Proc Natl Acad Sci U S A. 2004;101:14385–9. doi: 10.1073/pnas.0405913101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezza G, Nicoletti L, Angelini R, Romi R, Finarelli AC, Panning M, Cordioli P, Fortuna C, Boros S, Magurano F, Silvi G, Angelini P, Dottori M, Ciufolini MG, Majori GC, Cassone A. Infection with chikungunya virus in Italy: an outbreak in a temperate region. Lancet. 2007;370:1840–6. doi: 10.1016/S0140-6736(07)61779-6. [DOI] [PubMed] [Google Scholar]
- Sanchez-Vargas I, Travanty EA, Keene KM, Franz AW, Beaty BJ, Blair CD, Olson KE. RNA interference, arthropod-borne viruses, and mosquitoes. Virus Res. 2004;102:65–74. doi: 10.1016/j.virusres.2004.01.017. [DOI] [PubMed] [Google Scholar]
- Sheng G, Thouvenot E, Schmucker D, Wilson DS, Desplan C. Direct regulation of rhodopsin 1 by Pax-6/eyeless in Drosophila: evidence for a conserved function in photoreceptors. Genes Dev. 1997;11:1122–31. doi: 10.1101/gad.11.9.1122. [DOI] [PubMed] [Google Scholar]
- Shiao SH, Higgs S, Adelman Z, Christensen BM, Liu SH, Chen CC. Effect of prophenoloxidase expression knockout on the melanization of microfilariae in the mosquito Armigeres subalbatus. Insect Mol Biol. 2001;10:315–21. doi: 10.1046/j.0962-1075.2001.00268.x. [DOI] [PubMed] [Google Scholar]
- Tamang D, Tseng SM, Huang CY, Tsao IY, Chou SZ, Higgs S, Christensen BM, Chen CC. The use of a double subgenomic Sindbis virus expression system to study mosquito gene function: effects of antisense nucleotide number and duration of viral infection on gene silencing efficiency. Insect Mol Biol. 2004;13:595–602. doi: 10.1111/j.0962-1075.2004.00516.x. [DOI] [PubMed] [Google Scholar]
- Wang XH, Aliyari R, Li WX, Li HW, Kim K, Carthew R, Atkinson P, Ding SW. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452–4. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD. RNA silencing: genomic defence with a slice of pi. Nature. 2007;446:864–5. doi: 10.1038/446864a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.