Abstract
OBJECTIVE— Epidemiological and family studies have demonstrated that susceptibility genes play an important role in the etiology of diabetic nephropathy, defined as persistent proteinuria or end-stage renal disease (ESRD) in type 1 diabetes.
RESEARCH DESIGN AND METHODS— To efficiently search for genomic regions harboring diabetic nephropathy genes, we conducted a scan using 5,382 informative single nucleotide polymorphisms on 100 sibpairs concordant for type 1 diabetes but discordant for diabetic nephropathy. In addition to being powerful for detecting linkage to diabetic nephropathy, this design allows linkage analysis on type 1 diabetes via traditional affected sibpair (ASP) analysis. In weighing the evidence for linkage, we considered maximum logarithm of odds score (maximum likelihood score [MLS]) values and corresponding allelic sharing patterns, calculated and viewed graphically using the software package SPLAT.
RESULTS— Our primary finding for diabetic nephropathy, broadly defined, is on chromosome 19q (MLS = 3.1), and a secondary peak exists on chromosome 2q (MLS = 2.1). Stratification of discordant sibpairs based on whether disease had progressed to ESRD suggested four tertiary peaks on chromosome 1q (ESRD only), chromosome 20p (proteinuria only), and chromosome 3q (two loci 58 cm apart, one for ESRD only and another for proteinuria only). Additionally, analysis of 130 ASPs for type 1 diabetes confirmed the linkage to the HLA region on chromosome 6p (MLS = 9.2) and IDDM15 on chromosome 6q (MLS = 3.1).
CONCLUSIONS— This study identified several novel loci as candidates for diabetic nephropathy, none of which appear to be the sole genetic determinant of diabetic nephropathy in type 1 diabetes. In addition, this study confirms two previously reported type 1 diabetes loci.
Diabetic nephropathy is the major complication of type 1 diabetes. Clinically, diabetic nephropathy is manifested as persistent proteinuria that frequently progresses to end-stage renal disease (ESRD) (1). While hyperglycemia plays a major role in diabetic nephropathy (1), genetic predisposition has become apparent. Familial aggregation of diabetic nephropathy has been observed in all family studies with multiple type 1 diabetic siblings (2–6). The most comprehensive study, conducted at the Joslin Clinic (4), demonstrated that in comparison with a lifetime diabetic nephropathy risk of 35% among unrelated patients with type 1 diabetes, the risk to a second diabetic sibling increases to 72% or decreases to 25%, depending on whether the first diabetic sibling had diabetic nephropathy. Since familial clustering of glycemic control could not account for this large disparity, a major gene effect was proposed as a plausible explanation (4). To map such a gene, we showed that discordant sibpairs (DSPs) for diabetic nephropathy would be four times as efficient as affected sibpairs (ASPs) (7).
Previously, we applied the DSP strategy to a collection of 66 DSPs to test for linkage with genes of the renin-angiotensin system (8). Manual genotyping of microsatellites did not identify any evidence for linkage with AGT (chromosome 1q) and ACE (chromosome 17q); however, we obtained suggestive evidence for linkage with the region on chromosome 3q containing ATR1 (8). Subsequent sequencing of this gene and association studies, however, excluded this gene (8). This report features a larger sample size (100 DSPs from 83 families) and a more stringent definition of diabetic nephropathy.
RESEARCH DESIGN AND METHODS
We identified patients with type 1 diabetes and diabetic nephropathy attending the Joslin Clinic between 1991 and 2003. Of 900 of such patients, 714 (95% Caucasians) were enrolled into the Joslin Study on Genetics of Diabetic Nephropathy. Initially, we identified 125 families having a diabetic nephropathy proband with one or more living siblings with ≥10 years of type 1 diabetes. After consent, each participant was examined by a trained recruiter. For siblings not attending the Joslin Clinic, we obtained medical record information from their primary physicians. The committee on human subjects of the Joslin Diabetes Center approved the protocol and informed consent procedures.
Diagnosis of type 1 diabetes and diabetic nephropathy.
Type 1 diabetes was assumed if hyperglycemia was diagnosed before age 35 years and its control required insulin treatment within 1 year of diagnosis. The diabetic nephropathy status of each patient was determined on the basis of questionnaires, medical records, and measurements of the urinary albumin-to-creatinine ratio using a previously described method (9). Patients were classified as not having diabetic nephropathy if they had ≥10 years of diabetes and a urinary albumin-to-creatinine ratio <20 mg/g in two of three consecutive urine specimens. Patients were considered case subjects if they had persistent proteinuria, ESRD, or renal transplant. Persistent proteinuria was defined as two of three successive positive urinalyses determined either by a urinary albumin-to-creatinine ratio >250 mg/g (men) or >355 mg/g (women) or by a Multistix reagent strip (≥2; Bayer, Elkhart, IN).
Examined families.
Of 125 initial families, 42 were excluded because all siblings had either subclinical (microalbuminuria) or clinical (proteinuria or ESRD) evidence of diabetic nephropathy. The remaining 83 examined families each contained at least one type 1 diabetic sibling with diabetic nephropathy and one type 1 diabetic sibling without diabetic nephropathy. The distribution of diabetic siblings per family and the number of examined parents is shown in Table 1. Among these 83 families, we examined 80 parents, 96 siblings with type 1 diabetes but without diabetic nephropathy, 43 diabetic siblings with persistent proteinuria, and 44 diabetic siblings with ESRD. There were a total of 130 sibpairs concordant for type 1 diabetes and 100 sibpairs discordant for diabetic nephropathy.
TABLE 1.
Characteristics of families with type 1 diabetes used in the linkage study
| Number of examined diabetic siblings per family | Number of examined parents per family |
|||
|---|---|---|---|---|
| Two | One | None | Total families | |
| Number of families | ||||
| Two (control and proteinuria) | 11 | 11 | 13 | 35 |
| Two (control and ESRD) | 12 | 12 | 8 | 32 |
| Three or more (control and proteinuria or ESRD) | 0 | 11 | 5 | 16* |
| Total families | 23 | 34 | 26 | 83† |
Fifteen families with three siblings and one family with four siblings.
In these families, there were 130 sibpairs concordant for diabetes and 100 sibpairs discordant for diabetic nephropathy.
Clinical characteristics of siblings.
Clinical characteristics were obtained during the enrollment examination. This information was supplemented with data abstracted from the Joslin Clinic medical records or obtained from primary care physicians.
DNA extraction and genotyping.
DNA was extracted using a standard phenol-chloroform protocol. Samples were genotyped by Illumina Genotyping Services (San Diego, CA) with Illumina's high-throughput BeadArray Platform utilizing single nucleotide polymorphisms (SNPs) from their Linkage 4b Mapping Panel, which consists of 6,008 validated SNPs with an average heterozygosity >45% (in Caucasians) and an average interpolated genetic map distance of 0.62 cM. This panel was determined by simulation to allow for the extraction of most inheritance information.
Data management and cleaning.
Data were managed in SAS. The original dataset consisted of 5,984 SNPs. Five samples were genotyped as quality-control duplicates. Of nearly 30,000 replicated quality-control genotypes, not a single mismatched allele call was observed. The sample with the lowest quality genotyping had a 4.8% no-call rate and 86.8% GenCall >0.7; thus we rejected no samples based on genotyping quality. Using the Wiggington algorithm, we found only three SNPs that violated Hardy-Weinberg. None of these had a material impact on our results. All analyzed pairs were confirmed as full sibs with Prest (10).
Among the original 5,984 SNPs, 602 were removed before analysis: 343 localized to sex chromosomes, 20 were monomorphic, and 239 were not of sufficient quality (<98% of genotypes with a GenCall score >0.7). Thus, our autosomal genome scan utilized 5,382 high-quality, informative SNPs (mean information content of 84.9%). Mendelian errors were evaluated with PedCheck (11). Interestingly, nine of these occurred in a single sample at the chromosome 18p telomere. Upon further investigation, this sample was found to be homozygous across each of the first 37 SNPs, thus implying the existence of a large chromosomal deletion. Genotypes for this and another such region were removed, as were the remaining observed Mendelian errors. Finally, we used Merlin (12) to remove less obvious genotyping errors. In total, we removed 442 genotype errors identified by PedCheck and 334 genotypes flagged by Merlin.
Genetic analysis.
Since linkage disequilibrium can introduce bias (13), we used the linkage disequilibrium modeling capabilities of Merlin (14) in calculating IBD statistics. Simulation studies have shown that maximum likelihood score (MLS) inflation is negligible for r2 < 0.16 (15), so we used this threshold. Merlin identified all clusters for which r2 exceeded 0.16 and ran the expectation-maximization algorithm to calculate haplotype frequencies, thus reducing bias due to linkage disequilibrium. Two clusters with obligate recombinants were removed before analysis.
The resulting IBD files were combined and converted to Genehunter format then, in conjunction with a phenotype file, analyzed with SPLAT (16). SPLAT used the phenotype file to define DSPs and then implemented the expectation-maximization algorithm to calculate maximum-likelihood estimates of sharing and corresponding MLS values for each chromosomal position. For DSPs, diminished sharing (mean sharing <0.49 and z2 < 0.2) was required for declaring linkage (17). Unless otherwise indicated, the MLS values reflect unconstrained maximization and should be compared with reference distribution with two degrees of freedom. Contour plots of peak MLS values together with various constraint regions are available in an online appendix (available at http://dx.doi.org/10.2337/db07-1086). For comparison to Lander-Kruglyak criteria (17), we report the largest MLS value falling in the ASP/DSP triangle. For both ASPs and DSPs, triangle-constrained values >4.0 are considered significant and >2.6 are considered suggestive (17,18). Calculations underlying these thresholds assume all inheritance information is extracted, so there is no need to adjust further for the large number of SNPs genotyped.
Genetic maps.
All genetic positions refer to the deCODE genetic map, and findings from previous studies have been converted to this scale.
RESULTS
Characteristics of examined families.
Clinical characteristics are shown in Table 2. Siblings with diabetic nephropathy, particularly those with ESRD, had shorter diabetes duration, were younger, and the majority had retinopathy requiring laser treatment. About one-third of siblings without diabetic nephropathy had had severe diabetic retinopathy requiring laser treatment. All siblings were treated with insulin and had similar A1C levels at enrollment. In short, all siblings had typical type 1 diabetes and the different diabetic nephropathy status of these siblings could not be accounted for by differences in diabetes duration. Our families comprise 130 ASPs for type 1 diabetes and 100 DSPs for diabetic nephropathy.
TABLE 2.
Clinical characteristics of examined diabetic siblings according to renal status
| Characteristics | Control subjects with normoalbuminuria | Case subjects with proteinuria | Case subjects with ESRD |
|---|---|---|---|
| n | 96 | 42 | 44* |
| Men (%) | 42 | 57 | 48 |
| Age at diagnosis of diabetes (years) | 15 ± 10 | 12 ± 8 | 12 ± 8 |
| Duration of diabetes (years)† | 28 ± 10‡ | 28 ± 6 | 25 ± 9§ |
| Age (years)† | 43 ± 9 | 40 ± 8 | 37 ± 9§ |
| Total insulin dose (units)† | 48 ± 24 | 50 ± 24 | 38 ± 17 |
| A1C (%)† | 8.1 ± 1.7 | 8.3 ± 1.2 | 8.1 ± 2.2 |
| Retinopathy requiring laser treatment (%)† | 30 | 62 | 88 |
| Serum creatinine (mg/dl)† | 0.9 ± 0.2 | 1.6 ± 0.8 | NA |
This group consists of 14 patients with new-onset ESRD, 2 patients on hemodialysis, and 28 patients with kidney transplant.
At the time of the enrollment into the study. In control subjects 10% were treated with antihypertensive drugs (including ACE inhibitors).
In control subjects, 33, 35, and 32% had duration of diabetes for 10–19 years, 20–29 years, and 30–49 years, respectively.
Value at the time of initiation of renal replacement therapy.
Linkage scan for type 1 diabetes loci.
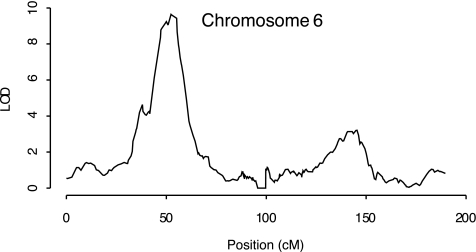
The 130 ASPs for type 1 diabetes were used to search for chromosomal regions with increased allele sharing (i.e., >25% sharing both chromosomes and mean allele sharing ≥50%). We present these results for two important reasons. First, the highly significant peak (MLS = 9.6 at 52 cM) location in the HLA region of chromosome 6p (Fig. 1) provides validation that our patient population is comprised of type 1 diabetic patients. Second, these results provide additional information throughout the genome to those studying the genetics of type 1 diabetes. Excluding HLA, the largest MLS occurs at a secondary peak on chromosome 6q (MLS = 3.1 at 141–143 cM) (Fig. 1). Here, the type 1 diabetic ASPs have >60% mean allele sharing and 42.6% share both alleles. The only other notable result from the ASP scan occurs on chromosome 5. However, the proportion of ASPs sharing both alleles at this location (19%) is uncharacteristically low for a true linkage result.
FIG. 1.
Chromosome 6 linkage results for 130 sibpairs concordant for type 1 diabetes. The computations and the plots were obtained using SPLAT (16).
Linkage scan for diabetic nephropathy susceptibility loci.
In contrast to the analysis carried out in 130 ASPs for type 1 diabetes, the 100 DSPs for diabetic nephropathy were analyzed to search for chromosomal regions with diminished sharing. This implies that >25% of the DSPs will share neither of their two chromosomes in the vicinity of the gene, leading to mean allele sharing <50%.
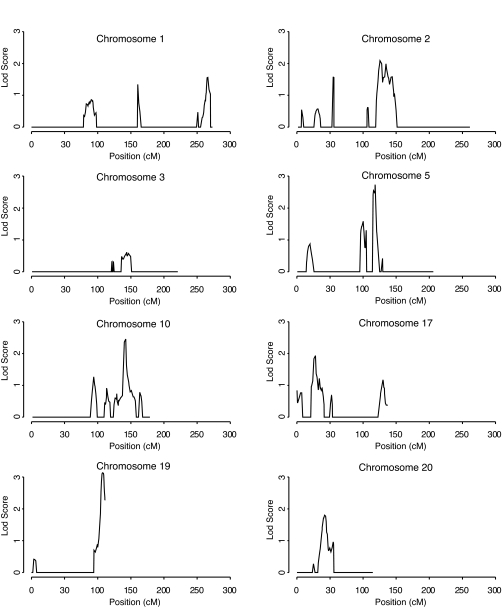
Fifteen chromosomes show no evidence of linkage (see online appendix). Figure 2 shows the remaining chromosomes and also chromosome 3, since we have previously reported linkage to this chromosome (8). The peak on chromosome 19 is the strongest (MLS = 3.1 at 107.2 cM). Three peaks have MLS >2: chromosome 5 (MLS = 2.7 at 118 cM), chromosome 10 (MLS = 2.5 at 142 cM), and chromosome 2 (MLS = 2.1 at 125 cM). Three others have MLS >1: chromosome 17 (MLS = 1.9 at 28 cM), chromosome 20 (MLS = 1.8 at 42 cM), and chromosome 1 (MLS = 1.6 at 266 cM). Three regions in Fig. 2 have a sharing pattern not reflective of linkage, characterized by an increased proportion of sharing one allele (67, 65, and 65% on chromosomes 5, 10, and 17, respectively) but a decreased proportion sharing neither allele (20, 25, and 21% on chromosomes 5, 10, and 17, respectively).
FIG. 2.
Linkage results for 100 sibpairs concordant for type 1 diabetes and discordant for nephropathy. Siblings considered unaffected had normoalbuminuria despite a minimum of 10 years duration of diabetes, while affected siblings had proteinuria or ESRD. The computations and the plots were obtained using SPLAT (16).
Progression from proteinuria to ESRD occurred in exactly half of the 100 DSPs, providing a natural partition of families for subanalysis (Table 3). The highest peak in the combined analysis (MLS = 3.1 on chromosome 19) shows strong consistency in both MLS values and sharing patterns in the two phenotypic subsets (MLS values of 1.9 [ESRD] and 1.8 [proteinuria] with zero allele sharing of 43 and 42%, respectively). The region on chromosome 2 shows similar consistency (MLS values of 1.3 [ESRD] and 1.4 [proteinuria] with zero allele sharing of 40 and 36%, respectively). In contrast, chromosome 20 results arise solely from the 50 DSPs with proteinuria (MLS = 2.8) and chromosome 1 results arise solely from the 50 DSPs with ESRD (MLS = 1.8). An additional twist emerges for chromosome 3. There, the rather modest peak in the set of all 100 DSPs (MLS = 0.6) partitions into two distinct peaks, one at 96 cM for the 50 DSPs with proteinuria (MLS = 1.5) and one at 154 cM for the 50 DSPs with ESRD (MLS = 1.1). Finally, of three chromosomes with sharing patterns not consistent with linkage in the overall dataset, the chromosome 10q region was consistent with linkage in the set of 50 DSPs with proteinuria (MLS = 1.7).
TABLE 3.
Linkage results (pattern of sharing) according to type of DSP*
| Chromosome | Rs no. | DSPs with proteinuria (n = 50) |
DSPs with ESRD (n = 50) |
All DSPs (n = 100) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cM† | z0 | z1 | z2 | MLS | Rs no. | cM | z0 | z1 | z2 | MLS | Rs no. | cM | z0 | z1 | z2 | MLS | ||
| 1q | — | — | — | — | — | — | rs987179 | 266 | 0.41 | 0.47 | 0.12 | 1.8 | rs987179 | 266 | 0.37 | 0.45 | 0.18 | 1.6 |
| 2q | rs1847694 | 126 | 0.39 | 0.49 | 0.13 | 1.4 | rs855004 | 122 | 0.36 | 0.51 | 0.12 | 1.3 | rs895415 | 125 | 0.35 | 0.51 | 0.14 | 2.1 |
| 3q | rs1002200 | 96 | 0.41 | 0.45 | 0.14 | 1.5 | rs1381768 | 154 | 0.39 | 0.45 | 0.16 | 1.1 | rs750543 | 144 | 0.28 | 0.54 | 0.18 | 0.6 |
| 5q | rs1501656 | 99 | 0.16 | 0.70 | 0.14 | 1.7 | rs27342 | 118 | 0.16 | 0.74 | 0.10 | 2.7 | rs27342 | 118 | 0.20 | 0.67 | 0.13 | 2.7 |
| 10q | rs1467813 | 142 | 0.35 | 0.57 | 0.08 | 1.7 | rs703422 | 140 | 0.15 | 0.73 | 0.12 | 1.8 | rs1467813 | 142 | 0.25 | 0.65 | 0.10 | 2.4 |
| 17p | rs1047365 | 28 | 0.18 | 0.65 | 0.16 | 0.8 | rs6503211 | 26 | 0.25 | 0.65 | 0.10 | 1.5 | rs1047365 | 28 | 0.21 | 0.66 | 0.13 | 1.9 |
| 19q | rs260462 | 111 | 0.43 | 0.44 | 0.13 | 1.9 | rs7478 | 105 | 0.42 | 0.46 | 0.12 | 1.8 | rs306450 | 107 | 0.39 | 050 | 0.11 | 3.1 |
| 20p | rs466243 | 41 | 0.35 | 0.59 | 0.06 | 2.8 | — | — | — | — | — | — | rs775133 | 42 | 0.31 | 0.56 | 0.13 | 1.8 |
Probabilities of sharing: z0 = 0 alleles, z1 = 1 allele, and z2 = 2 alleles.
cM according to the deCODE map.
Refinement of results based on sharing patterns.
Because siblings can share zero, one, or two alleles at a given chromosomal location, describing the sharing pattern requires two parameters, the proportion sharing zero alleles and the proportion sharing one allele. The remaining siblings share two alleles. Under the null hypothesis, siblings share zero, one, or two alleles with frequencies of 25, 50, and 25%, respectively.
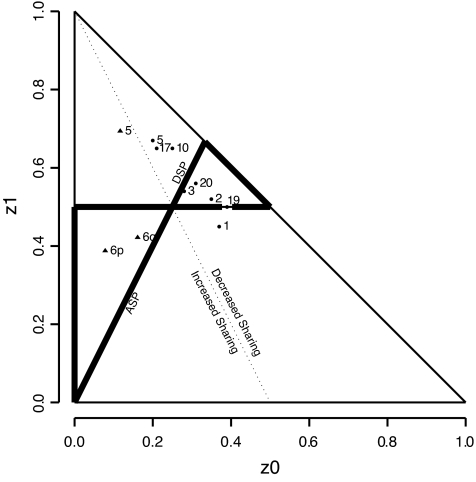
The two free parameters can be visualized on a two-dimensional graph with the constraint that the sum of the two parameters is one or more. However, not all patterns are consistent with those likely to occur in the vicinity of a susceptibility gene. Holmans (19) systematically characterized the patterns consistent with linkage in ASPs (Holmans’ “ASP Triangle,” see Fig. 3). We derived the analogous triangular region for DSPs (18), also shown in Fig. 3. The only point of overlap of the two triangles is the null hypothesis. Otherwise, the entire ASP triangle falls in the region of increased mean sharing and the entire DSP triangle falls in the region of decreased mean sharing.
FIG. 3.
Sharing patterns for ASP (▴) and DSP (•) linkage peaks. Note that both ASP results on chromosome 6 and the DSP peaks for chromosomes 2, 3, 19, and 20 reside within the triangles defined by biological consistency under the respective models. The sharing pattern for the ASP peak on chromosome 5, for example, though comparatively distant from null sharing of 0.25, 0.5, and 0.25, is close to the dotted line defining mean sharing of one-half and is therefore not consistent with true linkage. The computations and the plot were obtained using SPLAT (16).
Figure 3 shows that the two ASP results on chromosome 6 are in the ASP triangle, but the ASP result on chromosome 5 is far from it, reinforcing our observation that the sharing pattern on chromosome 5 is not typical of linkage. For the 100 DSPs, the results for chromosomes 19, 2, 20, and 3 are within the DSP triangle, while the chromosome 1 result is not. We also get a visual sense of the clustering of the chromosome 5, 10, and 17 results, which are outside of the DSP triangle and dissimilar to the null hypothesis primarily because of an excess of one allele sharing.
The sharing patterns in the subphenotypes of ESRD and proteinuria, reported in Table 3, can be viewed graphically in the online appendix. For chromosome 2, the peak for the proteinuria group is within the DSP triangle, while the peak for the ESRD group is nearby but just outside the triangle. The proximity of these two peaks to each other suggests a single linked locus, which is independent of subphenotype, located close to the lower boundary of the DSP triangle. That the actual sharing pattern for proteinuria falls just below the boundary is not concerning, since the corresponding point estimate of the location also has a rather wide confidence interval. A similar situation occurs for chromosome 19, where the peaks for ESRD and proteinuria are close to each other and to the peak in the overall sample. Here, the peaks for both subphenotypes fall just below the lower boundary of the DSP triangle, again likely due to sampling variation in a small sample. The chromosome 3 peaks for subphenotypes are also close to each other and just below the lower boundary of the DSP triangle. The chromosome 1 peak in ESRD is in a similar location to the overall peak, which is just below the DSP triangle.
DISCUSSION
Our primary finding for linkage to diabetic nephropathy is on chromosome 19q (triangle MLS = 3.1), with a secondary peak on chromosome 2q (triangle MLS = 2.1). The former, but not the latter, exceeds the Lander and Kruglyak criterion of triangle MLS ≥2.6 (17,18) for suggestive linkage. For reference, triangle MLS values of 3.3, 2.3, and 1.7 correspond to unadjusted P values of 0.0001, 0.001, and 0.005, respectively.
Stratification of DSPs based on proteinuria or ESRD suggested four tertiary peaks: linkage with ESRD on chromosome 1q (MLS = 1.8), linkage with proteinuria on chromosome 20p (MLS = 2.8), and linkage with two separate regions on chromosome 3q, one for proteinuria (MLS = 1.5) and another, 58 cM away, for ESRD (MLS = 1.1). We also found two chromosomal regions linked with type 1 diabetes. The most striking, not surprisingly, was on chromosome 6p (MLS = 9.2, 52 cM), confirming the well-established linkage with HLA. We also replicated IDDM15 on chromosome 6q (MLS = 3.1, 142 cM) (http://t1dbase.org/page/Loci/display/?species=Human).
Two previous publications have used the DSP study design developed by our group. The first was a pilot study done at Joslin Diabetes Center (8). Sixty-six DSPs from 52 families were used to test chromosomal regions containing genes of the renin-angiotensin system. We found no evidence for linkage with AGT (chromosome 1q) or with ACE (chromosome 17q); however, we did obtain suggestive evidence (MLS = 3.1 at 157 cM) on the chromosome 3q region containing ATR1. In the second study of 83 DSPs from 73 Finnish families with type 1 diabetes, suggestive evidence was found on chromosome 3q (MLS = 2.7 at 141 cM). The sharing patterns leading to the linkage effect were not presented (20).
Contrary to these studies, the strongest linkage signal in our current study is on chromosome 19q. Our enthusiasm for this finding stems not only from the magnitude of the linkage statistic but also from the sharing pattern, which occurs well within the DSP triangle. Moreover, this signal was detected in both phenotypic subsets (ESRD and proteinuria). The 1 logarithm of odds support interval around the combined ESRD/proteinuria DSP result encompasses 6.5 cm, within which there are 136 genes (94 known and 42 hypothetical or predicted) (see online appendix).
As with chromosome 19q, our secondary peak on chromosome 2q (MLS = 2.1) has a sharing pattern consistent with linkage, and the peak exists in both subsets of DSPs. The 1 logarithm of odds support interval around the combined ESRD/proteinuria DSP result encompasses 25 cM, within which there are 206 genes (100 known and 106 hypothetical or predicted) (see online appendix).
Our remaining findings stemmed from subset analysis of 50 DSPs defined by ESRD and 50 DSPs defined by proteinuria. Specifically, we found modest evidence for linkage with ESRD on chromosome 1q (MLS = 1.8). Conversely, there is evidence for linkage with proteinuria on chromosome 20p (MLS = 2.8). The results on chromosome 3q point toward two separate regions: one linked to proteinuria (MLS = 1.5) and another, 58 cM away, linked to ESRD (MLS = 1.1). One possible explanation is phenotypic heterogeneity, with some locus (or loci) related to abnormalities in urinary albumin excretion (UAE) and others related to progression to ESRD (or differences in survival rates once ESRD occurs). Recently, we demonstrated such phenotypic heterogeneity in extended families with type 2 diabetes. In particular, we performed a whole genome scan using variance components analysis to study two renal phenotypes, UAE and renal function estimated with serum cystatin C. We found strong or suggestive evidence for linkage to UAE on chromosome 5q, 7q, and 22p (21) and independently strong or suggestive evidence for linkage to renal function on chromosome 2q, 7p, 10q, and 18p (22).
Our primary finding on chromosome 19q is novel and does not overlap with any regions reported by other authors. In contrast, our secondary finding on chromosome 2q overlaps exactly with a recently reported linkage result from the Family Investigation of Nephropathy and Diabetes (FIND) study for UAE variation in several ethic groups (23). The region from FIND spanned markers D2S410–D2S1328 located at 127–138 cM. Among our tertiary results, chromosome 1q is novel, while there is some agreement with the results on chromosome 20p and 3q. Specifically, our finding on chromosome 20p seems to validate a finding reported in a genome scan of 59 Pima Indian families with 98 sibpairs concordant for both type 2 diabetes and diabetic nephropathy (21). An MLS = 1.8 was found near D20S115 (24.7 cM) and a two-point MLS = 1.9 was found near GATA65E01 (57.2 cM). Speaking broadly, the location of our tertiary peak on 3q is in agreement with seven previous studies (Table 4). However, the location of peaks varied between 95 and 210 cM. In our current study, we found two minor peaks 58 cM apart.
TABLE 4.
Summary of linkage results reported on chromosome 3q
| Authors | Study design | Phenotype | MLS | Markers | Genetic position* |
|---|---|---|---|---|---|
| Studies in type 1 diabetes | |||||
| Moczulski et al. (8) (Caucasians) | DSP (n = 66) | Proteinuria and ESRD | 3.1 | D3S1308 | 157.0 |
| Osterholm et al. (20) (Caucasians) | DSP (n = 83) | Proteinuria | 2.7 | D3S3606–D3S3694 | 134.6–148.8 |
| Current study (Caucasians) | DSP (n = 50) DSP (n = 50) | Proteinuria ESRD | 1.5 1.1 | rs1002200 rs1381768 | 95.6 154.2 |
| Studies in type 2 diabetes | |||||
| Imperatore et al. (24) (Pima Indians) | ASP (n = 90) | Proteinuria and ESRD | 1.5 | D3S3053 | 174 |
| Bowden et al. (25) (African Americans) | ASP (n = 206) | ESRD | 1.3 (4.6)† | D3S2460 | 126 |
| Krolewski et al. (21) Caucasians) | Quantitative trait loci using 5,656 relative pairs | UAE | 1.0 | D3S1744 | 153.4 |
| Placha et al. (22) (Caucasians) | Quantitative trait loci using 5,187 relative pairs | GFR estimated by cystatin C (nondiabetic relative pairs only) | 2.8 | D3S1744 | 153.4 |
| Chen et al. (26) (West Africans) | Quantitative trait loci using 360 relative pairs | Serum creatinine | 2.2 | D3S2418 | 210 |
cM according to the deCODE map.
Ordered subset analysis.
Phenotypic or genetic heterogeneity may underlie these discrepant results. For example, in our pilot study, we had 66 DSPs and we found a peak with MLS = 3.1 on chromosome 3q, close to our current ESRD peak (MLS = 1.1). The current study comprised 48 original and 52 new DSPs. In the 48 DSPs, the MLS was 2.6 at 154 cM. In the new 52 DSPs, we identified only modest evidence for linkage (MLS = 1.3) at position 103 cM. The noticeable difference between the groups was significantly shorter diabetes duration in diabetic nephropathy cases in the original DSPs. Due to the small number of DSPs, it is impossible to evaluate whether this phenotypic difference could explain the different linkage results. In our previous publication, however, we demonstrated an effect of diabetes duration at onset of complications on the results of genetic studies (27).
There are certain limitations to this study. First, DSP studies are prone to inflated MLS values if allele-sharing patterns are improperly estimated (e.g., due to missing parents). In our study, in 26 families, neither parent was genotyped. We attempted to minimize any bias by choosing a highly reliable genotyping platform and being diligent in removing markers failing to meet strict quality-control standards. Our confidence in the genotyping platform is based on previous validation as well as nearly 30,000 replicate quality-control genotypes from five samples in our study. As a further check, we reanalyzed the top peaks using subsets of every third marker and found consistency in each of the three subsets (data not shown). Such consistency would not be expected were the results due to genotyping errors. Inclusion of diabetic nephropathy ASPs would have been another way to mitigate the risk of misgenotyping, and methods to analyze DSPs and ASPs together exist (28,29). However, as documented (7), collection of diabetic nephropathy ASPs is extremely difficult and, being concordant for both diabetic nephropathy and type 1 diabetes, these siblings would present an additional challenge in terms of interpretation. Second, while 100 DSPs are sufficient to detect a major locus, the power to detect moderate/minor genetic players is more modest. Moreover, the subanalyses we performed by distinguishing ESRD from proteinuria was a post hoc analysis that should be viewed as exploratory. Third, mortality in ESRD is quite high (30), leading, potentially, to survival bias. This could manifest through reduced power among the 50 ESRD DSPs, which might be misinterpreted as phenotypic heterogeneity, or through drop-out of poor survivors with ESRD such that the genetic variants associated with good survival would appear to be related to diabetic nephropathy susceptibility. Fourth, while two loci (DSP triangle MLS = 3.1 on chromosome 19 and ASP triangle MLS = 3.1 on chromosome 6q) were “suggestive” (17), none except HLA achieved “significance” (triangle MLS ≥4.0). Therefore, our results must be considered largely as hypothesis generating. Given the difficulty in assembling collections of sibs concordant for type 1 diabetes and discordant for diabetic nephropathy, a more practical approach may be to now focus on detecting association using resources such as GoKinD (31).
In conclusion, our study has provided two sets of genome scan results: one for type 1 diabetes using 130 ASPs and another for diabetic nephropathy using 100 DSPs. The type 1 diabetes scan overwhelmingly confirmed the HLA region on chromosome 6p and it provided additional support for IDDM15. The diabetic nephropathy scan introduced a new candidate region on chromosome 19q and confirmed linkage to UAE on chromosome 2q reported by the FIND study. Using exploratory subset analysis based on the degree of diabetic nephropathy, we found a novel locus on chromosome 1q and confirmed a locus on chromosome 20p described in Pima Indians. Finally, we found evidence for two loci on chromosome 3q, adding to the list of positive studies on this chromosome. Taken together, the results from our diabetic nephropathy scan suggest several loci as candidates for susceptibility, none of which appear to the sole determinant of diabetic nephropathy.
Supplementary Material
Acknowledgments
This research has been supported by National Institutes of Health Grants DK 053534 and DK 077532. K.W. has been supported by Juvenile Diabetes Research Federation Fellowship no. 3-2005-908.
All supplemental tables and figures can be obtained online at http://www.joslinresearch.org/LabSites/Krolewski/t1dn.dsp/.
Published ahead of print at http://diabetes.diabetesjournals.org on 16 June 2008.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Parving HH, Mauer M, Ritz E: Diabetic nephropathy. In Brenner and Rector's The Kidney. 7th ed. Brenner BM, Ed. Philadelphia, Elsevier,2004. , p.1777 –1818
- 2.Seaquist ER, Goetz FC, Rich S, Barbosa J: Familial clustering of diabetic kidney disease: evidence for genetic susceptibility to diabetic nephropathy. N Engl J Med 320 :1161 –1165,1989 [DOI] [PubMed] [Google Scholar]
- 3.Borch-Johnsen K, Norgaard K, Hommel E, Mathiesen ER, Jensen JS, Deckert T, Parving HH: Is diabetic nephropathy and inherited complication? Kidney Int 41 :719 –722,1992 [DOI] [PubMed] [Google Scholar]
- 4.Quinn M, Angelico MC, Warram JH, Krolewski AS: Familial factors determine the development of diabetic nephropathy in patients with IDDM. Diabetologia 39 :940 –945,1996 [DOI] [PubMed] [Google Scholar]
- 5.The Diabetes Control and Complications Trial Research Group: Clustering of long-term complications in families with diabetes in the Diabetes Control and Complications Trial. Diabetes 46 :1829 –1839,1997 [PubMed] [Google Scholar]
- 6.Harjutsalo V, Katoh S, Sarti C, Tajima N, Tuomilehto J: Population-based assessment of familial clustering of diabetic nephropathy in type 1 diabetes. Diabetes 53 :2449 –2454,2004 [DOI] [PubMed] [Google Scholar]
- 7.Rogus JJ, Krolewski AS: Using discordant sib pairs to map loci for qualitative traits with high sibling recurrence risk. Am J Hum Genet 59 :1376 –1381,1996 [PMC free article] [PubMed] [Google Scholar]
- 8.Moczulski DK, Rogus JJ, Antonellis A, Warram JH, Krolewski AS: Major susceptibility locus for nephropathy in type 1 diabetes on chromosome 3q: results of novel discordant sibpair analysis. Diabetes 47 :1164 –1169,1998 [DOI] [PubMed] [Google Scholar]
- 9.Warram JH, Gearin G, Laffel L, Krolewski AS: Effect of duration of IDDM on the prevalence of stages of diabetic nephropathy defined by urinary albumin/creatinine ratio. J Am Soc Nephrol 7 :930 –937,1996 [DOI] [PubMed] [Google Scholar]
- 10.McPeek S: Statistical tests for detection of misspecified relationships by use of genome-screen data. Am J Hum Genet 66 :1076 –1094,2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Connell JR, Weeks DE: PedCheck: a program for identifying genotype incompatibilities in linkage analysis. Am J Hum Genet 63 :259 –266,1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abecasis GR, Cherny SS, Cookson WO, Cardon LR: Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30 :97 –101,2002 [DOI] [PubMed] [Google Scholar]
- 13.Huang Q, Shete S, Amos CI: Ignoring linkage disequilibrium among tightly linked markers induces false-positive evidence of linkage for affected sib pair analysis. Am J Hum Genet 75 :1106 –1112,2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abecasis GR, Wigginton JE: Handling marker-marker linkage disequilibrium: pedigree analysis with clustered markers. Am J Hum Genet 77 :754 –767,2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyles AL, Scott WK, Martin ER, Schmidt S, Li YJ, Ashley-Koch A, Bass MP, Schmidt M, Pericak-Vance MA, Speer MC, Hauser ER: Linkage disequilibrium inflates type I error rates in multipoint linkage analysis when parental genotypes are missing. Hum Hered 59 :220 –227,2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poznik GD, Adamska K, Xu X, Krolewski AS, Rogus JJ: A novel framework for sib pair linkage analysis. Am J Hum Genet 78 :222 –230,2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lander E, Kruglyak L: Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11 :241 –247,1995 [DOI] [PubMed] [Google Scholar]
- 18.Lunetta KL, Rogus JJ: Strategy for mapping minor histocompatibility genes involved in graft-versus-host disease: a novel application of discordant sib pair methodology. Genet Epidemiol 15 :595 –607,1998 [DOI] [PubMed] [Google Scholar]
- 19.Holmans P: Asymptotic properties of affected-sib-pair linkage analysis. Am J Hum Genet 52 :362 –374,1993 [PMC free article] [PubMed] [Google Scholar]
- 20.Osterholm AM, He B, Pitkaniemi J, Albinsson L, Berg T, Sarti C, Tuomilehto J, Tryggvason K: Genome-wide scan for type 1 diabetic nephropathy in the Finnish population reveals suggestive linkage to a single locus on chromosome 3q. Kidney Int 71 :140 –145,2007 [DOI] [PubMed] [Google Scholar]
- 21.Krolewski AS, Poznik DG, Placha GP, Canani L, Dunn J, Walker W, Smiles A, Krolewski B, Fogarty D, Moczulski D, Araki S, Makita Y, Ng DPK, Rogus JJ, Duggirala R, Rich SS, Warram JH: A genome-wide linkage scan for genes controlling variation in urinary albumin excretion in type 2 diabetes. Kidney Int 69 :129 –136,2006 [DOI] [PubMed] [Google Scholar]
- 22.Placha G, Poznik GD, Dunn J, Smiles A, Krolewski B, Glew T, Puppala S, Schneider J, Rogus JJ, Rich SS, Duggirala R, Warram JH, Krolewski AS: A genome-wide linkage scan for genes controlling variation in renal function estimated by serum cystatin C levels in extended families with type 2 diabetes. Diabetes 55 :3358 –3365,2006 [DOI] [PubMed] [Google Scholar]
- 23.Iyengar SK, Abboud He, Goddard KA, Saad MF, Adler SG, Arar NH, Bowden DW, Duggirala R, Elston RC, Hanson RL, Ipp E, Kao WH, Kimmel PL, Klag MJ, Knowler WC, Meoni LA, Nelson RG, Nicholas SB, Pahl MV, Parekh RS, Quade SR, Rich SS, Rotter JI, Scavini M, Schelling JR, Sedor JR, Sehgal AR, Shah VO, Smith MW, Taylor KD, Winkler CA, Zager PG, Freedman BI: Genome-wide scans for diabetic nephropathy and albuminuria in multiethnic populations: the Family Investigation of Nephropathy and Diabetes (FIND). Diabetes 56 :1577 –1585,2007 [DOI] [PubMed] [Google Scholar]
- 24.Imperatore G, Hanson RL, Pettitt DJ, Kobes S, Bennett PH, Knowler WC: Sib-pair linkage analysis for susceptibility genes for microvascular complications among Pima Indians with type 2 diabetes: Pima Diabetes Genes Group. Diabetes 47 :821 –830,1998 [DOI] [PubMed] [Google Scholar]
- 25.Bowden DW, Colicigno CJ, Langefeld CD, Sale MM, Williams A, Anderson PJ, Rich SS, Freedman BI: A genome scan for diabetic nephropathy in African Americans. Kidney Int 66 :1517 –1526,2004 [DOI] [PubMed] [Google Scholar]
- 26.Chen G, Adeyemo AA, Zhou J, Chen Y, Doumatey A, Lashley K, Huang H, Amoah A, Agyenim-Boateng K, Eghan BA, Okafor G, Acheampong J, Oli J, Fasanmade O, Johnson T, Rotimi C: A genome-wide search for linkage to renal function phenotypes in West Africans with type 2 diabetes. Am J Kidney Dis 49 :394 –400,2007 [DOI] [PubMed] [Google Scholar]
- 27.Rogus JJ, Warram JH, Krolewski AS: Genetics studies of late diabetic complications: the overlooked importance of diabetes duration before complication onset. Diabetes 51 :1655 –1662,2002 [DOI] [PubMed] [Google Scholar]
- 28.Shih PY, Wang T, Xing C, Sinha M, Song Y, Elston RC: Linkage analysis of alcohol dependence using both affected and discordant sib pairs. BMC Genet 6 (Suppl. 1):S36 ,2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xing C, Sinha R, Xing G, Lu Q, Elston RC: The affected-/discordant-sib-pair design can guarantee validity of multipoint model-free linkage analysis of incomplete pedigrees when there is marker-marker disequilibrium. Am J Hum Genet 79 :396 –401,2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adler AI, Stevens RJ, Manley SE, Bilous RW, Cull CA, Holman RR: Development and progression of nephropathy in type 2 diabetes: the United Kingdom Prospective Diabetes Study (UKPDS 64). Kidney Int 63 :225 –232,2003 [DOI] [PubMed] [Google Scholar]
- 31.Mueller PM, Rogus JJ, Cleary PA, Zhao Y, Smiles AM, Steffes MW, Bucksa J, Gibson TB, Cordovado SK, Krolewski AS, Nierras CR, Warram JH: The Genetics of Kidneys in Diabetes (GoKinD) Study: a genetics collection available for identifying the genetic susceptibility factors for diabetic nephropathy in type 1 diabetes mellitus. J Am Soc Neph 17 :1782 –1790,2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.