Abstract
OBJECTIVE— Loss of lipin 1 activity causes lipodystrophy and insulin resistance in the fld mouse, and LPIN1 expression and common genetic variation were recently suggested to influence adiposity and insulin sensitivity in humans. We aimed to conduct a comprehensive association study to clarify the influence of common LPIN1 variation on adiposity and insulin sensitivity in U.K. populations and to examine the role of LPIN1 mutations in insulin resistance syndromes.
RESEARCH DESIGN AND METHOD— Twenty-two single nucleotide polymorphisms tagging common LPIN1 variation were genotyped in Medical Research Council (MRC) Ely (n = 1,709) and Hertfordshire (n = 2,901) population-based cohorts. LPIN1 exons, exon/intron boundaries, and 3′ untranslated region were sequenced in 158 patients with idiopathic severe insulin resistance (including 23 lipodystrophic patients) and 48 control subjects.
RESULTS— We found no association between LPIN1 single nucleotide polymorphisms and fasting insulin but report a nominal association between rs13412852 and BMI (P = 0.042) in a meta-analysis of 8,504 samples from in-house and publicly available studies. Three rare nonsynonymous variants (A353T, R552K, and G582R) were detected in severely insulin-resistant patients. However, these did not cosegregate with disease in affected families, and Lipin1 protein expression and phosphorylation in patients with variants were indistinguishable from those in control subjects.
CONCLUSIONS— Our data do not support a major effect of common LPIN1 variation on metabolic traits and suggest that mutations in LPIN1 are not a common cause of lipodystrophy in humans. The nominal associations with BMI and other metabolic traits in U.K. cohorts require replication in larger cohorts.
Lipin 1, a multifunctional protein highly expressed in mouse and human adipose tissue, has been shown to influence adipose tissue development and function. Null mutations in the murine lipin 1 gene (Lpin1) result in impaired adipocyte differentiation leading to a severe reduction in adipose tissue mass, insulin resistance, and progressive peripheral neuropathy in the fld and fld2J mouse models (1). In contrast, transgenic mice with adipose tissue–specific overexpression of Lpin1 exhibit diet-induced obesity and enhanced insulin sensitivity compared with those seen in wild-type littermates (2). In humans, LPIN1 expression in adipose tissue appears to be inversely correlated with measures of adiposity such as BMI and positively correlated with insulin sensitivity (3–6).
The mechanism through which lipin 1 influences adiposity and insulin sensitivity in mice and humans is not entirely known. However, recent data indicate that lipin 1 is a magnesium-dependent phospatidate phosphatase responsible for catalyzing the penultimate step in triacylglyceride synthesis, explaining why Lpin1-deficient fld mice cannot accumulate fat in mature adipocytes (7). Lipin 1 is also thought to regulate transcription of genes involved in adipocyte differentiation (PPARγ and C/EBPα), fat synthesis and storage (DGAT, ACC-1, PEPCK, FAS, and SCD1), and fatty acid oxidation (CPT-1, AOX, and PPARα) (2,6–10).
There has only been one study sequencing LPIN1 in lipodystrophic patients (n = 15), with no pathogenic mutation reported (11). Furthermore, although a number of studies have evaluated the role of common variation in LPIN1 and metabolic quantitative phenotypes (12–14), the results have been inconsistent across studies and sometimes within the same study. For example, rs2716610 and a SNP in high linkage disequilibrium, rs2716609, were associated with BMI in a Finnish obesity case-control study and in the Quebec Family Study (12,14) but not in a German population-based cohort (the the MONItoring of trends and determinants in CArdiovascular disease [MONICA] study) (13). Moreover, LPIN1 haplotypes were strongly associated with traits underlying the metabolic syndrome in the MONICA study, but these haplotypes often had the opposite effect on the same traits in a replication cohort (13). This inconsistency suggests that further studies are needed to clarify the role of LPIN1 variation on human metabolic traits. In this study, we have taken complementary approaches to study the role of LPIN1 variation in human metabolic traits in U.K. populations: 1) we genotyped 22 SNPs that tag common LPIN1 variation (minor allele frequency >0.01) in two white U.K. population-based cohorts (n = 4,610) and tested for association with fasting serum insulin levels, BMI, and a number of additional metabolic traits with previously reported association with LPIN1, and 2) we sequenced LPIN1 in a cohort of patients with syndromes of severe insulin resistance (n = 135) and lipodystrophy (n = 23) to identify potentially pathogenic mutations.
RESEARCH DESIGN AND METHODS
Definition of cohorts
ELY cohort.
The MRC Ely Study is a population-based cohort study of the etiology and pathogenesis of type 2 diabetes and related metabolic disorders in the U.K. (15). The subject population is comprised of white men and women aged 35–79 years without diagnosed diabetes. Measurements of anthropometric and metabolic data analyzed in this study have previously been described (16). Informed consent was obtained from all participants, and ethical approval for the study was granted by the Cambridge Local Research Ethics Committee.
Hertfordshire cohort.
The study population comprising the Hertfordshire Cohort was recruited from the Hertfordshire population born between 1931 and 1939. The cohort details and measurements of metabolic traits analyzed in this study have previously been described (17).
European Prospective Investigation into Cancer and Nutrition–Obesity study.
The European Prospective Investigation into Cancer and Nutrition (EPIC)-Obesity study is nested within the EPIC-Norfolk study, a population-based cohort study of 25,663 white European men and women aged 39–79 and recruited in Norfolk, U.K., between 1993 and 1997 (18). Height and weight were measured using standard anthropometric techniques (18). All samples were genotyped using the Affymetrix GeneChip Human Mapping 500K Array Set, which contained genotype information for five of our LPIN1 tagSNPs, each of which had a call rate >90%. In total, 2,415 individuals with height and weight measures and quality-controlled genotype data were available for analyses.
Human genome diversity cell line panel and Centre d'Etude du Polymorphisme Humain.
The Human Genomse Diversity Cell Line Panel (HGDP) Centre d'Etude du Polymorphisme Humain (CEPH) is a resource of 1,064 DNA samples from individuals distributed around the world and has previously been described (19). The 48 unrelated individuals from CEPH families supplied by Coriell Cell Repositories (20) are control individuals of north and west European origin.
Severe insulin resistance cohort.
All patients had severe insulin resistance, defined as fasting insulin above 150 pmol/l or peak insulin on oral glucose tolerance testing >1,500 pmol/l in nondiabetic patients. Complete insulin deficiency was defined as an insulin requirement above 3 units · kg−1 · day−1. Most patients had a BMI <30 kg/m2, and at least 58 patients had BMI >30 kg/m2. Those with partial β-cell decompensation and clinical features including acanthosis nigricans and those with BMI >30 kg/m2 were included at the investigators’ discretion. All patients gave informed consent with approval of the local research ethics committee in Cambridge, U.K.
Selection of single nucleotide polymorphisms to common tag LPIN1 variation.
Tag single nucleotide polymorphisms (SNPs) were selected to cover variation with minor allele frequency (MAF) >1% present in LPIN1 and its flanking 4-kb regions (chromosome 2, coordinates 11800212–11889941 [NCBI B36 assembly]). The International HapMap (release 20, phase II) reports 61 SNPs for CEPH Utah (CEU) samples in this chromosomal region, while we identified five novel SNPs during resequencing of 48 CEPH individuals (supplementary Table 1, available in an online appendix at http://dx.doi.org/10.2337/db08-0422). Twenty-five tag SNPs were selected to cover all 66 novel and known HapMap polymorphisms with a pairwise r2 ≥ 0.8 using Tagger (21) as a stand-alone program in Haploview (22) (supplementary Fig. 1). Twenty-two of these tag SNPs passed assay design and prescreening (supplementary Table 2). The three failed SNPs tagged only five intronic SNPs. We calculated that we have >80% power to detect a per-allele effect on BMI of >1.33 kg/m2 with MAF 0.01 and >0.27 kg/m2 with MAF 0.5. For logged fasting insulin data, this range is >1.04 to >1.22.
Genotyping.
Genotyping was performed by the genotyping facility within the Genetics of Complex Traits in Humans team at the Wellcome Trust Sanger Institute. LPIN1 mutations A353T, R552K, and G582R were genotyped on the CEPH human diversity panel as stand-alone assays using the Sequenom MassArray hME platform ((Sequenom, San Diego, CA) according to the manufacturer's instructions. Twenty-two LPIN1 tagging SNPs (supplementary Table 2) were genotyped using the Sequenom MassArray iPlex platform according to the manufacturer's instructions. Details regarding primers, probes, and conditions are available on request. All SNPs, except for rs17603755, were successfully genotyped in >85% of samples (call rates for each SNP are presented in supplementary Table 2) and did not deviate from Hardy-Weinberg Equilibrium (P > 0.01). The average call rate across Ely and Hertfordshire samples pooled together was 92.1%.
Statistical analysis.
Deviation of LPIN1 tagSNP genotype from Hardy-Weinberg equilibrium was assessed using a goodness-of-fit χ2 test. Linear regression analysis was used to assess the association between individual SNPs and BMI, log-transformed fasting plasma insulin, and log-transformed additional metabolic traits (systolic and diastolic blood pressure, HDL and LDL cholesterol, plasma triglycerides, waist circumference, and A1C levels) in Ely and Hertfordshire cohorts using Stata (version 9; Stata, College Station, TX). All analyses were adjusted for age and sex and, in the case of the joint analysis, included an indicator term for study. Logistic regression in Stata was used to test for association between LPIN1 SNPs and risk of hypertension and diabetes. χ2 analysis was performed to test for significant differences (P < 0.01) in call rate between cases and controls. The joint Ely and Hertfordshire cohort analysis of additional traits underlying metabolic syndrome was comprised of 189 tests, so the P value threshold adjusted for multiple testing using the Bonferroni correction is 0.0003. Fixed-effects meta-analysis using the inverse variance method was performed by using the “metan” command in Stata (23). Heterogeneity among studies was assessed using the Q statistic. IMPUTE software (http://www.stats.ox.ac.uk/∼marchini/software/gwas/impute.html) was used to impute genotypes for rs17603420 in the EPIC cohort. Plink (http://pngu.mgh.harvard.edu/∼purcell/plink/) was used to perform hapotype analysis (24), and Ely and Hertfordshire cohorts were meta-analyzed using METAL (http://www.sph.umich.edu/csg/abecasis/metal/index.html).
PCR and sequencing.
Genomic DNA from patients was randomly preamplified in a GenomiPhi reaction (GE Healthcare UK, Chalfont St. Giles, U.K.) before amplification with gene-specific primers (designed using Primer3 software [http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi]) covering all coding exons, splice junctions, and 3′ untranslated region (sequences and cycling conditions available on request). PCR was performed using standard conditions, and products were purified using exonuclease I and shrimp alkaline phosphatase (USB, Cleveland, OH). Bidirectional sequencing was performed using the Big Dye Terminator 3.1 kit (Applied Biosystems, Foster City, CA). Sequencing reactions were run on ABI3730 capillary machines (Applied Biosystems) and analyzed using Mutation Surveyor (version 2.20; SoftGenetics, State College, PA). All nonsynonymous variants with MAF <0.01 were confirmed in a second PCR and sequencing reaction using patient genomic DNA. DNA from family members used for cosegregation analysis was genomic. Only one amplicon within the 3′ untranslated region failed sequencing, and all others passed on >85% of samples (with an average pass rate of 95%). PANTHER was used to predict the functional impact of nonsynonymous mutations (http://www.pantherdb.org/tools/csnpScoreForm.jsp). Sequencing of preamplified CEPH samples was done in the same way.
Western blotting.
Patient fibroblast cells were maintained in Dulbecco's modified Eagle's supplemented with 10% fetal bovine serum and 2 mmol/l l-glutamine in a humidified 37°C incubator with 5% CO2. All cells were routinely assessed for and protected against Macroplasma pulmonis infection using VenorGeM Mycoplasma detection kit (VGM-025; Minerva Biolabs, CamBio) and BM-cyclin (799050; Roche) respectively. Fibroblasts were collected by trypsin EDTA release, washed with PBS, and then lysed in a 50 mmol/l HEPES pH7.4 buffer containing 150 mmol/l NaCl; 1% Triton X-100; 100 μmol/l 4-(2-aminoethyl)-benzenesulfonyl fluoride, hydrochloride; protease inhibitor cocktail 1; phosphatase inhibitor cocktail II; 1 μg/ml Dnase; and 4 mmol/l MgCl2 chilled to 4°C. Each sample was homogensized by passing through a 25-gauge needle 10 times. Insoluble debris was removed by a 16,000g centrifugation step at 4°C. Sample protein concentration was measured by a comparative Bradford protein assay. Samples were then suspended in 1× SDS sample buffer and boiled at 95°C for 5 min, loaded onto 7% SDS-PAGE, transferred onto nitrocellulose, and blocked in PBS with 1% TX-100 and 5% milk. These were then probed with protein-specific primary antibodies: lipin 1, lipin 2, anti-Mab414 (MMS-120P; Covance), anti-laminB (sc-6217; Santa-Cruz;). Lipin 1 and 2 polyclonal antibody production will be described elsewhere (N.G. and S.S., unpublished observations). This was followed by species-specific secondary antibodies coupled to horseradish peroxidase: anti-rabbit IgG (211-032-171; Jackson ImmunoResearch), anti-goat IgG (NB 710-H; Novus Biologicals), anti-mouse IgG (H & L) highly cross-adsorbed (A11029; Molecular Probes). Proteins were then detected using standard electrochemiluminescence techniques (Amersham ECL reagents).
Indirect immunofluorescence by confocal microscopy.
Fibroblasts were fixed with 3% formaldehyde, permiablized with 0.1% Triton X-100, and blocked with 1 mg/ml BSA in PBS. Each coverslip was labeled with primary mouse −αMab414 (nuclear pore marker) and secondary anti-mouse conjugated to fluorescein isothiocyanate (green), primary rabbit endoplasmic reticulum calcium-binding protein (αCalreticulin, no. 208910; Calbiochem), and secondary anti-rabbit conjugated to Alexa fluor 594 (red) (A11037; Molecular Probes). DNA was stained with DAPI (blue). Each slide was mounted onto glass slides and then visualized with a 63× or 100× Plan Apochromat objective lens (numerical aperture 1.4) on a Ziess Axiovert 200M inverted microscope with an LSM 510 confocal laser-scanning attachment.
RESULTS
Association studies of LPIN1 tag SNPs.
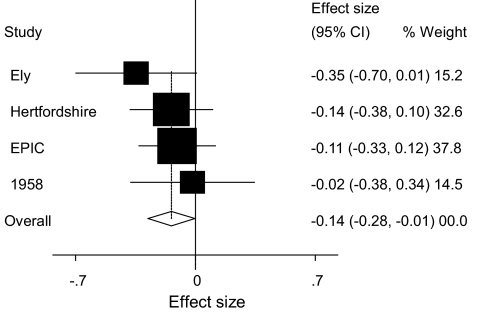
Twenty-one LPIN1 tag SNPs were successfully genotyped in two white U.K. population–based cohorts: the MRC Ely study (n = 1,709) and the Hertfordshire cohort study (n = 2,901). In the MRC Ely cohort, the minor allele of rs13412852 is nominally associated with lower fasting insulin levels (P = 0.041) and the minor allele of rs17603350 is nominally associated with higher BMI (P = 0.031), but these associations are not replicated in the Hertfordshire cohort (Table 1). Conversely, in the Hertfordshire cohort, and not in the MRC Ely cohort, rs17603420 and rs2577261 are nominally associated with BMI (P = 0.01 and P = 0.006, respectively) (Table 1). In a joint analysis of the pooled Ely and Hertfordshire cohorts (supplementary Table 3), no SNPs were associated with fasting insulin levels, but rs13412852, rs17603420 and rs2577261 were nominally associated with BMI (P ≤ 0.05). Two of these SNPs, rs13412852 and rs2577261, overlapped with SNPs on the Affymetrix 500k and Illumina 300k SNP chips, and rs17603420 could be imputed. Consequently, we were able to increase the power of our study to detect modest effects of these SNPs on BMI by performing meta-analyses with in-house data (EPIC-Obesity study; n = 2,415) and, in the case of rs13412852 and rs2577261, with data deposited by the Wellcome Trust Case Control Consortium and the Wellcome Trust Sanger Institute and published online from the British 1958 DNA collection (n = 1,479) (http://www.b58cgene.sgul.ac.uk/, accessed January 2008). SNPs rs2577261 and rs17603420 were not associated with BMI in the meta-analysis (P = 0.114 and 0.071, respectively). However, the association between rs13412852 and BMI remained marginally statistically significant (P = 0.042) in the meta-analysis (Fig. 1).
TABLE 1.
Mean fasting insulin levels and mean BMI of study participants by LPIN1 tagSNP genotype in the MRC Ely cohort and the Hertfordshire cohort studies
| SNP | Ely |
Hertfordshire | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Insulin (pmol/l) |
BMI (kg/m2) |
Insulin (pmol/l) |
BMI (kg/m2) |
|||||||||||||
| 0 | 1 | 2 | P | 0 | 1 | 2 | P | 0 | 1 | 2 | P | 0 | 1 | 2 | P | |
| rs893346 | 49.18 ± 1.02 | 47.65 ± 1.05 | 57.57 ± 1.08 | 0.609 | 27.23 ± 0.13 | 27.5 ± 0.39 | 26.3 ± 1.7 | 0.303 | 70.6 ± 1.01 | 72.33 ± 1.04 | 78.17 ± 1.2 | 0.486 | 27.36 ± 0.09 | 27.43 ± 0.24 | 26.14 ± 1.08 | 0.993 |
| rs4669778 | 49.99 ± 1.03 | 48.88 ± 1.02 | 47.23 ± 1.03 | 0.216 | 27.42 ± 0.24 | 27.41 ± 0.16 | 27.11 ± 0.27 | 0.109 | 71.28 ± 1.02 | 72.11 ± 1.02 | 69.06 ± 1.03 | 0.409 | 27.41 ± 0.16 | 27.37 ± 0.12 | 27.33 ± 0.16 | 0.719 |
| rs893345 | 48.17 ± 1.03 | 48.69 ± 1.02 | 49.76 ± 1.03 | 0.424 | 27.27 ± 0.28 | 27.75 ± 0.32 | 25.87 ± 1.57 | 0.967 | 71.15 ± 1.03 | 71.73 ± 1.02 | 69.14 ± 1.03 | 0.406 | 27.21 ± 0.17 | 27.38 ± 0.12 | 27.49 ± 0.17 | 0.234 |
| rs7595221 | 47.75 ± 1.03 | 50.08 ± 1.02 | 48.59 ± 1.04 | 0.55 | 27.11 ± 0.22 | 26.99 ± 0.24 | 26.09 ± 0.92 | 0.815 | 70.01 ± 1.02 | 71.45 ± 1.02 | 70.3 ± 1.03 | 0.708 | 27.34 ± 0.14 | 27.44 ± 0.12 | 27.23 ± 0.19 | 0.734 |
| Novel1 | 48.46 ± 1.02 | 49.79 ± 1.03 | 49.67 ± 1.08 | 0.409 | 27.15 ± 0.15 | 27.49 ± 0.21 | 27.01 ± 0.64 | 0.393 | 71.71 ± 1.02 | 70.63 ± 1.02 | 71.25 ± 1.06 | 0.575 | 27.34 ± 0.1 | 27.52 ± 0.16 | 27.72 ± 0.45 | 0.172 |
| rs16857866 | 49.1 ± 1.01 | 53.35 ± 1.08 | 47.7 ± 0 | 0.398 | 27.23 ± 0.12 | 28.02 ± 0.64 | 29.65 ± 0 | 0.215 | 70.96 ± 1.01 | 72.11 ± 1.06 | 0 ± 0 | 0.734 | 27.36 ± 0.09 | 27.8 ± 0.4 | 0 ± 0 | 0.306 |
| rs13412852 | 50.18 ± 1.02 | 48.63 ± 1.02 | 45.1 ± 1.05 | 0.042 | 27.42 ± 0.18 | 27.36 ± 0.18 | 26.44 ± 0.35 | 0.054 | 70.57 ± 1.02 | 71.63 ± 1.02 | 69.18 ± 1.04 | 0.89 | 27.47 ± 0.13 | 27.36 ± 0.12 | 27.15 ± 0.2 | 0.249 |
| rs2278513 | 47.41 ± 1.03 | 49.45 ± 1.02 | 48.82 ± 1.04 | 0.37 | 27.06 ± 0.22 | 27.28 ± 0.16 | 27.45 ± 0.26 | 0.467 | 69.47 ± 1.02 | 71.46 ± 1.02 | 72.34 ± 1.03 | 0.234 | 27.45 ± 0.14 | 27.32 ± 0.12 | 27.42 ± 0.2 | 0.753 |
| rs3795974 | 48.59 ± 1.02 | 49.57 ± 1.02 | 49.93 ± 1.04 | 0.441 | 27.21 ± 0.19 | 27.76 ± 0.96 | 27.86 ± 3.19 | 0.844 | 71.04 ± 1.02 | 70.16 ± 1.02 | 72.91 ± 1.04 | 0.602 | 27.31 ± 0.13 | 27.41 ± 0.13 | 27.42 ± 0.22 | 0.633 |
| rs33997857 | 48.95 ± 1.01 | 46.11 ± 1.1 | 56.18 ± 1.9 | 0.627 | 27.24 ± 0.12 | 27.7 ± 0.67 | 28.76 ± 0.57 | 0.401 | 70.82 ± 1.01 | 78.33 ± 1.07 | 73.83 ± 0 | 0.166 | 27.36 ± 0.08 | 27.91 ± 0.44 | 25.2 ± 0 | 0.259 |
| rs17603350 | 48.99 ± 1.02 | 47.89 ± 1.05 | 99.5 ± 1.37 | 0.986 | 27.34 ± 0.13 | 26.3 ± 0.36 | 27.52 ± 1.28 | 0.031 | 71.27 ± 1.01 | 67.78 ± 1.05 | 60.16 ± 2.08 | 0.282 | 27.4 ± 0.09 | 27.15 ± 0.36 | 30.9 ± 4.96 | 0.725 |
| rs17603420 | 50.63 ± 1.03 | 47.75 ± 1.02 | 49.14 ± 1.03 | 0.271 | 27.33 ± 0.2 | 27.39 ± 0.18 | 26.9 ± 0.25 | 0.269 | 70.43 ± 1.02 | 71.41 ± 1.02 | 70.26 ± 1.03 | 0.971 | 27.58 ± 0.16 | 27.42 ± 0.12 | 26.92 ± 0.17 | 0.01 |
| rs6729430 | 48.95 ± 1.02 | 45.77 ± 1.1 | 49.99 ± 1.47 | 0.61 | 27.24 ± 0.12 | 27.39 ± 0.34 | 29.79 ± 1.28 | 0.508 | 70.98 ± 1.01 | 71.68 ± 1.08 | 0 ± 0 | 0.849 | 27.34 ± 0.08 | 28.37 ± 0.53 | 0 ± 0 | 0.053 |
| rs2577264 | 48.61 ± 1.02 | 48.91 ± 1.02 | 50.6 ± 1.04 | 0.368 | 27.19 ± 0.2 | 27.37 ± 0.18 | 27.21 ± 0.3 | 0.733 | 71.27 ± 1.02 | 69.8 ± 1.02 | 73.51 ± 1.04 | 0.644 | 27.28 ± 0.13 | 27.39 ± 0.12 | 27.48 ± 0.23 | 0.462 |
| rs2577262 | 49.25 ± 1.02 | 48.96 ± 1.02 | 48.72 ± 1.05 | 0.639 | 27.24 ± 0.17 | 27.61 ± 0.28 | 27.99 ± 0.86 | 0.857 | 70.76 ± 1.02 | 71.58 ± 1.02 | 69.77 ± 1.04 | 0.95 | 27.47 ± 0.12 | 27.37 ± 0.13 | 26.98 ± 0.25 | 0.148 |
| rs2577261 | 49.52 ± 1.02 | 46.67 ± 1.03 | 48.09 ± 1.12 | 0.15 | 27.18 ± 0.12 | 27.38 ± 0.18 | 27.23 ± 0.3 | 0.256 | 71.08 ± 1.02 | 71.39 ± 1.03 | 73.5 ± 1.08 | 0.719 | 27.23 ± 0.09 | 27.67 ± 0.22 | 29.06 ± 0.72 | 0.006 |
| rs4669781 | 48.91 ± 1.02 | 49.56 ± 1.04 | 43.59 ± 1.21 | 0.844 | 27.22 ± 0.12 | 27.19 ± 0.18 | 27.25 ± 0.29 | 0.780 | 71.03 ± 1.01 | 69.44 ± 1.03 | 66.11 ± 1.11 | 0.426 | 27.41 ± 0.09 | 27.02 ± 0.24 | 26.81 ± 2.07 | 0.126 |
| rs2716609 | 48.99 ± 1.02 | 48.25 ± 1.05 | 44.85 ± 1.2 | 0.61 | 27.13 ± 0.15 | 27.34 ± 0.17 | 26.82 ± 0.24 | 0.073 | 71.19 ± 1.01 | 69.03 ± 1.04 | 78.39 ± 1.22 | 0.597 | 27.35 ± 0.1 | 27.42 ± 0.17 | 27.82 ± 0.66 | 0.465 |
| Novel2 | 48.11 ± 1.02 | 50.44 ± 1.04 | 52.4 ± 1.17 | 0.117 | 27.25 ± 0.12 | 27.25 ± 1.26 | 0 ± 0 | 0.987 | 71.65 ± 1.02 | 68.38 ± 1.03 | 76.59 ± 1.08 | 0.34 | 27.38 ± 0.08 | 25.71 ± 0.72 | 0 ± 0 | 0.103 |
| rs1050800 | 49.01 ± 1.01 | 47.15 ± 1.19 | 0 ± 0 | 0.728 | 27.21 ± 0.14 | 27.34 ± 0.23 | 26.55 ± 0.62 | 0.993 | 71.02 ± 1.01 | 69.3 ± 1.15 | 0 ± 0 | 0.848 | 27.4 ± 0.1 | 27.34 ± 0.16 | 27.14 ± 0.55 | 0.585 |
| rs2577256 | 48.81 ± 1.02 | 49.32 ± 1.03 | 49.09 ± 1.09 | 0.862 | 27.02 ± 0.24 | 27.41 ± 0.19 | 26.94 ± 0.38 | 0.208 | 70.48 ± 1.02 | 71.77 ± 1.02 | 74.09 ± 1.07 | 0.386 | 27.19 ± 0.16 | 27.42 ± 0.12 | 27.47 ± 0.18 | 0.219 |
Data are means ± SE. The P values indicate the results of a regression analysis assuming an additive model of gene action (nominally significant values, P < 0.05, are highlighted in bold). For fasting insulin, the analysis was performed on log-transformed data, and the table shows geometric means and SEs. 0, homozygous for the major allele (refer to Supplementary table 2); 1, heterozygous; and 2, homozygous for the minor allele.
FIG. 1.
Association between rs13412852 and BMI in individual studies and combined effect size (overall), P = 0.045. 1958, 1958 British Birth cohort; EPIC, EPIC Obesity cohort; Ely, MRC Ely study; and Hertfordshire, Hertfordshire cohort study.
In analyses of pooled Ely and Hertfordshire cohorts, a number of nominal associations (P < 0.05) were detected between LPIN1 tag SNPs and traits previously reported to be associated with LPIN1 variation (13). These data are presented in supplementary Tables 4 and 5. For SNPs overlapping with the Affymetrix 500k SNP chip, meta-analyses were performed on continuous traits with publicly available data from the Broad Institute (http://www.broad.mit.edu/diabetes/scandinavs/metatraits.html) (supplementary Table 4). For rs2577256, meta-analysis was performed with publicly available Wellcome Trust Case Control Consortium data to test for association with diabetes and hypertension (supplementary Table 5).
Mutation screening in the severe insulin resistance cohort.
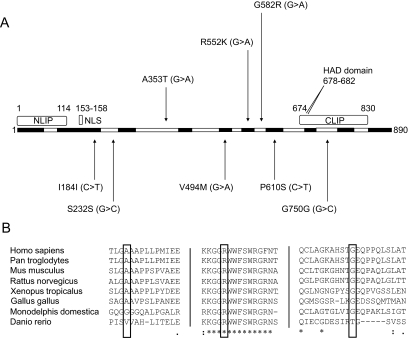
A total of 44 variants were detected in insulin-resistant or lipodystrophic patients (supplementary Table 1), eight of which were present in the coding sequence (Fig. 2A). Coding sequence variants that did not alter the amino acid sequence and/or that were also present in control subjects were considered unlikely to be pathogenic (shown below the schematic in Fig. 2A). This left three rare nonsynonymous variants detected to be heterozygous (shown above the schematic) that did not fall within any known functional domains within LPIN1.
FIG. 2.
A: Schematic of the lipin 1 protein showing exons in alternating black and white and known domains among lipin family proteins in gray. Arrows indicate the location of coding SNPs detected in LPIN1 by sequencing 23 patients with partial lipodystrophy and 135 patients with other syndromes of severe insulin resistance. Rare (MAF <1%) nonsynonymous mutations (above) were considered potentially pathogenic. CLIP (amino acids 674–830), COOH-terminal lipin domain, also referred to as the LNS2 (lipin/Ned1/Smp2) domain; HAD, haloacid dehalogenase domain; NLIP (amino acids 1–114), NH2-terminal lipin domain; and NLS (amino acids 153–158), nuclear localization signal. B: Multiple sequence alignments (using ClustalW) showing conservation of LPIN1 amino acids A353, R552, and G582. Straight lines indicate hidden sequence.
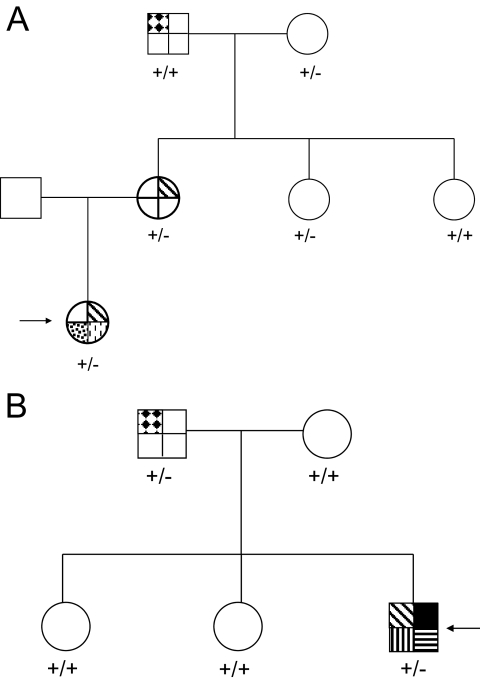
A353T was detected in a female patient with a Pakistani father and British white mother. She presented with clinical features of severe insulin resistance at 8 years old, which worsened with weight gain in the second decade, before improving dramatically with weight loss in adult life. She had no evidence of lipodystrophy. A353T was predicted to have no functional impact on the lipin 1 protein by PANTHER. DNA from the patient's mother, maternal aunts, and maternal grandparents was sequenced and demonstrated that the A353T variant did not segregate with the hallmarks of insulin resistance in the family (Fig. 3A). A353T was also genotyped in 1,064 participants of the HGDP-CEPH Human Genome Diversity Cell Line Panel (referred to below as the diversity panel) but was not detected.
FIG. 3.
A: A family pedigree demonstrating that the A353T mutation does not segregate with disease in a fully penetrant manner. The patient (indicated by the arrow) has hyperinsulinemia (diagonal stripes), hirsutism (spots), and acanthosis nigricans (dashes). The grandfather has diabetes (diamonds). There is no fasting insulin data for the grandmother or the youngest maternal aunt. ±, heterozygous genotype; +/+, wild-type genotype. B: A family pedigree demonstrating that the G582R mutation does not segregate with disease in a fully penetrant manner. The patient (indicated by the arrow) has hyperinsulinemia (diagonal stripes), severe peripheral neuropathy (black), previous bone marrow transplant for acute myeloid leukemia (horizontal stripes), and an intracerebral cavernous hemangioma (vertical stripes). His father has diabetes (diamonds). ±, heterozygous genotype; +/+, wild-type genotype.
R552K was detected in two unrelated white European female subjects but not in 1,064 control subjects from the diversity panel. The first proband presented with severe insulin resistance and femorogluteal lipodystrophy at 15 years old. The lipodystrophy progressed to become generalized in conjunction with the appearance of aggressive hemolytic anemia and autoimmune liver disease. Liver failure led to her death at 24 years old. The other proband was diagnosed with insulin-resistant diabetes at 32 years old and subsequently required in excess of 4 units/day exogenous insulin to maintain satisfactory glycemic control. She had no clinical evidence of lipodystrophy, and her BMI was sustained above 30 kg/m2. R552 is within a highly conserved tract (Fig. 2B), and mutation to lysine is predicted by PANTHER to have deleterious effects on lipin 1 function. Family DNA was not available for cosegregation analysis for either patient.
G582R was identified in a white European male with a complex syndrome. This included severe insulin resistance and severe early-onset sensorimotor neuropathy that confined him to a wheelchair, a combination reminiscent of lipodystrophy/insulin resistance and neuropathy in the fld mouse. This patient also underwent allogeneic bone marrow transplantation in childhood for acute lymphoblastic leukemia and had a cerebral cavernous hemangioma. All genomic analyses were undertaken on DNA extracted from cultured skin fibroblasts. G582 is a well-conserved residue within the protein (Fig. 2B), and mutation to arginine is predicted by PANTHER to have deleterious effects on lipin 1 function. Cosegregation analysis was performed using DNA from first-degree relatives of the patient (Fig. 3B). The father also carried the variant, but although he was diagnosed with diabetes at age 69 years, he had no peripheral neuropathy or clinical or biochemical evidence of insulin resistance/lipodystrophy. Subsequently, the G582R variant was genotyped in the Diversity Panel and detected in a Bedouin control subject from Nedev, Israel.
In summary, we identified three rare LPIN1 missense variants in a cohort of insulin-resistant patients. G582R and R552K are predicted to be deleterious by PANTHER, and the proband carrying the G582R variant had a syndrome reminiscent of the fld mouse. Thus, despite the failure of this variant to cosegregate with disease in the kindred, and despite the absence of available family members from the R552K kindred, we elected to investigate the possibility of impaired lipin 1 function in primary skin fibroblasts from the probands. As A353T was predicted to be benign, it did not segregate with disease in the family, and as no fibroblasts were available, this variant was not investigated further.
Assessing the functional impact of LPIN1 mutations.
To investigate the impact of R552K and G582R mutations on lipin 1 protein levels and phosphorylation status, total cell extracts from patient fibroblasts were probed with lipin 1, lipin 2, a nuclear pore marker (Mab414), and lamin B-specific primary antibodies to detect protein levels (supplementary Fig. 2A). The resulting Western blot, shown in supplementary Fig. 2A, shows similar intensities of all four proteins in patient fibroblasts compared with those in control cells. Immunocytochemistry was employed to detect changes in nuclear membrane morphology by staining a nuclear pore marker (supplementary Fig. 2B). There was no discernable difference in morphology between patient and control fibroblasts.
DISCUSSION
In this study, we performed a comprehensive analysis of LPIN1 variants and their effects on metabolic quantitative traits and syndromes of insulin resistance (including lipodystrophy). Analysis of LPIN1 common variation (MAF >0.01) in two U.K. population-based cohorts (n = 4,610) revealed nominal significant associations with BMI, and rs13412582 remained marginally associated with BMI (P = 0.042) in a meta-analysis of U.K. population-based samples from in-house and publicly available genome-wide studies (n = 8,504). We also detected nominal associations between our tag SNPs and metabolic traits previously reported to be associated with LPIN1 variation (13). Sequencing of 23 patients with lipodystrophy and 135 patients with syndromes of insulin resistance revealed that mutations in LPIN1 are not a common cause of these diseases in humans.
To our knowledge, neither rs13412582 nor any highly correlated SNPs have been tested in other association studies published to date. Further replication in larger cohorts will be required to confirm the association between rs13412582 and BMI.
Seven of our 21 tag SNPs were directly genotyped in at least one of the other association studies (12–14). No analyses, including our own, found an association between rs4669781, rs1050800, and rs2577256 and insulin levels and measures of adiposity. Results for the other four SNPs are inconsistent between studies. For example, rs2716610 was associated with BMI in lean Finnish men (12) and with quantitative measures of adiposity in French-Canadian families in the Quebec Family Study (14). Here, the highly correlated SNP rs2716609 (r2 = 1.0 in HapMap trios) was associated with skinfolds and waist circumference, and BMI showed the same trend. Given our sample size of 4,130 individuals with full rs2716609 genotype and BMI data, we had >80% power to detect the effect size observed in the Quebec Family study. Nevertheless, we did not replicate the association between rs2716609 and BMI or waist circumference in the Ely and Hertfordshire cohorts (Table 1). Our results agree with the MONICA study Augsburg (n = 1,416), a German population–based cohort, which found no association between rs2716610 and BMI in men or women (13).
Two other SNPs, rs893346 and rs2577262, were associated with BMI in lean Finnish men (12) but showed no statistical association with BMI in 1,873 lean men from the Ely and Hertfordshire cohort studies (P = 0.631 and 0.253, respectively). Similarly, rs2278513 and rs2577262 were associated with BMI in obese Finnish men but not in obese men from the U.K. (P = 0.780 and 0.676, respectively). Our data agree with the results of the MONICA study, which showed no association of rs893346 and two SNPs highly correlated with rs2577262 in HapMap CEU trios (r2 = 1.0 and 0.96 for rs6744682 and rs6708316, respectively) with BMI in men (13).
The MONICA study reported strong associations between haplotypes of rs33997857, rs6744682, and rs6708316 with hypertension-, obesity-, or diabetes-related traits (13). Several of the traits were also statistically associated with the same haplotypes in a replication study, but the effect was always in the opposite direction compared with the original cohort. To attempt replication of the MONICA study data, we tested haplotypes of rs33997857 and rs2577262 (highly correlated with rs6744682 [r2 = 1.0] and rs6708316 [r2 = 0.96] in HapMap CEU trios) against metabolic traits in the Ely and Hertfordshire cohorts but only found nominal associations with hypertension (supplementary Table 6). We did detect a number of nominal associations between these traits and other SNPs in our study (supplementary Tables 4 and 5). However, none of these reached statistical significance after adjustment of the P value threshold for multiple testing using the Bonferroni correction, and all require further replication.
There are several possible reasons why we could not replicate previously reported associations between LPIN1 variants and metabolic quantitative traits. Firstly, we may have reported false-negative results. However, where effect sizes were reported in previously published studies, we were able to calculate that our study had >80% power to detect them. Secondly, previous studies might have reported false-positive results. In particular, as a consequence of multiple testing, detection of false-positive associations becomes more likely when analyses are performed in subsets of samples and on many traits. Furthermore, one has to expect false-positive findings among previously reported disease associations given the low prior probability of detecting a true association with a complex trait (25). Alternatively, the discrepancy in results between studies may be due to genetic and/or environmental differences between the populations genotyped. For example, the degree of linkage disequilibrium between LPIN1 tag SNPs and the putative unmeasured true functional variant(s) may vary between the cohorts. Also, LPIN1 SNPs may interact with other genetic and/or environmental risk factors in different studies.
In the fld mouse model, Lpin1-null mutations cause lipodystrophy, insulin resistance, and peripheral neuropathy (1). However, of the three rare (MAF <0.01) nonsynonymous LPIN1 variants detected within our cohort of patients with syndromes of severe insulin resistance, none are likely to be pathogenic in isolation in heterozygous form; family cosegregation analysis showed that A353T and G582R did not segregate with disease in a fully penetrant manner, and G582R was also detected in one Bedouin control.
Western blotting of patient fibroblasts showed that G582R and R552K had no discernable impact on lipin 1 protein levels. Proteins orthologous to lipin 1 in yeast are proposed to be involved in nuclear membrane growth and morphology (26–28). However, staining of a nuclear pore marker in patient fibroblasts with R552K and G582R variants revealed no abnormalities in membrane morphology compared with control fibroblasts.
To date, our study and a previously published work (11), including 23 and 15 patients with lipodystophy screened, respectively, have demonstrated that LPIN1 coding mutations are unlikely to be a common cause of human lipodystrophy. However, we cannot rule out the possibility that LPIN1 mutations interact with other genetic defects to cause disease or that they are rarer causes of these disorders. The methods used to screen for mutations would not have detected copy number variations affecting large regions or mutations affecting regulatory regions; therefore, we cannot exclude these types of LPIN1 variations as causes of human lipodystrophy and insulin resistance. Furthermore, the in vitro assays used to assess the functional impact of LPIN1 nonsynonymous variants might have missed some functional effects, such as phosphatidic acid phosphatase activity.
We conclude that LPIN1 coding variants are not a common cause of lipodystrophy and severe insulin resistance in humans and that polymorphisms in LPIN1 are unlikely to importantly contribute to insulin sensitivity and waist circumference in U.K. populations. Nominal associations between LPIN1 variants and BMI, blood pressure, cholesterol, triglycerides, A1C, and risk of hypertension need replicating in larger cohorts.
Supplementary Material
Acknowledgments
We acknowledge use of genotype data from the British 1958 Birth Cohort DNA collection, funded by Medical Research Council Grant G0000934 and Wellcome Trust Grant 068545/Z/02. We also acknowledge use of genotype data from the Wellcome Trust Case Control Consortium and the Diabetes Genetics Initiative. K.F., E.W., A.D., and I.B. are funded by the Wellcome Trust. I.B. and E.W. also acknowledge support from EU FP6 funding (contract no. LSHM-CT-2003-503041). N.G. and S.S. are supported by a Wellcome Trust Career development fellowship. R.S., M.S., and S.O. are grateful for the support of the Wellcome Trust (Intermediate Clinical Fellowship 080952/Z/06/Z and Programme Grant 078986/Z/06/Z to R.S.) and the U.K. National Institute for Health Research Cambridge Biomedical Research Centre.
We thank Susannah Bumpstead and Andrew Keniry, members of the genotyping facility within the Genetics of Complex Traits in Humans team at the Wellcome Trust Sanger Institute, for genotyping LPIN1 variants in the Ely and Hertfordshire cohorts and the HGDP CEPH panel.
Published ahead of print at http://diabetes.diabetesjournals.org on 30 June 2008.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Peterfy M, Phan J, Xu P, Reue K: Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat Genet 27 :121 –124,2001 [DOI] [PubMed] [Google Scholar]
- 2.Phan J, Reue K: Lipin, a lipodystrophy and obesity gene. Cell Metab 1 :73 –83,2005 [DOI] [PubMed] [Google Scholar]
- 3.Croce MA, Eagon JC, LaRiviere LL, Korenblat KM, Klein S, Finck BN: Hepatic lipin 1β expression is diminished in insulin-resistant obese subjects and is reactivated by marked weight loss. Diabetes 56 :2395 –2399,2007 [DOI] [PubMed] [Google Scholar]
- 4.Donkor J, Sparks LM, Xie H, Smith SR, Reue K: Adipose tissue lipin-1 expression is correlated with peroxisome proliferator-activated receptor alpha gene expression and insulin sensitivity in healthy young men. J Clin Endocrinol Metab 93 :233 –239,2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindegaard B, Larsen LF, Hansen AB, Gerstoft J, Pedersen BK, Reue K: Adipose tissue lipin expression levels distinguish HIV patients with and without lipodystrophy. Int J Obes (Lond )31 :449 –456,2007 [DOI] [PubMed] [Google Scholar]
- 6.Yao-Borengasser A, Rasouli N, Varma V, Miles LM, Phanavanh B, Starks TN, Phan J, Spencer HJ 3rd, McGehee RE Jr, Reue K, Kern PA: Lipin expression is attenuated in adipose tissue of insulin-resistant human subjects and increases with peroxisome proliferator–activated receptor γ activation. Diabetes 55 :2811 –2818,2006 [DOI] [PubMed] [Google Scholar]
- 7.Han GS, Wu WI, Carman GM: The Saccharomyces cerevisiae lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J Biol Chem 281 :9210 –9218,2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finck BN, Gropler MC, Chen Z, Leone TC, Croce MA, Harris TE, Lawrence JC Jr, Kelly DP: Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell Metab 4 :199 –210,2006 [DOI] [PubMed] [Google Scholar]
- 9.Phan J, Peterfy M, Reue K: Lipin expression preceding peroxisome proliferator-activated receptor-gamma is critical for adipogenesis in vivo and in vitro. J Biol Chem 279 :29558 –29564,2004 [DOI] [PubMed] [Google Scholar]
- 10.Xu J, Lee WN, Phan J, Saad MF, Reue K, Kurland IJ: Lipin deficiency impairs diurnal metabolic fuel switching. Diabetes 55 :3429 –3438,2006 [DOI] [PubMed] [Google Scholar]
- 11.Cao H, Hegele RA: Identification of single-nucleotide polymorphisms in the human LPIN1 gene. J Hum Genet 47 :370 –372,2002 [DOI] [PubMed] [Google Scholar]
- 12.Suviolahti E, Reue K, Cantor RM, Phan J, Gentile M, Naukkarinen J, Soro-Paavonen A, Oksanen L, Kaprio J, Rissanen A, Salomaa V, Kontula K, Taskinen MR, Pajukanta P, Peltonen L: Cross-species analyses implicate lipin 1 involvement in human glucose metabolism. Hum Mol Genet 15 :377 –386,2006 [DOI] [PubMed] [Google Scholar]
- 13.Wiedmann S, Fischer M, Koehler M, Neureuther K, Riegger G, Doering A, Schunkert H, Hengstenberg C, Baessler A: Genetic variants within the LPIN1 gene, encoding lipin, are influencing phenotypes of the metabolic syndrome in humans. Diabetes 57 :209 –217,2007 [DOI] [PubMed] [Google Scholar]
- 14.Loos RJF, Rankinen T, Pérusse L, Tremblay A, Després J-P, Bouchard C: Association of lipin 1 gene polymorphisms with measures of energy and glucose metabolism. Obesity 15 :2723 –2732,2007 [DOI] [PubMed] [Google Scholar]
- 15.Wareham NJ, Hennings SJ, Byrne CD, Hales CN, Prentice AM, Day NE: A quantitative analysis of the relationship between habitual energy expenditure, fitness and the metabolic cardiovascular syndrome. Br J Nutr 80 :235 –241,1998 [DOI] [PubMed] [Google Scholar]
- 16.Ekelund U, Franks PW, Sharp S, Brage S, Wareham NJ: Increase in physical activity energy expenditure is associated with reduced metabolic risk independent of change in fatness and fitness. Diabetes Care 30 :2101 –2106,2007 [DOI] [PubMed] [Google Scholar]
- 17.Syddall HE, Aihie Sayer A, Dennison EM, Martin HJ, Barker DJ, Cooper C: Cohort profile: the Hertfordshire cohort study. Int J Epidemiol 34 :1234 –1242,2005 [DOI] [PubMed] [Google Scholar]
- 18.Day N, Oakes S, Luben R, Khaw KT, Bingham S, Welch A, Wareham N: EPIC-Norfolk: study design and characteristics of the cohort. European Prospective Investigation of Cancer. Br J Cancer 80 (Suppl 1):95 –103,1999 [PubMed] [Google Scholar]
- 19.Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, Piouffre L, Bodmer J, Bodmer WF, Bonne-Tamir B, Cambon-Thomsen A, Chen Z, Chu J, Carcassi C, Contu L, Du R, Excoffier L, Ferrara GB, Friedlaender JS, Groot H, Gurwitz D, Jenkins T, Herrera RJ, Huang X, Kidd J, Kidd KK, Langaney A, Lin AA, Mehdi SQ, Parham P, Piazza A, Pistillo MP, Qian Y, Shu Q, Xu J, Zhu S, Weber JL, Greely HT, Feldman MW, Thomas G, Dausset J, Cavalli-Sforza LL: A human genome diversity cell line panel. Science 296 :261 –262,2002 [DOI] [PubMed] [Google Scholar]
- 20.Dausset J, Cann H, Cohen D, Lathrop M, Lalouel JM, White R: Centre d'etude du polymorphisme humain (CEPH): collaborative genetic mapping of the human genome. Genomics 6 :575 –577,1990 [DOI] [PubMed] [Google Scholar]
- 21.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D: Efficiency and power in genetic association studies. Nat Genet 37 :1217 –1223,2005 [DOI] [PubMed] [Google Scholar]
- 22.Barrett JC, Fry B, Maller J, Daly MJ: Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21 :263 –265,2005 [DOI] [PubMed] [Google Scholar]
- 23.Bradburn MJ, Deeks JJ, Altman DG: Metan: an alternative meta-analysis command. Stata Technical Bulletin Reprints 8 :86 –100,1999 [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC: PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81 :559 –575,2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N: Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst 96 :434 –442,2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santos-Rosa H, Leung J, Grimsey N, Peak-Chew S, Siniossoglou S: The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. Embo J 24 :1931 –1941,2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siniossoglou S, Santos-Rosa H, Rappsilber J, Mann M, Hurt E: A novel complex of membrane proteins required for formation of a spherical nucleus. Embo J 17 :6449 –6464,1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tange Y, Hirata A, Niwa O: An evolutionarily conserved fission yeast protein, Ned1, implicated in normal nuclear morphology and chromosome stability, interacts with Dis3, Pim1/RCC1 and an essential nucleoporin. J Cell Sci 115 :4375 –4385,2002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.