Abstract
OBJECTIVE— We evaluated the impact on diabetes-related intermediary traits of common novel type 2 diabetes–associated variants in the JAZF1 (rs864745), CDC123/CAMK1D (rs12779790), TSPAN8 (rs7961581), THADA (rs7578597), ADAMTS9 (rs4607103), and NOTCH2 (rs10923931) loci, which were recently identified by meta-analysis of genome-wide association data.
RESEARCH DESIGN AND METHODS— We genotyped the six variants in 4,516 middle-aged glucose-tolerant individuals of the population-based Inter99 cohort who were all characterized by an oral glucose tolerance test (OGTT).
RESULTS— Homozygous carriers of the minor diabetes risk G-allele of the CDC123/CAMK1D rs12779790 showed an 18% decrease in insulinogenic index (95% CI 10–27%; P = 4 × 10−5), an 18% decrease in corrected insulin response (CIR) (8.1–29%; P = 4 × 10−4), and a 13% decrease in the ratio of area under the serum-insulin and plasma-glucose curves during an OGTT (AUC-insulin/AUC-glucose) (5.8–20%; P = 4 × 10−4). Carriers of the diabetes-associated T-allele of JAZF1 rs864745 had an allele-dependent 3% decrease in BIGTT-AIR (0.9–4.3%; P = 0.003). Furthermore, the diabetes-associated C-allele of TSPAN8 rs7961581 associated with decreased levels of CIR (4.5% [0.5–8.4]; P = 0.03), of AUC-insulin/AUC-glucose ratio (3.9% [1.2–6.7]; P = 0.005), and of the insulinogenic index (5.2% [1.9–8.6]; P = 0.002). No association with traits of insulin release or insulin action was observed for the THADA, ADAMTS9, or NOTCH2 variants.
CONCLUSIONS— If replicated, our data suggest that type 2 diabetes at-risk alleles in the JAZF1, CDC123/CAMK1D, and TSPAN8 loci associate with various OGTT-based surrogate measures of insulin release, emphasizing the contribution of abnormal pancreatic β-cell function in the pathogenesis of type 2 diabetes.
Recent discoveries using genome-wide association (GWA) studies have led to progression in the understanding of the molecular genetic background of type 2 diabetes, dramatically increasing the number of common validated type 2 diabetes loci with modest impact on relative diabetes risk (1–5). The Diabetes Genetics Replication and Meta-analysis (DIAGRAM) consortium recently reported the outcome of a meta-analysis of data from three GWA studies. Six additional type 2 diabetes loci reaching genome-wide significance levels were identified in the JAZF1, CDC123/CAMK1D, TSPAN8, THADA, ADAMTS9, and NOTCH2 loci; all were modestly affecting disease risk with odds ratios between 1.09 and 1.15 (6).
As for most other findings obtained from GWA studies, little is known about the function of the putative regional candidate genes thought to be affected by the at-risk variants. Recent studies have, however, shown that many validated type 2 diabetes risk variants confer an impaired pancreatic β-cell function, which seems to be the case for risk alleles in the CDKAL1, SLC30A8, HHEX/IDE, CDKN2A/2B, IGF2BP2, TCF7L2, and KCNJ11 loci (2,7–9). Indeed, only the PPARG Pro12Ala variant has so far displayed a diabetogenic potential through affecting peripheral insulin sensitivity (10) and variants in FTO by increasing fat accumulation (11). Of the six novel type 2 diabetes loci (6), the biological function of NOTCH2 points to an impact on pancreatic β-cell function because of its critical role in fetal pancreatic development (12), yet little or no prior implication in the pathogenesis of type 2 diabetes or diabetes-related phenotypes can be claimed for genes in the JAZF1, CDC123/CAMK1D, TSPAN8, THADA, or ADAMTS9 regions.
Given the sparse knowledge of the biological functions of the six novel type 2 diabetes–associated variants, we have characterized the influence of these variants on quantitative surrogate measures of oral glucose-stimulated insulin release, insulin sensitivity, and body fat accumulation in a population-based study of glucose-tolerant middle-aged Danes who all had undertaken an oral glucose tolerance test (OGTT).
RESEARCH DESIGN AND METHODS
Studies of quantitative metabolic traits were performed in the Inter99 cohort, which is a population-based, randomized, nonpharmacological intervention study of 6,784 middle-aged subjects for the prevention of ischemic heart disease, conducted at the Research Centre for Prevention and Health in Glostrup, Copenhagen (ClinicalTrials.gov ID-no: NCT00289237) (13). An OGTT was performed in all participants with measurements of plasma glucose and serum insulin at fasting and at 30 and 120 min, and 6,083 subjects with available DNA were subsequently classified as individuals with normal glucose tolerance (NGT) (n = 4,516), impaired fasting glycemia (n = 503), impaired glucose tolerance (n = 692), screen-detected and treatment-naïve type 2 diabetes (n = 253), or previously diagnosed type 2 diabetes (n = 119). In the analysis of quantitative diabetes-related phenotypes, we included 4,516 subjects with NGT (2,101 men/2,415 women, age 45.2 ± 7.9 years and BMI 25.5 ± 4.1 kg/m2 [mean ± SD]). Type 2 diabetes was diagnosed according to World Health Organization 1999 criteria.
Informed written consent was obtained from all participants. The study was conducted in accordance with the Declaration of Helsinki II and was approved by the local ethics committee of Copenhagen.
Biochemical and anthropometrical measures.
Height and weight were measured in light indoor clothing and without shoes. Waist circumference was measured in the upright position midway between the iliac crest and the lower costal margin. Blood samples were drawn after a 12-h overnight fast. Plasma glucose was analyzed by a glucose oxidase method (Granutest; Merck, Darmstadt, Germany). Serum insulin [excluding des (31,32)] and intact proinsulin) was measured using the AutoDELFIA insulin kit (Perkin-Elmer, Wallac, Turku, Finland).
Indexes of insulin release and insulin sensitivity.
Oral glucose-stimulated insulin release was reported as the insulinogenic index, the corrected insulin response (CIR), the ratio of the area under the curve (AUC) of insulin to the AUC of glucose during the OGTT (AUC-insulin/AUC-glucose), and the BIGTT-acute insulin response (AIR) index. The insulinogenic index was calculated as follows: (serum insulin30 min − serum insulin0 min [pmol/l])/plasma glucose30 min [mmol/l]. CIR was calculated as follows: 100 × serum insulin30 min/[plasma glucose30 min × (plasma glucose30 min − 3.89)] (14). Indexes of insulin sensitivity were reported as the insulin sensitivity index (ISI), calculated as the reciprocal of homeostasis model assessment of insulin resistance {22.5/[plasma glucose0 min (mmol/l) × serum insulin0 min (pmol/l)]} (15), and as the OGTT-derived BIGTT-SI. The BIGTT indexes apply information on sex and BMI combined with plasma glucose and serum insulin during an OGTT to provide indexes for AIR and SI that are highly correlated with indexes obtained during an intravenous glucose tolerance test and were calculated as reported (16). To construct OGTT-based disposition indexes, we multiplied the CIR index with ISI, since these measures are not intrinsically interdependent. Furthermore, we multiplied the BIGTT-SI index with the BIGTT-AIR index.
Genotyping.
The six gene variants (rs864745, rs12779790, rs7961581, rs7578597, rs4607103, and rs10923931) were genotyped by TaqMan allelic discrimination (KBiosciences, Hoddesdon, U.K). All genotyping success rates were above 96% and all mismatch rates were below 1% in 1,090 duplicate samples. The distributions of genotypes for all variants were in the Hardy-Weinberg equilibrium (all P > 0.05).
Statistical analysis.
A general linear statistical methodology was used to test quantitative traits in relation to genotype, applying additive, dominant, and recessive models while adjusting for the effect of age (BIGTT-SI and BIGTT-AIR), age and sex (BMI and waist), or age, sex, and BMI (all other traits). BMI and all values of plasma glucose and serum insulin and derived indexes of insulin release and insulin sensitivity were logarithmically transformed before analysis. In the main text, parameter estimates (95% CI) of associated quantitative traits are given, while data in tables are unadjusted medians or means. The multivariate method, Hotelling's T2, was applied to test the simultaneous effect of genotype on insulin release and insulin sensitivity. A P value of <0.05 was considered significant. All analyses were performed using RGui, version 2.6.1 (http://www.r-project.org).
Estimation of statistical power.
Statistical power for the quantitative traits was estimated using simulations. We assumed an additive genetic model for both the simulation of the data and for testing the data using a linear model. We used the empirical variance of the observed traits to simulate phenotypes from a normal distribution so that variance across genotypes is drawn from the estimated variance. Because we also include adjustment factors in our analysis, we estimated the variance from the residuals of a linear model containing the adjustment factors. Thus, we assume that the genotype and the adjustment factors are independent. The power was estimated using 5,000 simulations and a significance threshold of 0.05. Based on the allele frequencies of the six examined gene variants and a sample size of 4,516 subjects, we estimated the effect sizes per allele of quantitative traits for which we had 80 and 90% statistical power, respectively, to detect an association. Depending on allele frequency (range 9.5–48.0%) and assuming an additive model, we had 80% power to detect an allele-dependent difference of 0.8–1.4% in BMI, 2.2–3.8% in BIGTT-AIR, 3.2–5.4% in insulinogenic index, and 3.0–5.0% in ISI. Similarly, we had 90% statistical power to detect a 1.0–1.7% change per allele in BMI, 2.6–4.3% in BIGTT-AIR, 3.7–6.2% in insulinogenic index, and 3.4–5.9% in ISI, respectively.
RESULTS
We investigated the JAZF1 rs864745, CDC123/CAMK1D rs12779790, TSPAN8 rs7961581, THADA rs7578597, ADAMTS9 rs4607103, and NOTCH2 rs10923931 variants for association with type 2 diabetes–related quantitative traits in a population-based sample of 4,516 glucose-tolerant subjects. Assuming an additive genetic model, carriers of the major diabetes-associated T-allele of JAZF1 rs864745 had a 0.21 kg/m2 decreased BMI (0.048–0.39 kg/m2; P = 0.02), a 0.47 cm decreased waist circumference (0.03–0.90 cm; P = 0.04), and a 2.6% (0.9–4.3%; P = 0.003) decreased insulin release per allele as assessed by the BIGTT-AIR index. The variant did not associate with other measures of insulin release (Table 1). Homozygous carriers of the minor diabetes risk G-allele of the CDC123/CAMK1D rs12779790 showed a 15% decreased serum insulin at 30 min during an OGTT (7.8–23%, P = 8 × 10−5), an 18% decreased insulinogenic index (10–27%; P = 4 × 10−5), an 18% decreased CIR (8.1–29%; P = 4 × 10−4), and a 13% decreased AUC-insulin/AUC-glucose (5.8–20%; P = 4 × 10−4) (Table 2). When applying a dominant genetic model, the minor diabetes risk C-allele of the TSPAN8 rs7961581 associated with a modest decrease in serum insulin at 30 min during OGTT (4.9% [1.9–7.9]; P = 0.001), a decrease in CIR (4.5% [0.5–8.4]; P = 0.03), a decrease in AUC-insulin/AUC-glucose (3.9% [1.2–6.7]; P = 0.005), and a decrease in insulinogenic index (5.2% [1.9–8.6]; P = 0.002) (Table 3).
TABLE 1.
Unadjusted quantitative metabolic traits in the population-based Inter99 cohort including 4,377 middle-aged subjects with normal glucose tolerance stratified according to genotype of JAZF1 rs864745
| 11 (CC) | 12 (CT) | 22 (TT) | PADDITIVE | P22 + 12 VS. 11 | P22 VS. 12 + 11 | |
|---|---|---|---|---|---|---|
| n (men/women) | 996 (453/543) | 2,238 (1,056/1,182) | 1,143 (513/630) | |||
| Age (years) | 45.5 ± 8 | 45.2 ± 7.8 | 45.1 ± 7.7 | |||
| BMI (kg/m2) | 25.7 ± 4.3 | 25.6 ± 4.1 | 25.2 ± 3.9 | 0.02 | 0.2 | 0.008 |
| Waist (cm) | 84 ± 13 | 84 ± 12 | 83 ± 12 | 0.04 | 0.2 | 0.03 |
| Fasting serum insulin (pmol/l) | 33 (23–48) | 32 (23–46) | 31 (22–44) | 0.2 | 0.1 | 0.5 |
| Serum insulin at 30 min (pmol/l) | 246 (180–359) | 250 (182–347) | 235 (168–341) | 0.3 | 0.6 | 0.2 |
| Serum insulin at 120 min (pmol/l) | 142 (92–219) | 141 (87–212) | 134 (87–209) | 0.7 | 0.6 | 0.8 |
| Fasting plasma glucose (mmol/l) | 5.3 (5.0–5.6) | 5.3 (5.1–5.6) | 5.3 (5.1–5.6) | 0.08 | 0.2 | 0.1 |
| Plasma glucose at 30 min (mmol/l) | 8.2 (7.2–9.1) | 8.1 (7.2–9.2) | 8.2 (7.2–9.2) | 0.7 | 0.8 | 0.7 |
| Plasma glucose at 120 min (mmol/l) | 5.7 (4.9–6.4) | 5.6 (4.7–6.4) | 5.6 (4.8–6.3) | 0.9 | 0.6 | 0.4 |
| ISI | 0.13 (0.09–0.18) | 0.13 (0.09–0.19) | 0.14 (0.09–0.19) | 0.3 | 0.2 | 0.7 |
| BIGTT-SI | 10.2 ± 3.8 | 10.3 ± 3.6 | 10.5 ± 3.7 | 0.06 | 0.3 | 0.06 |
| AUC-insulin/AUC-glucose | 28.0 (20.8–38.3) | 27.6 (20.8–37.8) | 26.5 (19.3–36.9) | 0.2 | 0.5 | 0.2 |
| CIR | 760 (477–1,220) | 749 (487–1,210) | 747 (462–1,150) | 0.4 | 0.5 | 0.4 |
| Insulinogenic index | 26.1 (18.1–39.1) | 26.3 (18.5–38) | 25.5 (17–37) | 0.4 | 0.8 | 0.3 |
| BIGTT-AIR | 1,700 (1,370–2,150) | 1,690 (1,350–2,120) | 1,610 (1,320–2,060) | 0.003 | 0.03 | 0.007 |
Data are medians (25% to 75% range) or means ± SD (BMI, waist, and BIGTT-SI). Values of BMI, plasma glucose, serum insulin, and derived indices were logarithmically transformed before statistical analysis. Calculated P values were adjusted for age (BIGTT-SI and BIGTT-AIR), age and sex (BMI and waist), or age, sex, and BMI (all other traits), assuming an additive, dominant, or recessive model. Indices of insulin release and insulin sensitivity were calculated as described in research design and methods. 1, type 2 diabetes–protective allele; 2, diabetes-associated allele.
TABLE 2.
Unadjusted quantitative metabolic traits in the population-based Inter99 cohort including 4,395 middle-aged subjects with normal glucose tolerance stratified according to genotype of CDC123/CAMK1D rs12779790
| 11 (AA) | 12 (AG) | 22 (GG) | PADDITIVE | P22 + 12 VS. 11 | P22 VS. 12 + 11 | |
|---|---|---|---|---|---|---|
| n (men/women) | 2,859 (1,324/1,535) | 1,365 (620/745) | 171 (88/83) | |||
| Age (years) | 45.2 ± 7.8 | 45.3 ± 7.9 | 45.2 ± 8.1 | |||
| BMI (kg/m2) | 25.5 ± 4.1 | 25.5 ± 4.0 | 25.8 ± 4.6 | 0.8 | 0.9 | 0.5 |
| Waist (cm) | 84 ± 12 | 84 ± 12 | 85 ± 12 | 0.5 | 0.5 | 0.6 |
| Fasting serum insulin (pmol/l) | 32 (23–46) | 32 (23–47) | 31 (21–49) | 0.7 | 0.7 | 0.06 |
| Serum insulin at 30 min (pmol/l) | 246 (178–351) | 246 (180–347) | 217 (159–299) | 0.02 | 0.3 | 8 × 10−5 |
| Serum insulin at 120 min (pmol/l) | 138 (87–212) | 141 (92–216) | 139 (80–190) | 0.4 | 0.1 | 0.2 |
| Fasting plasma glucose (mmol/l) | 5.3 (5.0–5.6) | 5.4 (5.1–5.6) | 5.3 (5.1–5.6) | 0.1 | 0.07 | 1 |
| Plasma glucose at 30 min (mmol/l) | 8.2 (7.2–9.2) | 8.2 (7.2–9.1) | 8.2 (7.4–9.3) | 1 | 0.9 | 0.7 |
| Plasma glucose at 120 min (mmol/l) | 5.6 (4.7–6.3) | 5.7 (4.9–6.4) | 5.8 (4.8–6.4) | 0.01 | 0.01 | 0.3 |
| ISI | 0.132 (0.09–0.189) | 0.131 (0.09–0.188) | 0.131 (0.084–0.201) | 0.9 | 0.6 | 0.08 |
| BIGTT-SI | 10.4 ± 3.7 | 10.2 ± 3.7 | 10.4 ± 3.7 | 0.4 | 0.3 | 0.8 |
| AUC-insulin/AUC-glucose | 27.6 (20.5–37.7) | 27.2 (20.3–38.1) | 25.4 (18.7–31.7) | 0.1 | 0.6 | 4 × 10−4 |
| CIR | 753 (480–1190) | 752 (483–1240) | 614 (402–926) | 0.07 | 0.5 | 4 × 10−4 |
| Insulinogenic index | 26.0 (18.3–38.3) | 26.1 (18.2–37.7) | 23.1 (15.3–30.4) | 0.01 | 0.2 | 4 × 10−5 |
| BIGTT-AIR | 1,680 (1,350–2,120) | 1,670 (1,350–2,120) | 1,620 (1,310–2,040) | 0.3 | 0.5 | 0.2 |
Data are median (25% to 75% range) or means ± SD (BMI, waist, and BIGTT-SI). Values of BMI, plasma glucose, serum insulin, and derived indices were logarithmically transformed before statistical analysis. Calculated P values were adjusted for age (BIGTT-SI and BIGTT-AIR), age and sex (BMI and waist), or age, sex, and BMI (all other traits), assuming an additive, dominant, or recessive model. Indices of insulin release and insulin sensitivity were calculated as described in research design and methods. 1, type 2 diabetes–protective allele; 2, diabetes-associated allele.
TABLE 3.
Unadjusted quantitative metabolic traits in the population-based Inter99 cohort including 4,410 middle-aged subjects with normal glucose tolerance stratified according to genotype of TSPAN8 rs7961581
| 11 (TT) | 12 (TC) | 22 (CC) | PADDITIVE | P22 + 12 VS. 11 | P22 VS. 12 + 11 | |
|---|---|---|---|---|---|---|
| n (men/women) | 2,404 (1,129/1,275) | 1,686 (771/915) | 320 (147/173) | |||
| Age (years) | 45.3 ± 7.7 | 45.2 ± 7.9 | 44.6 ± 8.0 | |||
| BMI (kg/m2) | 25.5 ± 4.1 | 25.5 ± 4.1 | 25.6 ± 4.3 | 0.9 | 0.7 | 0.7 |
| Waist (cm) | 84 ± 12 | 84 ± 12 | 84 ± 12 | 0.9 | 0.9 | 0.7 |
| Fasting serum insulin (pmol/l) | 32 (23–47) | 32 (23–46) | 31 (22–44) | 0.05 | 0.2 | 0.03 |
| Serum insulin at 30 min (pmol/l) | 251 (181–352) | 238 (175–343) | 245 (173–359) | 0.003 | 0.001 | 0.3 |
| Serum insulin at 120 min (pmol/l) | 141 (88–217) | 137 (87–202) | 140 (90–225) | 0.2 | 0.2 | 0.6 |
| Fasting plasma glucose (mmol/l) | 5.3 (5.1–5.6) | 5.3 (5.0–5.6) | 5.3 (5.1–5.6) | 0.6 | 0.3 | 0.7 |
| Plasma glucose at 30 min (mmol/l) | 8.1 (7.2–9.1) | 8.2 (7.2–9.3) | 8.2 (7–9) | 0.3 | 0.7 | 0.08 |
| Plasma glucose at 120 min (mmol/l) | 5.6 (4.8–6.3) | 5.6 (4.8–6.4) | 5.6 (4.7–6.4) | 0.4 | 0.3 | 0.9 |
| ISI | 0.134 (0.089–0.192) | 0.133 (0.092–0.193) | 0.136 (0.095–0.211) | 0.05 | 0.2 | 0.04 |
| BIGTT-SI | 10.3 ± 3.7 | 10.4 ± 3.6 | 10.2 ± 3.6 | 0.4 | 0.2 | 0.7 |
| AUC-insulin/AUC-glucose | 27.9 (20.4–38.2) | 26.6 (20.3–36) | 28.2 (20.1–39.0) | 0.02 | 0.005 | 0.7 |
| CIR | 754 (494–1,210) | 738 (464–1,130) | 741 (469–1,330) | 0.1 | 0.03 | 0.4 |
| Insulinogenic index | 26.7 (18.3–38.6) | 25.1 (17.8–36.7) | 25.4 (18.3–39.3) | 0.01 | 0.002 | 1 |
| BIGTT-AIR | 1,670 (1,360–2,130) | 1,660 (1,330–2,080) | 1,720 (1,330–2,140) | 0.4 | 0.2 | 0.5 |
Data are medians (25% to 75% range) or means ± SD (BMI, waist, and BIGTT-SI). Values of BMI, plasma glucose, serum insulin, and derived indices were logarithmically transformed before statistical analysis. Calculated P values were adjusted for age (BIGTT-SI and BIGTT-AIR), age and sex (BMI and waist), or age, sex, and BMI (all other traits), assuming an additive, dominant, or recessive model. Indices of insulin release and insulin sensitivity were calculated as described in research design and methods. 1, type 2 diabetes–protective allele; 2, diabetes-associated allele.
The THADA rs7578597 did not associate with measures of obesity (BMI: P = 0.4), insulin response (insulinogenic index: P = 0.4), or insulin sensitivity (BIGTT-SI: P = 1) (Supplementary Table 1 [available in an online appendix at http://dx.doi.org/10.2337/db08-0436]). Similarly, the ADAMTS9 rs4607103 and NOTCH2 rs10923931 variants did not significantly associate with measures of oral glucose-stimulated insulin response (all P ≥ 0.5), insulin sensitivity (P ≥ 0.1), or obesity (P ≥ 0.1) in the Inter99 cohort (Supplementary Tables 2 and 3). Similar results were found when including all 5,964 treatment-naïve individuals from the Inter99 cohort (data not shown).
Because the insulin response to glucose is highly dependent on the level of insulin sensitivity, we constructed two OGTT-based disposition indexes by combining existing indexes of insulin response and insulin sensitivity and tested association with the six genotyped variants. Homozygous carriers of the CDC123/CAMK1D diabetes-associated G-allele showed a nominal association with a 13% decrease in a disposition index based on CIR and ISI (1.1–24%; P = 0.03). A disposition index based on BIGTT-AIR and BIGTT-SI did, however, not differ significantly between genotype groups for any of the six variants, although a tendency toward an allele-dependent decrease in minor G-allele carriers of the CDC123/CAMK1D variant was observed (P = 0.05).
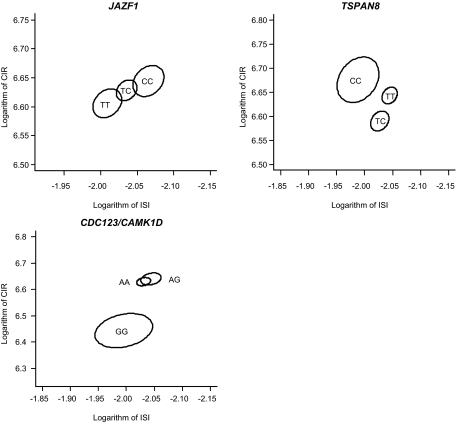
To further evaluate the relationship between insulin release, insulin sensitivity, and genetic predispositions of the type 2 diabetes–associated variants, we applied the multivariate Hotelling's T2 method to simultaneously test the effect of genotype on a combination of CIR and ISI as well as BIGTT-AIR and BIGTT-SI (Fig. 1). We demonstrated statistically significant multivariate associations of the JAZF1 and CDC123/CAMK1D variants with the combination of CIR and ISI (PADDITIVE = 0.04 and PRECESSIVE = 0.002, respectively). Furthermore, borderline association was observed for the TSPAN8 variant (PDOMINANT = 0.09 and PRECESSIVE = 0.05). The multivariate analysis did not show any influence of genotype on the combination of BIGTT-AIR and BIGTT-SI (data not shown).
FIG. 1.
Two-dimensional standard error of the mean of logarithm of the ISI and logarithm of CIR stratified according to genotype of JAZF1 rs864745, CDC123/CAMK1D rs12779790, and TSPAN8 rs7961581 in 4,516 subjects from the Inter99 cohort with normal glucose tolerance. Assuming bivariate normal distribution, we constructed standard error ellipses around the means of each genotype level. Numbers of subjects are: rs864745: TT: 1,039, TC: 2,020, CC: 898; rs12779790: AA: 2,588, AG: 1,229, GG: 153; rs7961581: TT: 2,181, TC: 1,518, CC: 289. The multivariate method, Hotelling T2, was applied to test the simultaneous effect of genotype on the two traits of insulin release and insulin sensitivity, and association was found for the JAZF1 rs864745 and CDC123/CAMK1D rs12779790 (PADDITIVE = 0.04 and PRECESSIVE = 0.002, respectively). Furthermore, borderline association was observed for TSPAN8 rs7961581 (PDOMINANT = 0.09 and PRECESSIVE = 0.05).
DISCUSSION
We report the association testing of six recently discovered type 2 diabetes risk variants (6) with intermediary diabetes-related phenotypes. Our results, if replicated in independent and statistically well-powered studies, suggest an impairment of pancreatic β-cell function for diabetes risk alleles in or near JAZF1, CDC123/CAMK1D, and TSPAN8, since these variants were associated with various surrogate measures of insulin release during an OGTT. Further support of the role of the CDC123/CAMK1D and TSPAN8 variants in altered pancreatic β-cell function was provided when analyzing an OGTT-based disposition index and for JAZF1 and CDC123/CAMK1D variants when doing multivariate analysis of estimates of insulin sensitivity and insulin release. The observed associations for all three variants are concordant with an impaired oral glucose-stimulated insulin release in subjects carrying the reported type 2 diabetes risk alleles (6).
In the analyses, we primarily focused on glucose-tolerant subjects to avoid the confounding influence of disturbances in glucose homeostasis and to circumvent the risk that associations with especially impaired insulin response were driven by the known association with type 2 diabetes. We did, however, observe similar results when including subjects with impaired fasting glycemia, impaired glucose tolerance, or screen-detected type 2 diabetes.
rs864745 resides in intron 1 of the JAZF1 (juxtaposed with another zinc finger gene 1) gene, which encodes a transcriptional repressor of the nuclear receptor subfamily 2, group C, member 2 (NR2C2) gene (17). NR2C2 (also known as TR4) is a member of the nuclear hormone receptor family and acts as a ligand-activated transcription factor (18). NR2C2 is widely expressed and Nr2c2−/− knockout mice display a phenotype of growth retardation, hypoglycemia, and reduced gluconeogenesis by decreased activation of PEPCK (19,20); however, no obvious involvement in pancreatic β-cell function has been demonstrated. Yet, since JAZF1 is expressed in the pancreas (17), one might speculate that a gain-of-function variant in JAZF1 may lead to postnatal growth restriction also affecting pancreatic β-cell mass and function.
rs12779790 is located ∼90 kb from CDC123 and ∼63.5 kb from CAMK1D. CDC123 (cell division cycle 123 homolog [S. cerevisiae]) encodes a protein involved in cell cycle regulation and nutritional control of gene transcription with no known relation to type 2 diabetes pathogenesis (21). Because CAMK1D (calcium/calmodulin-dependent protein kinase I delta) regulates granulocyte function (22), it is also possible that a causative variant in this region is related to CAMK1D and affects pancreatic β-cell function through increased apoptosis.
Lastly, rs7961581 resides ∼110 kb upstream of TSPAN8 (tetraspanin 8), which encodes a widely expressed cell surface glycoprotein known to form complexes with integrins to regulate cell motility in cancer cell lines (23). Because α6-integrin binding to laminin has been shown to negatively affect pancreatic β-cell mass maintenance (24), it is possible that variation in TSPAN8 biologically influences pancreatic β-cell function.
In this article, we have performed a thorough evaluation of a range of OGTT-based surrogate estimates of insulin release and insulin sensitivity. The associations of examined gene variants to various measures of pancreatic β-cell function highlight the need for cautious interpretation of outcomes. Variants in the CDC123/CAMK1D and TSPAN8 regions associate with the insulinogenic index, the corrected insulin response, and the ratio of AUC-insulin to AUC-glucose, which are widely used and well-documented estimates of insulin release (25,26), yet not with the recently described BIGTT-AIR index (16), and the opposite is true for the JAZF1 variant. These discrepancies may be caused by different accuracy and/or sensitivity of the applied surrogate indexes or the possibility that the different indexes capture particular and diverse roles of the encoded proteins in specific steps of insulin biosynthesis, insulin secretion, or insulin elimination. However, we cannot exclude that the associations to various measures are caused by statistical type I or II errors. Although we analyzed a range of OGTT-based surrogate indexes of insulin release, we acknowledge that application of more precise measures of insulin release, such as estimates based on an intravenous glucose tolerance test, may have modified the outcome of our analyses.
Type 2 diabetes–associated variants in the THADA, ADAMTS9, and NOTCH2 loci did not associate with metabolic traits in the Inter99 cohort. Lack of statistical power is a possible explanation, since these variants confer a modestly increased risk of type 2 diabetes. Based on 95% CIs of effect size estimates, we can with confidence exclude an allele-dependent effect in the current study on BMI, insulinogenic index, BIGTT-AIR, and ISI above 4.5% for THADA rs7578597, 3% for ADAMTS9 rs4607103, and 4% for NOTCH2 rs10923931. However, we are unable to estimate potential associations below these effect sizes.
We recognize that since no correction for multiple hypothesis testing was applied, the present results are of an explorative nature and call for validation in statistically powered and well-characterized cohorts. If, however, stringent Bonferroni correction for multiple testing (252 tests) was performed, only the associations of the CDC123/CAMK1D rs12779790 variant with measures of insulin response (insulinogenic index and serum insulin at 30 min during the OGTT) would remain statistically significant, underlining the need for replication. Based on the effect sizes of the current study, we estimate that ∼3,300, 6,100, and 3,900 subjects are needed for future studies to achieve 80% statistical power to replicate associations of JAZF1 rs864745 with BIGTT-AIR (additive model), CDC123/CAMK1D rs12779790 with insulinogenic index (recessive model), and TSPAN8 rs7961581 with insulinogenic index (dominant model), respectively.
In conclusion, we report data suggesting an impaired pancreatic β-cell function in glucose-tolerant carriers of novel type 2 diabetes risk alleles in the JAZF1, CDC123/CAMK1D, and TSPAN8 regions. No associations of common variants in THADA, ADAMTS9, and NOTCH2 with quantitative measures of insulin release or insulin sensitivity could be shown in the cohort of middle-aged people.
Supplementary Material
Acknowledgments
The study was supported by grants from the Lundbeck Foundation Centre of Applied Medical Genomics for Personalized Disease Prediction, Prevention and Care (LUCAMP); the Danish Health Research Council, The European Union (EUGENE2, grant no. LSHM-CT-2004-512013), Danish Council for Strategic Research (DanORC, grant no. 2101-06-0005), the Faculty of Health Sciences of Aarhus University, the Danish Clinical Intervention Research Academy, and the Danish Diabetes Association and Novo Nordisk.
We are thankful to Dr. Mark McCarthy, Dr. Michael Boehnke, Dr. David Altshuler, Dr. Leif Groop, and Dr. Francis Collins from the DIAGRAM consortium for prepublication information on the identity of novel type 2 diabetes genes and loci. The authors thank A. Forman, I.-L. Wantzin, and M. Stendal for technical assistance and G. Lademann for secretarial support.
We acknowledge all the members of the Inter99 team. The steering committee of the Inter99 study comprises: T. Jørgensen (principal investigator [PI]), K. Borch-Johnsen (co-PI), H. Ibsen, T. Thomsen, C. Pisinger, and C. Glümer. The study was financially supported by The Danish Medical Research Council, The Danish Centre for Health Technology Assessment, Novo Nordisk, Research Foundation of Copenhagen County, Ministry of Internal Affairs and Health, The Danish Heart Foundation, The Danish Pharmaceutical Association, The Augustinus Foundation, The Ib Henriksen Foundation, and The Becket Foundation.
Published ahead of print at http://diabetes.diabetesjournals.org on 20 June 2008.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P: A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445 :881 –885,2007 [DOI] [PubMed] [Google Scholar]
- 2.Steinthorsdottir V, Thorleifsson G, Reynisdottir I, Benediktsson R, Jonsdottir T, Walters GB, Styrkarsdottir U, Gretarsdottir S, Emilsson V, Ghosh S, Baker A, Snorradottir S, Bjarnason H, Ng MCY, Hansen T, Bagger Y, Wilensky RL, Reilly MP, Adeyemo A, Chen Y, Zhou J, Gudnason V, Chen G, Huang H, Lashley K, Doumatey A, So WY, Ma RCY, Andersen G, Borch-Johnsen K, Jorgensen T, Vliet-Ostaptchouk JV, Hofker MH, Wijmenga C, Christiansen C, Rader DJ, Rotimi C, Gurney M, Chan JCN, Pedersen O, Sigurdsson G, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K: A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet 39 :770 –775,2007 [DOI] [PubMed] [Google Scholar]
- 3.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL, Sprau AG, Tong M, White PP, Hetrick KN, Barnhart MW, Bark CW, Goldstein JL, Watkins L, Xiang F, Saramies J, Buchanan TA, Watanabe RM, Valle TT, Kinnunen L, Abecasis GR, Pugh EW, Doheny KF, Bergman RN, Tuomilehto J, Collins FS, Boehnke M: A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316 :1341 –1345,2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JRB, Rayner NW, Freathy RM, Barrett JC, Shields B, Morris AP, Ellard S, Groves CJ, Harries LW, Marchini JL, Owen KR, Knight B, Cardon LR, Walker M, Hitman GA, Morris AD, Doney ASF, The Wellcome Trust Case Control Consortium, McCarthy MI, Hattersley AT: Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 316 :1336 –1341,2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research; Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PIW, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson Bostrom K, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Rastam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjogren M, Sterner M, Surti A, Svensson M, Svensson M, Tewhey R, Blumenstiel B, Parkin M, DeFelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma Q, Parikh H, Richardson D, Ricke D, Purcell S: Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316 :1331 –1336,2007 [DOI] [PubMed] [Google Scholar]
- 6.Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, de Bakker PI, Abecasis GR, Almgren P, Andersen G, Ardlie K, Bostrom KB, Bergman RN, Bonnycastle LL, Borch-Johnsen K, Burtt NP, Chen H, Chines PS, Daly MJ, Deodhar P, Ding CJ, Doney ASF, Duren WL, Elliott KS, Erdos MR, Frayling TM, Freathy RM, Gianniny L, Grallert H, Grarup N, Groves CJ, Guiducci C, Hansen T, Herder C, Hitman GA, Hughes TE, Isomaa B, Jackson AU, Jorgensen T, Kong A, Kubalanza K, Kuruvilla FG, Kuusisto J, Langenberg C, Lango H, Lauritzen T, Li Y, Lindgren CM, Lyssenko V, Marvelle AF, Meisinger C, Midthjell K, Mohlke KL, Morken MA, Morris AD, Narisu N, Nilsson P, Owen KR, Palmer CN, Payne F, Perry JRB, Pettersen E, Platou C, Prokopenko I, Qi L, Qin L, Rayner NW, Rees M, Roix JJ, Sandbaek A, Shields B, Sjogren M, Steinthorsdottir V, Stringham HM, Swift AJ, Thorleifsson G, Thorsteinsdottir U, Timpson NJ, Tuomi T, Tuomilehto J, Walker M, Watanabe RM, Weedon MN, Willer CJ, Illig T, Hveem K, Hu FB, Laakso M, Stefansson K, Pedersen O, Wareham NJ, Barroso I, Hattersley AT, Collins FS, Groop L, McCarthy MI, Boehnke M, Altshuler D: Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 40 :638 –645,2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grarup N, Rose CS, Andersson EA, Andersen G, Nielsen AL, Albrechtsen A, Clausen JO, Rasmussen SS, Jørgensen T, Sandbæk A, Lauritzen T, Schmitz O, Hansen T, Pedersen O: Studies of association of variants near the HHEX, CDKN2A/B, and IGF2BP2 genes with type 2 diabetes and impaired insulin release in 10,705 Danish subjects: validation and extension of genome-wide association studies. Diabetes 56 :3105 –3111,2007 [DOI] [PubMed] [Google Scholar]
- 8.Florez JC, Jablonski KA, Bayley N, Pollin TI, de-Bakker PI, Shuldiner AR, Knowler WC, Nathan DM, Altshuler D: Diabetes Prevention Program Research Group: TCF7L2 polymorphisms and progression to diabetes in the Diabetes Prevention Program. N Engl J Med 355 :241 –250,2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nielsen EM, Hansen L, Carstensen B, Echwald SM, Drivsholm T, Glümer C, Thorsteinsson B, Borch-Johnsen K, Hansen T, Pedersen O: The E23K variant of Kir6.2 associates with impaired post-OGTT serum insulin response and increased risk of type 2 diabetes. Diabetes 52 :573 –577,2003 [DOI] [PubMed] [Google Scholar]
- 10.Deeb SS, Fajas L, Nemoto M, Pihlajamäki J, Mykkänen L, Kuusisto J, Laakso M, Fujimoto W, Auwerx J: A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat Genet 20 :284 –287,1998 [DOI] [PubMed] [Google Scholar]
- 11.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, Shields B, Harries LW, Barrett JC, Ellard S, Groves CJ, Knight B, Patch AM, Ness AR, Ebrahim S, Lawlor DA, Ring SM, Ben-Shlomo Y, Jarvelin MR, Sovio U, Bennett AJ, Melzer D, Ferrucci L, Loos RJ, Barroso I, Wareham NJ, Karpe F, Owen KR, Cardon LR, Walker M, Hitman GA, Palmer CN, Doney AS, Morris AD, Smith GD, Hattersley AT, McCarthy MI: A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316 :889 –894,2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Apelqvist A, Li H, Sommer L, Beatus P, Anderson DJ, Honjo T, Hrabe-de A, Lendahl U, Edlund H: Notch signalling controls pancreatic cell differentiation. Nature 400 :877 –881,1999 [DOI] [PubMed] [Google Scholar]
- 13.Jørgensen T, Borch-Johnsen K, Thomsen TF, Ibsen H, Glumer C, Pisinger C: A randomized non-pharmacological intervention study for prevention of ischaemic heart disease: baseline results Inter99 (1). Eur J Cardiovasc Prev Rehab 10 :377 –386,2003 [DOI] [PubMed] [Google Scholar]
- 14.Sluiter WJ, Erkelens DW, Reitsma WD, Doorenbos H: Glucose tolerance and insulin release, a mathematical approach I: assay of the beta-cell response after oral glucose loading. Diabetes 25 :241 –244,1976 [DOI] [PubMed] [Google Scholar]
- 15.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC: Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28 :412 –419,1985 [DOI] [PubMed] [Google Scholar]
- 16.Hansen T, Drivsholm T, Urhammer SA, Palacios RT, Vølund A, Borch-Johnsen K, Pedersen O: The BIGTT test: a novel test for simultaneous measurement of pancreatic β-cell function, insulin sensitivity, and glucose tolerance. Diabetes Care 30 :257 –262,2007 [DOI] [PubMed] [Google Scholar]
- 17.Nakajima T, Fujino S, Nakanishi G, Kim YS, Jetten AM: TIP27: a novel repressor of the nuclear orphan receptor TAK1/TR4. Nucleic Acid Res 32 :4194 –4204,2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang C, da Silva SL, Ideta R, Lee Y, Yeh S, Burbach JPH: Human and rat TR4 orphan receptors specify a subclass of the steroid receptor superfamily. Proc Natl Acad Sci U S A 91 :6040 –6044,1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collins LL, Lee YF, Heinlein CA, Liu NC, Chen YT, Shyr CR, Meshul CK, Uno H, Platt KA, Chang C: Growth retardation and abnormal maternal behavior in mice lacking testicular orphan nuclear receptor 4. Proc Natl Acad Sci U S A 101 :15058 –15063,2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu NC, Lin WJ, Kim E, Collins LL, Lin HY, Yu IC, Sparks JD, Chen LM, Lee YF, Chang C: Loss of TR4 orphan nuclear receptor reduces phosphoenolpyruvate carboxykinase-mediated gluconeogenesis. Diabetes 56 :2901 –2909,2007 [DOI] [PubMed] [Google Scholar]
- 21.Bieganowski P, Shilinski K, Tsichlis PN, Brenner C: Cdc123 and checkpoint forkhead associated with RING proteins control the cell cycle by controlling eIF2gamma abundance. J Biol Chem 279 :44656 –44666,2004 [DOI] [PubMed] [Google Scholar]
- 22.Verploegen S, Ulfman L, van-Deutekom HW, van-Aalst C, Honing H, Lammers JW, Koenderman L, Coffer PJ: Characterization of the role of CaMKI-like kinase (CKLiK) in human granulocyte function. Blood 106 :1076 –1083,2005 [DOI] [PubMed] [Google Scholar]
- 23.Gesierich S, Paret C, Hildebrand D, Weitz J, Zgraggen K, Schmitz-Winnenthal FH, Horejsi V, Yoshie O, Herlyn D, Ashman LK, Zöller M: Colocalization of the tetraspanins, CO-029 and CD151, with integrins in human pancreatic adenocarcinoma: impact on cell motility. Clin Cancer Res 11 :2840 –2852,2005 [DOI] [PubMed] [Google Scholar]
- 24.Kilkenny DM, Rocheleau JV: Fibroblast growth factor receptor-1 signaling in pancreatic islet beta-cells is modulated by the extracellular matrix. Mol Endocrinol 22 :196 –205,2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stumvoll M, Mitrakou A, Pimenta W, Jenssen T, Yki J, Van Haeften T, Renn W, Gerich J: Use of the oral glucose tolerance test to assess insulin release and insulin sensitivity. Diabetes Care 23 :295 –301,2000 [DOI] [PubMed] [Google Scholar]
- 26.Hanson RL, Pratley RE, Bogardus C, Narayan KM, Roumain JM, Imperatore G, Fagot-Campagna A, Pettitt DJ, Bennett PH, Knowler WC: Evaluation of simple indices of insulin sensitivity and insulin secretion for use in epidemiologic studies. Am J Epidemiol 151 :190 –198,2000 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.