Abstract
Background
Acquisition of virulence factors and antibiotic resistance by many clinically important bacteria can be traced to horizontal gene transfer (HGT) between related or evolutionarily distant microflora. Comparative genomic analysis has become an important tool for identifying HGT DNA in emerging pathogens. We have adapted the multi-genome alignment tool EvoPrinter to facilitate discovery of HGT DNA sequences within bacterial genomes and within their mobile genetic elements.
Principal Findings
EvoPrinter analysis of 13 different Staphylococcus aureus genomes revealed that one of the human isolates, the hospital epidemic methicillin-resistant MRSA252 strain, uniquely shares multiple putative HGT DNA sequences with different causative agents of bovine mastitis that are not found in the other human S. aureus isolates. MRSA252 shares over 14 different DNA sequence blocks with the bovine mastitis ET3 S. aureus strain RF122, and many of the HGT DNAs encode virulence factors. EvoPrinter analysis of the MRSA252 chromosome also uncovered virulence-factor encoding HGT events with the genome of Listeria monocytogenes and a Staphylococcus saprophyticus associated plasmid. Both bacteria are also causal agents of contagious bovine mastitis.
Conclusions
EvoPrinter analysis reveals that the human MRSA252 strain uniquely shares multiple DNA sequence blocks with different causative agents of bovine mastitis, suggesting that HGT events may be occurring between these pathogens. These findings have important implications with regard to animal husbandry practices that inadvertently enhance the contact of human and livestock bacterial pathogens.
Introduction
Staphylococcus aureus (S. aureus) infections in both man and domestic livestock present growing and formidable global challenges for human and animal health concerns. Methicillin-resistant S. aureus (MRSA) is now the leading cause of hospital- and community-acquired S. aureus infections [1], [2]. Likewise, among dairy herds S. aureus is one of the major causal agents of contagious bovine mastitis [3], [4]. A recent survey of mastitis outbreaks in Canadian dairy cows reported a total of 3,149 S. aureus infections in 106 farms over the course of a single year [5].
Comparative analysis of different S. aureus genomes has revealed that many strains have independently acquired genes from members of their surrounding microflora that confer antibiotic resistance and/or encode virulence factors [6], [7], [8]. Horizontal gene transfer (HGT) among bacteria and their mobile genetic elements (MGEs) is the primary mode for the spread of antibiotic resistance and virulence factors in clinically important pathogens [6], [9], [10]. Bacteriophage, plasmids, transposons and uptake of naked DNA have been shown to be involved in the movement of DNA between different bacteria [7], [11], [12], [13], [14].
HGT events have played a prominent role in the rapid acquisition of antibiotic resistance in S. aureus. Before the emergence of methicillin resistance in the early 1960s, a steady increase in penicillin resistance, mediated by plasmid transfer, was detected in hospital infections of S. aureus in both the UK and the USA [8], [15]. Soon after methicillin was used in 1960, methicillin-resistant isolates were reported, and by 1967, multidrug-resistant MRSA was reported in numerous countries. Although the origins of the mecA gene (the principle component of methicillin resistance in staphylococci) are unknown, a mecA homologue (88% similarity) is ubiquitous in the antibiotic-susceptible S. sciuri, and may be a possible evolutionary precursor of the mecA gene of the MRSA strains. Transfer of mecA is mediated by the Staphylococcal Cassette Chromosome mec (SCCmec) [16].
Bacterial co-infections are likely to be a major point of origin for HGT events. For example, the gene encoding the biofilm-associated protein (Bap), a surface protein implicated in formation of aggregates of microorganisms, is present in S. aureus that have been isolated from chronic bovine mastitis infections, and is thought to be acquired by HGT, since it is shared by other causative agents of mastitis [17]. HGT has also been documented between different bacterial genera; transfer of vancomycin resistance has been shown to occur between Enterococcus faecalis and S. aureus [18]. Other examples of transfer under conditions of co-infection include the transfer of antibiotic resistance from E. coli to the plague bacillis, Yersinia pestis [19], and transfer of vancomycin resistance from porcine to human Enterococcus faecium [20].
Multi-genome alignments play an important role in the identification of unique or uniquely shared HGT DNA sequences among related or evolutionary distant bacteria, and hence can be used to detect more recently acquired virulence factor genes in emerging pathogens [10]. To expedite the search for HGT sequences, we have adapted the web-accessed comparative genomics tool EvoPrinter for the rapid screening of chromosomal and MGE DNA [21], [22]. Evoprinter works rapidly: the alignment time for a 40 kb sequence to 17 staphylococcal genomes is accomplished in less than 20 sec. EvoPrinter algorithms serve as an initial search tool to help identify DNA sequences that are not uniformly shared by other related bacteria. Whereas BLAST sequence homology searches highlight sequence similarities, bacterial EvoPrinter algorithms highlight sequence differences between DNAs that may otherwise go unnoticed in a BLAST search, especially when large sequence files are searched. The uninterrupted EvoPrinter readouts allow for rapid visual screening of up to 40 kb of DNA, without having to sort through the multiple pairwise BLAST alignments that include both orthologs and less related sequence comparisons. EvoPrinter is currently formatted for the automated comparative analysis of 17 staphylococcal, 20 streptococcal and 22 enteric bacterial genomes and can be used to detect HGT sequences among bacterial chromosomes and their MGEs. Staphylococcal genomes included in the EvoPrinter automated analysis currently include 13 S. aureus, two S. epidermidis, one S. haemolyticus and one S. saprophyticus.
Our search for HGT DNA sequences within the genomes of different human MRSA isolates has led to the discovery that one of the human strains, the hospital-acquired epidemic MRSA252 [23], uniquely shares multiple DNA sequence blocks with three different causative agents of contagious bovine mastitis but not with other human isolates. These putative HGT sequences encode virulence factors that are also incorporated into the chromosomes or plasmids of the bovine S. aureus ET3 strain RF122, S. saprophyticus and Listeria monocytogenes. Analysis of the MRSA252 complete genome EvoUnique profile (compared to 12 other S. aureus isolates), identified over 20 different regions that were either unique to the MRSA252 genome or uniquely shared with just one other S. aureus isolate included in the analysis, the bovine RF122. Taken together, the multiple uniquely shared DNAs indicate that the human MRSA252 or another related epidemic MRSA strain may have undergone repeated HGT events with different bovine pathogens.
Results and Discussion
The comparative analysis of the bacterial chromosomes and their MGEs described in this study was performed using the EvoPrinterHD alignment algorithms. Evoprinter functions as a tool for the rapid discovery of unique DNA sequence differences among multiple bacterial genomes and their MGEs. Specifically, an EvoDifference profile identifies sequences present in a group of related genomes that are lacking in a single genome, and an EvoUnique profile highlights sequences that are either unique or uniquely shared by a subset of bacteria included in the analysis.
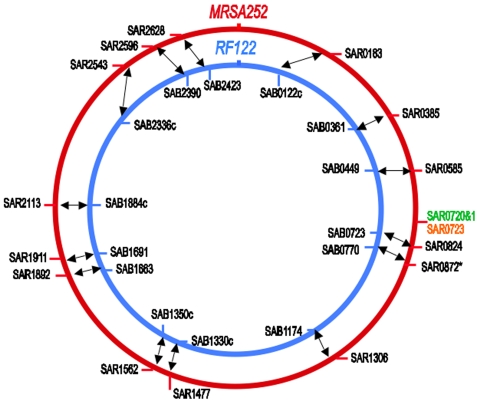
To identify uniquely shared DNA sequence blocks and unique single nucleotide polymorphisms (SNPs) among subsets of 13 different S. aureus genomes, we generated EvoUnique profiles of their chromosomes. Our initial MRSA252 EvoUnique profile alerted us to multiple instances of exclusive sequence sharing with other S. aureus chromosomes, indicated by green uppercase letters (Figure 1). To our surprise, the MRSA252 EvoUnique profile revealed that the highlighted DNAs were almost exclusively shared with a contagious bovine mastitis strain known as RF122. Notably, MRSA252 and RF122 chromosomes share 14 different unique DNA sequence blocks that are not present in any of the other S. aureus or in any of the other Staphylococci included in the EvoPrinter database (Figure 1, Table 1 and Table S1; see materials and methods for complete list of genomes). The different sequence blocks, which are from 144 to 4,950 bp in length, exhibit between 93% and 99% pairwise identity, with most having 97% or greater identity. Complete EvoUnique profiles of the MRSA252 and RF122 chromosomes are available at the EvoPrinter website.
Figure 1. Circular displays of the S. aureus human MRSA252 and bovine RF122 chromosomes showing the relative genomic positions of the uniquely shared DNAs.
Shown are the relative positions of the MRSA252 (SAR) and RF122 (SAB) shared genes. The chromosomal position of the MRSA252 - Listeria monocytogenes shared genes copper ATPase and copper oxidase is indicated in green. The chromosome position of the cadmium-transporting ATPase cadA gene shared with the S. saprophyticus pSSP2 plasmid is indicated in orange. Arrows indicate orthologous DNAs.
Table 1. MRSA252 uniquely shares multiple DNA sequences with other causative agents of bovine mastitis.
| Uniquely shared sequences | Sequence description | Flanking MGE or phage DNA | |
| MRSA252 | RF122 | ||
| SAR0183 | SAB0122c | Partial sequence of Acetylglutamate Kinase. | None detected |
| SAR0385 | SAB0361 | Similar to bovine pathogenicity island protein ORF3 | Phage |
| SAR0585 | SAB0449 | Hypothetical Phosphomethylpyrimidine kinase | None detected |
| SAR0824 | SAB0723 | Malolactic enzyme ORF plus flanking sequences. | None detected |
| SAR0872* | SAB0770* | Intragenic | None detected |
| SAR1306 | SAB1174 & SAB1175c | Hypothetical novel protein. | Transposon |
| SAR1477 & SAR1478 | SAB1330c & SAB1331c | Chorismate Synthase gene & Nucleoside Diphosphate Kinase | None detected |
| SAR1562* | SAB1350c & SAB1349c | Hypothetical novel proteins | Phage |
| SAR1892 & SAR1889 | SAB1663 & others | Hyaluronate lyase precursor 1 & other hypothetical proteins | None detected |
| SAR1911 | SAB1691 | Hypothetical novel protein | None detected |
| SAR2113 | SAB1884c | Hypothetical novel protein | Phage |
| SAR2543 | SAB2336c | ATP binding ABC transporter | None detected |
| SAR2596 | SAB2390 | Fructose 1,6 bisphosphatase | None detected |
| SAR2628 | SAB2423 | Putative ATP-dependent protease ATP-binding subunit ClpL | None detected |
| MRSA252 | Listeria monocytogenes | ||
| SAR0720 | FSL R2-503 | CopB (Copper cation ATPase transporter) | Transposon |
| SAR0721 | FSL R2-503 | Copper oxidase | Transposon |
| MRSA252 | S. saprophyticus PSSP2 plasmid | ||
| SAR0723 | SSPP217 | Cadmium-transporting ATPase | Transposon |
Sequence flanks the gene.
Considerable information has been acquired about the biology of these two pathogens [23], [24], [25], [27], [28]. Comparative analysis of the human and bovine S. aureus strains has revealed that they are phylogenetically distinct from one another and from other S. aureus strains [23], [25] and that their pathogenicity-associated genes have undergone significant genetic divergence [23], [24], [25]. MRSA252 is a sub-clone of the hospital epidemic EMRSA-16 clone [23]. The EMRSA-16 clone and its representative members are responsible for half of all MRSA infections in the U.K., and it is now considered one of the most clinically important global lineages within the U.S. [23]. The MRSA252 chromosome contains a 58.8 kb SCCmec element that carries multiple antibiotic resistance genes [23]. MRSA252 also contains the Tn552 transposon that harbors penicillin-resistance genes, which are components of the inducible S. aureus β-lactamase operon [26]. The RF122 isolate belongs to the ST151 sub-clone of the bovine ET3 clone [27], and members of this subclone display greater virulence than other ET3 sub-clones in a mouse model of mastitis [28]. In addition, comparative analysis of the different bovine ET3 clonal subtypes revealed that multiple episodes of HGT may have occurred within the ET3 lineage [28].
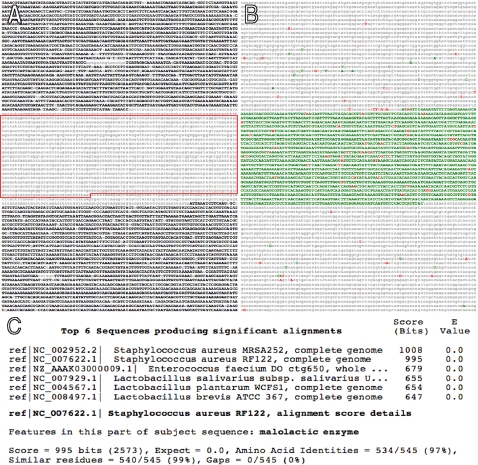
Database searches reveal that many of the MRSA252-RF122 unique sequence blocks span genes that encode virulence factors, metabolic enzymes or novel protein encoding sequences found in other bacteria (Table 1). For example, among the Staphylococcal genomes included in this analysis, a sequence encoding the malolactic enzyme gene (annotated respectively as SAR0824 and SAB0723 in the MRSA252 and RF122 genomes) is unique to MRSA252 and RF122 but present in many fermentation bacteria (Figure 2 and Table 1). The shared sequence, consisting of 1,981 bp, is located at synonymous genomic locations and exhibits 97.8% sequence identity (Figures 1 and 2). Malolactic enzyme is a component of the anaerobic respiration pathway and confers bacterial virulence by enabling survival in the anaerobic environment of deep tissue abscesses [29]. Given that abscess formation is an important aspect of human MRSA pathogenesis [30], the acquisition of this enzyme is most likely of clinical importance.
Figure 2. EvoPrint and EvoUnique comparative analysis of 13 different S. aureus isolates reveals that only the human MRSA252 and contagious bovine mastitis RF122 chromosomes contain the malolactic enzyme gene.
A) An EvoPrint generated from pairwise alignments of MRSA252 DNA (6,497 bp) and aligning regions from 12 other S. aureus genomes (S. aureus COL; S. aureus MSSA476, S. aureus Mu50; S. aureus MW2; S. aureus N315; S. aureus NCTC 8325; S. aureus RF122; S. aureus USA300; S. aureus JH1; S. aureus JH9; S. aureus Mu3 and S. aureus Newman) reveals high homology between all aligning regions except for a central 1,981 bp region. Uppercase black-colored letters identify MRSA252 sequences that align with all genomes and lowercase gray-colored bases indicate nucleotides that do not align with at least one of the genomes. The boxed ORF sequence encodes the 545 amino acid malolactic enzyme. B) Generated from the same analysis, the MRSA252 EvoUnique print indicates that only one of the 12 genomes included in the analysis contains homologous sequences that span the MRSA252 malolactic enzyme gene locus (SAR0824). Uppercase red-colored SNPs are unique to the MRSA252 sequence; green-colored letters represent bases that align with only one of the 12 genomes (S. aureus RF122) and lowercase gray-colored letters represent sequences that are common to two or more alignments. (C) Protein database homology searches identify the RF122 malolactic enzyme as sharing the highest identity with the MRSA252 enzyme. Note: no homologies to other malolactic enzyme encoding genes were detected in other S. aureus species.
The EvoUnique profile of the malolactic enzyme region also demonstrates the utility of this approach for the detection of single nucleotide polymorphisms (SNPs). For example, the red-colored bases in Figure 2B represent SNPs that are unique in MRSA252 and not present in any of the other genomes analyzed, including RF122. The EvoUnique profile of the first 50 kb of the MRSA252 chromosome identifies 229 SNPs that distinguishes it from the other isolates (data not shown). The speed and base-pair resolution of this approach should prove invaluable when markers are sought to distinguish between different MRSA isolates.
All of the MRSA252-RF122 shared DNAs exhibit features of HGT events [6]; the acquired DNA is not found in other closely related strains and the sequences flanking the unique DNA share greater identity with the closely-related genomes (that lack the unique DNA) than they do with the putative donor genome. For example, pairwise alignments reveal that the highest shared identity with 5 kb of MRSA252 DNA flanking either side of the malolactic enzyme gene (Figure 2) is not with RF122 but with the human community-acquired MRSA MW2 isolate (data not shown). Of all of the genomes included in the analysis, the RF122 flanking DNA is least homologous to the MRSA252 flanks. The multiplicity and synonymous genomic locations of many of the unique DNAs indicate that multiple HGT events have occurred, and that homologous recombination most likely played a role in many of the integration events.
The bovine RF122 isolate or related clones may have undergone HGT events with other human S. aureus isolates. For example, DNA sequences between bases 404,577 and 409,784 of RF122 are exclusively shared with two human isolates, MSSA476 and MW2, but not with MRSA252 or the other S. aureus isolates examined. MSSA476 and MW2 are representative of community-acquired S. aureus strains; MSSA476 is a hyper-virulent community-acquired methicillin-susceptible strain isolated in the United Kingdom [23] and the MW2 strain is one of the major MRSA pathogens causing community-acquired infections in the mid-western region of the USA [31]. The shared sequences contain ORFs but do not encode proteins of identifiable function (data not shown).
EvoPrinter analysis of the MRSA252 chromosome also identified additional DNA sequence blocks that are absent from the other S. aureus but shared with other causative agents of bovine mastitis: Listeria monocytogenes and S. saprophyticus [32], [33]. Most notable is a region (at position 754,883) that spans the copper cation ATPase transporter copB (SAR0720) and copper oxidase genes (SAR0721) (Figure 1 and 3A). The MRSA252 genomic region that spans these two genes is nearly identical to DNA present in the genome of Listeria monocytogenes, as revealed by a BLASTn alignment (data not shown). Pairwise alignments between the Listeria DNA and the aligning region of the MRSA252 chromosome reveal that the two DNAs share 99.8% identity over 3,131 bp (Figure 3A). Our analysis also uncovered another human MRSA isolate, USA300, that contains a copper ATPase transporter gene (SAUSA300_0078, identified as copA), however, its sequence homology to the MRSA252/Listeria genes is significantly less and it lacks the flanking copper oxidase gene (Figure 3B). The MRSA252 copB and copper oxidase genes were most likely transferred via a plasmid, as the flanking DNA in MRSA252 is homologous to plasmid sequences (data not shown). The copA and copper oxidase genes constitute a copper resistance operon (cop) associated with resistance to metal toxicity [34]. It is surprising that Listeria and MRSA252 have near identical copper resistance genes. Metal ion transporters play important roles in both nutrient uptake and in secretion of toxins [35].
Figure 3. S. aureus MRSA252 and Listeria monocytogenes chromosomes share near identical copper ATPase transporter and copper oxidase genes.
A) Shown is a Listeria monocytogenes DNA (4,680 bp) – MRSA252 pairwise enhanced BLAT (eBLAT) DNA alignment revealing near sequence identity that spans the ORFs for the copper ATPase transporter (SAR0720; upper box) and the copper oxidase genes (SAR0721; lower box) [23]. The Listeria genomic region of this eBLAT (GenBank accession AARR00000000), used as the reference sequence, was selected as ‘other species’ in the sequence input page of EvoPrinter. Use of this feature allows for the comparative analysis of any sequence with respect to the genomes in the EvoPrinter database. Red-colored letters indicate unique bases that not shared with MRSA252. Green-colored sequences are common to both Listeria and MRSA252. B) Listeria DNA (same sequence as in panel A) eBLAT alignment with the S. aureus USA300 genome identifies CopA (SAUSA300_0078) but with a significantly lower sequence homology when compared to the MRSA252 DNA (A).
Another MRSA252 unique sequence block (2,152 bp; SAR0723) is also shared with the Staphylococcus saprophyticus plasmid pSSP2 (locus tag: SSPP217, GenBank accession NC_007352) and it encodes the cadmium-transporting ATPase CadA protein (93.5% shared identity). Heavy metal cadmium resistance has been associated with S. aureus plasmids [36], although the current selective pressure for cadmium resistance is unknown [37].
Our comparative analysis identified additional putative HGT DNA sequences within the MRSA252 genome that have no counterpart in other Staphylococci examined in this study. For example, the unique DNA sequence found at nucleotides 303,347–306,097 contains an ORF coding for a 763 amino acid protein (Figure S1), annotated as a nitric oxide reductase (SAR0261) and identified as a region of inserted DNA [23]. A BLASTp database search revealed 51% amino acid identity to nitric oxide reductase encoding genes in Geobacillus species and Bacillus licheniformi (data not shown). In other bacterial pathogens, the nitric oxide reductase enzyme is considered a virulence factor that allows survival under very low oxygen tension and/or allows the organism to take advantage of de-nitrification to cope with nitric oxide production in macrophages [38], [39].
The presence of mobile genetic element-associated genes integrated into the MRSA252 genome and the sharing of sequences between MRSA252 and RF122 prompted us to examine sequenced S. aureus associated MGEs for the presence of genes that might be shared by these two isolates, thus suggesting a potential mode of transfer. EvoPrinter analysis of the 13 different S. aureus isolates was performed using the 35 kb pTZ2162 plasmid as the reference input sequence. This plasmid is widely distributed among healthcare-associated MRSA strains [40]. The pTZ2162 EvoUnique profile identified multiple regions that are uniquely shared with different S. aureus genomes (data not shown). An EvoDifference analysis of one such region identified two flanking sub-regions, one shared with MRSA252 and the other with RF122 (Figure S2). The shared pTZ2162-MRSA252 sequence contains a partial sequence that matches the blaZ antibiotic resistance gene, which encodes ß-lactamase [41]. blaZ has been identified in a significant fraction of clinical S. aureus isolates [42] and in S. aureus isolates from persistent bovine mastitis [43]. The RF122 shared sequence contains an ORF that encodes a quinone oxidoreductase/DT diaphorase, a member of a subfamily of alcohol dehydrogenases, annotated as SAB1296c [24]. In E. coli, oxidoreductases have been shown to be drug resistance factors [44]. The HGT events were most likely due to plasmid insertions, as both the MRSA252 (located at 866,929) and RF122 (located at 1,418,565 bp) sequences are flanked by plasmid sequences (data not shown). The pTZ2162 plasmid also shares near sequence identity to multiple genes in Staphylococcus haemolyticus and Staphylococcus epidermidis human pathogens (data not shown).
EvoUnique profiles of other S. aureus isolates also identified putative HGT sequences that are not part of the MRSA252 or RF122 chromosomes. For example, our analysis revealed sequences within the human USA300 chromosome that are shared with S. epidermidis, S. haemolyticus and S. saprophyticus, but not with other S. aureus isolates (data not shown). Additionally, the USA300 EvoUnique profile revealed sequences uniquely shared with the human MW2 and Col isolates, but no sequences were found uniquely shared between USA300 and MRSA252 or RF122.
Summary
The finding of multiple putative HGT DNAs that are shared between a human epidemic MRSA isolate and a contagious bovine mastitis S. aureus isolate, but that are absent from other S. aureus, indicates that these two phylogenetically distinct strains most likely have undergone multiple gene transfers either between themselves or with as yet unidentified additional bacteria. Clearly, comparisons between bacterial genomes establish only that they uniquely share DNA sequences, but the analysis does not establish a transfer mechanism or the identity of the exchanging partners. However, our analysis reveals multiple instances of HGT events between RF122 and MRSA252 that are adjacent to MGE and bacteriophage sequences. In most cases, the flanking vector sequences are in MRSA252 and not in RF122, suggesting that MRSA252 or one of its related clones was the recipient in the HGT exchange.
One likely scenario is that the DNA exchanges may have occurred during human and/or bovine co-infections. In light of the documented cases of direct transmission of MRSA between cows and humans [45] and the isolation in cows of a MRSA related to human epidemic strain Irish 01 [4], this possibility is highly probable. Clearly, more studies are required to determine how and where the putative HGT events took place. Nevertheless, the finding of unique virulence factor genes in a human pathogen whose potential source(s) may have originated from different causative agents of contagious bovine mastitis, including Listeria monocytogenes and S. saprophyticus, suggests that there may be a common epidemiological association between these bacteria and that co-infections are a likely point of origin for these exchanges. A similar concern regarding co-infection has been raised over the possibility of HGT of vancomycin resistance from enterococcus to S. aureus [46]. Taken together, these findings suggest that HGT events may be more prevalent between human and livestock bacteria than previously recognized and that animal husbandry practices that enhance contact between human and livestock pathogens should be avoided. Further work is required to elucidate the details and ramifications of these exchanges.
Materials and Methods
EvoPrinter analysis
EvoPrinter algorithms consist of a series of web-accessed tools for discovering and comparing conserved or uniquely shared sequences within orthologous DNAs [21], [22] (http://evoprinter.ninds.nih.gov/). Unlike other multi-genome comparative tools that display columns of aligning bases with gaps to optimize alignments, EvoPrinter displays, in a single uninterrupted view, DNA sequences that are either conserved, unique or uniquely shared, including single nucleotide polymorphisms (SNPs), as they exist in the genome or MGE of interest. Because EvoPrinter readouts show only the input reference DNA sequence and not the aligning regions of the multiple genomes included in the analysis, more sequence can be displayed in a single view than is possible with conventional multi-genome alignments.
The following algorithms were developed to help identify putative HGT DNA sequences: (1) an EvoUnique profile highlights unique or uniquely shared sequences among subsets of genomes that are otherwise absent from the other genomes included in the analysis; (2) a repeat finder detects putative MGE sequences based on the repetitive presence of their sequences within bacterial chromosomes; (3) an EvoDifferences profile portrays, in a single view, those sequences that are detected in all but one of the genomes included in the analysis, and (4) input reference DNA exchange allows for re-initiation of the comparative analysis using the aligning region of another genome, thus facilitating the search for unique differences among the genomes included in the analysis. EvoPrinterHD also includes algorithms that identify sequence rearrangements in the aligning regions of the test genomes.
For Staphylococcus chromosome analysis, after inserting the DNA sequence to be analyzed (the reference sequence) into the DNA sequence input window, the alignment algorithms automatically generate 9 eBLAT alignments for each of the 17 staphylococcal genomes and then assembles composite eBLAT (ceBLAT) alignments for the top three homology scoring regions of each genome. The alignment process takes seconds to complete and allows the user to examine the input DNA for repetitive sequences and to view the alignment results, in the form of an alignment scorecard [22]. The alignment scorecard gives information regarding the extent of homology to each of the test species for the top scoring alignment, and the second and third most significant alignments. Accessible from the alignment scorecard are the eBLAT alignments to the top three aligning regions for each pairwise analysis, and a ceBLAT that superimposes the top three alignments. From the scorecard, the user can select or deselect different alignments to be used in the EvoPrint analysis. The complete EvoUnique prints of the S. aureus MRSA252 and S. aureus RF122 genomes are also available online through links provided in the EvoPrinterHD bacterial genome resources section.
Genomic DNA sequence files
The 17 Staphylococcus genomes were curated from databases listed below. The following Staphylococcus genome sequence files were curated from the BacMap database of University of Alberta (http://wishart.biology.ualberta.ca/BacMap/): S. aureus COL, S. aureus JH1, S. aureus JH9, S. aureus MRSA252, S. aureus MSSA476, S. aureus Mu3, S. aureus Mu50, S. aureus MW2, S. aureus N315, S. aureus NCTC8325, S. aureus Newman, S. aureus RF122, S. aureus USA300, S. epidermidis ATCC12228, S. epidermidis RP62A and S. haemolyticus JCSC1435. The genome sequence files for S. aureus subsp. aureus JH1, S. aureus subsp. aureus JH9, S. aureus Mu3, and S. aureus subsp. aureus str. Newman were curated from the European Bioinformatics Institute of the European Molecular Biology Laboratory (http://www.ebi.ac.uk/genomes/bacteria.html). The pTZ2162 plasmid sequence was obtained from the NCBI GenBank nucleotide sequence database.
Genomic indexing
Each of the genomes was parsed into non-overlapping K-mers three different ways and held in memory. In addition to the original 11-mer index of BLAT [47], EvoPrinterHD indexes each genome into a second set of non-overlapping 11-mers, offset by four base pairs from the initial indexing, and into a third set of non-overlapping 9-mers. By performing three independent alignments using the staggered genomic indices and then superimposing the resulting alignments to show all aligning sequences, the enhanced-BLAT (eBLAT) detects as much as 75% more conserved sequences when evolutionary distant sequences are aligned [22].
NCBI BLAST database searches
Database homology BLAST searches against 988 microbial genomes were performed using the standard tBLASTn, BLASTn and BLASTp options at the National Center for Biotechnology Information web site (http://www.ncbi.nlm.nih.gov).
Supporting Information
MRSA252-RF122 uniquely shared chromosomal DNA sequence blocks.
(0.06 MB DOC)
An EvoUnique profile identifies a putative nitric oxide reductase gene in the human S. aureus MRSA252 genome that is not present in the genomes of 16 other Staphylococci. EvoUnique analysis of the human S. aureus MRSA252 genomic DNA (bases 303,347 to 306,097) with 16 other Staphylococcus genomes (S. aureus COL; S. aureus MSSA476, S. aureus Mu50; S. aureus MW2; S. aureus N315; S. aureus NCTC 8325; S. aureus RF122; S. aureus USA300; S. aureus JH1; S. aureus JH9; S. aureus Mu3; S. aureus Newman; S. epidermidis ATCC 12228; S. epidermidis RP62; S. haemolyticus JCSC1435; and S. saprophyticus) reveals a unique 2,750 bp region that contains a 763 amino acid protein encoding ORF identified as encoding a nitric oxide reductase (SAR0261). Upper case, red-colored letters represent sequences that are unique to S. aureus MRSA252 and were not found in the other 16 Staphylococci genomes analyzed; green bases are shared with one other isolate and lowercase gray-colored bases are common to three or more of the test species aligning regions. Protein database homology searches reveal that the predicted protein shares 52% homology with the Geobacillis kaustophilus HTA426 nitric oxide reductase enzyme.
(4.89 MB TIF)
Identification of HGT exchanges between plasmid and bacterial genomes. EvoPrinter comparative analysis of the Staphylococcus pTZ2162 plasmid with the S. aureus COL; S. aureus MRSA252; S. aureus MSSA476, S. aureus Mu50; S. aureus MW2; S. aureus N315; S. aureus NCTC 8325; S. aureus RF122; S. aureus USA300; S. aureus JH1; S. aureus JH9; S. aureus Mu3 and S. aureus Newman genomes identifies HGT sequences that are uniquely shared with different bacterial genomes. Shown is a pTZ2162 plasmid DNA EvoDifferences profile (25,815 to 30,799 bp) that highlights two different putative HGT events. Highlighted with green-colored letters, pTZ2162 uniquely shares a 766 bp sequence with the human MRSA252 genome that includes a partial match to the blaZ gene (ORF boxed). Flanking the MRSA252 - pTZ2162 shared homology is a 2,121 bp fragment (red-colored sequences) that is uniquely shared with the bovine RF122 chromosome that contains the quinone oxidoreductase / DT diaphorase gene (ORF boxed).
(5.06 MB TIF)
Acknowledgments
We are grateful to Howard Nash, Alan Koretsky, Story Landis, Michael Otto and Frank DeLeo for helpful discussions during the preparation of the manuscript. We also thank Ken Weeks and Jack Bishop for their technical expertise and acknowledge the editorial expertise of Judith Brody.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by the Intramural Research Program of the NIH, NINDS.
References
- 1.Klevens RM, Edwards JR, Tenover FC, McDonald LC, Horan T, et al. Changes in the epidemiology of methicillin-resistant Staphylococcus aureus in intensive care units in US hospitals, 1992–2003. Clin Infect Dis. 2006;42:389–91. doi: 10.1086/499367. [DOI] [PubMed] [Google Scholar]
- 2.Jarvis WR, Schlosser J, Chinn RY, Tweeten S, Jackson M. National prevalence of methicillin-resistant Staphylococcus aureus in inpatients at US health care facilities, 2006. Am J Infection Control. 2006;35:631–637. doi: 10.1016/j.ajic.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Barkema HW, Schukken YH, Zadoks RN. Invited Review: The role of cow, pathogen, and treatment regimen in the therapeutic success of bovine Staphylococcus aureus mastitis. J Dairy Sci. 2006;89:1877–95. doi: 10.3168/jds.S0022-0302(06)72256-1. [DOI] [PubMed] [Google Scholar]
- 4.Monecke S, Kuhnert P, Hotzel H, Slickers P, Ehricht R. Microarray based study on virulence-associated genes and resistance determinants of Staphylococcus aureus isolates from cattle. Vet Microbiol. 2007;125:128–40. doi: 10.1016/j.vetmic.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 5.Olde Riekerink RG, Barkema HW, Kelton DF, Scholl DT. Incidence rate of clinical mastitis on Canadian dairy farms. J Dairy Sci. 2008;91:1366–77. doi: 10.3168/jds.2007-0757. [DOI] [PubMed] [Google Scholar]
- 6.Ochman H, Lawrence JG, Grolsman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 7.Lindsay JA, Holden MT. Understanding the rise of the superbug: investigation of the evolution and genomic variation of Staphylococcus aureus. Functional Integrative Genomics. 2006;6:186–201. doi: 10.1007/s10142-005-0019-7. [DOI] [PubMed] [Google Scholar]
- 8.de Lencastre H, Oliveira D, Tomasz A. Antibiotic resistant Staphylococcus aureus: a paradigm of adaptive power. Curr Opin Microbiol. 2007;10:428–35. doi: 10.1016/j.mib.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diep BA, Carleton HA, Chang RF, Sensabaugh GF, Perdreau-Remington F. Roles of 34 virulence genes in the evolution of hospital- and community-associated strains of methicillin-resistant Staphylococcus aureus. J Infect Dis. 2006;193:1495–503. doi: 10.1086/503777. [DOI] [PubMed] [Google Scholar]
- 10.Zaneveld JR, Nemergut DR, Knight R. Are all horizontal gene transfers created equal? Prospects for mechanism-based studies of HGT patterns. Microbiology. 2008;154:1–15. doi: 10.1099/mic.0.2007/011833-0. [DOI] [PubMed] [Google Scholar]
- 11.Boyd EF, Davis BM, Hochhut B. Bacteriophage-bacteriophage interactions in the evolution of pathogenic bacteria. Trends Microbiol. 2001;9:137–44. doi: 10.1016/s0966-842x(01)01960-6. [DOI] [PubMed] [Google Scholar]
- 12.Hughes AL, Friedman R. Nucleotide substitution and recombination at orthologous loci in Staphylococcus aureus. J Bacteriology. 2005;187:2698–2704. doi: 10.1128/JB.187.8.2698-2704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pallen MJ, Wren BW. Bacterial pathogenomics. Nature. 2007;449:835–42. doi: 10.1038/nature06248. [DOI] [PubMed] [Google Scholar]
- 14.Nwaneshiudu AI, Mucci T, Pickard DJ, Okeke IN. A second large plasmid encodes conjugative transfer and antimicrobial resistance in O119:H2 and some typical O111 enteropathogenic Escherichia coli strains. J Bacteriol. 2007;189:6074–9. doi: 10.1128/JB.00349-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, et al. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc Natl Acad Sci USA. 2002;99:7687–92. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deurenberg RH, Vink C, Kalenic S, Friedrich AW, Bruggeman CA, et al. The molecular evolution of methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect. 2007;13:222–35. doi: 10.1111/j.1469-0691.2006.01573.x. [DOI] [PubMed] [Google Scholar]
- 17.Tormo MA, Knecht E, Götz F, Lasa I, Penadés JR. Bap-dependent biofilm formation by pathogenic species of Staphylococcus: evidence of horizontal gene transfer? Microbiology. 2005;151:2465–75. doi: 10.1099/mic.0.27865-0. [DOI] [PubMed] [Google Scholar]
- 18.Noble WC, Virani Z, Cree RG. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiol Lett. 1992;72:195–8. doi: 10.1016/0378-1097(92)90528-v. [DOI] [PubMed] [Google Scholar]
- 19.Hinnebusch BJ, Rosso ML, Schwan TG, Carniel E. High-frequency conjugative transfer of antibiotic resistance genes to Yersinia pestis in the flea midgut. Mol Microbiol. 2002;46:349–54. doi: 10.1046/j.1365-2958.2002.03159.x. [DOI] [PubMed] [Google Scholar]
- 20.Moubareck C, Bourgeois N, Courvalin P, Doucet-Populaire F. Multiple antibiotic resistance gene transfer from animal to human enterococci in the digestive tract of gnotobiotic mice. Antimicrob Agents Chemother. 2003;47:2993–6. doi: 10.1128/AAC.47.9.2993-2996.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Odenwald WF, Rasband W, Kuzin A, Brody T. EVOPRINTER, a multigenomic comparative tool for rapid identification of functionally important DNA. Proc Natl Acad Sci USA. 2005;102:14700–5. doi: 10.1073/pnas.0506915102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yavatkar AS, Lin Y, Ross J, Fann Y, Brody T, et al. Rapid detection and curation of conserved DNA via enhanced-BLAT and EvoPrinterHD analysis. BMC Genomics. 2008;9:106. doi: 10.1186/1471-2164-9-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holden MTG, Feil EJ, Lindsay JA, Peacock SJ, Day NP, et al. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci USA. 2004;101:9786–91. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herron LL, Chakravarty R, Dwan C, Fitzgerald JR, Musser JM, et al. Genome sequence survey identifies unique sequences and key virulence genes with unusual rates of amino acid substitution in bovine Staphylococcus aureus. Infection Immunity. 2002;70:3978–81. doi: 10.1128/IAI.70.7.3978-3981.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J Bacteriol. 2008;190:300–10. doi: 10.1128/JB.01000-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rowland SJ, Dyke KG. Tn552, a novel transposable element from Staphylococcus aureus. Mol Microbiol. 1990;4:961–75. doi: 10.1111/j.1365-2958.1990.tb00669.x. [DOI] [PubMed] [Google Scholar]
- 27.Herron-Olson L, Fitzgerald JR, Musser JM, Kapur V. Molecular correlates of host specialization in Staphylococcus aureus. PLoS ONE. 2007;2:e1120. doi: 10.1371/journal.pone.0001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guinane CM, Sturdevant DE, Herron-Olson L, Otto M, Smyth DS, et al. Pathogenomic analysis of the common bovine Staphylococcus aureus clone (ET3): emergence of a virulent subtype with potential risk to public health. J Infectious Diseases. 2008;197:205–13. doi: 10.1086/524689. [DOI] [PubMed] [Google Scholar]
- 29.Field J, Rosenthal B, Samuelson J. Early lateral transfer of genes encoding malic enzyme, acetyl-CoA synthetase and alcohol dehydrogenases from anaerobic prokaryotes to Entamoeba histolytica. Mol Microbiol. 2000;38:446–55. doi: 10.1046/j.1365-2958.2000.02143.x. [DOI] [PubMed] [Google Scholar]
- 30.Saiman L, O'Keefe M, Graham PL, Wu F, Saïd-Salim B, et al. Hospital transmission of community-acquired methicillin-resistant Staphylococcus aureus among postpartum women. Clin Infect Dis. 2003;37:1313–9. doi: 10.1086/379022. [DOI] [PubMed] [Google Scholar]
- 31.Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–1827. doi: 10.1016/s0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 32.Bourry A, Poutrel B, Rocourt J. Bovine mastitis caused by Listeria monocytogenes: characteristics of natural and experimental infections. J Med Microbiol. 1995;43:125–32. doi: 10.1099/00222615-43-2-125. [DOI] [PubMed] [Google Scholar]
- 33.Moon JS, Lee AR, Kang HM, Lee ES, Joo YS, et al. Antibiogram and coagulase diversity in staphylococcal enterotoxin-producing Staphylococcus aureus from bovine mastitis. J Dairy Sci. 2007;90:1716–24. doi: 10.3168/jds.2006-512. [DOI] [PubMed] [Google Scholar]
- 34.Cooksey DA. Molecular mechanisms of copper resistance and accumulation in bacteria. FEMS Microbiol Rev. 1994;14:381–6. doi: 10.1111/j.1574-6976.1994.tb00112.x. [DOI] [PubMed] [Google Scholar]
- 35.Stähler FN, Odenbreit S, Haas R, Wilrich J, Van Vliet AHM, et al. The novel Helicobacter pylori CznABC metal efflux pump is required for cadmium, zinc, and nickel resistance, urease modulation, and gastric colonization. Infect Immun. 2006;74:3845–52. doi: 10.1128/IAI.02025-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novick RP, Roth C. Plasmid-linked resistance to inorganic salts in Staphylococcus aureus. J Bacteriol. 1968;95:1335–42. doi: 10.1128/jb.95.4.1335-1342.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Porter FD, Silver S, Ong C, Nakahara H. Selection for mercurial resistance in hospital settings. Antimicrob Agents Chemother. 1982;22:852–858. doi: 10.1128/aac.22.5.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loisel-Meyer S, Jiménez de Bagüés MP, Bassères E, Dornand J, Köhler S, et al. Requirement of norD for Brucella suis virulence in a murine model of in vitro and in vivo infection. Infect Immun. 2006;74:1973–6. doi: 10.1128/IAI.74.3.1973-1976.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pittman MS, Elvers KT, Lee L, Jones MA, Poole RK, et al. Growth of Campylobacter jejuni on nitrate and nitrite: electron transport to NapA and NrfA via NrfH and distinct roles for NrfA and the globin Cgb in protection against nitrosative stress. Mol Microb. 2006;63:575–590. doi: 10.1111/j.1365-2958.2006.05532.x. [DOI] [PubMed] [Google Scholar]
- 40.Nakaminami H, Noguchi N, Nishijima S, Kurokawa I, So H, et al. Transduction of the plasmid encoding antiseptic resistance gene qacB in Staphylococcus aureus. Biol Pharm Bull. 2007;30:1412–5. doi: 10.1248/bpb.30.1412. [DOI] [PubMed] [Google Scholar]
- 41.Brown DF, Reynolds PE. Intrinsic resistance to beta-lactam antibiotics in Staphylococcus aureus. FEBS Lett. 1980;122:275–78. doi: 10.1016/0014-5793(80)80455-8. [DOI] [PubMed] [Google Scholar]
- 42.Zhu LX, Zhang ZW, Wang C, Yang HW, Jiang D, et al. Use of a DNA microarray for simultaneous detection of antibiotic resistance genes among staphylococcal clinical isolates. J Clin Microbiol. 2007;45:3514–21. doi: 10.1128/JCM.02340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haveri M, Roslöf A, Rantala L, Pyörälä S. Virulence genes of bovine Staphylococcus aureus from persistent and nonpersistent intramammary infections with different clinical characteristics. J Appl Microbiol. 2007;103:993–1000. doi: 10.1111/j.1365-2672.2007.03356.x. [DOI] [PubMed] [Google Scholar]
- 44.Adams MA, Jia Z. Modulator of drug activity B from Escherichia coli: Crystal Structure of a prokaryotic homologue of DT-diaphorase. J Molec Biol. 2006;359:455–65. doi: 10.1016/j.jmb.2006.03.053. [DOI] [PubMed] [Google Scholar]
- 45.Juhász-Kaszanyitzky E, Jánosi S, Somogyi P, Dán A, van der Graaf-van Bloois L, et al. MRSA transmission between cows and humans. Emerg Infect Dis. 2007;13:630–2. doi: 10.3201/eid1304.060833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sigurdardottir B, Berg JV, Hu J, Alamu J, McNutt, et al. Descriptive epidemiology and case-control study of patients colonized with vancomycin-resistant enterococcus and methicillin-resistant Staphylococcus aureus. Infect Control Hosp Epidemiol. 2006;27:913–9. doi: 10.1086/507278. [DOI] [PubMed] [Google Scholar]
- 47.Kent WJ. BLAT-the BLAST-like alignment tool. Genome Res. 2002;12:656–64. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MRSA252-RF122 uniquely shared chromosomal DNA sequence blocks.
(0.06 MB DOC)
An EvoUnique profile identifies a putative nitric oxide reductase gene in the human S. aureus MRSA252 genome that is not present in the genomes of 16 other Staphylococci. EvoUnique analysis of the human S. aureus MRSA252 genomic DNA (bases 303,347 to 306,097) with 16 other Staphylococcus genomes (S. aureus COL; S. aureus MSSA476, S. aureus Mu50; S. aureus MW2; S. aureus N315; S. aureus NCTC 8325; S. aureus RF122; S. aureus USA300; S. aureus JH1; S. aureus JH9; S. aureus Mu3; S. aureus Newman; S. epidermidis ATCC 12228; S. epidermidis RP62; S. haemolyticus JCSC1435; and S. saprophyticus) reveals a unique 2,750 bp region that contains a 763 amino acid protein encoding ORF identified as encoding a nitric oxide reductase (SAR0261). Upper case, red-colored letters represent sequences that are unique to S. aureus MRSA252 and were not found in the other 16 Staphylococci genomes analyzed; green bases are shared with one other isolate and lowercase gray-colored bases are common to three or more of the test species aligning regions. Protein database homology searches reveal that the predicted protein shares 52% homology with the Geobacillis kaustophilus HTA426 nitric oxide reductase enzyme.
(4.89 MB TIF)
Identification of HGT exchanges between plasmid and bacterial genomes. EvoPrinter comparative analysis of the Staphylococcus pTZ2162 plasmid with the S. aureus COL; S. aureus MRSA252; S. aureus MSSA476, S. aureus Mu50; S. aureus MW2; S. aureus N315; S. aureus NCTC 8325; S. aureus RF122; S. aureus USA300; S. aureus JH1; S. aureus JH9; S. aureus Mu3 and S. aureus Newman genomes identifies HGT sequences that are uniquely shared with different bacterial genomes. Shown is a pTZ2162 plasmid DNA EvoDifferences profile (25,815 to 30,799 bp) that highlights two different putative HGT events. Highlighted with green-colored letters, pTZ2162 uniquely shares a 766 bp sequence with the human MRSA252 genome that includes a partial match to the blaZ gene (ORF boxed). Flanking the MRSA252 - pTZ2162 shared homology is a 2,121 bp fragment (red-colored sequences) that is uniquely shared with the bovine RF122 chromosome that contains the quinone oxidoreductase / DT diaphorase gene (ORF boxed).
(5.06 MB TIF)