Summary
The regulation of gene expression requires a wide array of protein factors that can modulate chromatin structure, act at enhancers, function as transcriptional coregulators, or regulate insulator function. Poly(ADP-ribose) polymerase-1 (PARP-1), an abundant and ubiquitous nuclear enzyme that catalyzes the NAD+-dependent addition of ADP-ribose polymers on a variety of nuclear proteins, has been implicated in all of these functions. Recent biochemical, genomic, proteomic, and cell-based studies have highlighted the role of PARP-1 in each of these processes and provided new insights about the molecular mechanisms governing PARP-1-dependent regulation of gene expression. In addition, these studies have demonstrated how PARP-1 functions as an integral part of cellular signaling pathways that culminate in gene regulatory outcomes.
Introduction
Poly(ADP-ribose) polymerase-1 (PARP-1) is an abundant (as many as 1 to 2 million copies per cell [1]) and ubiquitous nuclear enzyme with biochemical properties that make it ideally suited for the regulation of nuclear processes. Although originally characterized as a key factor in DNA repair pathways, a wealth of studies over the past decade have demonstrated a role for PARP-1 in the regulation of gene expression under basal, signal-activated, and stress-activated conditions [1–3]. Recent studies using a variety of experimental approaches have highlighted the role of PARP-1 in at least four distinct modes of transcriptional regulation (see below) and provided new insights about the cellular signaling systems that interface with PARP-1 in the nucleus.
PARP-1, the founding member of the PARP superfamily [4], has a carboxyl-terminal catalytic domain that polymerizes linear or branched chains of ADP-ribose (ADPR) from donor nicotinamide adenine dinucleotide (NAD+) molecules on target proteins (Fig. 1) [1,2]. Poly(ADP-ribosyl)ation (PARylation) by PARP-1 is likely the major source of poly(ADP-ribose) (PAR) production in the cell [1]. PARP-1 also has an amino-terminal DNA binding domain (DBD) containing two zinc finger motifs, as well as a central automodification domain (AMD) that functions as the target of direct covalent automodification [1,4] (Fig. 1a). Together, these domains allow PARP-1 to interact with genomic DNA and chromatin, poly(ADP-ribosyl)ate relevant nuclear targets, and regulate gene expression. Studies from the past few years, which will be the focus of this review, have begun to elucidate the underlying mechanisms and consequences of gene regulation by PARP-1.
Figure 1. Schematic representation of PARP-1's structural and functional organization and the reactions in the PARP-1-dependent protein PARylation pathway.
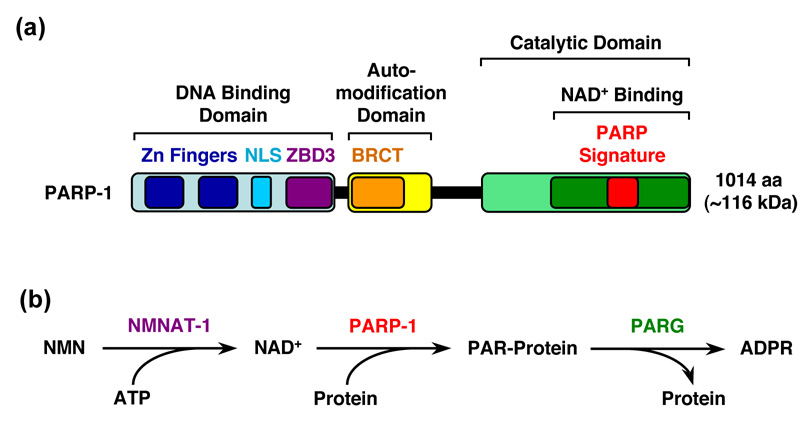
(a) PARP-1 has three major structural and functional domains: (1) an amino-terminal DNA binding domain containing two zinc finger motifs (Zn fingers), a nuclear localization signal (NLS), and a recently identified third zinc-binding domain (ZBD3), (2) a central automodification domain containing a BRCA1 C-terminus-like (BRCT) motif, and (3) a carboxyl-terminal catalytic domain containing an NAD+-binding domain and a highly conserved "PARP signature" motif that defines the PARP superfamily of proteins. (b) The following reactions occur in the PARP-1-dependent protein PARylation pathway: (1) NMNAT-1 synthesizes NAD+ from nicotinamide mononucleotide (NMN) and ATP, (2) PARP-1 polymerizes linear or branched chains of ADP-ribose (ADPR) from donor NAD+ molecules on target proteins, and (3) PARG hydrolyzes the chains of poly(ADP-ribose) (PAR) to release free ADPR.
PARP-1 activities and interactions
The biochemical activities of PARP-1 provide clues to how it might function in gene regulation in vivo. High affinity binding of PARP-1 to certain forms of DNA (double strand breaks, cruciforms, crossovers) and nucleosomes are mediated by the DBD [1,3,5–8]. PARP-1's enzymatic activity is low in basal or unstimulated conditions, but is potently allosterically activated by the binding of PARP-1 to a variety of interaction partners, including protein interaction partners, nucleosomes, and the aforementioned forms of DNA [1,3,5,7,9]. A newly identified third zinc binding domain located between the second zinc finger and the AMD (Fig. 1a) couples DNA binding to the allosteric activation of PARP-1's catalytic activity [10•]. Numerous nuclear targets for PARP-1 enzymatic activity have been identified, including core histones, the linker histone H1, and a variety of transcription factors, but PARP-1 is the major acceptor for PARP-1-dependent poly(ADP-ribosyl)ation (PARylation) reactions in vivo [1,3].Automodification blocks the ability of PARP-1 to bind to both DNA and nucleosomes [1,5,6•].
In addition to its interactions with DNA and substrates, PARP-1 associates with complexes containing a variety of other nuclear proteins, including transcription-related factors [11]. Recent studies have demonstrated functional associations of PARP-1 with a variety of regulatory complexes, including a TLE (Transducin-like enhancer of Split) corepressor complex [12], a Mediator coregulator complex [13], a condensin I/XRCC1 repair complex [14], a macroH2A1.1 nucleosome complex [15••], and a CTCF insulator complex [16,17]. As highlighted below, interactions with these protein partners dictate the location and actions of PARP-1 in gene regulation.
PARP-1 genomics
Understanding the role of PARP-1 in gene regulation requires knowledge of where PARP-1 binds in the genome and which genes are directly regulated by its actions. Recent genomic studies have begun to provide answers to these questions. Chromatin immunoprecipitation coupled to hybridization to genomic microarrays (i.e., ChIP-chip) has shown that PARP-1 binding is enriched at the promoters of perhaps as many as 90 percent of expressed RNA polymerase II (Pol II)-transcribed promoters in MCF-7 cells [18••]. Interestingly, the enrichment of PARP-1 at these promoters correlates with the depletion of the linker histone H1, and a high PARP-1/H1 ratio specifies genes that are actively transcribed. This does not, however, imply a stimulatory role for PARP-1 at all of these promoters, but rather indicates that PARP-1 localizes to sites of ongoing transcription, where it may in fact exert stimulatory or inhibitory effects [18••]. Although PARP-1 localizes to actively transcribed promoters, its localization pattern may not completely overlap that of active (i.e., phosphorylated) Pol II in a given gene [5] since PARP-1 generally peaks just upstream of the transcription start site (i.e., approximately −250 bp), whereas active Pol II localizes to the transcription start site and the body of the gene [18••].
Microarray expression analyses using cells and tissues derived from PARP-1-deficient (Parp-1−/−) mice have begun to yield information about the PARP-1-regulated transcriptome [19–21]. In a recent study exploring gene expression profiles in embryonic stem cells and livers from Parp-1−/− mice, ~3.5% of the transcriptome was regulated by PARP-1, with approximately 60 to 70% of the genes being positively regulated by PARP-1 [19]. The regulated gene sets play roles in critical cellular processes such as metabolism, stress responses, signal transduction, cell cycle control, and transcription, fitting well with the known functions of PARP-1 [19], but perhaps a bit surprising given the mild overall phenotype of Parp-1−/− mice [22].
The difference between the large number of PARP-1-bound promoters from ChIP-chip analyses and the limited number of PARP-1-regulated genes from expression analyses is striking, but perhaps not unexpected. A number of genomic analyses have revealed more genomic binding sites than regulated genes for a variety of factors. In the case of PARP-1, this may suggest that other factors, including related nuclear PARPs (e.g., PARP-2 ) [4], play redundant regulatory roles at many PARP-1-bound promoters and mask the effects of PARP-1 depletion or inhibition [2]. Alternatively, PARP-1-dependent regulation of some PARP-1-bound promoters may only occur in certain cell types or in response to appropriate cellular signals. In contrast, the localization of PARP-1 to some actively transcribed promoters may occur as a consequence of the transcription process (e.g., histone modification; cleavage or oxidation of promoter DNA [23••,24]) without PARP-1 playing a specific role in the regulation of those promoters. In this regard, PARP-1 may have an alternate role in promoter-localized transcription-coupled DNA repair distinct from the regulation of transcription initiation [14,23••–25], although such speculation requires additional proof.
Modulation of chromatin structure and composition by PARP-1
The earliest characterized effects of PARP-1 on the genome were the modulation of chromatin structure and the PARylation of histones [1,3,26,27] (Fig. 2a). The effects of PARP-1 on chromatin structure have been elaborated and elucidated in more recent biochemical and in vivo studies [5,6•,28,29.]. For example, biochemical assays have shown that PARP-1 binds to nucleosomes at the dyad axis with a stoichiometry of one [5]. Saturated PARP-1 binding to nucleosomes in the absence of NAD+ promotes the compaction of nucleosomal arrays into higher order structures [5], as recently visualized by atomic force microscopy [6•] (Fig. 3a). These PARP-1-dependent higher order structures are refractory to in vitro transcription [5,6•]. In the presence of saturating amounts of NAD+, PARP-1 automodifies and releases from the chromatin, leading to decompaction and the restoration of transcription [5,6•] (Fig. 3c). At first glance, these results may seem at odds with the genomic studies described above [18••]. How is PARP-1 binding at promoters associated with actively transcribed genes in vivo when it promotes chromatin compaction and transcriptional repression in vitro? The reason may be the NAD+ concentrations, with the former occurring at a physiological nuclear NAD+ concentration and the latter occurring in the non-physiological absence of NAD+ [5,6•,18••] (Fig. 3). Thus, although PARP-1 can bind to nucleosomes and promote saturating levels of chromatin compaction in vitro, more studies are needed to determine the extent to which PARP-1 compacts chromatin at promoters in vivo.
Figure 2. Multiple modes of transcriptional regulation by PARP-1.
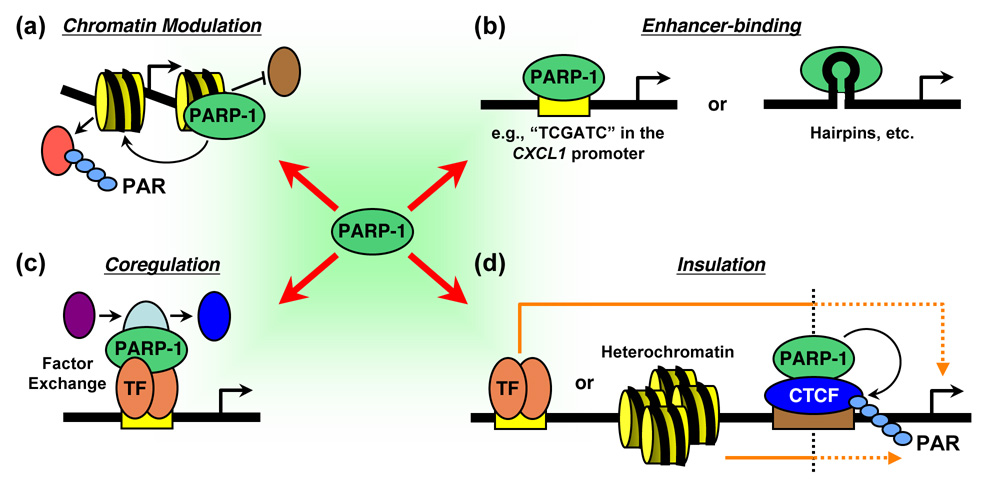
PARP-1 regulates transcription in perhaps as many as four modes, as indicated. (a) PARP-1 can modulate chromatin structure by binding to nucleosomes, modifying histone proteins, or regulating the composition of chromatin. (b) PARP-1 can act as an enhancer-binding factor that functions in a manner similar to classical sequence-specific DNA-binding activators or repressors. In this mode, PARP-1 may bind to specific sequences or structures in the DNA. (c) PARP-1 can function as a transcriptional coregulator in a manner similar to classical coactivators and corepressors. In this mode, PARP-1 may function as a promoter-specific "exchange factor" that promotes the release of inhibitory factors and the recruitment of stimulatory factors during signal-regulated transcriptional responses. TF, DNA-binding transcription factor (d) PARP-1 can function as a component of insulators, which act to limit the effects of enhancers on promoters or by preventing the spread of heterochromatin. In this mode, the PARylation of CTCF by PARP-1 is likely to play a role in the maintenance of insulator function.
Figure 3. Regulation of chromatin structure by PARP-1.
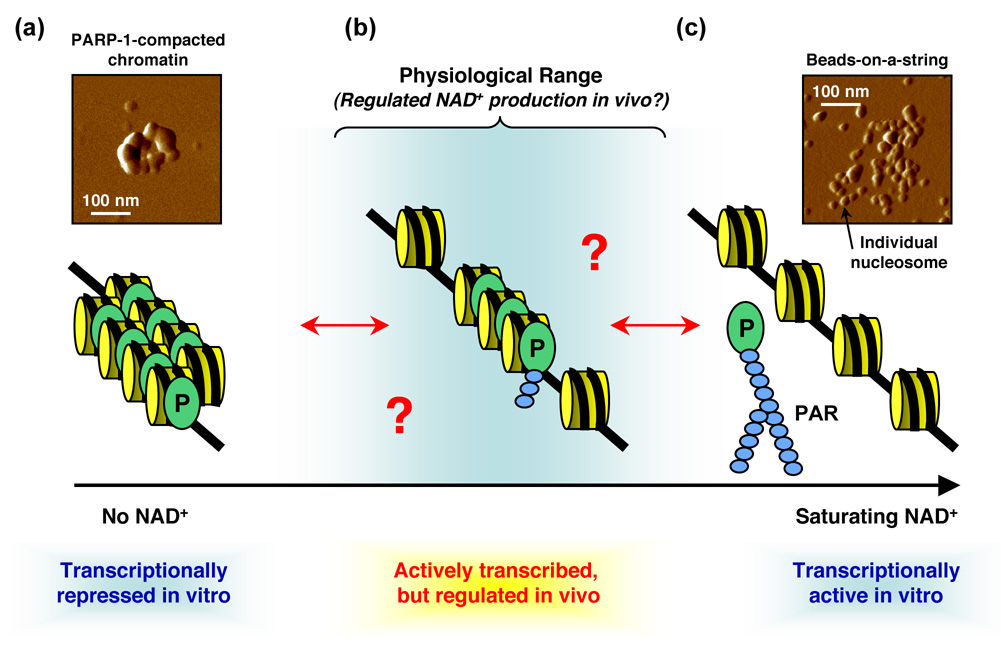
Schematic representation of the effects of PARP-1 on chromatin structure in the presence of different levels of NAD+. (a) In biochemical assays, saturated binding of PARP-1 (P) to nucleosomes in the absence of NAD+ and promotes the compaction of nucleosomal arrays into higher order structures. Inset: PARP-1-dependent compaction of a single molecule of reconstituted chromatin assembled from an ~11 kb plasmid, as visualized by atomic force microscopy (similar to the images described by Wacker et al., 2007 [6•]). A length scale bar is shown. (b) Hypothetical model of an intermediate chromatin state at physiological levels of NAD+ that might be found in vivo, although the actual structure is currently unknown. In this state, oligo(ADP-ribosyl)ation of PARP-1 may have a regulatory effect without promoting the dissociation of PARP-1 from chromatin. (c) In biochemical assays with saturating amounts of NAD+, PARP-1 automodifies and releases from the chromatin, leading to decompaction. Inset: NAD+-dependent decompaction ("beads-on-a-string" conformation) of a single molecule of PARP-1-containing chromatin assembled from an ~11 kb plasmid, as visualized by atomic force microscopy (similar to the images described by Wacker et al., 2007 [6•]). A length scale bar is shown. PAR, poly(ADP-ribose).
Recent studies have suggested a role for PARP-1 in regulating the composition of chromatin (Fig. 2a). For example, PARP-1 acts to exclude H1 from the promoters of some PARP-1-regulated genes [18••], possibly by competing with H1 for binding to nucleosomes [5] or by PARylating it [27]. Furthermore, during estrogen-induced transcription of the TFF1 (a.k.a. pS2) gene, PARP-1 not only promotes the removal of H1, but also increases the levels of HMGB1, a chromatin architectural protein that enhances transcription [23••]. In addition, PARP-1-dependent PARylation of DEK, an abundant and ubiquitous component of chromatin, promotes the release of DEK from chromatin, the loading of the Mediator coregulatory complex, and the enhancement of transcription [30•]. PARP-1-dependent PARylation may also affect the genomic actions of SIRT1, an NAD+-dependent histone deacetylase [2].
Two recent studies have suggested that interactions between PARP-1 and the histone variant macroH2A, an H2A variant containing a large carboxyl-terminal non-histone region called a "macro" domain, may provide another means for bringing PARP-1 to chromatin and regulating PARP-1 enzymatic activity [15••,31•]. The common observations from these studies are: (1) binding of PARP-1 to the "non-histone" domain of macroH2A1 (splice variants 1.1 or 1.2) and (2) inhibition of PARP-1 enzymatic activity by macroH2A (full length 1.1 [15••] or the non-histone domain from 1.1, 1.2, or 2 [31•]). In spite of these similarities, RNAi-mediated depletion of macroH2A1 or PARP-1 differentially affects the expression of target genes in the two systems studied. Specifically, depletion of macroH2A1 or PARP-1 blocks heat shock-induced expression of the HSP70.1 gene in HeLa cells [15••], but reactivates expression of an inactive X (Xi)-linked GFP transgene in mouse embryo fibroblast cells [31•]. Further studies are required to fully understand the functional link between PARP-1 and macroH2A.
Enhancer-binding actions of PARP-1
Many of the initial studies describing direct effects of PARP-1 on the transcriptional regulation of target genes focused on the binding of PARP-1 to specific DNA sequences or structures in the regulatory regions of the genes, allowing PARP-1 to function like a classical enhancer-binding factor [32–36] (Fig. 2b). In fact, direct binding of PARP-1 to hairpins may underlie an autoregulatory mechanism governing the expression of the PARP-1 gene itself [37]. Two recent studies have examined the role of direct DNA binding by PARP-1 in the regulation of the PARP-1 target genes CXCL1 [38•] and BCL6 [39•]. PARP-1 binds to specific sequences immediately upstream of the CXCL1 promoter and in the first intron of BCL6 to repress transcription. For CXCL1, the binding of PARP-1 inhibits expression by preventing the binding of NF-κB to an adjacent element, an effect that is reversed upon PARP-1 activation and automodification, resulting in a loss of PARP-1 binding to the promoter [38•]. The generality of enhancer-binding as a mode of transcriptional regulation is unknown. As the determinants of PARP-1 binding to DNA are elucidated, bioinformatic analyses may aid in the identification of new PARP-1 target genes subject to this type of control.
Transcriptional coregulation by PARP-1
Roles for PARP-1 as a promoter-specific coregulator (either a coactivator or a corepressor) for a number of different sequence-specific DNA-binding transcriptional regulators, such as NF-κB, nuclear receptors, HES1, B-Myb, Oct-1, HTLV Tax-1, Sp1, NFAT, Elk1, and others, have been reported [2,3,12,13,40–43••] (Fig. 2c). In most of these cases, the DNA-binding factor is thought to recruit PARP-1 to relevant target promoters. Yet, ChIP-chip analyses have shown that many peaks of promoter-proximal PARP-1 binding are quite broad (i.e., as much as 3 kb or more shoulder to shoulder) [18••]. How such a genomic localization pattern relates to recruitment by a DNA-binding factor to a specific site is not clear. In some cases, PARP-1 enzymatic activity is not required for its coregulatory activity (e.g., with NF-κB, B-Myb, and HTLV Tax-1) [2,3,40], while in others it is required (e.g., HES1, Sp1, NFAT, and Elk1) [12,41–43••]. In many of the latter cases, the DNA-binding factor or other components of the coregulatory complex are targets for PARP-1-dependent PARylation [12,41,42].
A key question regarding PARP-1 coregulatory activity is the effect that it has on the transcription complexes assembled at target promoters. Recent studies have shown that PARP-1 can function as a promoter-specific "exchange factor" that promotes the release of inhibitory factors and the recruitment of stimulatory factors during signal-regulated transcriptional responses [12,13,23••]. For example, PARP-1 has been shown to promote the exchange of a TLE1 corepressor complex for a HAT-containing coactivator complex with HES1 during signal-dependent activation in neuronal cells [12]. Likewise, PARP-1 can promote the exchange of an inactive cdk8-positive Mediator for an active cdk8-negative Mediator during retinoic acid-regulated activation [13].
More recently, a study describing perhaps the most intriguing actions of PARP-1 at promoters has added a new twist to the exchange factor model [23••]. In this study, PARP-1 was shown to promote the recruitment of topoisomerase IIβ (TopoIIβ) to hormone-regulated promoters, leading to concomitant promoter DNA cleavage, factor exchange, and transcriptional activation [23••,44,45]. The study focused on the estrogen-regulated TFF1 gene promoter, which is bound by a PARP-1 corepressor complex containing the corepressor NCoR and the histone deacetylase HDAC3 prior to activation. Estrogen exposure rapidly promotes an exchange of the corepressor complex for a PARP-1 coactivator complex containing TopoIIβ, the coregulator ASC2, and the DNA repair proteins Ku86/70 and DNA-PK. This results in a transient TopoIIβ-dependent cleavage of the promoter DNA near the estrogen receptor binding site, which may resolve a topological barrier and allow for favorable structural changes at the promoter. Recruitment of the PARP-1 coactivator complex also promotes the release of histone H1, the recruitment of HMGB1/2, changes in chromatin architecture, and ultimately increased transcription of the gene [23••,44,45]. Together, these studies highlight the diverse mechanisms of PARP-1 coregulator function, which are likely to vary in an activator-and gene-specific manner.
Insulator functions of PARP-1: CTCF, the nuclear matrix, and DNA methylation
Insulators are DNA elements that help to organize the genome into discrete regulatory units by limiting the effects of enhancers on promoters or by preventing the spread of heterochromatin [46]. Recent studies have implicated PARP-1-dependent PARylation of CTCF, a ubiquitous DNA-binding protein that functions at insulators, in the preservation of insulator function [16,17,47] (Fig. 2d). In this regard, the general PARP inhibitor 3-aminobenzamide blocks insulator function in cell-based assays [16,17,47]. PARP-1 actions at insulators may also involve associations with the nuclear matrix and DNA methylation, both of which have been implicated in insulator function [46,47]. Note, for example, that CTCF, PARP-1, and PARP-1-associated proteins (e.g., nucleophosmin, topoisomerase II, Ku, DNA-PK) have been shown to associate with components of the nuclear matrix [17,48–51]. In addition, PARP-1 has been shown to regulate DNA methylation [52,53], perhaps through the regulation of the DNA methyltransferase DNMT1 [54], and DNA methylation can modulate the binding of CTCF to insulators [55]. Additional studies are needed to clarify the role of PARP-1 in insulator function, especially as it relates to the regulation of PARP-1 target genes.
Signaling and regulation in PARP-1-dependent gene expression pathways
A number of cellular signaling pathways culminate with the regulation of PARP-1-dependent transcriptional processes (Fig. 4). Mediating signals include small molecules (e.g., steroid and vitamin hormones) [13,23••], heat shock [15••,29], and kinases (e.g., CaM kinase IIδ and ERK2) [12,43••]. Cellular signaling may result in the post-translational modification of PARP-1 through autoPARylation [43••], acetylation [56], or phosphorylation [57•,58], which alter PARP-1 activity for gene-specific or other regulatory outcomes.
Figure 4. Signaling and regulation in PARP-1-dependent gene expression pathways.
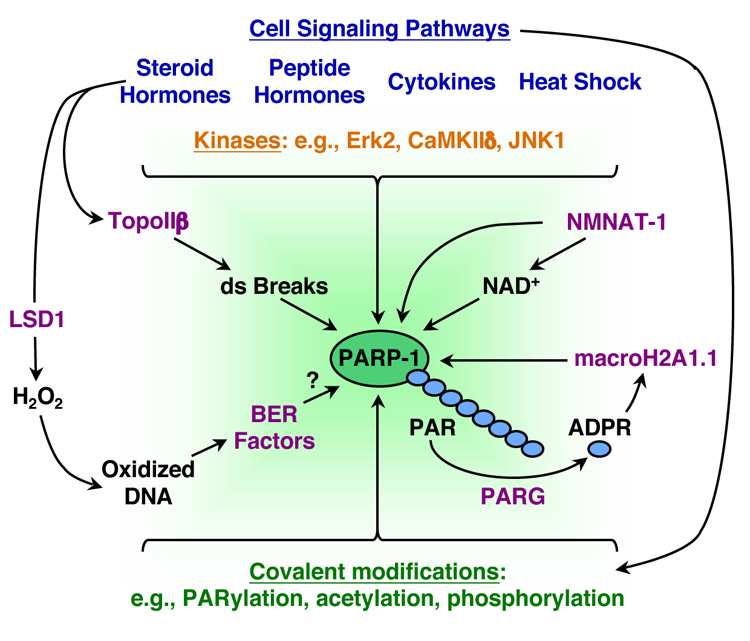
Schematic representation of some of the many signaling and regulatory inputs that affect PARP-1-dependent transcriptional outcomes. Mediating signals include hormones, cytokines, heat shock, and intracellular kinases (e.g., CaM kinase IIδ and Erk2). One endpoint of these signaling pathways is the post-translational modification of PARP-1 by autoPARylation, acetylation, and phosphorylation. NAD+ synthesis by NMNAT-1 and PAR catabolism by PARG represent additional points of control. In addition to producing NAD+ for use by PARP-1, NMNAT-1 also stimulates PARP-1 catalytic activity. The degradation of PAR polymers by PARG yields monomers of ADP-ribose (ADPR), a ligand for the macro domain of macroH2A1.1, which may have signaling functions in the nucleus. Transcription-induced DNA damage may also affect PARP-1's gene-regulatory activities at target promoters. For example, estrogen signaling can induce transient TopoIIβ-dependent double strand (ds) DNA breaks, which may promote the recruitment and activation of PARP-1. Likewise, estrogen-induced demethylation of histones by LSD1 generates H2O2 that can oxidize promoter DNA and promote the recruitment of components of the base excision repair (BER) machinery and TopoIIβ, which may in turn promote the recruitment and activation of PARP-1.
A recent study has revealed an interesting alternate signal-dependent mechanism for the activation of PARP-1 enzymatic activity that does not require DNA binding by PARP-1 [43••]. The binding of PARP-1 by ERK2 potently stimulates PARP-1 enzymatic activity and, as a result, increases PARP-1 autoPARylation. Upon activation, PARP-1 promotes the ERK2-dependent phosphorylation of a downstream effector, the DNA-binding transcription factor Elk1, resulting in an increase in histone acetylation and target gene expression [43••]. Direct phosphorylation of PARP-1 by ERK1/2 may also enhance PARP-1 activity, but the effects on transcription have not been determined [57•]. Another recent study has suggested a means by which promoter-localized effects of signaling might promote DNA oxidation-dependent PARP-1 actions at promoters [24]. Estrogen-induced demethylation of histones by LSD1 at the BCL-2 gene generates H2O2 that can oxidize the promoter DNA, leading to the recruitment of components of the base excision repair machinery (e.g., OGG1) and TopoIIβ [24], which could conceivably recruit and activate PARP-1 (Fig. 4). The latter is reminiscent of the estrogen-induced recruitment of TopoIIβ to the TFF1 gene promoter [23••]. Whether such a mechanism also involves the recruitment of PARP-1, however, has not been determined.
The regulation of nuclear NAD+ synthesis and PAR catabolism represent additional points of control for PARP-1-dependent gene regulation (Fig. 4). The production of NAD+ in the nucleus is controlled by the nuclear NAD+ synthase nicotinamide mononucleotide adenylyltransferase-1 (NMNAT-1), whereas the degradation of PAR is mediated by poly(ADP-ribose) glycohydrolase (PARG) [2,59]. NMNAT-1 not only produces NAD+ for use by PARP-1, but it also stimulates PARP-1 activity and binds to PAR, thereby regulating PARP-1-dependent outcomes [60•]. The catabolism of PAR polymers by PARG occurs rapidly in vivo to yield monomers of ADP-ribose (ADPR). This may: (1) inhibit PAR-dependent processes, (2) re-set PAR-dependent processes so that continued regulation can occur, or (3) generate ADPR, which may have signaling functions in the nucleus [1–3,61]. With respect to the latter, ADPR has recently been shown to be a ligand for the macro domain of macroH2A1.1, but not the 1.2 splice variant [62]. Although the functional consequences of this are unknown, it may represent a mode of chromatin-dependent nuclear signaling.
Conclusions
The available data indicate that PARP-1 regulates transcription in perhaps as many as four modes: (1) as a modulator of chromatin structure by binding to nucleosomes, modifying histone proteins, or regulating the composition of chromatin, (2) as an enhancer-binding factor that functions in a manner similar to classical sequence-specific DNA-binding activators or repressors, (3) as a transcriptional coregulator that functions in a manner similar to classical coactivators and corepressors, and (4) as a component of transcriptional insulators. Although recent studies have clarified some of the molecular details of these different modes of regulation, a unified model for the regulation of gene expression by PARP-1 remains elusive. Further studies examining the generality of these types of regulation, their interrelationships, and how they are regulated by cellular signaling pathways are needed.
Acknowledgements
The author thanks Matthew Gamble, Kristine Frizzell, and Raga Krishnakumar for critical comments and helpful suggestions. The author's laboratory is supported by funding from the National Institute of Diabetes, Digestive, and Kidney Disorders.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.D'Amours D, Desnoyers S, D'Silva I, Poirier G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 2.Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: 'PAR-laying' NAD+ into a nuclear signal. Genes Dev. 2005;19:1951–1967. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- 3.Kraus WL, Lis J. PARP goes transcription. Cell. 2003;113:677–683. doi: 10.1016/s0092-8674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- 4.Amé JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 5.Kim MY, Mauro S, Gevry N, Lis J, Kraus WL. NAD+-dependent modulation of chromatin structure and tanscription by nucleosome binding properties of PARP-1. Cell. 2004;119:803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Wacker DA, Ruhl DD, Balagamwala EH, Hope KM, Zhang T, Kraus WL. The DNA binding and catalytic domains of poly(ADP-ribose) polymerase 1 cooperate in the regulation of chromatin structure and transcription. Mol Cell Biol. 2007;27:7475–7485. doi: 10.1128/MCB.01314-07.This paper provides direct visual evidence for PARP-1-dependent compaction of chromatin by using atomic force microscopy of individual molecules of chromatin assembled in vitro. In addition, this paper demonstrates that: (1) the DBD of PARP-1 is necessary and sufficient for binding to nucleosomes, but alone is unable to promote chromatin compaction and (2) the catalytic domain of PARP-1, which does not bind nucleosomes on its own, cooperates with the DBD to promote chromatin compaction in a manner independent of its enzymatic activity. Thus, the catalytic domain of PARP-1 plays a previously uncharacterized role in chromatin compaction
- 7.Lonskaya I, Potaman VN, Shlyakhtenko LS, Oussatcheva EA, Lyubchenko YL, Soldatenkov VA. Regulation of poly(ADP-ribose) polymerase-1 by DNA structure-specific binding. J Biol Chem. 2005;280:17076–17083. doi: 10.1074/jbc.M413483200. [DOI] [PubMed] [Google Scholar]
- 8.Potaman VN, Shlyakhtenko LS, Oussatcheva EA, Lyubchenko YL, Soldatenkov VA. Specific binding of poly(ADP-ribose) polymerase-1 to cruciform hairpins. J Mol Biol. 2005;348:609–615. doi: 10.1016/j.jmb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 9.Kun E, Kirsten E, Mendeleyev J, Ordahl CP. Regulation of the enzymatic catalysis of poly(ADP-ribose) polymerase by dsDNA, polyamines, Mg2+, Ca2+, histones H1 and H3, and ATP. Biochemistry. 2004;43:210–216. doi: 10.1021/bi0301791. [DOI] [PubMed] [Google Scholar]
- 10.Langelier MF, Servent KM, Rogers EE, Pascal JM. A third zinc-binding domain of human poly(ADP-ribose) polymerase-1 coordinates DNA-dependent enzyme activation. J Biol Chem. 2008;283:4105–4114. doi: 10.1074/jbc.M708558200.Using spectroscopic and crystallographic analyses, this paper identifies and characterizes a previously unidentified third zinc-binding domain in PARP-1. This new zinc binding domain, which is located between the amino-terminal zinc fingers in the DBD and the BRCT motif in the AMD, mediates interdomain contacts important for DNA-dependent enzyme activation. The crystal structure reveals a zinc ribbon fold and points to conserved residues that could form interdomain contacts that may help to couple the DNA binding and catalytic functions of PARP-1. Together, the results described in this paper provide a structural and mechanistic basis for the DBD-dependent allosteric activation of PARP-1 enzymatic activity.
- 11.Droit A, Hunter JM, Rouleau M, Ethier C, Picard-Cloutier A, Bourgais D, Poirier GG. PARPs Database: A LIMS systems for protein-protein interaction data mining or Laboratory Information management system. BMC Bioinformatics. 2007;8:483. doi: 10.1186/1471-2105-8-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ju BG, Solum D, Song EJ, Lee KJ, Rose DW, Glass CK, Rosenfeld MG. Activating the PARP-1 sensor component of the groucho/ TLE1 corepressor complex mediates a CaMKinase IIdelta-dependent neurogenic gene activation pathway. Cell. 2004;119:815–829. doi: 10.1016/j.cell.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 13.Pavri R, Lewis B, Kim TK, Dilworth FJ, Erdjument-Bromage H, Tempst P, de Murcia G, Evans R, Chambon P, Reinberg D. PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol Cell. 2005;18:83–96. doi: 10.1016/j.molcel.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 14.Heale JT, Ball AR, Jr, Schmiesing JA, Kim JS, Kong X, Zhou S, Hudson DF, Earnshaw WC, Yokomori K. Condensin I interacts with the PARP-1-XRCC1 complex and functions in DNA single-strand break repair. Mol Cell. 2006;21:837–848. doi: 10.1016/j.molcel.2006.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ouararhni K, Hadj-Slimane R, Ait-Si-Ali S, Robin P, Mietton F, Harel-Bellan A, Dimitrov S, Hamiche A. The histone variant mH2A1.1 interferes with transcription by down-regulating PARP-1 enzymatic activity. Genes Dev. 2006;20:3324–3336. doi: 10.1101/gad.396106.Together with [31], this paper describes interactions between PARP-1 and the "non-histone" domain of the histone variant macroH2A1 (specifically the 1.1 splice variant in this case),which may help to bring PARP-1 to chromatin and inhibit its enzymatic activity. These interactions between PARP-1 and macroH2A1 play a role in regulating gene expression. Specifically, this paper shows that RNAi-mediated depletion of macroH2A1 or PARP-1 blocks heat shock-induced expression of the HSP70.1 gene. Together, these studies suggest an interesting functional link between PARP-1 and macroH2A.
- 16.Yu W, Ginjala V, Pant V, Chernukhin I, Whitehead J, Docquier F, Farrar D, Tavoosidana G, Mukhopadhyay R, Kanduri C, et al. Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat Genet. 2004;36:1105–1110. doi: 10.1038/ng1426. [DOI] [PubMed] [Google Scholar]
- 17.Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 18.Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, Kraus WL. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319:819–821. doi: 10.1126/science.1149250.Using ChIP-chip, this paper provides the first high-resolution map of PARP-1 and histone H1 localization across a mammalian genome. The data show that PARP-1 and H1 exhibit a reciprocal localization pattern at many Pol II-transcribed promoters, with PARP-1 enriched and H1 depleted. This high PARP-1/H1 ratio is associated with actively transcribed genes. Gene-specific studies showed that PARP-1 acts to exclude H1 from a subset of PARP-1-stimulated promoters, suggesting a functional interplay between PARP-1 and H1 at the level of nucleosome binding reminiscent of the results from the biochemical studies presented in [5]. Together, these data indicate that PARP-1 and H1 have opposing actions at many Pol II-transcribed promoters.
- 19.Ogino H, Nozaki T, Gunji A, Maeda M, Suzuki H, Ohta T, Murakami Y, Nakagama H, Sugimura T, Masutani M. Loss of Parp-1 affects gene expression profile in a genome-wide manner in ES cells and liver cells. BMC Genomics. 2007;8:41. doi: 10.1186/1471-2164-8-41. see Erratum in BMC Genomics 2007, 8:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simbulan-Rosenthal CM, Ly DH, Rosenthal DS, Konopka G, Luo R, Wang ZQ, Schultz PG, Smulson ME. Misregulation of gene expression in primary fibroblasts lacking poly(ADP-ribose) polymerase. Proc Natl Acad Sci U S A. 2000;97:11274–11279. doi: 10.1073/pnas.200285797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zingarelli B, Hake PW, O'Connor M, Denenberg A, Kong S, Aronow BJ. Absence of poly(ADP-ribose)polymerase-1 alters nuclear factor-kappa B activation and gene expression of apoptosis regulators after reperfusion injury. Mol Med. 2003;9:143–153. doi: 10.2119/2003-00011.zingarelli. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang ZQ, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, Wagner EF. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 1995;9:509–520. doi: 10.1101/gad.9.5.509. [DOI] [PubMed] [Google Scholar]
- 23.Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIβ-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196.This paper describes intriguing actions of PARP-1 at target gene promoters, adding a new twist to the exchange factor model. Specifically, this paper uses the estrogen-regulated TFF1 gene to study hormone-induced gene activation in the context of chromatin. Prior to activation, the TFF1 promoter is bound by a PARP-1 corepressor complex containing NCoR and the histone deacetylase HDAC3. Estrogen exposure rapidly promotes an exchange of the corepressor complex for a PARP-1 coactivator complex containing TopoIIβ, which transiently cleaves the promoter DNA. Recruitment of the PARP-1 coactivator complex also promotes the release of histone H1, the recruitment of HMGB1/2, changes in chromatin architecture, and ultimately increased transcription of the gene. This novel mechanism links the actions of two DNA-associated enzymes, namely PARP-1 and TopoIIβ, in the regulation of gene expression.
- 24.Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 25.Malanga M, Althaus FR. The role of poly(ADP-ribose) in the DNA damage signaling network. Biochem Cell Biol. 2005;83:354–364. doi: 10.1139/o05-038. [DOI] [PubMed] [Google Scholar]
- 26.Poirier G, de Murcia G, Jongstra-Bilen J, Niedergang C, Mandel P. Poly(ADP-ribosyl) ation of polynucleosomes causes relaxation of chromatin structure. Proc. Natl. Acad. Sci. USA. 1982;79:3423–3427. doi: 10.1073/pnas.79.11.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huletsky A, de Murcia G, Muller S, Hengartner M, Menard L, Lamarre D, Poirier GG. The effect of poly(ADP-ribosyl)ation on native and H1-depleted chromatin. A role of poly(ADP-ribosyl)ation on core nucleosome structure. J Biol Chem. 1989;264:8878–8886. [PubMed] [Google Scholar]
- 28.Tulin A, Stewart D, Spradling AC. The Drosophila heterochromatic gene encoding poly(ADP-ribose) polymerase (PARP) is required to modulate chromatin structure during development. Genes Dev. 2002;16:2108–2119. doi: 10.1101/gad.1003902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- 30.Gamble MJ, Fisher RP. SET and PARP1 remove DEK from chromatin to permit access by the transcription machinery. Nat Struct Mol Biol. 2007;14:548–555. doi: 10.1038/nsmb1248.This paper uses in vitro reconstitutions with chromatin templates to examine the functional interplay between PARP-1, the histone chaperone SET, and the chromatin-associated factor DEK. In the absence of SET and NAD+, PARP-1 and DEK repress transcription by restricting access of the transcriptional machinery to chromatin. SET enhances transcription in the absence of NAD+ by promoting the eviction of PARP-1 and DEK. In the presence of NAD+, PARP1 PARylates and evicts DEK and itself from chromatin to permit the loading of the transcriptional machinery in a SET-independent manner. Thus, these studies point to a role for PARP-1 in regulating the composition of chromatin.
- 31.Nusinow DA, Hernandez-Munoz I, Fazzio TG, Shah GM, Kraus WL, Panning B. Poly(ADP-ribose) polymerase 1 is inhibited by a histone H2A variant, macroH2A, and contributes to silencing of the inactive X chromosome. J Biol Chem. 2007;282:12851–12859. doi: 10.1074/jbc.M610502200.Together with [15], this paper describes interactions between PARP-1 and the "non-histone" domain of the histone variant macroH2A1 (specifically the 1.2 splice variant in this case), which may help to bring PARP-1 to chromatin and inhibit its enzymatic activity. RNAi-mediated depletion of macroH2A1 or PARP-1 reactivates expression of an inactive X-linked GFP transgene in mouse embryo fibroblast cells, suggesting that PARP-1 participates in the maintenance of silencing. Together, these studies suggest an interesting functional link between PARP-1 and macroH2A.
- 32.Huang K, Tidyman WE, Le KU, Kirsten E, Kun E, Ordahl CP. Analysis of nucleotide sequence-dependent DNA binding of poly(ADP-ribose) polymerase in a purified system. Biochemistry. 2004;43:217–223. doi: 10.1021/bi0301800. [DOI] [PubMed] [Google Scholar]
- 33.Butler AJ, Ordahl CP. Poly(ADP-ribose) polymerase binds with transcription enhancer factor 1 to MCAT1 elements to regulate muscle-specific transcription. Mol Cell Biol. 1999;19:296–306. doi: 10.1128/mcb.19.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nirodi C, NagDas S, Gygi SP, Olson G, Aebersold R, Richmond A. A role for poly(ADP-ribose) polymerase in the transcriptional regulation of the melanoma growth stimulatory activity (CXCL1) gene expression. J Biol Chem. 2001;276:9366–9374. doi: 10.1074/jbc.M009897200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Z, Hildebrandt EF, Simbulan-Rosenthal CM, Anderson MG. Sequence-specific binding of poly(ADP-ribose) polymerase-1 to the human T cell leukemia virus type-I tax responsive element. Virology. 2002;296:107–116. doi: 10.1006/viro.2002.1385. [DOI] [PubMed] [Google Scholar]
- 36.Akiyama T, Takasawa S, Nata K, Kobayashi S, Abe M, Shervani NJ, Ikeda T, Nakagawa K, Unno M, Matsuno S, et al. Activation of Reg gene, a gene for insulin-producing beta-cell regeneration: poly(ADP-ribose) polymerase binds Reg promoter and regulates the transcription by autopoly(ADP-ribosyl)ation. Proc Natl Acad Sci U S A. 2001;98:48–53. doi: 10.1073/pnas.240458597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soldatenkov VA, Chasovskikh S, Potaman VN, Trofimova I, Smulson ME, Dritschilo A. Transcriptional repression by binding of poly(ADP-ribose) polymerase to promoter sequences. J Biol Chem. 2002;277:665–670. doi: 10.1074/jbc.M108551200. [DOI] [PubMed] [Google Scholar]
- 38.Amiri KI, Ha HC, Smulson ME, Richmond A. Differential regulation of CXC ligand 1 transcription in melanoma cell lines by poly(ADP-ribose) polymerase-1. Oncogene. 2006;25:7714–7722. doi: 10.1038/sj.onc.1209751.This paper and [39] provide new examples of gene regulation through sequence-specific binding of PARP-1 to DNA regulatory elements. This paper shows that unactivated PARP-1 binds to a DNA element in the CXCL1 gene promoter and acts to block the binding of NF-κB to an adjacent element. Activation of PARP-1 leads to autoPARylation, loss of PARP-1 binding, increased NF-κB binding, and enhanced CXCL1 expression.
- 39.Ambrose HE, Papadopoulou V, Beswick RW, Wagner SD. Poly-(ADP-ribose) polymerase-1 (Parp-1) binds in a sequence-specific manner at the Bcl-6 locus and contributes to the regulation of Bcl-6 transcription. Oncogene. 2007;26:6244–6252. doi: 10.1038/sj.onc.1210434.This paper and [38] provide new examples of gene regulation through sequence-specific binding of PARP-1 to DNA regulatory elements. This paper identifies a conserved regulatory element in the first intron of the BCL6 gene to which PARP-1 binds. PARP inhibitors and PARP-1 depletion by RNAi induce BCL6 expression, suggesting that PARP-1 activation plays a role in inhibiting BCL6 expression.
- 40.Hassa PO, Hottiger MO. The functional role of poly(ADP-ribose)polymerase 1 as novel coactivator of NF-kappaB in inflammatory disorders. Cell Mol Life Sci. 2002;59:1534–1553. doi: 10.1007/s00018-002-8527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olabisi OA, Soto-Nieves N, Nieves E, Yang TT, Yang XY, Yu RY, Suk HY, Macian F, Chow CW. Regulation of Transcription Factor NFAT by ADP-Ribosylation. Mol Cell Biol. 2008 doi: 10.1128/MCB.01746-07. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaniolo K, Desnoyers S, Leclerc S, Guerin SL. Regulation of poly(ADP-ribose) polymerase-1 (PARP-1) gene expression through the post-translational modification of Sp1: a nuclear target protein of PARP-1. BMC Mol Biol. 2007;8:96. doi: 10.1186/1471-2199-8-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cohen-Armon M, Visochek L, Rozensal D, Kalal A, Geistrikh I, Klein R, Bendetz-Nezer S, Yao Z, Seger R. DNA-independent PARP-1 activation by phosphorylated ERK2 increases Elk1 activity: a link to histone acetylation. Mol Cell. 2007;25:297–308. doi: 10.1016/j.molcel.2006.12.012.Together with [57], this paper describes a functional interplay between PARP-1 and ERK (ERK2 in this paper), although the outcomes of the interplay are different than those shown in [57]. The results of this paper reveal an alternate signal-dependent mechanism for the activation of PARP-1 enzymatic activity that does not require DNA binding by PARP-1. Specifically, the binding of PARP-1 by ERK2 potently stimulates PARP-1 enzymatic activity and, as a result, increases PARP-1 autoPARylation. PARP-1 activated in this manner dramatically increases ERK2- dependent phosphorylation of its downstream effector, the DNA-binding transcription factor Elk1, resulting in an increase in histone acetylation and target gene expression. Thus, this study brings together the ERK2 signaling pathway and the gene regulatory actions of PARP-1
- 44.Ju BG, Rosenfeld MG. A breaking strategy for topoisomerase IIbeta/PARP-1-dependent regulated transcription. Cell Cycle. 2006;5:2557–2560. doi: 10.4161/cc.5.22.3497. [DOI] [PubMed] [Google Scholar]
- 45.Lis JT, Kraus WL. Promoter cleavage: a topoIIbeta and PARP-1 collaboration. Cell. 2006;125:1225–1227. doi: 10.1016/j.cell.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 46.Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr Opin Genet Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klenova E, Ohlsson R. Poly(ADP-ribosyl)ation and epigenetics. Is CTCF PARt of the plot? Cell Cycle. 2005;4:96–101. doi: 10.4161/cc.4.1.1398. [DOI] [PubMed] [Google Scholar]
- 48.Yusufzai TM, Felsenfeld G. The 5'-HS4 chicken beta-globin insulator is a CTCF-dependent nuclear matrix-associated element. Proc Natl Acad Sci U S A. 2004;101:8620–8624. doi: 10.1073/pnas.0402938101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Galande S, Kohwi-Shigematsu T. Poly(ADP-ribose) polymerase and Ku autoantigen form a complex and synergistically bind to matrix attachment sequences. J Biol Chem. 1999;274:20521–20528. doi: 10.1074/jbc.274.29.20521. [DOI] [PubMed] [Google Scholar]
- 50.Vidakovic M, Grdovic N, Quesada P, Bode J, Poznanovic G. Poly(ADP-ribose) polymerase-1: association with nuclear lamins in rodent liver cells. J Cell Biochem. 2004;93:1155–1168. doi: 10.1002/jcb.20289. [DOI] [PubMed] [Google Scholar]
- 51.Vidakovic M, Koester M, Goetze S, Winkelmann S, Klar M, Poznanovic G, Bode J. Co-localization of PARP-1 and lamin B in the nuclear architecture: a halo-fluorescenc-eand confocal-microscopy study. J Cell Biochem. 2005;96:555–568. doi: 10.1002/jcb.20516. [DOI] [PubMed] [Google Scholar]
- 52.Althaus FR. Poly(ADP-ribose): a co-regulator of DNA methylation? Oncogene. 2005;24:11–12. doi: 10.1038/sj.onc.1208382. [DOI] [PubMed] [Google Scholar]
- 53.Zardo G, Reale A, De Matteis G, Buontempo S, Caiafa P. A role for poly(ADPribosyl)ation in DNA methylation. Biochem Cell Biol. 2003;81:197–208. doi: 10.1139/o03-050. [DOI] [PubMed] [Google Scholar]
- 54.Reale A, Matteis GD, Galleazzi G, Zampieri M, Caiafa P. Modulation of DNMT1 activity by ADP-ribose polymers. Oncogene. 2005;24:13–19. doi: 10.1038/sj.onc.1208005. [DOI] [PubMed] [Google Scholar]
- 55.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 56.Hassa PO, Haenni SS, Buerki C, Meier NI, Lane WS, Owen H, Gersbach M, Imhof R, Hottiger MO. Acetylation of poly(ADP-ribose) polymerase-1 by p300/CREB-binding protein regulates coactivation of NF-kappaB-dependent transcription. J Biol Chem. 2005;280:40450–40464. doi: 10.1074/jbc.M507553200. [DOI] [PubMed] [Google Scholar]
- 57.Kauppinen TM, Chan WY, Suh SW, Wiggins AK, Huang EJ, Swanson RA. Direct phosphorylation and regulation of poly(ADP-ribose) polymerase-1 by extracellular signal-regulated kinases 1/2. Proc Natl Acad Sci U S A. 2006;103:7136–7141. doi: 10.1073/pnas.0508606103.Together with [43], this paper describes functional interplay between PARP-1 and ERK (ERK2 in this paper), although the outcomes of the interplay are different than those shown in [43]. This paper (1) demonstrates that direct phosphorylation of PARP-1 by ERK1/2 can enhance PARP-1 activity and (2) identifies ERK1/2 phosphorylation sites on PARP-1 by mass spectrometry and site-directed mutagenesis. Although ERK1/2 pathway inhibitors blocked PARP-1 activation and PARP-1-mediated neuronal death, the effects on PARP-1-dependent gene expression were not determined
- 58.Zhang S, Lin Y, Kim YS, Hande MP, Liu ZG, Shen HM. c-Jun N-terminal kinase mediates hydrogen peroxide-induced cell death via sustained poly(ADP-ribose) polymerase-1 activation. Cell Death Differ. 2007;14:1001–1010. doi: 10.1038/sj.cdd.4402088. [DOI] [PubMed] [Google Scholar]
- 59.Bonicalzi ME, Haince JF, Droit A, Poirier GG. Regulation of poly(ADP-ribose) metabolism by poly(ADP-ribose) glycohydrolase: where and when? Cell Mol Life Sci. 2005;62:739–750. doi: 10.1007/s00018-004-4505-1. [DOI] [PubMed] [Google Scholar]
- 60.Berger F, Lau C, Ziegler M. Regulation of poly(ADP-ribose) polymerase 1 activity by the phosphorylation state of the nuclear NAD biosynthetic enzyme NMN adenylyl transferase 1. Proc Natl Acad Sci U S A. 2007;104:3765–3770. doi: 10.1073/pnas.0609211104.This paper describes a role for the nuclear NAD+ synthase NMNAT-1 in the regulation of PARP-1 enzymatic activity. Specifically, the results show that NMNAT-1 stimulates PARP-1 automodification, as well as binds to PAR. Phosphorylation of NMNAT-1 by protein kinase C reduces its binding to PAR, suggesting that NMNAT-1 may be an endpoint of cell signaling pathways. Together, these results demonstrate that NMNAT-1 can enhance the activity of PARP-1 and, thus, amplify PARylation
- 61.Tulin A, Naumova NM, Menon AK, Spradling AC. Drosophila poly(ADP-ribose) glycohydrolase mediates chromatin structure and SIR2-dependent silencing. Genetics. 2006;172:363–371. doi: 10.1534/genetics.105.049239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kustatscher G, Hothorn M, Pugieux C, Scheffzek K, Ladurner AG. Splicing regulates NAD metabolite binding to histone macroH2A. Nat Struct Mol Biol. 2005;12:624–625. doi: 10.1038/nsmb956. [DOI] [PubMed] [Google Scholar]


