Abstract
The role of lipid domain size and protein-lipid interfaces in the thermotropic phase transition of dipalmitoyl phosphatidylcholine (DPPC) and dimyristoyl phosphatidylcholine (DMPC) bilayers in Nanodiscs was studied using small-angle X-ray scattering (SAXS), differential scanning calorimetry (DSC), and generalized polarization (GP) of the lipophilic probe Laurdan. Nanodiscs are water-soluble, monodisperse self-assembled lipid bilayers encompassed by a helical membrane scaffold protein (MSP). MSPs of different lengths were used to define the diameter of the Nanodisc lipid bilayer from 76 to 108 Å and the number of DPPC molecules from 164 to 335 per discoidal structure. In Nanodiscs of all sizes, the phase transitions were broader and shifted to higher temperatures relative to those observed in vesicle preparations. The size dependences of the transition enthalpies and structural parameters of Nanodiscs reveal the presence of a boundary lipid layer in contact with the scaffold protein encircling the perimeter of the disc. The thickness of this annular layer was estimated to be approximately 15 Å, or two lipid molecules. SAXS was used to measure the lateral thermal expansion of Nanodiscs and a steep decrease of bilayer thickness during the main lipid phase transition was observed. These results provide basis for the quantitative understanding of cooperative phase transitions in membrane bilayers in confined geometries at the nanoscale.
Keywords: Phase transition, Nanodisc, Phospholipid bilayer, Nanodomain, Differential scanning calorimetry, Small-angle X-ray scattering
Introduction
Biological membranes are complex systems with a hierarchy of structural elements consisting of lipids and proteins which can partition to form different domains1-3. The structural and dynamic properties of membranes are essential for the function of a diverse set of protein assemblies involved in important cellular phenomena such as signaling and membrane enzymatic reactions. The influence of the lipid bilayer phase state on isolated membrane proteins as well as the behavior of whole living cells has been the subject of many investigations4-6. Conclusions are often derived in the framework of the fluid mosaic model1, which describes the biological membrane as a sea of lipids populated with mobile proteins. However, this model is too simple to completely describe the properties of a heterogeneous multi-component membrane system that may contain a large variety of lipid and protein types7,8. The protein weight percentage in membranes can vary from 20% to 80%2, suggesting that a significant fraction of the lipid is in direct contact with protein. Numerous experimental studies have shown that such interfacial, or boundary lipids, are perturbed by these interactions with proteins, and the character and magnitude of this perturbation depend on protein and lipid structure as well as on the bilayer phase state9-15. These concepts have been the subject of several recent reviews16-19. Currently, it is commonly assumed that the main structural elements of biological membranes are segregated microdomains, usually formed by different lipids and often including proteins20. The properties of such domains are now the subject of intensive experimental and theoretical studies.
Several experimental methods are able to directly monitor the properties of a lipid bilayer domain smaller than 1 μm in diameter20. Nevertheless, such domains contain up to 105 - 106 lipid molecules, numbers still beyond the capabilities of even the fastest molecular dynamics simulations. Therefore, many recent efforts have focused on smaller systems, as the experimental and theoretical works indicate the typical correlation length in lipid bilayers corresponding to domains of 10 - 30 nm containing 102 – 103 lipid molecules21-24. Similar sizes for pure lipid domains in biological membranes have been obtained from simple estimates based on the protein/lipid ratio2,25. Such small domains have also been detected experimentally26-29. These findings suggest that up to half of all lipids may be in contact with protein and, by definition, belong to the boundary lipid layers. The impact of such observations on biomembrane function reinforces the importance of direct experimental studies of bilayer phase state on the scale of 5 – 50 nm, corresponding to 102 – 104 lipid molecules. While improved methods in vesicle preparation have made it possible to prepare small unilamellar vesicles (SUVs) under 100 nm in diameter, it has been shown that vesicles less than 50 nm in diameter have drastically different thermodynamic properties due to the high curvature of the surface and an overall lack of stability30. In contrast, the recently developed Nanodisc technology has been shown to produce stable water-soluble lipid bilayers measuring ∼10 nm in diameter31.
Nanodiscs (Figure 1) are formed through self-assembly from a detergent solubilized mixture of lipids and membrane scaffold proteins (MSPs)31,33-40. The amphipathic helices of the MSP render the phospholipid bilayer soluble in aqueous solution. The diameter of a Nanodisc is determined by the length of the two amphipathic helical MSPs surrounding the discoidal lipid bilayer. The stoichiometry of lipids can be optimized for each MSP to obtain monodisperse Nanodiscs of different size38. Herein we describe the study of the main phase transition of DPPC and DMPC lipid bilayers in Nanodiscs with diameters 9.7 nm and 12.5 nm and containing 164±2 and 335±12 DPPC molecules respectively38. We used small-angle X-ray scattering (SAXS), DSC and Laurdan fluorescence to monitor structural and dynamic changes of the lipid bilayer as a function of temperature. In addition, we compare our experimental results with the relevant structural parameters obtained in a recent molecular dynamics study of Nanodiscs formed with DPPC32. These studies of the thermotropic phase transition in Nanodiscs extend our preliminary investigation40 by providing quantitative characterization of size dependent structural changes of the bilayer and of the role of boundary lipids in the observed cooperative behavior.
Figure 1.

A crossection of MD simulated Nanodisc with DPPC 32. The numbers designate different cylindrical layers in the core – shell models used in the fitting of experimental SAXS data: (1) MSP double belt at the disc circumference, (2) phosphatidylcholine polar groups, (3) hydrocarbon acyl chains, (4) methyl end group layer in the middle of the lipid bilayer. D represents the overall diameter of the Nanodisc and H the bilayer height. The bar shown on top is a 100 Å scale.
Materials and methods
Membrane scaffold proteins
The design, expression and purification of MSPs was described previously31,38. Smaller Nanodiscs, 9.7 nm in diameter, were formed using MSP1 scaffold protein which contains 200 amino acids and a N-terminal hexa-histidine tag with a protease recognition site. Larger Nanodiscs, 12.5 nm in diameter, were formed with MSP1E3 which contains an additional 66 helical amino acids in the middle of the MSP1 sequence as previously described38.
Preparation of the disc samples
The general procedure for the self-assembly of Nanodiscs has been published31,38. Briefly, a solution of purified MSP at 0.15 – 0.3 mM concentration was combined with cholate and phospholipid (either DPPC or DMPC). After incubation at 38° C (DPPC) or at room temperature (DMPC), the self-assembly process is initiated by dialysis against 1000-fold excess buffer (20 mM Tris, pH 7.4, 100 mM NaCl, 1 mM NaN3) for 24 hours with fresh buffer exchange every 8 hours at the same temperature using 10,000 MW cut-off membranes. Alternatively, detergent removal can be accomplished through incubation with hydrophobic BiobeadsTM (Pierce Chemicals, Rockford, IL) for 3-4 hours. After this process, the main fractions of self-assembled Nanodiscs were isolated by size exclusion chromatography as described38. Monodisperse fractions (2 – 3 % variation of lipid/protein ratio) were used in experimental phase transition studies.
Analytical procedures
The concentration of lipids in Nanodisc samples was measured using 3H enriched lipids of the same chemical structure and scintillation counting of the HPLC column fractions31. The specific activity of each batch of radioactive lipids was determined by quantitation of phosphate41 and scintillation counting using standard procedures. For calibration we used standard phosphorous solutions (Sigma, P3869) and [1,2-3H] hexadecanol (Moravec Biochemicals, Bria, CA). Concentrations of scaffold proteins were determined using molar absorption coefficients calculated for the known amino acid sequences according to published methods42.
SAXS measurements and analysis
Small angle X-ray scattering (SAXS) was measured at the DND-CAT Sector 5 at the Advanced Photon Source (Argonne National Lab, IL) at a 2.012 m sample - 2D detector distance and a nominal photon energy of 15 keV (wavelength 0.826 Å). The Nanodisc solutions were sealed in glass capillaries with 1.5 mm diameter (Charles Supper Company, Natick, MA) and placed into an aluminum sample holder with temperature controlled by Peltier elements. Vacuum chambers with Mylar windows were used along the beam path before and after the sample holder to minimize scattering by air. Silver behenate with 58.38 Å spacing43 was used for calibration, and reference buffer solvents were used for background correction. The data collection was executed after equilibration of the sample at each temperature for three minutes; usually two data frames with 100 s exposure were collected at each temperature to ensure complete equilibration of the sample. After warming to temperatures above the lipid main phase transition, each sample was cooled to 22° C (DPPC) or 15° C (DMPC), and SAXS data were collected again to check for reversibility. In all cases the main lipid transition in Nanodiscs monitored by SAXS was fully reversible. In two experiments with prolonged incubation of Nanodiscs at 55° C for 1 – 2 hours, the beginning of irreversible degradation of Nanodiscs was detected by incomplete recovery of the SAXS curves after cooling (data not shown). The raw data were processed using the program FIT2D44,45 to give the scattering curves in the form log(I/I0) vs. Q = 4πsin(θ)/λ. Analysis of SAXS data was performed using the programs GNOM46 and CRYSOL47, as well as in-house subroutines written in MATLAB (MathWorks, Natick, MA). The MATLAB fitting programs used analytical expressions derived for cylindrical core-shell models48.
The models for data fitting were constructed using information on the size and composition of Nanodiscs obtained independently, as described earlier38. Four layers of different electron densities were used in these models, as shown in Figure 1, one for the protein and three for the lipid bilayer, the phosphocholine polar heads, the acyl chains and the methyl end groups at the center of the bilayer. Changes in the electron density of water at different temperatures were neglected. Experimental curves for each type of Nanodisc were fitted using six parameters, the four electron densities previously listed, the bilayer thickness, and the radius of the disc. The composition of Nanodiscs was determined as described38. All Nanodiscs contained two MSP1 or MSP1E3 molecules and 164±2 or 335±12 DPPC molecules. As has been shown before38, approximately 180 amino acids in each MSP1 molecule (240 – 250 amino acids in each MSP1E3 molecule) form the helical belt around the cylindrical fragment of the lipid bilayer in 9.7 nm or 12.5 nm Nanodiscs, as illustrated in Figure 1. The molecular volumes for these fragments of MSP were determined using CRYSOL from the published X-ray structure of the truncated human apolipoprotein A149 (Protein Data Bank file 1AJV), the sequence of which is similar to MSP. The total number of electrons in each protein molecule was calculated from the known amino acid sequence, and the electron densities of the protein in Nanodiscs of different sizes were obtained from these numbers divided by the volume of a corresponding cylindrical shell (layer 1 in Figure 1). A similar approach was used to calculate the electron densities for the layers modeling the lipid phase (i.e. layers 2, 3, and 4 in Figure 1). The total number of electrons in each layer was calculated from the experimentally determined lipid/protein ratio for each sample. The average electron densities in each layer were then calculated assuming a plane going through the glycerol backbone separating the lipid monolayer into a polar head layer and acyl chains. The end group region was composed of the terminal methyl groups and two penultimate methylenes, (-CH2CH2CH3). Thus, layer 2 (polar heads) contained 138 electrons per lipid molecule, layer 3 (acyl chains) containing 216 or 184 electrons per DPPC or DMPC correspondingly, and layer 4 (end groups) containing 52 electrons per lipid. The hydration water was also included in calculation of the electron density of layer 2, so that the volume of this layer per one lipid molecule was assumed to contain water in the amount corresponding to 27 Å3 per hydration water. We used 27 Å3 instead of 30 Å 3 as the molecular volume to take into account the higher density of hydration water50. As shown in Figure 1, the diameters for layers 2 – 4 are the same, and the radial thickness of protein layer 1 was fixed at 10 Å. The height of layer 1 was then calculated using the known volume and radial size. The height of layer 4, H4, was also fixed as 0.265H3, where H3 is the hydrophobic thickness of the lipid bilayer. These constraints were based on the available structural information and were necessary to optimize the convergence of the fitting algorithm. Structure factors were assumed to be unity because of the low sample concentrations, i.e., the volume fraction of Nanodiscs was never higher than 3.5 %. As a result, only three free parameters were fitted for each SAXS curve, one diameter and two bilayer heights. All other parameters, including electron densities, could be calculated from the known compositions and structures.
Differential Scanning Calorimetry
Differential scanning calorimetry (DSC) experiments were performed using a Microcal MCS differential scanning calorimeter (Microcal, North Hampton, MA). The disc samples were prepared as previously described with the exception that the buffer system used was 20 mM sodium phosphate pH 7.4, 100 mM sodium chloride, 1 mM sodium azide. The lipid/protein stoichiometry of each sample was determined by first exchanging the sample into 20 mM Tris HCl Buffer pH 7.4, 100 mM sodium chloride, 1 mM sodium azide and then measuring protein concentration and total phosphorous in each sample41. The lipid/protein stoichiometry was then used to calculate the lipid concentration from the protein concentration of each DSC sample. Samples contained 1 – 2 mM lipid concentration. DSC thermograms were generated using a scan rate of 35 degrees/hour. DMPC samples were scanned from 10°C to 50°C and DPPC samples were scanned from 10°C to 60°C. Enthalpies of the main transitions were determined by fitting the data to a non-two state transition using the software package Origin (Microcal, Natick, MA).
Laurdan fluorescence
Nanodiscs produced for fluorescence measurements were made as described except Laurdan (6-dodecanoyl-2-dimethylaminonaphthalene, Molecular Probes Eugene, OR) was added in a 1:200 probe:lipid molar ratio prior to detergent solubilization of the lipids. Fluorescence measurements utilized a Hitachi F-3010 spectrofluorimeter (Tokyo, Japan) with excitation at 340 nm, 5 nm spectral bandwidths for both excitation and emission monochromators, and scanning speed 1nm/s. The concentration of Nanodiscs in all samples was approximately 1 μM, as determined using molar absorption coefficients of scaffold proteins after correction for the absorption of Laurdan. Sample temperature was controlled by a Peltier element and was read directly from the sample by a submersible thermocouple.
Results
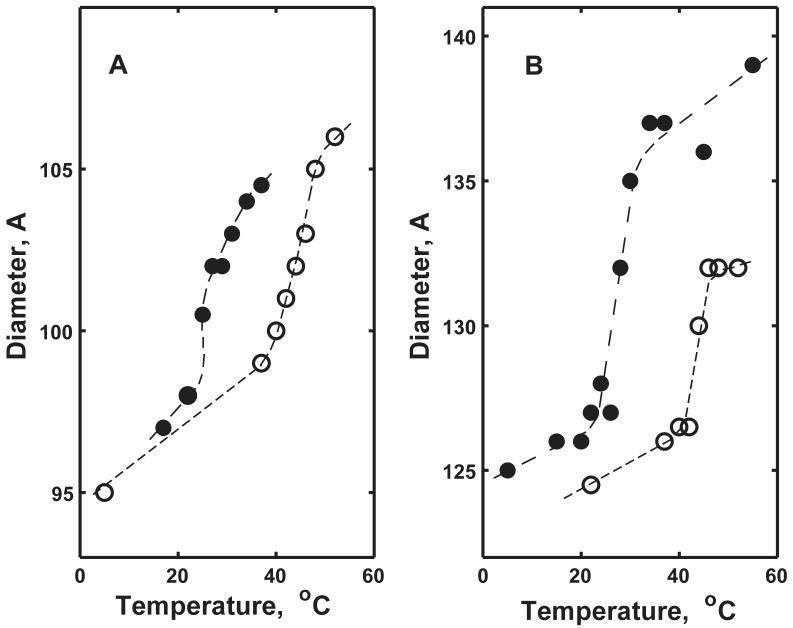
SAXS is one of the most rigorous direct experimental methods for structural studies of macromolecules in solution, especially when additional information about the composition of the system is available51-53. Our analysis of SAXS experimental results relies on the fitting of multiple data sets obtained on the same sample with known composition at different temperatures. The representative SAXS curves measured at different temperatures from Nanodiscs of different sizes are shown in Figure 2. For all samples, similar tendencies are observed with increasing temperature, namely: (i) the position of the first minimum on SAXS curves shifts to lower angles; (ii) the depth of this minimum decreases; (iii) the intensity at zero angle decreases. Analysis of these curves with GNOM46 provides the corresponding Nanodisc diameters and the density distribution functions. The results of this analysis are presented in Figures 3 and 4. The main phase transition of the lipid bilayer in Nanodiscs is clearly identified as a steep increase of Nanodisc diameter in all samples. The transition temperatures (Tm) estimated from the SAXS data in Figure 3 are in good agreement with those obtained using DSC and Laurdan fluorescence (see below), although the precision of the main transition position determined from SAXS data is lower. From Figure 3 it is clear that SAXS can directly detect changes in Nanodisc size on the order of 1 Å. The gradual increase of Nanodisc diameter with temperature was also observed below and above the phase transition range. These changes can be attributed to the lateral thermal expansion of the lipid bilayer54. The temperature dependent SAXS curves provide information concerning changes in all components of the Nanodisc, including the protein and boundary lipid layer. This may explain why the lipid phase transitions detected by SAXS in Nanodiscs are apparently broader and less cooperative than the same transitions monitored by calorimetry and Laurdan fluorescence (see below). Nevertheless, the size changes of Nanodiscs as a result of the cooperative melting of the bulk lipid are clearly observed for all samples in Figure 3. To our knowledge, this is the first direct observation of size changes of lipid nanoscale bilayers induced by a thermotropic phase transition.
Figure 2.
Temperature dependence of SAXS measured from Nanodiscs of different sizes formed with DPPC. Experimental data – (•), data fits obtained by GNOM are indicated by full lines. Temperatures, from top to bottom are: left panel: (1) 5°C; (2) 22°C; (3) 37°C; (4) 42°C; (5) 46°C; (6) 48°C; (7) 52°C ; and the right panel: (1) 22°C; (2) 37°C; (3) 42°C; (4) 46°C; (5) 48°C; (6) 52°C. Scattering vector Q = 4πsin(θ)/λ. The data obtained with DMPC Nanodiscs reveal a similar pattern (not shown).
Figure 3.
Temperature dependence of Nanodisc diameters determined from experimental SAXS curves using GNOM 46. (A) Smaller Nanodiscs formed with MSP1. (B) Larger Nanodiscs formed with MSP1E3. Filled symbols correspond to DMPC and open symbols to DPPC.
Figure 4.
Density distribution functions p(R) at different temperatures determined from experimental SAXS curves using GNOM 46. (A) Smaller Nanodiscs formed with DMPC (top) and DPPC (bottom). (B) Larger Nanodiscs formed with DMPC (top) and DPPC (bottom). The arrows show the direction of p(R) changes with temperature increase. The temperatures for each data set are the same as in Figures 3 and 5.
More detailed information about the structure of the lipid bilayer in Nanodiscs can be derived from the density distribution functions p(R)51 at different temperatures which are shown in Figure 4. For all samples, the lipid phase transition reveals itself as a decrease of p(R) values at intermediate scattering lengths. As a result, the integrated total scattering density of Nanodiscs also decreases with temperature (data not shown). This effect is almost totally due to the thermal expansion of the lipid bilayer and concomitant decrease of lipid phase scattering density, because the specific volume changes of proteins are known to be small53,55. Comparison of Nanodiscs of the same size but formed with different lipids shows the expected difference in positions of maxima, which approximately correspond to the bilayer thickness. For Nanodiscs formed with DPPC, the maximum of p(R) is observed at 56 – 58 Å below the phase transition, and shifts to 52 – 53 Å at higher temperatures. The position of the same maximum for Nanodiscs formed with DMPC is at 51-53 Å at low temperatures and shifts to 46 – 48 Å at high temperatures. These results are consistent with the expected decrease in the bilayer thickness at temperatures higher than Tm54 and provide an additional support for the cooperative melting of the lipid bilayer in Nanodiscs.
The changes in bilayer thickness with temperature obtained using a fitting program based on the analytical solution of a cylindrical core-shell model (Figure 1) are presented in Figure 5. Comparison of the results obtained for Nanodiscs of different size formed with the same lipid shows that the average bilayer thickness is slightly higher for bigger Nanodiscs. This difference of approximately 3 Å for both DPPC and DMPC can be explained by the significantly higher fraction of structurally perturbed boundary lipids at the protein-lipid interface in smaller Nanodiscs. Since these boundary lipids are expected to have larger surface area per lipid, and thus smaller bilayer height (see Figure 1 for illustration), this difference may be considered as the main factor responsible for the systematic changes in bilayer thickness with Nanodisc size. The thickness of DMPC bilayers is smaller by 7 – 8 Å than that of DPPC bilayers for both Nanodisc sizes. This difference is higher than 4 – 5 Å, which would be expected from the difference in the length of alkyl chains, C14 vs. C16 and therefore, other factors are contributing. Direct measurements of lipid/protein stoichiometry consistently show that Nanodiscs of the same size contain more molecules of DPPC than DMPC. This means that the surface area per lipid in Nanodiscs formed with DMPC is higher, and the bilayer height is lower, as compared with unperturbed thickness corresponding to the gel phase. The bilayer thickness determined by SAXS (Figure 5) in dilute solutions also includes the layer of hydration water molecules. Thus, the values obtained in our experiments are slightly higher than bilayer thicknesses determined by SAXS from multilamellar DPPC and DMPC samples at full hydration54,56.
Figure 5.
The bilayer thickness of Nanodiscs as a function of temperature for Nanodiscs formed with DMPC (filled symbols) and DPPC (open symbols). (A) Smaller Nanodiscs formed with MSP1, (B) larger Nanodiscs formed with MSP1E3.
The lipid phase transitions in Nanodiscs detected by DSC are shown in Figure 6. Each experimental curve was fitted with two Gaussian contours, the dominant strong band corresponding to the main lipid phase transition, and the second minor broad band representing the background. The sharp minor spike in Figure 6A indicates a small vesicular contamination in the Nanodisc sample. The parameters corresponding to the main transition are shown in Table 1 along with the same parameters determined from Laurdan fluorescence analysis. Note, that the size of the Nanodisc does not affect the main transition temperature. However, in all cases the observed transition temperatures are higher than those with unperturbed bilayers, 41.5°C and 23.5°C correspondingly57, by 2°C for DPPC and 5°C for DMPC. The good agreement between the Tm values determined experimentally using different methods indicates that the same phase transition is being monitored.
Figure 6.
DSC scans of Nanodiscs of different sizes formed with DPPC (A) and DMPC (B). On both panels (1) indicates the smaller Nanodiscs formed with MSP1 and (2) the larger Nanodiscs formed with MSP1E3.
Table 1.
Positions, Tm, and widths, ΔT, of the main phase transitions of lipid bilayer in Nanodiscs of different sizes formed with DPPC and DMPC. Data from Laurdan fluorescence and DSC (parentheses) are shown.
| Membrane Scaffold Protein |
DPPC | DMPC | ||
|---|---|---|---|---|
| Tm, °C | ΔT, °C | Tm, °C | ΔT, °C | |
| MSP1 | 43.7 (44.5) | 7 (7) | 29.0 (28.5) | 10 (7) |
| MSP1E3 | 43.8 (44.6) | 5 (4) | 28.6 (27.5) | 9 (6) |
An important feature of the lipid phase transition in Nanodiscs is the drastic increase of the transition enthalpy with Nanodisc size indicating the perturbation of the boundary lipid layer at the protein-lipid interface. The boundary lipids can be detected by structural and dynamic perturbation of the lipid bilayer at the protein-lipid interface10,58-62, as well as by changes in the parameters of the gel — liquid crystal phase transition, the transition temperature Tm and the decrease in the transition enthalpy9,11-13. The latter effect is usually interpreted as the absence of a cooperative phase transition at the boundary lipid layer9,11-14,63. This means that the transition enthalpy calculated per mole of total lipid is expected to increase with increasing Nanodisc size due to the decrease of the fraction of boundary lipid. With Nanodiscs we have precise control over particle composition and geometry, and have tested this hypothesis.
An essential advantage of the Nanodisc system is the similar geometry at all temperatures studied, and thus the fraction of boundary lipids at the protein-lipid interface does not depend on temperature. The values of ΔH are calculated from the experimental DSC curves as molar enthalpies per lipid, where the lipid concentrations correspond to the total lipid content in the Nanodisc sample, including boundary lipids. In previous studies it was usually assumed that the boundary lipid layer does not contribute to the observed heat effect 5,19, and the transition enthalpy is due only to the bulk unperturbed lipid domains. Within such simple models, all lipid molecules belong to the bulk lipid bilayer cooperative domains, or to the boundary lipid layer which does not melt cooperatively. In this case the transition enthalpy, ΔH, is proportional to the surface area of the unperturbed lipid domain at the center of the Nanodisc and can be estimated using equation 1:
| 1 |
where R is the radius of the cooperative domain at the center of the Nanodisc undergoing the phase transition, b is the thickness of the boundary layer and ΔHo is the transition enthalpy for pure, unperturbed lipid bilayer. If we let Z = R + b, the radius of the lipid surface, and rearrange, we obtain:
| 2 |
therefore, fitting the observed enthalpy to equation 2 yields both the thickness of the boundary layer and the transition enthalpy of pure lipids (the value at infinite radius). DSC experiments were performed on DMPC Nanodiscs with a bilayer radius ranging from 3.8 nm to 5.4 nm (Table 2). As a result of fitting the data obtained for DMPC discs to equation 2, we find that the thickness of the boundary lipid layer is approximately 1.5 nm, which corresponds to a perturbation propagated through two lipid layers in the vicinity of the scaffold protein. The transition enthalpy of this pure lipid phase is found to be 5.7 kcal/mole. Both values agree well with earlier preliminary analysis 64. Note that a similar dependence on the size of discoidal reconstituted high density lipoprotein formed with DMPC was detected for the proton NMR chemical shifts of choline N-methyl groups 65. This signal was used by the same authors as a sensitive probe of lipid packing order in vesicles of different size 66 and interpreted as the direct measure of the boundary lipid fraction in that system.
Table 2.
Observed enthalpies and lipid bilayer radii of DMPC Nanodiscs from SAXS 38.
| Membrane Scaffold Protein |
ΔH (kcal/mole) | Bilayer radius (nm) |
|---|---|---|
| MSP1 | 1.96 | 3.8 |
| MSP1E1 | 2.01 | 4.2 |
| MSP1E2 | 2.38 | 4.9 |
| MSP1E3 | 2.72 | 5.4 |
Laurdan fluorescence is sensitive to dipolar relaxation of the solvent molecules in the nanosecond timescale and is commonly used as a probe to detect changes in dynamic properties of lipid bilayers near the main phase transition 58-60. The temperature dependence of Laurdan general polarization (GP) in Nanodiscs of different sizes is shown in Figure 7. GP is defined by the formula GP = (I440-I490)/(I440+I490) where I440 and I490 are the fluorescence intensities at 440 nm and 490 nm respectively60. As the temperature increases, the steep decrease of GP indicates the cooperative phase transition of the lipid bilayer in all Nanodisc samples 59,60,67. With both lipids, the amplitude of this transition is higher for larger Nanodiscs, presumably because of the relatively smaller fraction of the boundary lipids which are structurally and dynamically perturbed at the protein-lipid interface 68. The positions of the main transition determined from GP following background correction are given in Table 1.
Figure 7.
Laurdan GP values for MSP1 Nanodiscs (triangles) and MSP1E3 Nanodiscs (circles) formed with DPPC (empty symbols) and DMPC (filled symbols) as a function of temperature. The lines are added only to guide the eye.
The cooperativity of phase transitions observed by DSC is slightly higher as evidenced by the transitions widths presented in Table 1. This difference may be due to the presence of Laurdan in the boundary lipid layer, as the latter does not show a cooperative melting transition and thus is not detected by DSC 9,11,13,14,63. Possible redistribution of Laurdan label between bulk and boundary lipid phases with the increase of temperature would result in the apparent broadening of the phase transition observed by GP. Another source of difference may be the fact that the Laurdan GP values in general can be only approximately represented as a linear combination of corresponding GP values of different species in the range of phase coexistence 58. As such, they can be treated quantitatively as a signal proportional to the species concentration only when the sum of fluorescence intensities at analytical wavelengths (I440+ I490) does not change with temperature 58, which is not usually the case in general.
Discussion
In this work we describe the results of an experimental study of thermotropic phase transition of lipid Nanodiscs containing from 140 to 340 lipid molecules. Earlier we have shown that Nanodiscs can be assembled in aqueous solution as highly monodisperse particles with similar structure, with the amphipathic helical membrane scaffold protein at the circumference of a circular fragment of lipid bilayer (Figure 1). Nanodiscs are stable for weeks at ambient conditions and persist as individual particles at temperatures up to 55 – 60° C, above which slow aggregation can be detected on a time scale of several hours. Nanodiscs are thus an ideal model system for studies of the lipid bilayer phase properties at the level of single cooperative domains.
Our results are in a good agreement with earlier studies of the thermotropic lipid transition in reconstituted high density lipoproteins (rHDL) which used DSC 13,69, fluorescent spectroscopy 68,70-77, Raman 10,78 and IR spectroscopic methods 74. The broad main transition of the lipid bilayer in rHDL is at the same or slightly higher temperature. However, in general, the rHDL particles in the previous studies were not homogeneous and their lipid-protein stoichiometry was not precisely characterized. As a result, the estimates for the configuration and size of the boundary lipid layer were heavily dependent on model assumptions and varied greatly
Comparison of bilayer height and mean area per lipid in Nanodiscs obtained from SAXS with the similar characteristics of lamellar DMPC and DPPC 54,56,79,80 preparations shows important differences, especially at lower temperatures (Figures 3 and 5). In Nanodiscs the values of A, the mean surface area per lipid below Tm, calculated from the data in Figure 5, are from 52 to 58 Å2. These are significantly higher than those typical for gel phase, A = 47 – 48 Å2, measured for DPPC at 20°C 79 and for DMPC at 10°C 56. This difference can be explained by the presence of perturbed boundary lipids with higher mean surface area, or by the fact that Nanodiscs are formed at temperatures near the main transition temperature of the unperturbed bilayer, 38°C for DPPC and 21 – 22°C for DMPC. At these temperatures the values of mean area per lipid are significantly higher than the ones at lower temperatures, where they are usually measured to characterize the gel phase. The helical belt of the scaffold protein can encompass a smaller number of lipids than one could estimate from the given circumference of MSP and the surface area per lipid molecule typical for the gel phase. At lower temperatures, the number of lipid molecules in the once formed Nanodiscs does not change, and thermal contraction of the bilayer will result in gradual decrease of the surface area of the bilayer fragment. The latter may be realized without significant changes in the length of the protein helix through continuous structural relaxation of the scaffold protein at the lipid-protein interface, which may be accompanied by the gradual transformation of the circular cylinder into an elliptical structure 32. However, because of hydrophobic mismatch and other structural distortions, the circular shape of the Nanodisc is expected to be roughly maintained at all temperatures to minimize the area of protein-lipid contact. Indeed, all experimental SAXS curves shown in Figure 2 are well fitted with circular discoidal models. Thus the protein conformation will be forced to accommodate the smaller protein-lipid interface area. In any of these scenarios a free energy penalty must be paid for such structural rearrangement, which may also result in perturbation of the ideal lipid packing in the gel phase.
Another important parameter, the thickness of the lipid bilayers, is also shown to be higher in Nanodiscs (see Figure 5) than in corresponding lamellar structures 54,56,79,80. From Figure 5 it is clear that for both lipids the observed bilayer thickness is higher by 2 – 3 Å for the larger Nanodiscs. This fact could be expected as an effect of the boundary lipids. The average bilayer height of the boundary lipid layer must be lower because of hydrophobic mismatch at the protein-lipid interface and the packing distortions at the protein – lipid interface. As a result, the bilayer thickness averaged over all Nanodiscs, as in the structural model shown in the Figure 1, will be higher in larger Nanodiscs. The absolute values of bilayer thickness are, however, systematically higher for Nanodiscs, as compared with unperturbed lamellar lipid samples 56,81. This comparison refers mostly to the gel phase region. Preliminary interpretation of this difference may be based on the assumption that the lipids in the gel phase in Nanodiscs are not tilted with respect to the bilayer plane (as shown in Figure 1), contrary to the well established tilt of 32° of DPPC and DMPC in the Lβ phase 82,83. This assumption is inspired by the axial symmetry of Nanodiscs and finds confirmation in the recent molecular dynamics simulations of Nanodiscs formed with DPPC 32. The direct consequence of this packing difference will be a substantially higher bilayer thickness in Nanodiscs, as compared with the similar thickness in liposomes. Note that the thickness of Nanodiscs formed with DPPC was measured as 56 – 59 Å by scanning force microscopy on mica surfaces 31. Slightly higher values calculated from SAXS from bigger discs (Figure 5, 61 – 62 Å with DPPC, and 55 Å with DMPC at low temperatures) may be explained by taking into account the hydration water layers, which add effectively to the scattering density at both bilayer surfaces. The contribution to the total scattering from hydration water layer, which has higher density than the bulk water, may increase the apparent bilayer thickness estimated from SAXS in dilute Nanodisc solution by 4 – 5 Å. This number is sufficient to explain the difference between the data shown at Figure 5 and those reported for multilamellar samples at full hydration, where there is little or no bulk water between bilayers 58.
DSC thermograms also display a shifted Tm and a broad transition when compared to pure lipid vesicle preparations. The cooperativity of the main transition, indicated by the width of the peaks shown in Table 1, is much lower than that seen in pure lipid vesicles 64 because of the number of lipids available to participate in the cooperative transition. It was estimated that the cooperative domains in multilamellar vesicles involve up to 1700 lipid molecules 64 and thus the intrinsic cooperativity within a Nanodisc containing 150 to 330 lipids is much lower. The size of cooperatively melting lipid domain as estimated from the ratio of van’t Hoff enthalpy and calorimetric transition enthalpy 64 is within 60 – 120 molecules for all Nanodisc samples. In addition, in the smaller Nanodiscs, more than 50% of the total lipids are present in the boundary layer with concomitant loss of cooperativity. With the ability to control the Nanodisc size, we used the observed enthalpies to estimate that an approximately 1.5 nm layer of lipid molecules near the scaffold protein is excluded from the cooperative phase transition. Consistent with this interpretation is the value obtained for the enthalpy of transition at infinite radius, 5.7 kcal/mole, which is in a good agreement with the enthalpy of transition observed for pure DMPC vesicles, 5.6 kcal/mol 64,84.
The increase in the melting temperature and the stabilization of the gel phase in Nanodiscs shown by all methods may be explained by the additional lateral pressure provided by the scaffold protein. As we have shown recently 38, the lipid packing in monodisperse Nanodiscs formed at the optimum lipid/protein ratio corresponds to a mean surface area of approximately 52 Å2 per DPPC molecule, characteristic for the gel state. This value is slightly higher than 47 Å2, measured at 20°C by Nagle and coworkers 54. The difference may be attributed to the presence of the structurally perturbed boundary lipids in Nanodiscs, or also due to the fact that the DPPC discs are self-assembled at 38°C where the unperturbed mean surface area per lipid molecule is higher than at 20°C. As a result of the thermal expansion of the lipid bilayer during the thermotropic transition in Nanodiscs, both the surface area of the thermodynamically unfavorable lipid/protein interface and the length of the scaffold protein helical belt must increase. These two factors provide the additional lateral pressure on the lipid bilayer against its expansion.
We have estimated the lateral thermal expansion coefficients, CT=(1/A)·(dA/dT), for DPPC and DMPC in Nanodiscs from the data shown in Figure 3, where A is the mean surface area per lipid. Below Tm for both lipids the average lateral thermal expansion coefficient is (2.2 – 2.9)•10-3 K-1, i.e. approximately a half of those measured for the giant unilamellar DMPC vesicles 85. This value of CTcorresponds to 0.4 – 0.55 Å/K increase of the perimeter of smaller and bigger Nanodiscs respectively, with the lengths of α-helical MSPs surrounding the lipid bilayer increase by the same amount. From the elastic modulus of α-helix estimated as 250 pN/Å 86, we calculate the stretching energy in MSP helices as 100 pN•Å (or 1.4 kcal/mol) per 1 Kelvin increase in temperature. This pressure is not enough to generate the compression energy of the lipid bilayer calculated as 300 pN•Å using the known elastic area compressibility modulus of DMPC in gel phase 85, Kcomp=0.35 N/m, and thermal expansion of the lipid phase estimated from Figure 3 as 11 Å2/K. The remainder of confinement is provided by thermodynamic coupling of thermal expansion with the concomitant increase of the area of hydrophobic mismatch at the protein-lipid interface, approximately 10 Å2/K. Such calculations show that the free energy penalty for the thermal expansion of this interface is 3 kcal/mol per Kelvin, similar to the solvation free energy of one methane molecule in water 87.
An additional feature of the scaffold protein influence on the transition temperature is the fact that the increase in Tm of lipid bilayers in Nanodiscs, as compared to corresponding transition temperatures of the same lipids in vesicles, is higher in case of DMPC (5 °C), than with DPPC (2 °C), even though the relative changes in diameter caused by the main phase transition of these lipids are very similar. These differences in Tm are similar for all Nanodiscs regardless of size. Most likely this is due to higher stability of the protein helical conformation at 29 °C than at 44 °C, the transition temperatures of DMPC and DPPC discs respectively. As the lipid phase transition results in the increase of Nanodisc diameter, the length of the scaffold protein helical belt must increase as well, which in turn will result in partial extension or unfolding of one or more of protein helices. The energetic cost of such structural perturbation is higher, if it happens at lower temperature, far from the unfolding temperature of the protein, as it is in the case of DMPC Nanodiscs. On the contrary, the transition in DPPC discs is at 44 °C, which may be in the initial region of gradual unfolding of the scaffold protein. Although we are not aware of any direct studies of conformation and stability of these or similar scaffold proteins in Nanodiscs or rHDL, according to many published results for rHDL and other apolipoproteins, the amphipathic α-helices are folded at temperatures below 50 °C and are predominantly unfolded at higher temperatures 88,89. These conclusions clearly depend on the character of lipid-protein interactions or on the presence of tertiary and quaternary structural interactions, which may also stabilize helical conformation of the protein. Thus, although the straightforward thermodynamic analysis of this system is complex, the observed features of the lipid phase transition in Nanodiscs are consistent with the known properties of the lipid and protein components.
Laurdan generalized polarization (GP) is a convenient probe for nanosecond relaxation changes correlated with the phase behavior of lipid bilayers 58-60. From the results shown in Table 2 it can be seen that the parameters of the phase transition detected by this method are similar for Nanodiscs of different sizes. Slightly higher GP values at the temperatures below Tm and lower GP values above Tmare observed for both lipids in the larger Nanodiscs (Figure 7). This increase in the amplitude of the transition detected by fluorescence may be attributed to the better average packing of lipids in the larger Nanodiscs, although the difference is small and cannot be rationalized in terms of the difference in the fraction of boundary lipids. In the case of equal distribution of Laurdan between bulk and boundary lipid fractions, approximately 65% of the fluorescent probe molecules would be incorporated in the boundary lipid layer in smaller Nanodiscs, and 50% in large discs. In this case it would be necessary to conclude that the boundary lipid layer undergoes the same, or a very similar thermotropic transition, at least in a dynamic sense, because Laurdan fluorescence probes the relaxation properties of the polar molecules in the time window corresponding to the lifetime of the excited state of the naphthalene fluorophore. Although this possibility cannot be excluded on the basis of our data, there are many evidences against this assumption 68,76,90-95. More probable is a preferential distribution of Laurdan into the bulk lipid fraction, at least during the phase transition and at higher temperatures. The equal distribution of the Laurdan probe between the bulk and boundary lipid fractions at low temperatures cannot be excluded because both lipid phases may have relatively low mobility despite the structural perturbation and the lack of cooperative melting for boundary lipids as compared to unperturbed lipid bilayer. The source of this perturbation is the significant hydrophobic mismatch between the average diameter of the membrane scaffold protein helix of 10 – 11 Å, and the hydrophobic thickness of the adjacent lipid monolayer, 14 – 19 Å for DMPC and DPPC, depending on the phase state 54,79. The average number of gauche isomers per DPPC molecule calculated in MD simulations at 27 °C 32 is two for the bulk lipids and almost five for the boundary lipids. These are comparable to the numbers characteristic for gel and LC lipid phases estimated from spectroscopic measurements 96, and detected by FTIR spectroscopy in reconstituted lipoproteins 97. Thus, the acyl chains in boundary lipids in MD model of Nanodisc equilibrated at 27 °C, i.e. below Tm, in average have conformations typical for LC state, while the lipid molecules in the middle of Nanodisc are not perturbed by interaction with the scaffold protein. The result of statistical analysis of MD simulation does not depend on the assumed thickness of boundary lipid layer within 13 – 18 Å, where 15 Å is the average thickness of two lipid layers, if the mean area per lipid is 56 Å2. Concomitantly, the thickness of the bilayer is higher in the center of the Nanodisc, as can be seen in Figure 1. While the strict quantitative comparison of all structural parameters derived from MD simulation with experimental results may be questioned, the analysis suggests the specific mode of structural perturbation of the lipid bilayer at the protein-lipid interface in Nanodiscs.
In conclusion, we have characterized the main lipid phase transition in monodisperse Nanodiscs formed with DPPC and DMPC as a function of disc diameter. The position of this transition Tm does not depend on the disc size and is shifted to higher temperatures as compared to pure lamellar lipid systems. This can be explained by the additional lateral pressure provided by the scaffold protein at the circumference of the Nanodisc. For both lipids, the apparent transition enthalpies and cooperativity are higher in larger discs because of the lower fraction of boundary lipids. Using SAXS we were able to demonstrate the changes in Nanodisc dimensions during the phase transition, namely, the increase of diameter and decrease of bilayer thickness. These parameters are in qualitative agreement with the known geometry of lamellar lipid bilayers, although the structural perturbations at the protein – lipid interface alter the mean surface area per lipid and the average bilayer thickness.
Acknowledgements
Portions of this work were performed at the DuPont-Northwestern-Dow Collaborative Access Team (DND-CAT) Synchrotron Research Center located at Sector 5 of the Advanced Photon Source. DND-CAT is supported by the E.I. DuPont de Nemours & Co., The Dow Chemical Company, the U.S. National Science Foundation through Grant DMR-9304725 and the State of Illinois through the Department of Commerce and the Board of Higher Education Grant IBHE HECA NWU 96. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Energy Research under Contract No. W-31-102-Eng-38. We gratefully acknowledge the help and support provided by Dr. J. Quintana, Dr. D. Keane and Dr. S. Weigand while working at Argonne. We thank A. Shih for the help in SAXS data collection and for atomic coordinates from MD simulations and Dr. T. Bayburt for helpful discussions. This work was supported by the Nanoscale Science and Engineering Initiative of the National Science Foundation under NSF Award Number EEC-0118025, and the NIH Grant GM33775.
References
- (1).Singer SJ, Nicolson GL. Science. 1972;175:720–731. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- (2).Gennis RB. Biomembranes. Molecular structure and function. Springer-Verlag; New York: 1989. [Google Scholar]
- (3).Yeagle PL, editor. The structure of biological membranes. 2nd CRC Press; 2005. p. 540 pp. [Google Scholar]
- (4).Jost PC, Griffith OH, editors. Lipid-Protein Interactions. Wiley; New York: 1982. [Google Scholar]
- (5).McElhaney RN. Biochim. Biophys. Acta. 1986;864:361–421. doi: 10.1016/0304-4157(86)90004-3. [DOI] [PubMed] [Google Scholar]
- (6).Bach D. Calorimetric studies of model and natural membranes. In: Chapman D, editor. Biomembrane structure and function. Vol. 4. Verlag Chemie; Weinheim: 1984. pp. 1–42. [Google Scholar]
- (7).Jost PC, Griffith OH, Capaldi RA, Vanderkooi G. Proc. Natl. Acad. Sci. U S A. 1973;70:480–484. doi: 10.1073/pnas.70.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Jost P, Griffith OH, Capaldi RA, Vanderkooi G. Biochim. Biophys. Acta. 1973;311:141–152. doi: 10.1016/0005-2736(73)90261-7. [DOI] [PubMed] [Google Scholar]
- (9).Alonso A, Restall CJ, Turner M, Gomez-Fernandez JC, Goni FM, Chapman D. Biochim. Biophys. Acta. 1982;689:283–289. [Google Scholar]
- (10).Edwards WL, Bush SF, Mattingly TW, Weisgraber KH. Spectrochim. Acta. 1993;49A:2027–2038. [Google Scholar]
- (11).Freire E, Markello T, Rigell C, Holloway PW. Biochemistry. 1983;22:1675–1680. doi: 10.1021/bi00276a024. [DOI] [PubMed] [Google Scholar]
- (12).Massey JB, Gotto AM, Jr., Pownall HJ. Biochemistry. 1981;20:1575–1584. doi: 10.1021/bi00509a026. [DOI] [PubMed] [Google Scholar]
- (13).Tall AR, Small DM, Deckelbaum RJ, Shipley GG. J. Biol. Chem. 1977;252:4701–4711. [PubMed] [Google Scholar]
- (14).Boggs JM, Moscarello MA. Biochemistry. 1978;17:5734–5739. doi: 10.1021/bi00619a021. [DOI] [PubMed] [Google Scholar]
- (15).Borbat PP, Costa-Filho AJ, Earle KA, Moscicki JK, Freed JH. Science. 2001;291:266–269. doi: 10.1126/science.291.5502.266. [DOI] [PubMed] [Google Scholar]
- (16).Lee AG. Biochim. Biophys. Acta. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- (17).Jensen MO, Mouritsen OG. Biochim. Biophys. Acta. 2004;1666:205–226. doi: 10.1016/j.bbamem.2004.06.009. [DOI] [PubMed] [Google Scholar]
- (18).Marsh D, Pali T. Biochim. Biophys. Acta. 2004;1666:118–141. doi: 10.1016/j.bbamem.2004.08.006. [DOI] [PubMed] [Google Scholar]
- (19).Lewis RNAH, McElhaney RN. In: The Structure of Biological Membranes. 2nd Yeagle PL, editor. CRC Press; Boca Raton, FL: 2005. pp. 53–120. [Google Scholar]
- (20).Binder WH, Barragan V, Menger FM. Angew. Chem. Int. Ed. 2003;42:5802–5827. doi: 10.1002/anie.200300586. [DOI] [PubMed] [Google Scholar]
- (21).Binder WH, Einzmann M, Knapp M, Kohler G. Monat. Chemie. 2004;135:13–21. [Google Scholar]
- (22).Xie AF, Yamada R, Gewirth AA, Granick S. Phys. Rev. Lett. 2002;89:246103/246101–246103/246104. doi: 10.1103/PhysRevLett.89.246103. [DOI] [PubMed] [Google Scholar]
- (23).Tokumasu F, Jin AJ, Dvorak JA. J. El. Microsc. 2002;51:1–9. doi: 10.1093/jmicro/51.1.1. [DOI] [PubMed] [Google Scholar]
- (24).Schrader W, Halstenberg S, Behrends R, Kaatze U. J. Phys. Chem. 2003;107B:14457–14463. [Google Scholar]
- (25).Marsh D, Watts A. In: Lipid-protein interactions. Jost PC, Griffith OH, editors. Vol. 2. Wiley; New York: 1982. pp. 53–126. [Google Scholar]
- (26).Damjanovich S, Gaspar R, Jr., Pieri C. Quart. Rev. Biophys. 1997;30:67–106. doi: 10.1017/s0033583596003307. [DOI] [PubMed] [Google Scholar]
- (27).Laude AJ, Prior IA. Mol. Membr. Biol. 2004;21:193–205. doi: 10.1080/09687680410001700517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Kusumi A, Koyama-Honda I, Suzuki K. Traffic. 2004;5:213–230. doi: 10.1111/j.1600-0854.2004.0178.x. [DOI] [PubMed] [Google Scholar]
- (29).Lommerse PHM, Spaink HP, Schmidt T. Biochim. Biophys. Acta. 2004;1664:119–131. doi: 10.1016/j.bbamem.2004.05.005. [DOI] [PubMed] [Google Scholar]
- (30).Lichtenberg D, Freire E, Schmidt CF, Barenholz Y, Felgner PL, Thompson TE. Biochemistry. 1981;20:3462–3467. doi: 10.1021/bi00515a024. [DOI] [PubMed] [Google Scholar]
- (31).Bayburt TH, Grinkova YV, Sligar SG. Nanoletters. 2002;2:853–856. [Google Scholar]
- (32).Shih AY, Denisov IG, Phillips JC, Sligar SG, Schulten K. Biophys. J. 2005;88:548–556. doi: 10.1529/biophysj.104.046896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Bayburt TH, Carlson JW, Sligar SG. Langmuir. 2000;16:5993–5997. [Google Scholar]
- (34).Bayburt TH, Sligar SG. Proc. Natl. Acad. Sci. U S A. 2002;99:6725–6730. doi: 10.1073/pnas.062565599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Bayburt TH, Sligar SG. Protein Sci. 2003;12:2476–2481. doi: 10.1110/ps.03267503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Bayburt TH, Carlson JW, Sligar SG. J. Struct. Biol. 1998;123:37–44. doi: 10.1006/jsbi.1998.4007. [DOI] [PubMed] [Google Scholar]
- (37).Civjan NR, Bayburt TH, Schuler MA, Sligar SG. Bio Techniques. 2003;35:556–558. 560, 562–563. doi: 10.2144/03353rr02. [DOI] [PubMed] [Google Scholar]
- (38).Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. J. Am. Chem. Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- (39).Baas BJ, Denisov IG, Sligar SG. Arch. Biochem. Biophys. 2004;430:218–228. doi: 10.1016/j.abb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- (40).Shaw AW, McLean MA, Sligar SG. FEBS Lett. 2004;556:260–264. doi: 10.1016/s0014-5793(03)01400-5. [DOI] [PubMed] [Google Scholar]
- (41).Chen PS, Toribara TY, Warner H. Analyt. Chem. 1956;28:1756–1759. [Google Scholar]
- (42).Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Huang TC, Toraya H, Blanton TN, Wu YJ. Appl. Cryst. 1993;26:180–184. [Google Scholar]
- (44).Hammersley AP. 1998 http://www.esrf.fr/computing/scientific/FIT2D
- (45).Hammersley AP, Svensson SO, Hanfland M, Fitch AN, Hausermann D. High Pressure Res. 1996;14:235–248. [Google Scholar]
- (46).Svergun DI. J. Appl. Cryst. 1992;25:493–503. [Google Scholar]
- (47).Svergun D, Barberato C, Koch MHJ. J. Appl. Cryst. 1995;28:768–773. [Google Scholar]
- (48).Pedersen JS. Adv. Coll. Int. Sci. 1997;70:171–210. [Google Scholar]
- (49).Borhani DW, Rogers DP, Engler JA, Brouillette CG. Proc. Natl. Acad. Sci. U S A. 1997;94:12291–12296. doi: 10.1073/pnas.94.23.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Svergun DI, Richard S, Koch MHJ, Sayers Z, Kuprin S, Zaccai G. Proc. Natl. Acad. Sci. U S A. 1998;95:2267–2272. doi: 10.1073/pnas.95.5.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Svergun DI, Koch MHJ. Rep. Prog. Phys. 2003;66:1735–1782. [Google Scholar]
- (52).Svergun DI. J. Appl. Cryst. 1997;30:792–797. [Google Scholar]
- (53).Semisotnov GV, Kihara H, Kotova NV, Kimura K, Amemiya Y, Wakabayashi K, Serdyuk IN, Timchenko AA, Chiba K, et al. J. Mol. Biol. 1996;262:559–574. doi: 10.1006/jmbi.1996.0535. [DOI] [PubMed] [Google Scholar]
- (54).Tristram-Nagle S, Nagle JF. Chem. Phys. Lipids. 2004;127:3–14. doi: 10.1016/j.chemphyslip.2003.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Harpaz Y, Gerstein M, Chothia C. Structure. 1994;2:641–649. doi: 10.1016/s0969-2126(00)00065-4. [DOI] [PubMed] [Google Scholar]
- (56).Tristram-Nagle S, Liu Y, Legleiter J, Nagle JF. Biophys. J. 2002;83:3324–3335. doi: 10.1016/S0006-3495(02)75333-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Cevc G, editor. Phospholipids Handbook. Marcel Dekker, Inc.; New York: 1993. [Google Scholar]
- (58).Parasassi T, De Stasio G, Ravagnan G, Rusch RM, Gratton E. Biophys. J. 1991;60:179–189. doi: 10.1016/S0006-3495(91)82041-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Parasassi T, Krasnowska EK, Bagatolli L, Gratton E. J. Fluorescence. 1998;8:365–373. [Google Scholar]
- (60).Parasassi T, De Stasio G, D’Ubaldo A, Gratton E. Biophys. J. 1990;57:1179–1186. doi: 10.1016/S0006-3495(90)82637-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Gilman K, Kauffman JW, Pownall HJ. Biochemistry. 1981;20:656–661. doi: 10.1021/bi00506a032. [DOI] [PubMed] [Google Scholar]
- (62).Ge M, Freed JH. Biophys. J. 1999;76:264–280. doi: 10.1016/S0006-3495(99)77195-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Faucon JF, Dufourcq J, Bernard E, Duchesneau L, Pezolet M. Biochemistry. 1983;22:2179–2185. doi: 10.1021/bi00278a019. [DOI] [PubMed] [Google Scholar]
- (64).Knoll W. Thermochim. Acta. 1984;77:35–47. [Google Scholar]
- (65).Brouillette CG, Jones JL, Ng TC, Kercret H, Chung BH, Segrest JP. Biochemistry. 1984;23:359–367. doi: 10.1021/bi00297a027. [DOI] [PubMed] [Google Scholar]
- (66).Brouillette CG, Segrest JP, Ng TC, Jones JL. Biochemistry. 1982;21:4569–4575. doi: 10.1021/bi00262a009. [DOI] [PubMed] [Google Scholar]
- (67).Bagatolli LA, Parasassi T, Fidelio GD, Gratton E. Photochem. Photobiol. 1999;70:557–564. [PubMed] [Google Scholar]
- (68).Dergunov AD, Dobretsov GE. Chem. Phys. Lipids. 2000;104:161–173. doi: 10.1016/s0009-3084(99)00125-5. [DOI] [PubMed] [Google Scholar]
- (69).Massey JB, She HS, Gotto AM, Pownall HJ. Biochemistry. 1985;24:7110–7116. doi: 10.1021/bi00346a014. [DOI] [PubMed] [Google Scholar]
- (70).Jonas A, Krajnovich DJ, Patterson BW. J. Biol. Chem. 1977;252:2200–2205. [PubMed] [Google Scholar]
- (71).Matz CE, Jonas A. J. Biol. Chem. 1982;257:4535–4540. [PubMed] [Google Scholar]
- (72).Zorich NL, Kezdy KE, Jonas A. Biochim. Biophys. Acta. 1987;919:181–189. doi: 10.1016/0005-2760(87)90206-2. [DOI] [PubMed] [Google Scholar]
- (73).Jonas A, Wald JH, Toohill KL, Krul ES, Kezdy KE. J. Biol. Chem. 1990;265:22123–22129. [PubMed] [Google Scholar]
- (74).Wald JH, Coormaghtigh E, De Meutter J, Ruysschaert JM, Jonas A. J. Biol. Chem. 1990;265:20044–20050. [PubMed] [Google Scholar]
- (75).Dergunov AD, Dobretsov GE, Taveirne J, Caster H, Vanloo B, Rosseneu M. Abstr. European Conference on the Spectroscopy of Biological Molecules, Enschede, Netherlands; 1999. pp. 347–348. [Google Scholar]
- (76).Dergunov AD, Taveirne J, Vanloo B, Hans C, Rosseneu M. Biochim. Biophys. Acta. 1997;1346:131–146. doi: 10.1016/s0005-2760(97)00025-8. [DOI] [PubMed] [Google Scholar]
- (77).Leroy A, Toohill KL, Fruchart JC, Jonas A. J. Biol. Chem. 1993;268:4798–4805. [PubMed] [Google Scholar]
- (78).Levin IW. Adv. Infrared Raman Spectr. 1984;11:1–48. [Google Scholar]
- (79).Nagle JF, Tristram-Nagle S. Biochim. Biophys. Acta. 2000;1469:159–195. doi: 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Nagle JF, Tristram-Nagle S. Curr. Opin. Struct. Biol. 2000;10:474–480. doi: 10.1016/s0959-440x(00)00117-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Wiener MC, Suter RM, Nagle JF. Biophys. J. 1989;55:315–325. doi: 10.1016/S0006-3495(89)82807-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Sun WJ, Tristram-Nagle S, Suter RM, Nagle JF. Biophys. J. 1996;71:885–891. doi: 10.1016/S0006-3495(96)79290-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Tristram-Nagle S, Zhang R, Suter RM, Worthington CR, Sun WJ, Nagle JF. Biophys. J. 1993;64:1097–1109. doi: 10.1016/S0006-3495(93)81475-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Knoll W, Hoehne GWH. Ber. Buns.-Gesellschaft. 1984;88:517–524. [Google Scholar]
- (85).Needham D, Evans E. Biochemistry. 1988;27:8261–8269. doi: 10.1021/bi00421a041. [DOI] [PubMed] [Google Scholar]
- (86).Rohs R, Etchebest C, Lavery R. Biophys. J. 1999;76:2760–2768. doi: 10.1016/S0006-3495(99)77429-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Ben-Naim A, Marcus Y. J. Chem. Phys. 1984;81:2016–2027. [Google Scholar]
- (88).Tall AR, Shipley GG, Small DM. J. Biol. Chem. 1976;251:3749–3755. [PubMed] [Google Scholar]
- (89).Gursky O, Atkinson D. Proc. Natl. Acad. Sci. U S A. 1996;93:2991–2995. doi: 10.1073/pnas.93.7.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Dergunov AD, Kaprel’yants AS, Kabishev AA, Simakova IM, Molchanov MI, Ostrovskii DN. Biokhimiya (Moscow) 1982;47:296–304. [PubMed] [Google Scholar]
- (91).Dergunov AD, Kabishev AA, Kaprelyants AS, Ostrovskii DN. FEBS Lett. 1981;131:181–185. [Google Scholar]
- (92).Kaprel’yants AS, Dergunov AD, Ostrovskii DN. Biokhimiya (Moscow) 1983;48:2049–2055. [PubMed] [Google Scholar]
- (93).Dobretsov GE, Dergunov AD, Taveirne J, Caster H, Vanloo B, Rosseneu M. Chem. Phys. Lipids. 1998;97:65–77. [Google Scholar]
- (94).Dergunov AD, Dobretsov GE, Visvikis S, Siest G. Chem. Phys. Lipids. 2001;113:67–82. doi: 10.1016/s0009-3084(01)00176-1. [DOI] [PubMed] [Google Scholar]
- (95).Kimelman D, Tecoma ES, Wolber PK, Hudson BS, Wickner WT, Simoni RD. Biochemistry. 1979;18:5874–5880. doi: 10.1021/bi00593a018. [DOI] [PubMed] [Google Scholar]
- (96).Mendelsohn R, Davies MA, Brauner JW, Schuster HF, Dluhy RA. Biochemistry. 1989;28:8934–8939. doi: 10.1021/bi00448a037. [DOI] [PubMed] [Google Scholar]
- (97).Lins L, Brasseur R, Rosseneu M, Vanloo B, Ruysschaert JM. Biochim. Biophys. Acta. 1993;1149:267–277. doi: 10.1016/0005-2736(93)90210-q. [DOI] [PubMed] [Google Scholar]