Abstract
Genomic medicine research requires substantial time and resources to obtain phenotype data. The electronic health record offers potential efficiencies in addressing these temporal and economic challenges, but few studies have explored the feasibility of using such data for genetics research. The main objective of this study was to determine the association of two genetic variants located on chromosome 9p21 conferring susceptibility to coronary heart disease and type 2 diabetes with a variety of clinical phenotypes derived from the electronic health record in a population of morbidly obese patients. Data on more than 100 clinical measures including diagnoses, laboratory values, and medications were extracted from the electronic health records of a total of 709 morbidly obese (body mass index (BMI) ≥ 40 kg/m2) patients. Two common single nucleotide polymorphisms located at chromosome 9p21 recently linked to coronary heart disease and type 2 diabetes (McPherson et al. Science 316:1488–1491, 2007; Saxena et al. Science 316:1331–1336, 2007; Scott et al. Science 316:1341-1345, 2007) were genotyped to assess statistical association with clinical phenotypes. Neither the type 2 diabetes variant nor the coronary heart disease variant was related to any expected clinical phenotype, although high-risk type 2 diabetes/coronary heart disease compound genotypes were associated with several coronary heart disease phenotypes. Electronic health records may be efficient sources of data for validation studies of genetic associations.
Keywords: SNP, Morbid obesity, Electronic health record, Cardiovascular disease, Type 2 diabetes
Introduction
Genomic medicine research requires substantial resources and time to assemble study populations, collect phenotypic data and biological samples, and to address specific research questions (Service et al. 2003). Moreover, the need for large sample sizes (Eberle et al. 2007) and increasingly precise definition of clinical phenotypes (Cupples et al. 2007) to study complex disorders, such as coronary heart disease (CHD) and type 2 diabetes (T2D), exacerbates demands on increasingly scarce research resources. Use of electronic health record (EHR) data on patient populations seeking care in large integrated delivery systems offers one potential solution to mitigate these challenges (Gerhard et al. in press).
Integrated delivery systems with EHRs offer several significant advantages over traditional approaches to genomic medicine research by simplifying logistics, reducing time lines, and reducing the overall costs through efficient data acquisition (Powell and Buchan 2005). Large numbers of patients can be readily identified and phenotyped using the EHR. Clinical infrastructure can be used to recruit patients, acquire biological samples (e.g., blood), and obtain supplemental data. However, few previous studies in genomic medicine research have used EHR data.
We examined the effectiveness of this model using comprehensive EHR data and biological samples on patients from the Geisinger Clinic Center for Nutrition and Weight Management. We performed a validation study on T2D and CHD genetic variants with a specific focus on patients with morbid obesity (BMI ≥ 40 kg/m2; Flegal et al. 2002). Few genetic studies of obesity-related disorders, such as T2D and CHD, have been conducted with morbidly obese populations (Koumanis et al. 2002). A large clinical database was constructed using data extracted from the EHR and evaluated through analysis of expected clinical associations. Two single nucleotide polymorphisms (SNPs) located in the same region of chromosome 9p21 which has previously been associated with T2D (Saxena et al. 2007; Scott et al. 2007; Zeggini et al. 2007) and CHD (Helgadottir et al. 2007; Larson et al. 2007; McPherson et al. 2007; O’Donnell et al. 2007; Samani et al. 2007) in genome wide association studies, were genotyped and associations with clinical variables determined. The resources and timeline required for these studies were considerably less than would have been required by traditional approaches.
Materials and methods
Center for Nutrition and Weight Management
The Center is an integrated practice model for weight management that seamlessly incorporates research as core to the practice. All patients who were enrolled in the Bariatric Surgery Program were recruited into a clinical research program in obesity (Still et al. 2007). Patients undergo a pre-operative assessment and preparation period during which a comprehensive set of clinical and laboratory measures were obtained along with blood samples for serum and DNA isolation. The Institutional Review Board of the Geisinger Clinic approved the research protocol and all participants provided written informed consent.
Patients
Patients from the Geisinger Clinic Center for Nutrition and Weight Management’s Bariatric Surgery program were recruited between October 2004 and August 2007. A comprehensive medical history and physical examination was performed during the initial visit. Standard of care laboratory tests were obtained pre-operatively, most approximately three weeks prior to surgery.
Biological samples
EDTA anti-coagulated blood samples for DNA isolation were obtained for the study as part of a clinical blood draw. For a small number of patients, blood was not obtained. DNA was then isolated from preserved liver tissue that was obtained from an intra-operative liver biopsy performed as standard of care for the bariatric surgery.
EHR
Geisinger Health System is an integrated delivery system with a significant presence in central and northeastern Pennsylvania. Installation of an EHR (EpicCare) began in 1996 in the current 40 community practice clinics and in specialty clinics in two hospitals and was completed (i.e., completely paperless operations) by 2001. The EHR is used for a variety of practice-based tasks including viewing test results, clinical messaging, dictation authorization, and order entry. Essentially all clinical notes are recorded in the EHR along with clinical measures, demographics, orders, diagnoses (based upon the International Classification of Diseases, Clinical Modification or ICD-9 codes), and data from other sources, including digital imaging and lab measures.
EHR data acquisition
Data were extracted from the EpicCare EHR (Verona, WI) using imbedded routines (known as Clarity), provided by the software vendor. Clarity tables can be manipulated using standard queries in SQL (standardized query language) applications. The Clarity tables containing the specific elements in the data dictionary for a particular domain (e.g., physical measurements, or lab measure) were identified, and those fields were extracted into a text file. Typically, all instances of an element were extracted (e.g., all lab results for a specific analyte). The extracted file was read into SAS/STAT software (SAS Institute Inc., Cary, NC).
Specific data elements from the EHR, summarized in Table 1, were selected because of their potential relevance to obesity and related complications, and their relative frequency among the morbidly obese population. For example, the co-morbidities (n = 25) and current medication subclasses (n = 67) extracted were found in at least 2% of the cohort at the initial visit; data elements not present in at least 2% of the cohort were omitted from the study database. The aggregate data obtained from the EHR were queried to extract those specific values (Table 1 variables) that were used to populate the study database. The resulting data file was merged with genotype data using a medical record number.
Table 1.
Variables extracted from the EHR
| Demographics | Laboratory values | |
| • Age at surgery | • Date of initial visit | • Triglycerides |
| • Gender | • Date of surgery | • High density lipoprotein cholesterol (HDL) |
| • Race | • Length of stay | • Low density lipoprotein cholesterol-calculated (LDL) |
| Clinical measurements | • HbA1c | |
| • Weight | • BMI | • Insulin |
| • Height | • Waist circumference | • Albumin |
| Diagnoses | • Total Bilirubin | |
| • Most common co-morbid conditions identified | • Alkaline Phosphatase | |
| • Hypertension | • Disorder of back | • Aspartate aminotransferase (AST) |
| • Diabetes | • Hypoglycemia | • Alanine aminotransferase (ALT) |
| • Hypercholesterolemia | • Ovarian dysfunction | • Protein |
| • General symptoms | • Chronic IHD | • Blood urea nitrogen (BUN) |
| • Depression | • Allergic rhinitis | • Creatinine |
| • Osteoarthrosis | • Invertebral disc disorder | • Sodium |
| • Hypothyroidism | • Respiratory symptoms | • Potassium |
| • Asthma | • Neurotic disorders | • Chloride |
| • Affective psychoses | • Nondependent drug abuse | • CO2 |
| • Disorder of soft tissue | • Adjustment reaction | • Glucose |
| • Disease of esophagus | • Migraine | • Calcium |
| Medications | • Glomerular filtration rate-estimated (GFR) | |
| • Most common medication subclasses identified included | • Thyroid stimulating hormone (TSH) | |
| • SSRIsa | • Biguanides | • Iron |
| • NSAIDsb | • Proton pump inhibitors | • Iron binding capacity |
| • ACE Inhibitorsc | • Statins | • Transferrin |
| • Loop Diuretics | • Salicylates | • Ferritin |
| • Thyroid hormones | • Sympathomimetics | • White blood cell count |
| • Beta blockers | • Opioid combinations | • Red blood cell count |
| • Sulfonylureas | • Insulin | • Hemoglobin |
| • Thiazides | • Insulin sensitizing agents | • Hematocrit |
| • Antihistamines | • Benzodiazepines | • Mean cell volume (MCV) |
| • Antihypertensive combos | • Nasal steroids | • Mean cell hemoglobin (MCH) |
| • Chol absorption inhibitor | • Multivitamins | • Mean cell hemoglobin concentration (MCHC) |
| • Calcium channel blocker | • Cough/Cold/Allergy combo | • Platelet count |
| • Steroid inhalants | • ARBse | • Mean platelet volume (MPV) |
| • SNRIsd | • Anticonvulsants | • Red cell distribution width (RDW) |
| • Central muscle relaxants | • H-2 Antagonistsf | |
| • Nitrates | • Fibric acid | |
| • Anti-platelet aggregation | • Coumarin anticoagulant | |
| • Non-narcotic analgesics | • Tricyclic agents | |
aSelective serotonin reuptake inhibitors
bNon-steroidal anti-inflammatory drugs
cAngiotensin converting enzyme inhibitors
dSelective serotonin norepinephrine reuptake inhibitors
eAngiotensin receptor blockers
fHistamine H2 receptor antagonists
DNA isolation
DNA was extracted from 0.35 ml of EDTA anti-coagulated whole blood using the Qiagen MagAttract DNA Blood Midi M48 Kit and Qiagen BioRobot M48 Workstation (Qiagen, Valencia, CA) according the manufacturer’s directions. The final elution volume was 200 μl. For a small number of patients, blood was not available so DNA was extracted from fixed liver tissue. Liver was first treated with proteinase K (1 μg/μl) in 350 μl Qiagen Tissue Lysis Buffer and incubated at 55°C overnight. Following digestion, samples were loaded to Qiagen BioRobot M48 Workstation and extracted for DNA as described above for blood samples. Quantification of DNA extracted was performed using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE).
Genotype analysis
Single nucleotide polymorphism (SNP) genotyping was performed on an Applied Biosystems 7500 real-time PCR System (Applied Biosystems, Foster City, CA). Assay reagents for each SNP were obtained from Applied Biosystems (rs10811661, Assay ID: C_31288917_10; rs2383206, Assay ID: C_1754669_10). DNA was genotyped according to the manufacturer’s protocol. Briefly, the reaction components for each genotyping reaction were as follows: 10 ng of DNA, 5 μl of TaqMan Genotyping Master Mix (Applied Biosystems, Foster City, CA), 0.25 μl of assay mix (40×), and water up to a total volume of 10 μl. The thermocycler conditions were as follows: 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 60 s. The reaction was then analyzed by Applied Biosystems Sequence Detection Software.
Statistical analysis
The HelixTree (Golden Helix, MT, USA) software package was used to analyze relationship of clinical variables using split-prediction methodology to either partition the data into subgroups or perform logistic regression on a predictor variable. For a binomial predictor (e.g., diagnosis code), all the observations with “0” as the predictor variable (i.e., lacking a diagnosis) are placed in one group, and all of the observations with a “1” as the predictor variable (i.e., carrying the diagnosis) are placed in a second group. A two-sample t-test is used to determine the probability that the two groups have the same mean. For a continuous-ordinal predictor (e.g., numeric lab value), observations are segmented into k subgroups, each with a different mean. The k − 1 cut-points that optimally split the data in a maximum likelihood sense are reduced by minimizing the sum of squared deviations of the subgroup means from the observations. An F-test was used to generate a raw P-value. An adjusted P-value (aP) was calculated by curve-fitting thousands of simulations. A Bonferroni corrected P-value (bP) was also calculated. A conservative threshold of a bP-value of <0.05 was used for all analyses. HelixTree was also used to determine differences in genotype and allele frequencies, estimate deviation from Hardy–Weinberg equilibrium, and to examine the association of SNPs with database variables. Graphical representation of data was performed using the KaleidaGraph software application (Syngergy Sofware, PA).
Results
More than 100 clinical variables (Table 1) were extracted from the EHR on a total of 824 patients who were consented as part of a bariatric surgery clinical research program on the genetics of obesity and related co-morbidities. Data in the EHR was obtained from a comprehensive history and physical examination performed on the initial visit, with laboratory measurements obtained within one month prior to surgery.
To define a population of morbidly obese patients for study, 49 patients (5.9%) whose body mass index (BMI) was <40, as well as 16 patients (1.9%) whose height and/or weight data were missing, were excluded from the analysis leaving 759 patients. Genotyping was then performed on available DNA from 709 of these patients. Gender, age, race, diagnoses, and medication use were obtained from the EHR on all patients. Values for laboratory measurements were obtained on at least 98% of patients for glomerular filtration rate, glucose, bun, sodium, potassium, chloride, CO2, calcium, and creatinine; on at least 97% of patients for white blood cell count (wbc), red blood cell count (rbc), hemoglobin (hgb), hematocrit (hct), mean cell volume (mcv), mean cell hemoglobin (mch), mean cell hemoglobin concentration (mchc), and red cell distribution width (rdw); on at least 96% of patients for triglycerides, cholesterol, high density lipoprotein cholesterol (hdl), alanine aminotransferase (alt), aspartate aminotransferase (ast), alkaline phosphatase, total bilirubin, and thyroid stimulating hormone (tsh); and on at least 94% of patients for low density lipoprotein cholesterol (ldl calculated), insulin, and hemoglobin A1c. Values were obtained on lower percentages of patients for iron (81%), iron binding capacity (81%), ferritin (81%), platelet count (86%), mean platelet volume (86%) albumin (33%), and total protein (33%). An “iron panel” (iron, iron binding capacity, and ferritin) was added to the clinical protocol after recruitment had begun, which accounts for the lower percentage of patients for those values. A platelet count and mean platelet volume were not reported if a hemoglobin and hematocrit was ordered rather than a complete blood count, which likely accounts for the lower percentage for these patients. Total protein and albumin were ordered only if nutritional status was deemed clinically necessary to evaluate.
Patients
The cohort consisted of 709 patients with BMI measurements of 40 or greater with a 97.5% self reported/clinically verified Caucasian ethnicity. Other demographic and relevant clinical data are shown in Table 2.
Table 2.
Demographic and selected clinical data on patient cohort (n = 709)
| Age (years) | BMIa (kg/m2) | T2Db (%) | CHDc (%) | HGB A1Cd (%) |
|---|---|---|---|---|
| 45.9 | 51.2 | 37 | 2.4 | 6.4 |
aBody mass index
bType II diabetes
cCoronary heart disease
dHemoglobin A1c
Clinical correlates of T2D and CHD
The database was used to determine whether expected relationships could be found with diabetes (i.e., ICD-9 code 250), defined as a binary variable for both split prediction analysis and regression analysis using the Golden Helix statistical software package. Of the more than 150 variables examined, the diagnosis of diabetes was associated with 35 following Bonferroni correction (bP < 0.05). The top ten statistically related measures to ICD-9 code 250 in the database are shown in Table 3 (3 variables represented by both split prediction and regression analyses). All can be directly related to diabetes. Pre-operative hemoglobin A1C was the most highly correlated (by regression) followed by the diabetes medication biguanides, hemoglobin A1c (split prediction), and insulin. The use of the statin class of lipid lowering drugs was also related, as was age (by both split prediction and regression). All of the relationships are expected based upon the clinical findings in diabetes.
Table 3.
Clinical variables with highest statistical relationship to diagnosis code for diabetes (ICD-9 250)
| Variable | P-value | aP-value | bP-value |
|---|---|---|---|
| HEMOGLOBIN_A1C | 3.90E-61 | 3.90E-61 | 8.00E-59 |
| MED BIGUANIDES | 4.54E-54 | 4.54E-54 | 9.31E-52 |
| HEMOGLOBIN_A1C | 8.61E-67 | 1.26E-51 | 2.59E-49 |
| MED INSULIN | 4.19E-41 | 4.19E-41 | 8.59E-39 |
| GLUCOSE | 1.66E-36 | 1.66E-36 | 3.41E-34 |
| MED INS SENS AGENTa | 6.92E-35 | 6.92E-35 | 1.42E-32 |
| MED SULFONYLUREAS | 5.47E-32 | 5.47E-32 | 1.12E-29 |
| GLUCOSE | 4.60E-43 | 5.21E-30 | 1.07E-27 |
| MED STATINS | 2.07E-22 | 2.07E-22 | 4.25E-20 |
| AGE | 1.89E-21 | 1.89E-21 | 3.87E-19 |
| GLUCOSE MONITOR | 3.10E-21 | 3.10E-21 | 6.35E-19 |
| AGE | 1.46E-19 | 2.17E-14 | 4.46E-12 |
| BUNb | 7.67E-14 | 7.67E-14 | 1.57E-11 |
Hemoglobin A1c, glucose, and age found by both split prediction and regression analyses
aMedication-Insulin Sensitizing Agent
bBlood Urea Nitrogen
A similar analysis was completed for CHD (i.e., defined as ICD-9 code 414 by clinical staff) as a dependent variable (Table 4). A total of 13 of the database variables were found to be statistically significant following Bonferroni correction (bP < 0.05). CHD medications including nitrates, beta blockers, platelet aggregation inhibitors, aspirin, statins and fibric acid derivatives, age (regression and split prediction), and gender were all statistically related, as was the diagnosis of hypercholesterolemia.
Table 4.
Clinical variables with highest statistical relationship to diagnosis code for ischemic heart diseases (ICD-9 code 414)
| Variable | P-value | aP-value | bP-value |
|---|---|---|---|
| MED NITRATES | 1.11E-30 | 1.11E-30 | 2.28E-28 |
| MED CARDIO BETA BLOCKER | 4.12E-22 | 4.12E-22 | 8.44E-20 |
| MED PLT AGG INHa | 1.36E-12 | 1.36E-12 | 2.79E-10 |
| MED FIBRIC ACID | 1.86E-11 | 1.86E-11 | 3.81E-09 |
| MED ASAb | 1.87E-09 | 1.87E-09 | 3.82E-07 |
| MED STATIN | 3.00E-08 | 3.00E-08 | 6.15E-06 |
| AGE | 3.06E-07 | 3.06E-07 | 6.28E-05 |
| MED COUMARIN | 4.12E-07 | 4.12E-07 | 8.46E-05 |
| PARENTERAL | 1.77E-06 | 1.77E-06 | 0.000361909 |
| MED VITAMINS | 5.65E-06 | 5.65E-06 | 0.001158545 |
| AGE | 1.66E-08 | 2.94E-05 | 0.006024747 |
| GENDER | 3.34E-05 | 3.34E-05 | 0.006848651 |
| DX HYPERCHOLc | 0.000159867 | 0.00015987 | 0.032772772 |
Age was found by both split prediction and regression analyses
aMedication-Platelet Aggregation Inhibitor
bMedication-Acetyl Salicylic Acid (aspirin)
cDiagnosis-Hypercholesterolemia
Genotypic correlates of T2D and CHD
A total of 709 patient DNA samples were genotyped for the chromosome 9p21 T2D SNP (r10811661) and CHD SNP (rs2383206) SNP variants (Table 5). Patients were defined as carriers of the “C” and/or “T” DNA sequences at the T2D SNP and the “G” and/or “A” DNA sequences at the CHD SNP. The T2D “T” SNP and the CHD “G” SNP are considered the high risk SNPs. The frequencies of the minor alleles of the T2D SNP and the CHD SNP (0.49 vs. 0.48) reported for control populations (McPherson et al.2007; Saxena et al. 2007) are in good agreement with the results here (0.17 vs. 0.17 for T2D and 0.49 vs. 0.48 for CHD).
Table 5.
Frequencies of the SNP DNA sequences
| Marker | Allele | Allele count | Allele freq. |
|---|---|---|---|
| rs10811661 | C | 239 | 0.17 |
| rs10811661 | T | 1179 | 0.83 |
| rs2383206 | A | 685 | 0.48 |
| rs2383206 | G | 733 | 0.52 |
To determine whether the population was genetically skewed through inbreeding or strong founder effects, a statistical test for Hardy–Weinberg equilibrium was performed. Both SNPs were found to be well within Hardy–Weinberg equilibrium (T2D P > 0.19; CHD P > 0.81). The frequency of the SNP alleles is thus consistent with an outbred mixed Caucasian/European population.
Because the SNPs are located within 20,000 bases of each other on chromosome 9, the extent of linkage disequilibrium between them was determined. No significant linkage disequilibrium was observed (LD Correlation R = 0.034), consistent with their presence in two distinct two haplotype blocks.
The diploid SNP sequences or genotypes (i.e., T2D “CC”, “CT”, and “TT”; CHD “AA”, “AG”, and “GG”), of each patient for each gene were also analyzed (Table 6). The T2D homozygous high risk “TT” genotype was present in ~70% of the population and the CHD homozygous high risk “GG” genotype was present in ~27%, consistent with previous studies. The T2D heterozygous “CT” and the CHD heterozygous “AG” genotypes were present at ~27% and ~50%, respectively. The low risk T2D genotype “CC” was present in ~3.5% of the population and the low risk CHD genotype “GG” was present in ~24%.
Table 6.
Frequencies of T2D and CHD SNP genotypes
| Marker | Genotype | Count | Freq. |
|---|---|---|---|
| rs10811661 | C_C | 25 | 0.035 |
| rs10811661 | C_T | 189 | 0.267 |
| rs10811661 | T_T | 495 | 0.698 |
| rs2383206 | A_A | 167 | 0.236 |
| rs2383206 | A_G | 351 | 0.495 |
| rs2383206 | G_G | 191 | 0.269 |
The relationship of the T2D and CHD SNP genotypes to the approximate 100 clinical variables obtained from the EHR was analyzed using the HelixTree Genetics Analysis Software. The initial analysis was performed using the individual T2D and CHD SNP genotypes (i.e., T2D “CC”, “CT”, and “TT”; CHD “AA”, “AG”, and “GG”). For T2D SNP rs10811661, two variables were found to be significantly different (bP < 0.05); the percentage of patients with the diagnoses of polycystic ovary syndrome (PCOS) and the diagnosis of hypertension (HTN). Interestingly, no patients with the CC genotype were diagnosed with PCOS and, correspondingly, a lower percentage had the diagnosis of HTN (Fig. 1). The mechanism by which this gene variant is related to PCOS and HTN is not clear.
Fig. 1.
Association of T2D SNP rs10811661 with the diagnosis of polycystic ovary syndrome (PCOS) and the diagnosis of hypertension (HTN). No patient with a “CC” genotype was diagnosed with PCOS
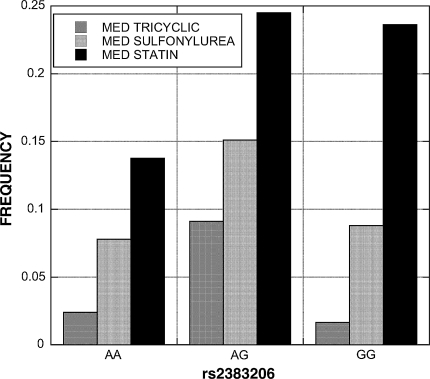
For CHD SNP rs2383206, 3 variables met the bonferroni corrected P-value threshold of 0.05; the percentage of patients on tricyclic antidepressants and sulfonylureas, as well as the laboratory value creatine kinase (CK). A fourth variable, the percentage of patients on statins, had a bP-value of 0.064. The genotype distribution patterns for tricyclic antidepressant and sulfonylurea use were different than for CK and statins. The AG heterozygotes had the highest use of tricyclics and sulfonylureas relative to AA and GG homozygotes (Fig. 2). The AG and GG genotypes had higher statin use. The GG CHD high risk genotype had CK levels that were over 2-fold higher than the non-GG genotypes (GG = 196 vs. AG = 86 vs. AA = 92).
Fig. 2.
Association of CHD SNP rs2383206 with tricyclic antidepressant, sulfonylurea, and statin use. The “AG” heterozygotes had the highest tricyclic and sulfonylurea use, while statin use was higher in patients carrying a “G” allele
Recognizing that each patient inherits the T2D and CHD risk alleles independently, we tested for compound genotype (i.e. T2D/CHD “CC”/“AA”, “CC”/“AG”, “CC”/“GG”, “CT”/“AA”, “CT”/“AG”, “CT”/“GG”, “TT”/“AA”, “TT”/“AG”, and “TT”/“GG”) associations. Each T2D and CHD genotype was classified as low (L), medium (M), and high (H) risk based upon the predicted risk group from previous studies (McPherson et al. 2007; Saxena et al. 2007). Thus, each patient could be categorized as T2D LOW/CHD LOW or L/L (“CC”/“AA”) through T2D High/CHD High or H/H (“TT”/“GG”).
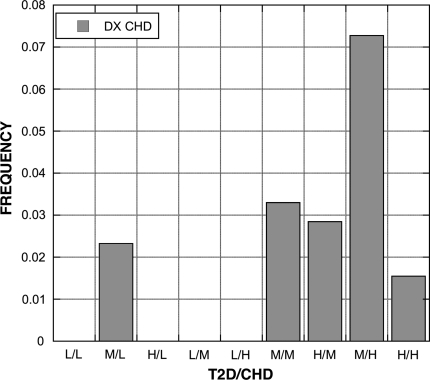
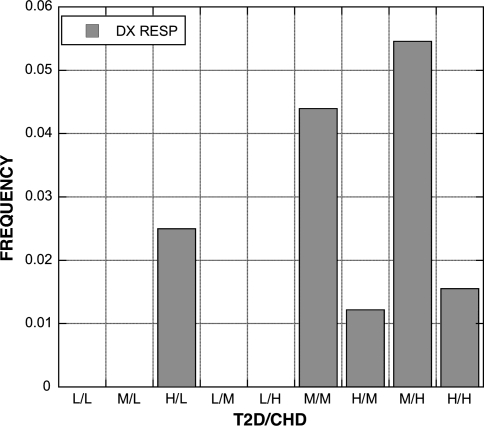
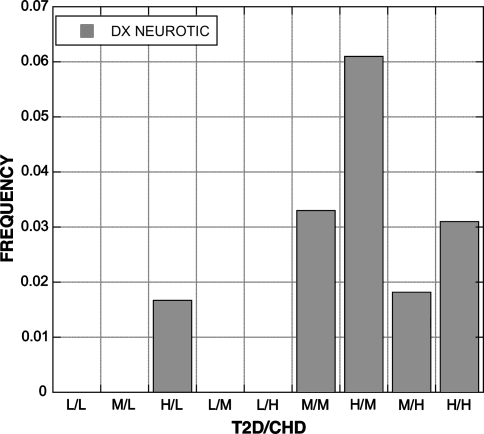
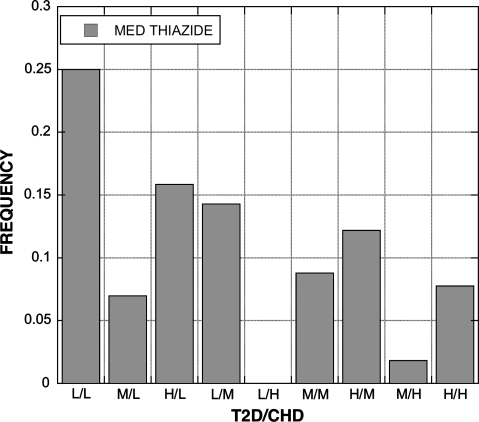
A total of 19 EHR derived variables (Table 7) were found to be statistically significant among the groups (bP < 0.05). The percentage of patients diagnosed with CHD was influenced by both SNPs; 4 of 5 compound genotypes with a low risk genotype had no patients diagnosed with CHD (Fig. 3). A similar pattern was present for the diagnoses of respiratory disorders (Fig. 4) and neurotic disorders (Fig. 5). The distribution of patients on thiazide diuretics was skewed toward low risk T2D/CHD alleles (Fig. 6). Patterns for the other associated variables were more complex and did not trend toward low or high risk genotypes.
Table 7.
Clinical variables with statistical association with type 2 diabetes/coronary heart disease compound genotypes
| Variable | Total | L/L | M/L | H/L | L/M | L/H | M/M | H/M | M/H | H/H |
|---|---|---|---|---|---|---|---|---|---|---|
| DX HTNa | 0.46 | 0.75 | 0.40 | 0.45 | 0.29 | 0.14 | 0.42 | 0.50 | 0.33 | 0.48 |
| DX ERDb | 0.25 | 0.00 | 0.37 | 0.23 | 0.07 | 0.14 | 0.27 | 0.25 | 0.16 | 0.28 |
| DX DJDc | 0.22 | 0.50 | 0.28 | 0.17 | 0.21 | 0.57 | 0.26 | 0.17 | 0.25 | 0.24 |
| DX LOTHYRd | 0.13 | 0.25 | 0.07 | 0.13 | 0.00 | 0.00 | 0.10 | 0.17 | 0.11 | 0.11 |
| DX SOFTISSe | 0.04 | 0.00 | 0.07 | 0.01 | 0.00 | 0.14 | 0.05 | 0.05 | 0.02 | 0.01 |
| DX CHDf | 0.02 | 0.00 | 0.02 | 0.00 | 0.00 | 0.00 | 0.03 | 0.03 | 0.07 | 0.02 |
| DX BACKg | 0.02 | 0.00 | 0.02 | 0.03 | 0.00 | 0.00 | 0.01 | 0.02 | 0.00 | 0.06 |
| DX RESPh | 0.02 | 0.00 | 0.00 | 0.03 | 0.00 | 0.00 | 0.04 | 0.01 | 0.05 | 0.02 |
| DX NEUROTICi | 0.04 | 0.00 | 0.00 | 0.02 | 0.00 | 0.00 | 0.03 | 0.06 | 0.02 | 0.03 |
| MED NITRATESj | 0.05 | 0.00 | 0.05 | 0.01 | 0.07 | 0.00 | 0.04 | 0.07 | 0.09 | 0.04 |
| MED BENZOk | 0.08 | 0.25 | 0.00 | 0.06 | 0.00 | 0.14 | 0.09 | 0.10 | 0.05 | 0.10 |
| MED STER INHl | 0.05 | 0.25 | 0.02 | 0.05 | 0.21 | 0.00 | 0.02 | 0.08 | 0.04 | 0.03 |
| MED SNRIsm | 0.06 | 0.25 | 0.09 | 0.02 | 0.00 | 0.00 | 0.08 | 0.06 | 0.04 | 0.09 |
| MED TRICYCLICn | 0.06 | 0.00 | 0.05 | 0.02 | 0.07 | 0.00 | 0.05 | 0.11 | 0.02 | 0.02 |
| MED STATINSo | 0.22 | 0.25 | 0.21 | 0.11 | 0.29 | 0.14 | 0.30 | 0.22 | 0.27 | 0.22 |
| MED INT CHOLp | 0.02 | 0.00 | 0.02 | 0.04 | 0.00 | 0.14 | 0.00 | 0.01 | 0.00 | 0.02 |
| MED ANTI-HTNq | 0.12 | 0.25 | 0.05 | 0.11 | 0.00 | 0.00 | 0.18 | 0.11 | 0.09 | 0.14 |
| MED COLDr | 0.04 | 0.00 | 0.00 | 0.07 | 0.07 | 0.29 | 0.04 | 0.02 | 0.00 | 0.04 |
| MED THIAZIDEs | 0.11 | 0.25 | 0.07 | 0.16 | 0.14 | 0.00 | 0.09 | 0.12 | 0.02 | 0.08 |
L = low risk genotype, M = medium risk genotype, H = high risk genotype. Numbers represent proportion of patients with compound genotype that were diagnosed with indicated condition or were on indicated medication
aDiagnosis-Hypertension
bDiagnosis-Esophageal Reflux Disease
cDiagnosis-Degenerative Joint Disease
dDiagnosis-Hypothyroidism
eDiagnosis-Soft Tissue Disorder
fDiagnosis-Coronary Heart Disease
gDiagnosis-Back Disorder
hDiagnosis-Respiratory Disorder
iDiagnosis-Neurotic Disorder
jMedication-Nitrates
kMedication-Benzodiazapines
lMedication-Steroid Inhalants
mMedication-Selective Serotonin Norepinephrine Reuptake Inhibitors
nMedication-Tricyclic Antidepressants
oMedication-HMG CoA Reductase Inhibitor Drugs
pMedication-Intestinal Cholesterol Absorption Inhibitors
qMedication-Anti-Hypertensives
r Medication-Cold Remedies
sMedication-Thiazide Diuretics
Fig. 3.
Frequency of diagnosis of CHD among patients with each T2D and CHD SNP compound genotype. The pattern is skewed toward those carrying the medium (M) and high (H) risk genotypes
Fig. 4.
Frequency of diagnosis of respiratory disorders (RESP) among patients with each T2D and CHD compound genotype. The pattern is skewed toward those carrying the medium (M) and high (H) risk genotypes, similar to the pattern with CHD
Fig. 5.
Frequency of diagnosis of neurotic disorders among patients with each T2D and CHD compound genotype. The pattern is skewed toward those carrying the medium (M) and high (H) risk genotypes, similar to the pattern with CHD
Fig. 6.
Frequency of thiazide medication use among patients with each T2D and CHD compound genotype. The distribution is skewed toward those with low risk genotypes
Discussion
The resource intensive nature of genomic medicine research stems both from costs related to DNA analysis, i.e., genotyping, as well as the costs and logistical challenges related to acquiring clinical data, i.e., phenotyping. While significant gains have been made in recent years in the cost-effectiveness of genotyping technologies, the methods used to recruit and profile clinical phenotypes have not changed and continue to rely on labor intensive processes. The advent of EHRs, pioneered by large integrated health delivery systems, may now provide a potentially rich source of phenotypic data for genomic medicine research (Gerhard et al. in press). Use of patient populations served by integrated delivery systems, related biobanked samples, and EHR data can substantially reduce the labor and time required to complete such studies. The use of EHRs to acquire phenotype data does not alter the essential nature of phenotyping; rather, it provides access to data that has already been gathered, and paid for, during the course of clinical care. The conversion of disparate clinical data sources (e.g., laboratory data, diagnostic coding, survey data, etc.) into an electronic format allows for efficient data extraction and database construction.
A potential limitation to the use EHR-derived data for genetics research is data quality (Thiru et al. 2003). Variation in completeness and quality of EHR data may be affected by different practices among staff and clinicians (Treweek 2003), potentially impacting consistency and accuracy of phenotypic definitions. The extent to which these and other issues impact data collection will vary from institution to institution, depending upon the capability of the specific EHR, how it is used in clinical operations, and which data domains are used (Persell et al. 2006). For example, laboratory values provide a relatively objective source of EHR data, while consistency of clinical definitions may vary if derived from a variety of clinicians (de Lusignan 2006) or if data must be extracted from free text (Voorham and Denig 2007). These concerns are mitigated to a large degree in this study through the acquisition of most data from a single clinic using a care delivery process that was optimized for obtaining from the EHR for research. All diagnosis and medication codes were derived from the initial comprehensive examination performed by the same staff using a common process, equivalent to a single visit data collection interview with a research participant. A standard set of laboratory values was measured as part of the clinical evaluation, and the testing was performed at the same laboratory using consistent methods. The true potential of the EHR for genomic medicine may exist in the ability to modify existing clinical processes to allow for research-grade data collection. The data presented here support the feasibility of this approach and represents one of the first examples of EHR-based genomic medicine research.
The depth and breadth of the data extracted from the EHR may also be useful for unraveling the complex interactions involving obesity, T2D, and CHD which are likely caused by a combination of genetic susceptibility and environmental effects. Substantially increasing the number of EHR variables extracted and analyzed carries only small incremental costs but greatly increases the potential to identify new genotype–phenotype correlations. For example, while an association with T2D was not found, the T2D SNP was related to the diagnoses of polycystic ovary syndrome (PCOS) and hypertension (HTN). PCOS has been associated with metabolic syndrome, a greatly increased risk of impaired glucose tolerance and type 2 diabetes mellitus, potential cardiovascular risks, and has a substantial genetic component (Norman et al. 2007). A number of other SNP-phenotype associations were identified that remain to be replicated and further explored. Unfortunately, little is known about the biological impact the SNPs. They are both located within about 20,000 bp of each other in an inter-genic region on chromosome 9p21 upstream of cyclin-dependent kinase inhibitors CDKN2A and CDKN2B, but it is not known whether the SNPs have a long-range effect on one of these genes or influence another gene(s).
A unique aspect of the cohort analyzed here was the level of obesity. The range of BMI values in the population studied here, 40–88 kg/m2, is more than double the range in most other studies, i.e., 20–40 kg/m2. The T2D and CHD SNPs were identified using non-obese, overweight, and/or mildly obese populations. For example, the CHD SNP rs2383206 was identified in populations of predominantly Caucasian men who had severe, premature CHD and was replicated in a much larger prospective study of CHD risk in Caucasian men and women (McPherson et al. 2007). We did not replicate these findings in a population consisting of primarily Caucasian, middle aged, morbidly obese women. Age and gender may also be important factors that may account for the lack of association with the CHD SNP. The average age of the morbidly obese population was less than 50 years and approximately 80% were female, thus many patients with genetic susceptibility to CHD may not yet have manifested any clinical evidence of the disease. In addition, statistical power may not have been sufficient given the low prevalence of clinically documented CHD. A 3–4-fold increase in CHD would need to be present in order to detect an influence of the homozygous CHD genotype given a prevalence of about 2%.
The T2D SNP rs10811661 was identified by two groups (Saxena et al. 2007; Scott et al. 2007) using populations of predominantly male non-obese patients from Finland and Sweden. We could not replicate these findings either, although no association of this SNP was found with several anthropometric traits, glucose tolerance and insulin secretion, lipids and apolipoproteins, and blood pressure, similar to our findings of no association with any lipid or diabetes related parameters. With the high frequency of the at risk T2D “AA” genotype and the high prevalence of T2D in our population, the analyses were sufficiently powered (>0.8) to detect a ~1.3 increased risk of T2D.
The results reported here represent studies of SNPs initially identified using genome wide association approaches. In addition to serving as a rapid and efficient means of evaluating the findings of such genome wide association studies, EHR data may also be useful as the primary source of phenotypes for genome wide association studies.
Acknowledgments
The corresponding author Glenn S. Gerhard had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. This work was supported by the Geisinger Clinical Research Fund.
References
- Cupples LA, Arruda HT, Benjamin EJ, D’Agostino RB Sr, Demissie S, DeStefano AL, Dupuis J, Falls KM, Fox CS, Gottlieb DJ, Govindaraju DR, Guo CY, Heard-Costa NL, Hwang SJ, Kathiresan S, Kiel DP, Laramie JM, Larson MG, Levy D, Liu CY, Lunetta KL, Mailman MD, Manning AK, Meigs JB, Murabito JM, Newton-Cheh C, O’Connor GT, O’Donnell CJ, Pandey M, Seshadri S, Vasan RS, Wang ZY, Wilk JB, Wolf PA, Yang Q, Atwood LD (2007) The Framingham Heart Study 100 K SNP genome-wide association study resource: overview of 17 phenotype working group reports. BMC Med Genet 8(Suppl 1):S1 [DOI] [PMC free article] [PubMed]
- de Lusignan S (2006) The optimum granularity for coding diagnostic data in primary care: report of a workshop of the EFMI Primary Care Informatics Working Group at MIE 2005. Inform Prim Care 14:133–137 [DOI] [PubMed]
- Eberle MA, Ng PC, Kuhn K, Zhou L, Peiffer DA, Galver L, Viaud-Martinez KA, Lawley CT, Gunderson KL, Shen R, Murray SS (2007) Power to detect risk alleles using genome-wide tag SNP panels. PLoS Genet 3:1827–1837 [DOI] [PMC free article] [PubMed]
- Flegal K, Carroll M, Ogden C, Johnson C (2002) Prevalence and trends in obesity among US adults. JAMA 288:1723–1727 [DOI] [PubMed]
- Gerhard G, Langer R, Carey D, Stewart W (in press) Electronic medical records in genomic medicine practice and research. In: Willard H, Ginsburg G (eds) Handbook of genomic medicine. Elsevier
- Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, Masson G, Gudbjartsson DF, Magnusson KP, Andersen K, Levey AI, Backman VM, Matthiasdottir S, Jonsdottir T, Palsson S, Einarsdottir H, Gunnarsdottir S, Gylfason A, Vaccarino V, Hooper WC, Reilly MP, Granger CB, Austin H, Rader DJ, Shah SH, Quyyumi AA, Gulcher JR, Thorgeirsson G, Thorsteinsdottir U, Kong A, Stefansson K (2007) A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 316:1491–1493 [DOI] [PubMed]
- Koumanis DJ, Christou NV, Wang XL, Gilfix BM (2002) Pilot study examining the frequency of several gene polymorphisms in a morbidly obese population. Obes Surg 12:759–764 [DOI] [PubMed]
- Larson MG, Atwood LD, Benjamin EJ, Cupples LA, D’Agostino RB Sr, Fox CS, Govindaraju DR, Guo CY, Heard-Costa NL, Hwang SJ, Murabito JM, Newton-Cheh C, O’Donnell CJ, Seshadri S, Vasan RS, Wang TJ, Wolf PA, Levy D (2007) Framingham Heart Study 100 K project: genome-wide associations for cardiovascular disease outcomes. BMC Med Genet 8(Suppl 1):S5 [DOI] [PMC free article] [PubMed]
- McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, Boerwinkle E, Hobbs HH, Cohen JC (2007) A common allele on chromosome 9 associated with coronary heart disease. Science 316:1488–1491 [DOI] [PMC free article] [PubMed]
- Norman RJ, Dewailly D, Legro RS, Hickey TE (2007) Polycystic ovary syndrome. Lancet 370:685–697 [DOI] [PubMed]
- O’Donnell CJ, Cupples LA, D’Agostino RB, Fox CS, Hoffmann U, Hwang SJ, Ingellson E, Liu C, Murabito JM, Polak JF, Wolf PA, Demissie S (2007) Genome-wide association study for subclinical atherosclerosis in major arterial territories in the NHLBI’s Framingham Heart Study. BMC Med Genet 8(Suppl 1):S4 [DOI] [PMC free article] [PubMed]
- Persell SD, Wright JM, Thompson JA, Kmetik KS, Baker DW (2006) Assessing the validity of national quality measures for coronary artery disease using an electronic health record. Arch Intern Med 166:2272–2277 [DOI] [PubMed]
- Powell J, Buchan I (2005) Electronic health records should support clinical research. J Med Internet Res 7:e4 [DOI] [PMC free article] [PubMed]
- Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, Barrett JH, Konig IR, Stevens SE, Szymczak S, Tregouet DA, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H (2007) Genomewide association analysis of coronary artery disease. N Engl J Med 357:443–453 [DOI] [PMC free article] [PubMed]
- Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson Bostrom K, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Rastam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjogren M, Sterner M, Surti A, Svensson M, Svensson M, Tewhey R, Blumenstiel B, Parkin M, Defelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma Q, Parikh H, Richardson D, Ricke D, Purcell S (2007) Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316:1331–1336 [DOI] [PubMed]
- Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL, Sprau AG, Tong M, White PP, Hetrick KN, Barnhart MW, Bark CW, Goldstein JL, Watkins L, Xiang F, Saramies J, Buchanan TA, Watanabe RM, Valle TT, Kinnunen L, Abecasis GR, Pugh EW, Doheny KF, Bergman RN, Tuomilehto J, Collins FS, Boehnke M (2007) A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316:1341–1345 [DOI] [PMC free article] [PubMed]
- Service SK, Sandkuijl LA, Freimer NB (2003) Cost-effective designs for linkage disequilibrium mapping of complex traits. Am J Hum Genet 72:1213–1220 [DOI] [PMC free article] [PubMed]
- Still CD, Benotti P, Wood GC, Gerhard G, Petrick A, Reed M, Strodel W (2007) Outcomes of preoperative weight loss in high-risk patients undergoing gastric bypass surgery. Arch Surg 142:994–998; discussion 999 [DOI] [PubMed]
- Thiru K, Hassey A, Sullivan F (2003) Systematic review of scope and quality of electronic patient record data in primary care. BMJ 326:1070 [DOI] [PMC free article] [PubMed]
- Treweek S (2003) The potential of electronic medical record systems to support quality improvement work and research in Norwegian general practice. BMC Health Serv Res 3:10 [DOI] [PMC free article] [PubMed]
- Voorham J, Denig P (2007) Computerized extraction of information on the quality of diabetes care from free text in electronic patient records of general practitioners. J Am Med Inform Assoc 14:349–354 [DOI] [PMC free article] [PubMed]
- Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW, Freathy RM, Barrett JC, Shields B, Morris AP, Ellard S, Groves CJ, Harries LW, Marchini JL, Owen KR, Knight B, Cardon LR, Walker M, Hitman GA, Morris AD, Doney AS, McCarthy MI, Hattersley AT (2007) Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 316:1336–1341 [DOI] [PMC free article] [PubMed]