Abstract
Single Nucleotide Polymorphisms (SNPs) are being intensively studied to understand the biological basis of complex traits and diseases. The Genetics of human phenotype variation could be understood by knowing the functions of SNPs. In this study using computational methods, we analyzed the genetic variations that can alter the expression and function of the CFTR gene responsible candidate for causing cystic fibrosis. We applied an evolutionary perspective to screen the SNPs using a sequence homology-based SIFT tool, which suggested that 17 nsSNPs (44%) were found to be deleterious. The structure-based approach PolyPhen server suggested that 26 nsSNPS (66%) may disrupt protein function and structure. The PupaSuite tool predicted the phenotypic effect of SNPs on the structure and function of the affected protein. Structure analysis was carried out with the major mutation that occurred in the native protein coded by CFTR gene, and which is at amino acid position F508C for nsSNP with id (rs1800093). The amino acid residues in the native and mutant modeled protein were further analyzed for solvent accessibility, secondary structure and stabilizing residues to check the stability of the proteins. The SNPs were further subjected to iHAP analysis to identify htSNPs, and we report potential candidates for future studies on CFTR mutations.
Keywords: CFTR gene, SIFT, PolyPhen, UTR, Modeled structure, Haplotype
Introduction
Cystic fibrosis (CF) is one of the most common life-threatening autosomal recessive diseases. It is a complex multisystem disorder, caused by mutations of the gene encoding for the cystic fibrosis transmembrane conductance regulator (CFTR), located on chromosome region 7q31. CFTR is made up of five domains: two membrane-spanning domains (MSD1 and MSD2) that form the chloride ion channel, two nucleotide-binding domains (NBD1 and NBD2) that bind and hydrolyze ATP (adenosine triphosphate), and a regulatory (R) domain. CFTR is localized in the apical membrane of epithelial cells and confers cAMP-activatable transport of chloride, bicarbonate and glutathione (Gabriela et al. 2007). One study reported that the basic defect in CF impairs apical permeability for the chloride ion, and is assessed in humans by increased chloride concentrations in sweat (Gibson and Cooke 1959). More recent studies report low chloride conductance of upper airway epithelium (Schuler et al. 2004), and lower chloride secretory response of the intestinal epithelium to secretagogues (De Jonge et al. 2004). The major disease causing mutation of the cystic fibrosis (CF) transmembrane conductance regulator (CFTR) protein occurs in the DNA sequence that codes for the first nucleotide-binding domain (NBD1). Approximately 70% of CF patients (Collins 1992) are homozygous for the F508 cftr and 90% carry at least one F508 cftr allele (compound heterozygotes). People who are homozygous for delta F508 mutation tend to have the most severe symptoms of cystic fibrosis due to critical loss of chloride ion transport.
Understanding the genomic differences in the human population is one of the major challenges in the field of current genomics research. The recent sequencing of the human genome (Venter et al. 2001; Lander et al. 2001) together with the large number of SNPs present in the human population (Sherry et al. 2001; Hinds et al. 2005; The International Hapmap Consortium 2003) opens the way for the development of a detailed understanding of the mechanisms by which genetic variation results in phenotype variation. The most common type of genome variation is single nucleotide polymorphisms (SNPs), which occur in the genome by the substitution of one single base, and account for 90% of all polymorphisms in the human genome (Sachidanandam et al. 2001). In addition, there are many common one base insertion and deletion polymorphisms. There are now several databases with these variations of SNPs, such as the human genome variation database, HGVBase (Fredman et al. 2002) and the National Center for Biotechnology Information (NCBI) database, dbSNP (Smigielski et al. 2000). Among the various types of SNPs, nonsynonymous SNPs (nsSNPs) are believed to have the greatest impact on protein function because they often lead to mutation of the encoded amino acids, which can have a deleterious effect on the structure and/or function of the proteins (Chasman and Adams 2001; Dryja et al. 1990; Smith et al. 1994). Recent studies show that SNPs may have functional effects on transcriptional regulation, by affecting transcription factor binding sites in promoter or intronic enhancer regions (Prokunina and Alarcn-Riquelme 2004; Prokunina et al. 2002), or alternatively splicing regulation by disrupting exonic splicing enhancers or silencers (Cartegni and Krainer 2002).
Over the past few years, several studies have attempted to predict the functional consequences of an nsSNP whether it is disease-related or neutral, based on sequence information and structural attributes (Richard et al. 2006). Currently, most of the diseases represented by the genes in the databases like OMIM, HGMD, and Swiss-Prot segregate in a Mendelian manner, which suggests that they are caused by single deleterious lesions.
Computational tools like SIFT and PolyPhen are able to predict 90% of damaging SNPs. These prediction methods can help us to narrow down candidate nsSNPs to identify the causative lesion within a large genomic region implicated in disease by linkage studies (Ng and Henokoff 2006). Several groups have tried to evaluate the deleterious nsSNPs based on 3-dimensional (3D) structure information of proteins by in-silico analysis. Karchin et al. considered that the strongest predicting signals in the lac repressor/lysozyme set were solvent accessibility and superfamily-level evolutionary conservation (Karchin et al. 2005a, b). Sunyaev et al. and Chen et al. also indicated that the residue solvent accessibility, which could identify the buried residues, was confidently proposed as predictors of deleterious substitutions (Sunyaev et al. 2001; Chen et al. 2005). However, the theoretical prediction methods for deleterious nsSNPs are still in their infancy because the 3D structural information of most proteins is not yet available (Bao and Cui 2006; Wagner et al. 2005; Nguyen 2006). Therefore, it is an inevitable trend to predict the deleterious variations in proteins using sequence-based and position-specific evolutionary information (Sunyaev et al. 2001; Saunders and Baker 2002; Balasubramanian et al. 2005). As a next step in the study of genetic variation, current interest is focused on disease-gene association, that is, identifying which DNA variation or set of DNA variations is highly associated with a specific disease. Recently, haplotype analysis has been successfully applied to the identification of the DNA variations relevant to several common and complex diseases and is now considered the most promising method for studying complex disease-gene association (Stumpf 2004).
Deleterious nsSNPs analyses for the CFTR gene have not been estimated computationally until now, although they have been the focus for experimental researchers. Therefore, in this work, the computational algorithms namely SIFT, PolyPhen, PupaSuite, FASTSNP, ASA View, DSSP and SRide were used to identify the deleterious nsSNPs that are likely to affect the function and structure of the protein and showed the htSNPs which are in the haplotype blocks using iHAP analysis. Based on SIFT and PolyPhen, we identified the possible mutation, proposed a modeled structure for the mutant proteins and compared this with the native protein in the 3-D modeled structure of the CFTR gene. We further analyzed native and mutant modeled proteins for solvent accessibility, secondary structure analysis and stabilizing residues. Our computational study also demonstrates the presence of other deleterious mutations in the CFTR gene that may affect the expression and function of proteins with possible roles in Cystic fibrosis. Moreover, our present study is well supported and documented by an in vivo experimental protocol in CFTR gene (Tsui 1992; Ghanem et al.1994; Bienvenu et al. 1998).
Materials and methods
Datasets
The SNPs and their related protein sequence of CFTR gene were retrieved from the Human genome variation database, HGVBase (http://hgvbase.cgb.ki.se) and National Center for Biotechnology Information (NCBI) database of SNPs, dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP) for our computational analysis.
Functional analysis of coding nsSNPs by SIFT
Sorting Intolerant From Tolerant (SIFT) is a sequence homology-based tool that sorts intolerant from tolerant amino acid substitutions and predicts whether an amino acid substitution in a protein will have a phenotypic effect. SIFT (Ng and Henikoff 2003) is based on the premise that protein evolution is correlated with protein function. We used SIFT to detect the deleterious coding non synonymous SNPs and submitted the query in the form of either SNPids or as protein sequences. SIFT analysis was performed by allowing the algorithm to search for homologous sequences (i.e., without inputting known homologs) and using the default settings (SWISS-PROT 45 and TrEMBL 28 databases, median conservation score 3.00, remove sequences >90% identical to query sequence). The underlying principle of this program is that it generates alignments with a large number of homologous sequences and assigns scores to each residue, ranging from zero to one. Scores close to zero indicate evolutionary conservation and intolerance to substitution, while scores close to one indicate tolerance to substitution. SIFT scores <0.05 are predicted by the algorithm to be intolerant or deleterious amino acid substitutions, whereas scores >0.05 are considered tolerant (Ng and Henikoff 2001). The higher a tolerance index, the less functional impact a particular amino acid substitution is likely to have.
Simulation for functional change in coding nsSNPs
PolyPhen (Polymorphism Phenotyping) is an automatic tool for prediction of possible impact of an amino acid substitution on the structure and function of a human protein available at http://coot.embl.de/PolyPhen/. This prediction is based on straightforward empirical rules which are applied to the sequence, phylogenetic and structural information characterizing the substitution. Input options for PolyPhen server (Ramensky et al. 2002) is protein sequence or SWALL database ID or accession number together with sequence position with two amino acid variants. We submitted the query in the form of protein sequence with mutational position and two amino acid variants. Basically, PolyPhen searches for 3D protein structures, multiple alignments of homologous sequences and amino acid contact information in several protein structure databases, calculates position-specific independent counts (PSIC) scores for each of two variants, and then computes the PSIC scores difference of two variants. The higher a PSIC score difference, the higher functional impact a particular amino acid substitution is likely to have. A PSIC score difference of 1.5 and above is considered to be damaging.
Analyzing the molecular phenotypic effects of SNPs
The SNPeffect (Reumers et al. 2006) and PupaSuite (Conde et al. 2006) are now synchronized to deliver annotations for both noncoding and coding SNP, as well as annotations for the SwissProt set of human disease mutations. In this approach, the input consists of a list of genes (genes belonging to a given pathway, involved in a particular biological function, etc.) and the user must specify the type of gene identifiers by selecting either Ensembl or an external database (which include GenBank, Swissprot/TrEMBL and other gene ids supported by Ensembl). PupaSuite is a unique and more integrated interface of PupaSNP (Conde et al. 2004) and PupasView (Conde et al. 2005) accessible at http://pupasuite.bioinfo.cipf.es and through http://www.pupasnp.org. PupasView retrieves SNPs that could affect conserved regions that the cellular machinery uses for the correct processing of genes (intron/exon boundaries or exonic splicing enhancers). It uses algorithms like Tango (β-aggregation regions in protein sequences) and FoldX (stability change caused by the single amino acid variation) to predict the effect of coding nonsynonymous SNPs on several phenotypic properties such as structure and dynamics, functional sites and cellular processing of human proteins using either sequence-based or structural bioinformatics tools.
Functional significance of noncoding SNPs in regulatory untranslated regions
Recent studies show that SNPs have functional effects on protein structure by a single change in the amino acid (Cargill et al. 1999; Sunyaev et al. 2000) and on transcriptional regulation. We used the FastSNP (Yuan et al. 2006) for predicting the functional significance of the 5′ and 3′ UTRs of the CFTR gene and also to identify the polymorphism involving the intron which may lead to defects in RNA and mRNA processing. The FastSNP server (http://fastsnp.ibms.sinica.edu.tw) follows the decision tree principle with external web service access to TFSearch, which predicts whether a noncoding SNP alters the transcription factor-binding site of a gene. The score will be given on the basis of levels of risk with a ranking of 0, 1, 2, 3, 4, or 5. This signifies the levels of no, very low, low, medium, high, and very high effect, respectively.
Modeling nsSNP locations on protein structure and their RMSD difference
Structure analysis was performed for evaluating the structural stability of native and mutant protein. We used the web resource SAAPdb (Cavallo and Martin 2005) and dbSNP to identify the protein coded by CFTR gene (PDB id 1nbd). We also confirmed the mutation positions and the mutation residues from this server. These mutation positions and residues were in complete agreement with the results obtained with SIFT and PolyPhen programs. The mutation was performed using SWISSPDB viewer, and energy minimization for 3D structures was performed using NOMAD-Ref server (Lindahl et al. 2006). This server use Gromacs as default forcefield for energy minimization based on the methods of steepest descent, conjugate gradient and L-BFGS methods (Delarue and Dumas 2004). We used the conjugate gradient method for optimizing the 3D structures. Deviation between the two structures was evaluated by their RMSD values.
Computation analysis of solvent accessibility, secondary structure and stabilizing residues
Solvent accessibility is the ratio between the solvent accessible surface area of a residue in a three dimensional structure and in an extended tripeptide conformation. We obtained the solvent accessibility information using NetASA view (Shander et al. 2004). The entire implementation of ASA View for all PDB proteins, as a whole or for an individual chain may be accessed at http://www.netasa.org/asaview/. Requirements for the use are simply the PDB code or the coordinate file. Solvent accessibility was divided into three classes, buried, partially buried and exposed indicating, respectively, low, moderate and high accessibility of the amino acid residues to the solvent (Gilis and Rooman 1996; Gilis and Rooman 1997). For a successful analysis of the relation between amino acid sequence and protein structure, an unambiguous and physically meaningful definition of secondary structure is essential. We obtained the information about secondary structures of the proteins using the program DSSP (Kabsch and Sander 1983).
In order to check the stability for the native and mutant modeled structures, identification of the stabilizing residues will be useful. We used the server SRide (Magyar et al. 2005) for identifying the stabilizing residues in native protein and mutant models. Stabilizing residues were computed using parameters such as surrounding hydrophobicity, long-range order, stabilization center and conservation score (Magyar et al. 2005).
Analysis of htSNPs
We used iHAP analysis (Song et al. 2006) to analyse optimal subsets of SNPs, commonly known as “haplotype tagging SNPs” (htSNPs), to capture most of the haplotype diversity of each haplotype block or gene-specific region. We submitted gene name, the iHAP resource determines the chromosomal region of interest using the UCSC Genome Browser Database. The setup of the analysis job is then defined according to parameters such as the HapMap population, allele frequency threshold, block definitions, tag SNP definitions, permutation test settings, as well as SNPs to be “force included” as tags. We selected only nsSNPs and SNPs in untranslated regions for iHAP analysis in three different populations namely CEU-CEPH (northern and western Europe), JPH (Japanese) and CHB (Chinese) respectively.
Results
SNP dataset
We selected (i) nonsynonymous coding SNPs (ii) 5′ and 3′ UTR region SNPs (iii) introns for our investigation. Out of 764 SNPs, 39 were nonsynonymous SNPs (nsSNPs) and 39 SNPs in coding synonymous region. Noncoding region is comprised of 2 SNPs in 5′ UTR region, 7 SNPs in 3′ UTR region and 677 SNPs were in the intronic region. Further it was observed that the number of nsSNPs in the coding region is much higher compared to the SNPs in the 5′and 3′ untranslated regions.
Deleterious nsSNP by SIFT program
SIFT predicts the functional importance of amino acid substitutions based on the alignment of orthologous and/or paralogous protein sequences. The protein sequences of 39 nsSNPs were submitted independently to the SIFT program to check its tolerance index. Among the 39 nsSNPs, 17 nsSNPs (44%) were identified to be deleterious with a tolerance index score of =0.05 as shown in Table 1. SIFT scores were classified as intolerant (0.00–0.05), potentially intolerant (0.051–0.10), borderline (0.101–0.20), or tolerant (0.201–1.00) according to the classification proposed by Ng et al. and Xi et al. The higher the tolerance index, the less functional impact a particular amino acid substitution is likely to have, and vice versa. 4 nsSNPs with ids (rs1800092, rs1800093, rs1800120 and rs4148725) showed a highly deleterious tolerance index score of 0.00 and could affect the protein function in the CFTR gene.
Table 1.
List of nsSNPs that were predicted to be deleterious by SIFT and PolyPhen
| SNPs ID | Alleles | AA change | Tolerance index | PSIC |
|---|---|---|---|---|
| rs1800072 | G/A | V11C | 1.00 | 0.150 |
| rs1800073 | C/T | R31C | 0.18 | 2.288 |
| rs1800074 | A/T | D44V | 0.01 | 2.532 |
| rs1800076 | G/A | R75Q | 0.03 | 1.754 |
| rs1800078 | T/C | L138P | 0.01 | 2.192 |
| rs35516286 | T/C | I148T | 0.41 | 1.743 |
| rs1800079 | G/A | R170H | 0.05 | 1.968 |
| rs1800080 | A/G | S182G | 0.03 | 1.699 |
| rs1800086 | C/G | T351S | 0.30 | 1.600 |
| rs1800087 | A/C | Q353H | 0.03 | 2.093 |
| rs4727853 | C/A | N417K | 1.00 | 0.015 |
| rs11531593 | C/A | F433L | 0.65 | 0.694 |
| rs1800089 | C/T | L467F | 0.15 | 1.568 |
| rs213950 | G/A | V470M | 0.17 | 1.432 |
| rs1800092 | C/A/G | I506M | 0.00 | 1.574 |
| rs1801178 | A/G | I507V | 0.38 | 0.314 |
| rs1800093 | T/G | F508C | 0.00 | 3.031 |
| rs35032490 | A/G | K532E | 1.00 | 1.525 |
| rs1800097 | G/A | V562I | 0.13 | 0.345 |
| rs41290377 | G/C | G576A | 0.33 | 1.262 |
| rs766874 | C/T | S605F | 0.03 | 2.147 |
| rs1800099 | A/G | S654G | 0.03 | 1.611 |
| rs1800100 | C/T | R668C | 0.01 | 2.654 |
| rs1800101 | T/C | F693L | 0.61 | 0.895 |
| rs1800103 | A/G | I807M | 0.01 | 1.554 |
| rs1800106 | T/C | Y903H | 0.52 | 0.183 |
| rs1800107 | G/T | S909I | 0.10 | 1.624 |
| rs1800110 | T/C | L967S | 0.07 | 1.683 |
| rs1800111 | G/C | L997F | 0.24 | 1.000 |
| rs1800112 | T/C | I1027T | 0.03 | 1.860 |
| rs1800114 | C/T | A1067V | 0.04 | 1.542 |
| rs36210737 | T/A | M1101K | 0.05 | 2.637 |
| rs35813506 | G/A | R1102K | 0.52 | 1.589 |
| rs1800120 | G/T | R1162L | 0.00 | 2.038 |
| rs1800123 | C/T | T1220I | 0.22 | 0.059 |
| rs34911792 | T/G | S1235R | 0.45 | 1.483 |
| rs11971167 | G/A | D1270N | 0.12 | 1.739 |
| rs4148725 | C/T | R1453W | 0.00 | 2.513 |
Highly deleterious by SIFT and damaging by PolyPhen are indicated as bold
Damaged nsSNP by PolyPhen server
The structural levels of alteration were determined by applying the PolyPhen program. It predicts the functional effect of amino acid changes by considering evolutionary conservation, the physiochemical differences, and the proximity of the substitution to predicted functional domains and/or structural features. All the 39 protein sequences of nsSNPs submitted to SIFT were also submitted as input to the PolyPhen server. 26 nsSNPs (66%) listed in Table 1 were considered to be damaging and exhibited a range of PSIC score difference between 1.52 and 3.03. 5 nsSNPs with ids (rs1800074, rs1800093, rs1800100, rs36210737 and rs4148725) with a PSIC score greater than 2.5 may have an affect on the tertiary structure of proteins and their functionality. 17 nsSNPs that were observed to be deleterious by the SIFT program also were damaging according to PolyPhen. To date, data on the validity of these algorithms has come from benchmarking studies based on the analysis of “known” deleterious substitutions annotated in databases, such as SwissProt, shown to successfully predict the effect of over 80% of amino acid substitutions (Savas et al. 2004; Sunyaev et al. 2000; Xi et al. 2004; Ng and Henikoff 2002). Experimental studies of individual proteins have also confirmed the accuracy of SIFT (Brooks-Wilson et al. 2004; Zhang et al.2004; Kanetsky et al. 2004). Hence, we could infer that the results obtained by the evolutionary-based approach (SIFT) correlated well with the results obtained by structural-based approach (PolyPhen), as can be seen from Table 1. The nsSNP with an id (rs1800093) showed a SIFT tolerance index of 0.00 and PSIC score difference 3.0 at position F508C and was selected for modeling analysis.
Predictions of potential phenotypic effect in SNPs
The effect of nonsynonymous coding SNPs can be analyzed by means of the physico-chemical properties of the affected proteins. Pupasuite tries to pinpoint the exact effect of a mutation to a specific structural or physico-chemical property, ranging from protein aggregation to the disruption of protein-protein interactions, or from changes in protein turnover rate to subcellular (mis) localisation. In-silico methods provide a useful tool for an initial approach to any mutation suspected of causing aberrant RNA processing. These mutations can result in either complete skipping of the exon, retention of the intron or the introduction of a new splice site within an exon or intron. In rare cases, mutations that do not disrupt or create a splice site, activate preexisting pseudo splice sites consistent with the proposal that introns contain splicing inhibitory sequences (Baralle 2005). Nonsense and missense mutations can disrupt exonic splicing enhancers (ESEs) and cause the splicing machinery to skip the mutant exon, with dramatic effects on the structure of the gene product (Cartegni et al. 2002). ESEs are common in alternative and constitutive exons, where they act as binding sites for Ser/Arg-rich proteins (SR proteins), a family of conserved splicing factors that participate in multiple steps of the splicing pathway (Graveley 2000). ESSs are sequence elements that are known to regulate alternative splicing and also play a role in splice site selection (Fairbrother and Chasin 2000). Out of 39 nsSNPs, 16 nsSNPs disrupted the exonic splicing enhancers, 3 SNPs in mRNA disrupted the exonic splicing enhancers, 3 nsSNPs disrupted the exonic splicing silencers, 6 nsSNPs (Pathological SNPs) were involved in cellular processing, 5 nsSNPs were involved in protein structure and dynamics and 3 nsSNPs were involved in functional sites as depicted in Table 2. Varied levels of alternative splicing have been detected for some of the splicing mutations in CFTR gene (Aznarez et al. 2003; Baralle 2005). The nsSNPs which were predicted to be deleterious in causing an effect in the structure and function of the protein by SIFT, PolyPhen and Pupasuite correlated well with experimental studies (Tsui 1992; Ghanem et al. 1994; Bienvenu et al. 1998) (Table 3).
Table 2.
List of nsSNPs that were predicted to be of functional significance by PupaSuite
| SNPs ID | Alleles | Region | Functional significance |
|---|---|---|---|
| rs1800072 | A/G | Coding nonsynonymous | Cellular processing |
| rs35516286 | C/T | Coding nonsynonymous | Cellular processing |
| rs1800080 | A/G | Coding nonsynonymous | Cellular processing |
| rs34911792 | G/T | Coding nonsynonymous | Cellular processing |
| rs1800092 | A/C/G | Coding nonsynonymous | Prot.structure and dynamics |
| Functional Sites | |||
| rs1800093 | G/T | Coding nonsynonymous | Prot.structure and dynamics |
| rs35813506 | A/G | Coding nonsynonymous | Prot.structure and dynamics |
| rs1801178 | C/T | Coding nonsynonymous | Prot.structure and dynamics |
| rs11531593 | A/C | Coding nonsynonymous | Prot.structure and dynamics |
| rs1800112 | C/T | Coding nonsynonymous | Functional Sites |
| rs766874 | A/G | Coding nonsynonymous | Cellular processing |
| Functional Sites | |||
| Exonic splicing enhancers | |||
| rs1800073 | C/T | Coding nonsynonymous | Exonic splicing enhancers |
| rs1800074 | A/T | Coding nonsynonymous | Exonic splicing enhancers |
| rs1800078 | C/T | Coding nonsynonymous | Exonic splicing enhancers |
| rs1800079 | A/G | Coding nonsynonymous | Exonic splicing enhancers |
| rs1800086 | C/G | Coding nonsynonymous | Exonic splicing enhancers |
| rs35032490 | A/G | Coding nonsynonymous | Exonic splicing enhancers |
| rs1800097 | A/G | Coding nonsynonymous | Exonic splicing enhancers |
| rs1800100 | C/T | Coding nonsynonymous | Exonic splicing enhancers |
| rs1800103 | A/G | Coding nonsynonymous | Exonic splicing enhancers |
| rs1800107 | G/T | Coding nonsynonymous | Exonic splicing enhancers |
| rs1800110 | C/T | Coding nonsynonymous | Exonic splicing enhancers |
| rs1800120 | G/T | Coding nonsynonymous | Exonic splicing enhancers |
| rs1800123 | C/T | Coding nonsynonymous | Exonic splicing enhancers |
| rs11971167 | A/G | Coding nonsynonymous | Exonic splicing enhancers |
| rs4148725 | C/T | Coding nonsynonymous | Exonic splicing enhancers |
| s1800501 | C/G | mRNA | Exonic splicing enhancers |
| rs1042166 | A/T | mRNA | Exonic splicing enhancers |
| rs1800501 | C/G | mRNA | Exonic splicing enhancers |
| rs1800080 | A/G | Coding nonsynonymous | Cellular processing |
| Exonic splicing silencers | |||
| rs1800099 | A/G | Coding nonsynonymous | Exonic splicing silencers |
| rs1800101 | C/T | Coding nonsynonymous | Exonic splicing silencers |
Table 3.
List of SNPs (UTR mRNA) predicted to be functionally significant by FastSNP
| SNPs ID | Alleles | UTR Position | Level of risk | Possible function effect |
|---|---|---|---|---|
| rs34255446 | A/C | 3′ UTR | Medium–High (3–4) | Splicing site |
| rs1800070 | A/G | 5′ UTR | Low–Medium (1–3) | Promoter/regulat ory region |
Functional SNPs in noncoding SNPs
Polymorphism in the 3′UTR region affects the gene expression by affecting the ribosomal translation of mRNA or by influencing the RNA half-life (Deventer 2000). The 5′ and 3′ UTRs are involved in various biological processes such as posttranscriptional regulatory pathways, stability, and translational efficiency (Sonenberg 1994; Nowak 1994). We found that out of 8 UTR SNPs, 1 SNPs in 3′and another in 5′UTR region with ids rs34255446 and rs1800070, respectively were predicted to be damaging by FAST SNP server as depicted in Table 3.
Modeling and analysis of mutant structure
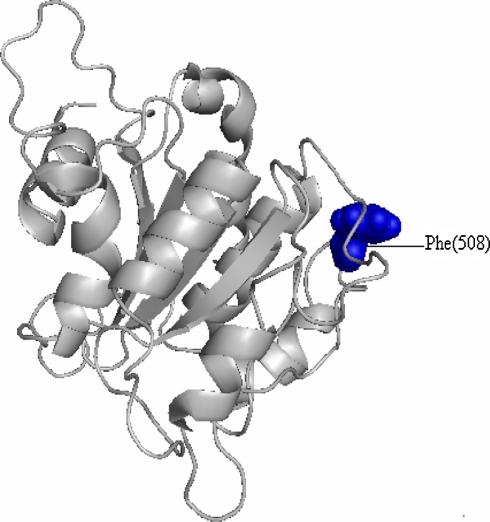
Single amino acid mutations can significantly change the stability of a protein structure. So, the knowledge of a protein’s three-dimensional (3D) structure is essential for a full understanding of its functionality. Mapping the deleterious nsSNPs into protein structure information was obtained from dbSNP and SAAPdb. The available structure for the CFTR gene is reported to have a PDB id (1nbd). Mutation analysis for the CFTR gene was performed based on the results obtained from highest SIFT and PolyPhen scores. It is noted that rs1800093 showed the highest deleterious (SIFT) and damaging (PolyPhen) scores, 0.00 and 3.031, respectively. According to this, the mutation occurred for native protein (1nbd) at position F508C with an SNP id namely (rs1800093), based on SIFT and PolyPhen results. The mutation for 1nbd at the corresponding position was performed by SWISS-PDB viewer independently to achieve modeled structures. Then, energy minimizations were performed by NOMAD-Ref server for the native type protein (1nbd) and the mutant type structures. It can be seen that total energy for the native (1nbd) and mutant type structure F508C were found to be −9786.37 and −9902.49 Kcal/mol respectively. RMSD values between the native amino acid phenylalanine and mutant amino acid cysteine at position 508 were found to be 1.75 Å. The superimposed structures of the native (1nbd) with mutant type protein F508C are shown in Figs. 1 and 2, respectively.
Fig. 1.
Structure of native protein 1nbd (grey) of CFTR gene
Fig. 2.
Superimposed structure of native amino acid phenylalanine (red) with mutant amino acid cysteine (grey) at 508 position in 1nbd
The prediction of residue solvent accessibility can help in better understanding the relationship between sequence and structure. Solvent accessibility of all the residues in the native protein and mutant proteins were computed with NetASA. It is interesting to note that the residues Ser (434), Tyr (512) and Ser (557) showed a change in solvent accessibility from a buried to exposed state and Ala (566) from an exposed to buried state in the mutant protein F508C. The native amino acid phenylalanine and mutant amino acid cysteine are hydrophobic in nature. Many studies have suggested that hydrophobic core residues are likely sites of deleterious mutations. Hence, change in solvent accessibility from an exposed to buried state could be considered functionally significant in the mutant protein at structural level (Chen and Zhou 2005). The occurrence of weak interactions has been observed at the terminus of the secondary structural units, in particular a-helix and β-sheets (Fabiola et al. 1997; Babu et al. 2002). These interactions play a definitive role in stabilizing these structures of proteins. The propensity of the amino acid residues to favor a particular conformation has been well documented. Such conformational preference is not dependent on the amino acid alone but is also dependent on the local amino acid sequence. We analyzed the secondary structure of each amino acid residue in the native and mutant structures of the protein. We found that the residue Met (498), Pro (499), Gly (500), Thr (507) and Leu (636) changed from helix in the native protein to turn conformation in the mutant protein.
SRide server was used for identifying the stabilizing residues of native type and mutant modeled structure. We obtained 12 amino acids which act as a stabilizing residue in the native, as well as in the mutant structure. Of these, ten residues were common in both the native and mutant structure. Interestingly, on mutation at position F508C two residues, namely Leu (453) and Cys (491), in the native protein were replaced with the residues Glu (542) and Gly (543), respectively. The change in the stabilizing residues on mutation at 508 results in increased stability of the mutant structure.
Identification of htSNPs
Sets of nearby SNPs on the same chromosome are inherited in blocks. The minimal informative subsets of SNPs associated with the limited number of haplotypes in a block are often referred to as htSNPs. We analyzed htSNPs in the coding region of CFTR gene by selecting the force tagSNPs selection in iHAP. We selected the htSNPs in different populations based on the proportion of haplotype diversity, haplotype entropy and minimax of pair wise LD measure r2 between tag and untagged SNPs. Base on these strategies we identified htSNPs in coding regions and untranslated regions of CFTR gene with ids rs766784 and rs1800100 in CEU-CEPH, and rs4148725 and rs1042180 in JPH and CHB populations, respectively. More specifically, tagSNPs chosen in one population are not appropriate for genotyping in a different population (Fullerton2004). Interestingly, the ids (rs766784, rs1800100 and rs4148725) were found to be deleterious and damaging by SIFT and PolyPhen.
Discussion
Understanding the functional impacts of inherited variations between individuals is an important goal of human genetics. Given that hundreds of thousands of SNPs are estimated to exist in the human population, only a small subset of variants that affect the phenotype will confer a disease risk. Among these variations, nsSNPs that lead to an amino acid change in the protein product are of particular interest for their close relevance to human inherited diseases and drug sensitivity (Yue and Moult 2006; Wang and Moult 2001). Therefore, the identification of nsSNPs that affect protein function and relate to disease will be a challenge in the coming years (Karchin et al. 2005a, b). The effect of many nsSNPs will probably be neutral as natural selection will have removed mutations on essential positions. Assessment of nonneutral SNPs is mainly based on phylogenetic information (i.e. correlation with residue conservation) extended to a certain degree with structural approaches (PolyPhen). However, there is increasing evidence that many human disease genes are the result of exonic or noncoding mutations affecting regulatory regions (Hudson 2003; Yan et al. 2002). Much attention has been focused on modeling by different methods the possible phenotypic effect of SNPs that cause amino acid changes, and only recently has interest focused on functional SNPs affecting regulatory regions or the splicing process.
Out of 39 nsSNPs in the CFTR gene, 17 of them were found to be deleterious (SIFT) and 26 of them found to be damaging (PolyPhen). 30 nsSNPs and 3 SNPs in mRNA region showed molecular phenotypic variation by PupaSuite. 1 SNP in the 3′ and another SNP in 5′ UTR region were found to be functionally significant by FASTSNP. We mapped the deleterious mutation for (1nbd) at position F508C with an SNP id (rs1800093) based on SIFT and PolyPhen results. Structural significance of native and mutant models of the CFTR gene at position F508C were further investigated in this work by solvent accessibility, secondary structure analysis and stabilizing residues. Solvent accessibility, considered as a discriminating feature in disease associated nsSNPs, tended to occur at buried sites; benign substitutions tended to occur at solvent accessible sites (Sunyaev et al. 2000; Ferrer-Costa 2002). Residues that form the hydrophobic core of a protein are critical for its stability. In the folded structure of a protein, polar and charged side chains have higher solvent accessibility than nonpolar side chains suggesting that formation of a hydrophobic core is a strong driving force in protein folding (Chan and Dill 1990). The mutation occurring at position 508 in CFTR gene provides hydrophobic contacts for domain–domain interactions that are crucial for the post-translation folding mechanism of NBD2 (Du et al. 2005). We further analyzed the CFTR gene by haplotype analysis and identified htSNPs. Our results from this study suggests that the application of computational algorithms, namely SIFT, PolyPhen, PupaSuite, FASTSNP, ASA view, SRide and iHAP analysis might provide an alternative approach to select target SNPs by understanding the effect of SNPs on the functional attributes or molecular phenotype of a protein. The models built in this work would be applicable for predicting the deleterious nsSNPs which would be helpful for further genotype–phenotype research as well as pharmacogenetics studies. The functional analysis in this study may be a good model for further research in genetically inherited disease.
Conclusion
Our results from this study suggest that the application of computational tools including SIFT, PolyPhen, Pupasiute, FASTSNP, ASA view, SRide and haplotype analysis might provide an alternative approach to select target SNPs in association studies. Our result also endorses a study with an in vivo experimental protocol. Importantly, the applications of these computational algorithms in association studies will greatly strengthen our understanding of the inheritance of complex human phenotype. Therefore, our analysis will provide useful information in selecting SNPs that are likely to have potential functional impact and ultimately contribute to an individual’s susceptibility to cystic fibrosis by the CFTR gene.
Acknowledgments
The authors thank the management of Vellore Institute of Technology for providing the facilities to carry out this work. The authors take this opportunity to thank the reviewers for their invaluable comments and suggestions to make this manuscript more readable and meaningful.
References
- Aznarez I, Chan EM, Zielenski J et al (2003) Characterization of disease-associated mutations affecting an exonic splicing enhancer and two cryptic splice sites in exon 13 of the cystic fibrosis transmembrane conductance regulator gene. Hum Mol Genet 12(16):2031–2040 [DOI] [PubMed]
- Babu MM, Singh KS, Balaram P et al (2002) C-H...O hydrogen bond stabilized polypeptide chain reversal motif at the C terminus of helices in proteins. J Mol Biol 322:871–880 [DOI] [PubMed]
- Balasubramanian S, Xia Y, Freinkman E et al (2005) Sequence variation in G- Protein-coupled receptors: analysis of single nucleotide polymorphisms. Nucleic Acids Res 33:1710–1721 [DOI] [PMC free article] [PubMed]
- Bao L, Cui Y (2006) Functional impacts of non-synonymous single nucleotide polymorphisms: selective constraint and structural environments. FEBS Lett 580:1231–1234 [DOI] [PubMed]
- Baralle D, Baralle M (2005) Splicing in action: assessing disease causing sequence changes. J Med Genet 42:737–748 [DOI] [PMC free article] [PubMed]
- Bienvenu T, Bousquet S, Vidaud D et al (1998) A novel missense mutation D513G in exon 10 of the cystic fibrosis transmembrane conductance regulator (CFTR) gene identified in a French CBAVD patient. Hum Mutat 12(3):213–214 [PubMed]
- Brooks-Wilson AR, Kaurah P, Suriano G (2004) Germline Ecadherin mutations in hereditary diffuse gastric cancer: assessment of 42 new families and review of genetic screening criteria. J Med Genet 41:508–517 [DOI] [PMC free article] [PubMed]
- Cargill M, Altshuler D, Ireland J et al (1999) Characterization of single nucleotide polymorphisms in coding regions of human genes. Nat Genet 22:231–238 [DOI] [PubMed]
- Cartegni L, Krainer AR (2002) Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nature Genet 30:377–384 [DOI] [PubMed]
- Cartegni L, Chew SL, Krainer AR (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 3:285–298 [DOI] [PubMed]
- Cavallo A, Martin AC (2005) Mapping SNPs to protein sequence and structure data. Bioinformatics 8:1443–1450 [DOI] [PubMed]
- Chasman D, Adams RM (2001) Predicting the functional consequences of non-synonymous single nucleotide polymorphisms: structure-based assessment of amino acid variation. J Mol Biol 307:683–706 [DOI] [PubMed]
- Chan HS, Dill KA (1990) Origins of structure in globular proteins. Proc Natl Acad Sci USA 87:6388–6392 [DOI] [PMC free article] [PubMed]
- Chen H, Zhou HX (2005) Prediction of solvent accessibility and sites of deleterious mutations from protein sequence. Nucleic Acids Res 33:3193–3199 [DOI] [PMC free article] [PubMed]
- Collins FS (1992) Cystic fibrosis: molecular biology and therapeutic implications. Science 256:774–779 [DOI] [PubMed]
- Conde L, Vaquerizas MJ, Santoyo J et al (2004) PupaSNP Finder: a web tool for finding SNPs with putative effect at transcriptional level. Nucleic Acids Res 32:W242–W248 [DOI] [PMC free article] [PubMed]
- Conde L, Vaquerizas JM, Ferrer-Costa C et al (2005) PupasView: a visual tool for selecting suitable SNPs, with putative pathological effect in genes, for genotyping purposes. Nucleic Acids Res 33:W501–W505 [DOI] [PMC free article] [PubMed]
- Conde L, Vaquerizas MJ, Dopazo H et al (2006) PupaSuite: findingfunctional single nucleotide polymorphisms for large-scale genotyping purposes. Nucleic Acids Res 34:W621–W625 [DOI] [PMC free article] [PubMed]
- De Jonge HR, Ballmann M, Veeze H et al (2004) Ex vivo CF diagnosis by intestinal current measurements (ICM) in small aperture, circulating Using chambers. J Cyst Fibros 3:159–163 [DOI] [PubMed]
- Delarue M, Dumas P (2004) On the use of low-frequency normal modes to enforce collective movements in refining macromolecular structural models. Proc Natl Acad Sci 101:6957–6962 [DOI] [PMC free article] [PubMed]
- Deventer SV (2000) Cytokine and cytokine receptor polymorphisms in infectious disease. Intensive Care Med 26:S98–S102 [DOI] [PubMed]
- Dryja TP, Mcgee TL, Halu LB et al (1990) Mutations within the rhodopsin gene in patients with autosomal dominant retinis pigmentosa. N Engl J Med 323:1302–1307 [DOI] [PubMed]
- Du K, Sharma M, Lukacs GL (2005) The F508 cystic fibrosis mutation impairs domain-domain interactions and arrests post-translational folding of CFTR. Nat Struct Mol Biol 12:17–25 [DOI] [PubMed]
- Fabiola GF, Krishnaswamy S, Nagarajan V et al (1997) C-H O Hydrogen bonds in βsheets. Acta Crystallogr D53:316–320 [DOI] [PubMed]
- Fairbrother WG, Chasin LA (2000) Human genomic sequences that inhibit splicing. Mol Cell Biol 20:6816–6825 [DOI] [PMC free article] [PubMed]
- Ferrer-Costa C, Orozco M, De la Cruz X (2002) Characterization of disease-associated single acid polymorphisms in terms of sequence and structure properties. J Mol Biol 315:771–786 [DOI] [PubMed]
- Fredman D, Siegfried M, Yuan YP et al (2002) HGVbase: a human sequence variation database emphasizing data quality and a broad spectrum of data sources. Nucleic Acids Res 30(1):387–391 [DOI] [PMC free article] [PubMed]
- Fullerton SM, Buchanan AV, Sonpar VA et al. (2004) The effects of scale: variation in the APOA1/C3/A4/A5 gene cluster. Hum Gene 115:36–56 [DOI] [PubMed]
- Gabriela MR, Alonso RP, Iris D (2007) XV-2c and KM.19 haplotype analysis in Chilean patients with cystic fibrosis and unknown CFTR gene mutations. Biol Res 40:223–229 [DOI] [PubMed]
- Ghanem N, Costes B, Girodon E et al (1994) Identification of eight mutations and three sequence variations in the cystic fibrosis tran membrane conductance regulator (CFTR) gene. Genomics 21:434–436 [DOI] [PubMed]
- Gibson LE, Cooke RE (1959) A test for concentration of electrolytes in sweat in cystic fibrosis of the pancreas utilizing pilocarpine by iontophoresis. Pediatrics 23:545–549 [PubMed]
- Gilis D, Rooman M (1996) Stability changes upon mutation of solvent accessible residues in proteins evaluated by database derived potentials. J Mol Biol 257:1112–1126 [DOI] [PubMed]
- Gilis D, Rooman M (1997) Predicting protein stability changes upon mutation using database-derived potentials: solvent accessibility determines the importance of local versus non-local interactions along the sequence. J Mol Biol 272:276–290 [DOI] [PubMed]
- Graveley BR (2000) Sorting out the complexity of SR protein functions. RNA 6:1197–1211 [DOI] [PMC free article] [PubMed]
- Hinds DA, Stuve LL, Nilsen GB et al (2005) Whole-genome patterns of common DNA variation in three human populations. Science 307:1072–1079 [DOI] [PubMed]
- Hudson TJ (2003) Wanted: regulatory SNPs. Nat Genet 33:439–440 [DOI] [PubMed]
- Kabsch W, Sander C (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577–2637 [DOI] [PubMed]
- Kanetsky PA, Ge F, Najarian D et al (2004) Assessment of polymorphic variants in the melanocortin-1 receptor gene with cutaneous pigmentation using an evolutionary approach. Cancer Epidemiol Biomarkers Prev 13:808–819 [PubMed]
- Karchin R, Kelly L, Sali A (2005a) Improving functional annotation of non-synonymous SNPs with information theory. Pac Symp Biocomput 10:397–408 [DOI] [PubMed]
- Karchin R, Diekhans M, Kelly L et al (2005b) LS-SNP: large-scale annotation of coding nonsynonymous SNPs based on multiple information sources. Bioinformatics 21(12):2814–2820 [DOI] [PubMed]
- Lander ES, Linton LM, Birren B et al (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921 [DOI] [PubMed]
- Lindahl E, Azuara C, Koehl P et al (2006) NOMAD-Ref: visualization, deformation and refinement of macromolecular structures based on all-atom normal mode analysis. Nucleic Acids Res 34:W52–W56 [DOI] [PMC free article] [PubMed]
- Magyar C, Gromiha MM, Pujadas G et al (2005) SRide: a server for identifying stabilizing residues in proteins. Nucleic Acids Res 33:W303–305 [DOI] [PMC free article] [PubMed]
- Ng PC, Henikoff S (2001) Predicting deleterious amino acid substitutions. Genome Res 11:863–874 [DOI] [PMC free article] [PubMed]
- Ng PC, Henikoff S (2002) Accounting for human polymorphisms predicted to affect protein. Genome Res 12:436–446 [DOI] [PMC free article] [PubMed]
- Ng PC, Henikoff S (2003) SIFT: predicting amino acid changes that affect protein function. Nucl Acids Res 31:3812–3814 [DOI] [PMC free article] [PubMed]
- Ng PC, Henikoff S (2006) Predicting the effects of amino acid substitutions on protein function. Annu Rev Genomics Hum Genet 7:61–80 [DOI] [PubMed]
- Nguyen MN, Rajapakse JC (2006) Two-stage support vector regression approach for predicting accessible surface areas of amino acids. Proteins 63:542–550 [DOI] [PubMed]
- Nowak R (1994) Mining treasures from ‘junk DNA’. Science 263:608–610 [DOI] [PubMed]
- Prokunina L, Alarcon-Riquelme ME (2004) Regulatory SNPs in complex diseases: their identification and functional validation. Expert Rev Mol Med:1–15 [DOI] [PubMed]
- Prokunina L, Castillejo-Lopez C, Oberg F et al (2002) A regulatory polymorphism in PDCD1 is associated with susceptibility to systemic lupus erythematosus in humans. Nat Genet 32:666–669 [DOI] [PubMed]
- Ramensky V, Pork P, Sunyaev S (2002) Human non-synonymous SNPs: server and survey. Nucleic Acids Res 30:3894–3900 [DOI] [PMC free article] [PubMed]
- Reumers J, Maurer-Stroh S, Schymkowitz J et al (2006) SNPeffect v2.0: a new step in investigating the molecular phenotypic effects of human non-synonymous SNPs. Bioinformatics 22(17):2183–2185 [DOI] [PubMed]
- Richard JD, Patricia BM, Mark JC et al (2006) Predicting deleterious nsSNPs: an analysis of sequence and structural attributes. BMC Bioinformatics 7:217 [DOI] [PMC free article] [PubMed]
- SachidanandamR, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry S, Mullikin JC, Mortimore BJ (2001) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409:928–933 [DOI] [PubMed]
- Saunders CT, Baker D (2002) Evaluation of structural and evolutionary contributions to deleterious mutation prediction. J Mol Biol 322:891–901 [DOI] [PubMed]
- Savas S, Kim DY, Ahmad MF et al (2004) Identifying functional genetic variants in DNA repair pathway using protein conservation analysis. Cancer Epidemiol Biomarkers 13:801–807 [PubMed]
- Schu¨ler D, Sermet-Gaudelus I, Wilschanski M et al (2004) Basic protocol for transepithelial nasal potential difference measurements. J Cyst Fibros 3:151–155 [DOI] [PubMed]
- Shander A et al (2004) ASA view: solvent accessibility graphics for proteins. Bioinformatics 51:51
- Sherry ST, Ward MH, Kholodov et al (2001) dbSNP: the NCBI database of genetic variation. Nucl Acids Res 29:308–311 [DOI] [PMC free article] [PubMed]
- Smigielski EM, Sirotkin K, Ward M et al (2000) dbSNP: a database of single nucleotide polymorphisms. Nucleic Acids Res 28(1):352–355 [DOI] [PMC free article] [PubMed]
- Smith EP, Boyd J, Frank GR et al (1994) Estrogen resistance caused by a mutation in the estrogen-receptor gene in a man. N Engl J Med 331:1056–1061 [DOI] [PubMed]
- Sonenberg N (1994) mRNA translation: influence of the 5′ and 3′ untranslated regions. Curr Opin Genet Dev 4:310–315 [DOI] [PubMed]
- Song CM, Yeo BH, Tantoso E et al (2006) iHAP – integrated haplotype analysis pipeline for characterizing the haplotype structure of genes. BMC Bioinformatics 7:525 [DOI] [PMC free article] [PubMed]
- Stumpf MPH (2004) Haplotype diversity and SNP frequency dependence in the description of genetic variation. Eur J Hum Genet 12:469–477 [DOI] [PubMed]
- Sunyaev S, Ramensky V, Bork P (2000) Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends Genet 16:198–200 [DOI] [PubMed]
- Sunyaev S, Ramensky V, Koch I et al (2001) Prediction of deleterious human alleles. Hum Mol Genet 10:591–597 [DOI] [PubMed]
- The International Hapmap Consortium (2003) The International HapMap Project. Nature 426:789–796 [DOI] [PubMed]
- Tsui L-C (1992) Mutations and sequence variations detected in the cystic fibrosis transmembrane conductance regulator (CFTR) gene: a report from the Cystic Fibrosis Genetic Analysis. Hum Mutat 1(3):197–203 [DOI] [PubMed]
- Venter JC, Adams MD, Myers EW et al (2001) The sequence of the human genome. Science 291:1304–1351 [DOI] [PubMed]
- Wagner M, Adamczak R, Porollo A et al (2005) Linear regression models for solvent accessibility prediction in proteins. J Comput Biol 12:355–369 [DOI] [PubMed]
- Wang Z, Moult J (2001) SNPs, protein structure, and disease. Hum Mutat 17(4):263–270 [DOI] [PubMed]
- Xi T, Jones IM, Mohrenweiser HW (2004) Many amino acid substitution variants identified in DNA repair genes during human population screenings are predicted to impact protein function. Genomics 83:970–979 [DOI] [PubMed]
- Yan H, Yuan W, Velculescu VE et al (2002) Allelic variation in human gene expression. Science 297:1143 [DOI] [PubMed]
- Yuan H, Chiou J, Tseng W et al (2006) FASTSNP: an always upto-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res 34:W635–W641 [DOI] [PMC free article] [PubMed]
- Yue P, Moult J (2006) Identification and analysis of deleterious human SNPs. J Mol Biol 356(5):1263–1274 [DOI] [PubMed]
- Zhang EY, Fu D-J, Pak YA et al (2004) Genetic polymorphisms in human proton-dependent dipeptide transporter PEPT1: implications for the functional role of Pro586. J Pharmacol Exp Ther 310:437–445 [DOI] [PubMed]