Abstract
BBDR rats develop autoimmune diabetes mellitus only after challenge with environmental perturbants. These include polyinosinic:polycytidylic acid (poly I:C, a ligand of toll-like receptor 3), agents that deplete regulatory T cell populations (Tregs), and a non-beta-cell-cytopathic parvovirus (Kilham rat virus, KRV). The dominant diabetes susceptibility locus Iddm4 is required for diabetes induced by treatment with poly I:C plus Treg depletion. Iddm4 is penetrant in congenic heterozygote rats on the resistant WF background, and is 79% sensitive and 80% specific as a predictor of induced diabetes. Surprisingly, an analysis of 190 (BBDR × WF)F2 rats treated with KRV after brief exposure to poly I:C revealed that the BBDR origin allele of Iddm4 is necessary but not entirely sufficient for diabetes expression. A genome scan identified a locus on chromosome 17, designated Iddm20, that is also required for susceptibility to diabetes after exposure to KRV and poly I:C (LOD score 3.7). These data suggest that the expression of autoimmune diabetes is a complex process that requires both major histocompatibility complex (MHC) genes that confer susceptibility and additional genes like Iddm4 and Iddm20 that operate only in the context of specific environmental perturbants, amplifying the immune response and the rate of disease progression.
Introduction
Type 1 diabetes results from inflammatory infiltration of pancreatic islets and selective beta cell destruction. It is thought to be caused by environmental factors operating in a genetically susceptible host (1,2). Susceptibility loci include the MHC, a promoter polymorphism of the insulin gene, and an allelic variant of CTLA4 (3). Among candidate environmental perturbants, viral infection is among the most likely (4). How genes interact with the environment to transform diabetes susceptibility into overt disease is unknown.
BBDR rats model virus-induced autoimmune diabetes remarkably well. (5). They are phenotypically normal and, in clean housing, never develop diabetes. They do, however, become diabetic when challenged with environmental perturbants, among them poly I:C in combination with depletion of regulatory T cells (Tregs) (6). Diabetes can also be induced in BBDR rats with KRV, a non-beta-cell cytopathic parvovirus (7). Naturally occurring KRV infection induces diabetes in ∼1% of animals; intentional infection with 107 plaque forming units (PFU) induces diabetes in ∼30% of BBDR rats (7). Infection with KRV following brief pre-treatment with a low, sub-diabetogenic dose of poly I:C (1 μg/gm daily ×3) leads to diabetes in 100% of animals (8). The effect is virus-specific; H-1, which is 98% sequence identical, uniformly fails to induce diabetes (8).
In analyses of (BBDP × WF) × WF rats, we used poly I:C plus Treg depletion to map a locus on chromosome 4 (Iddm4) with significant linkage to diabetes (6,9), and we recently positioned Iddm4 in a 2.8 cM region (10). The BB-origin allele of Iddm4 is dominant and 79% sensitive and 80% specific as a predictor of diabetes induced by Treg depletion and poly I:C. A radiation hybrid map has assigned Iddm4 to a 6.3-Mb segment between PTN and ZYX at 7q32 in the human genome, and to a 5.7-Mb segment between Ptn and Zyx in the mouse genome (11).
We now report a linkage analysis of 190 (BBDR × WF)F2 rats. It reveals that the BBDR-origin allele of Iddm4 is necessary but not entirely sufficient for diabetes expression in response to KRV infection. An additional gene or genes on chromosome 17 are necessary.
Methods
Animals
BBDR/Wor, WF.Iddm4, and WF.ART2a rats (all RT1u/u, ART2a) were obtained from colonies maintained by us. WF.Iddm4 congenic rats were generated by repetitive (BBDR/Wor × WF) × WF backcrosses using a marker-assisted selection protocol as described (10). They were studied at the N6 generation. WF.ART2a congenic rats were also developed by us and differ from ordinary WF animals in that they express the “a” rather than the “b” allotype of the ART2 T cell alloantigen on chromosome 1 (10). For simplicity, we refer to them here as WF rats. An F2 intercross was bred from (BBDR × WF)F1 hybrids. Animals were housed in viral antibody free conditions, confirmed monthly to be serologically free of rat pathogens (10), and maintained in accordance with recommendations in the Guide for the Care and Use of Laboratory Animals (National Academy of Sciences, 1996).
Microsatellite and Mapping Analyses
Genomic DNA was prepared as described (10 and Appendix, Figure 3). Microsatellite markers were placed evenly throughout the 20 autosomes. The source of primers and positions of markers on the genetic map are given in Appendix Figure 3. Primers were end-labeled using 32Pγ-ATP, used in a PCR reaction, and resolved by polyacrylamide gel electrophoresis as described (6). The position of markers on the genetic map was established by inspection of the dataset and conventional calculation methods to establish meiotic map distances, which are expressed in cM or megabases (Mb) according to the rat genome sequence, June 2003 build (genome.ucsu.edu).
Linkage of diabetes with segregation of BBDR-origin alleles was evaluated by composite interval mapping (CIM) using model 6 of the Zmapqtl program in Windows QTL Cartographer v1.30 (statgen.ncsu.edu/qtlcart/cartographer.html). CIM combines classical interval mapping with multiple regression analysis, allowing for more precise QTL localization than classical interval mapping (12).
Treatment Protocols
KRV-UMass was propagated in NRK cells grown in Dulbecco's minimal essential medium. Poly I:C (Sigma, St. Louis, MO) was dissolved in Dulbecco's PBS, sterile filtered, and stored at -20°C until used. Contaminating endotoxin concentration was <50 units/mg (Charles River Endosafe, Charleston, SC). In studies of KRV alone, rats of either sex 22-28 days old were injected intraperitoneally with 107 PFU in a volume of 1 ml. In other experiments, rats 21-25 days of age of either sex were injected intraperitoneally with poly I:C 1 (μg/gm body weight on three consecutive days) and either not treated further or injected on the following day with KRV. Pretreatment with poly I:C was used because it increases the frequency of diabetes in KRV-treated BBDR rats from ∼30% to 100% (8). Animals were screened three times weekly for glycosuria (Tes-Tape® Eli Lilly, Indianapolis, IN). Diabetes was diagnosed on the basis of a plasma glucose concentration >11.1 mM (OneTouch Ultra Glucometer, LifeScan, Milpitas, CA). For study of insulitis, pancreata were removed, fixed in formalin, and stained with hematoxylin and eosin. Pancreata were graded by a qualified pathologist on a scale of increasing intensity 0 to 4+ as described (10).
Results
Autoimmune Diabetes Induced by KRV and TLR3 Ligation
We first confirmed (8) that a significant fraction (41%) of parental BBDR rats become diabetic after infection with KRV alone, that none become diabetic in response to a 3 day course of poly I:C alone, and that 100% become diabetic in response to KRV after poly I:C (Table 1). We also confirmed (13) that WF rats resist diabetes induction in response to either KRV or KRV plus poly I:C (Table 1).
Table 1. Frequency of Diabetes in Rats Treated with KRV, Poly I:C, and Treg Depletion.
Frequency of induced diabetes. Male and female rats were entered into the indicated treatment protocols when 21-28 days old. In groups 1 and 2, KRV-UMass was given intraperitoneally at a dose of 107 PFU. In group 2, poly I:C was given at a dose of 1 μg/g body weight intraperitoneally on days -3, -2, and -1 relative to KRV. In group 3 poly I:C (1 μg/g) was given 3 times/week for 40 days and anti-ART2.1 mAb (25 μg) was given 5 times/week for 40 days as described (9). Rats in group 4 received only poly I:C (1 μg/g) for 3 consecutive days. Diabetes was defined as a plasma glucose concentration > 11.1 mM (250 mg/dl). The WF.Iddm4d congenic rats were from the N6 generation and bear ∼2.8 cM of the genetically dominant BBDR rat-derived Iddm4 region on chromosome 4 and at least one BBDR-origin “a” allele of the ART2 T cell alloantigen on chromosome 1 (10). Diabetic rats in all groups had severe insulitis or “end-stage” islets.
| Groups | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Protocols | KRV | KRV + poly I:C | Anti-ART2.1 | Poly I:C |
| Rat Strains | mAb + poly I:C | alone | ||
| BBDR | 11/27 | 27/27 | 12/12 | 0/6 |
| WF | 0/3 | 0/8 | 3/53 | — |
| WF.Iddm4d (N6) | 0/4 | 0/18 | 7/12 | — |
| (BBDR × WF)F1 | 0/6 | 5/13 | 11/11 | — |
| (WF × BBDR)F2 | — | 59/190 | — | — |
We next tested N6 generation WF.Iddm4 congenic rats (10) for disease susceptibility. To our surprise, we observed WF.Iddm4d rats to be uniformly resistant to diabetes in response to KRV infection either alone or after poly I:C, despite maintaining the expected degree of susceptibility to diabetes after treatment with poly I:C and Treg depletion (Table 1).
Autoimmune Diabetes Induced by KRV and TLR Ligation Segregates with Iddm4 and a Second Locus in (BBDR × WF)F2 rats
To determine if Iddm4 acts only in the presence of additional BBDR origin genes, we generated (BBDR × WF)F1 progeny. We observed that all F1 rats were resistant to KRV alone, but 38% were susceptible to treatment with KRV plus poly I:C. To identify susceptibility genes, we then generated (BBDR × WF) F2 progeny and treated them with KRV plus poly I:C. Diabetes occurred in 59 of 190 animals (31%) in this segregating population and affected both males and females (Table 1). Looking first at the Iddm4 interval, we observed that diabetes occurred almost exclusively in animals with at least one BBDR-origin allele of Iddm4 (58 of 59 diabetic animals, Table 2). The presence of the BBDR allele of Iddm4 was 98% sensitive but only 31% specific in predicting susceptibility to diabetes, implying that at least one gene of BBDR origin is required for the expression of diabetes in response to infection. We therefore performed a genome-wide scan on this F2 cohort. The remaining genome was assessed for linkage using 144 markers on the 20 autosomes (Appendix, Figure 3).
Table 2. Frequency of Diabetes in (BBDR × WF)F2 Rats as a Function of Iddm4 Genotype.
All (BBDR × WF)F2 rats in Table 1 were genotyped as described in Methods. They are grouped according to the presence of the WF and BBDR alleles of the microsatellite marker D4Arb9 used as the genotype for Iddm4 as described (10). Overall chi2 = 22.2, df=2, p<0.001.
| Iddm4 Genotype | N | Number Diabetic (%) | |
|---|---|---|---|
| BBDR/BBDR | 49 | 23 | (47%) |
| BBDR/WF | 100 | 35 | (35%) |
| WF/WF | 41 | 1 | (2%) |
Composite interval analysis of the linkage data is shown in Figure 1. Iddm4, as expected, showed strong linkage to diabetes (LOD >6.0 at ∼36 cM on chromosome 4) A second locus on chromosome 17 was linked to the diabetes in the F2 population with a LOD score of 3.7. This locus has been designated Iddm20 by the curators of the Rat Genome Database (www.rgd.mcw.edu).
Figure 1.
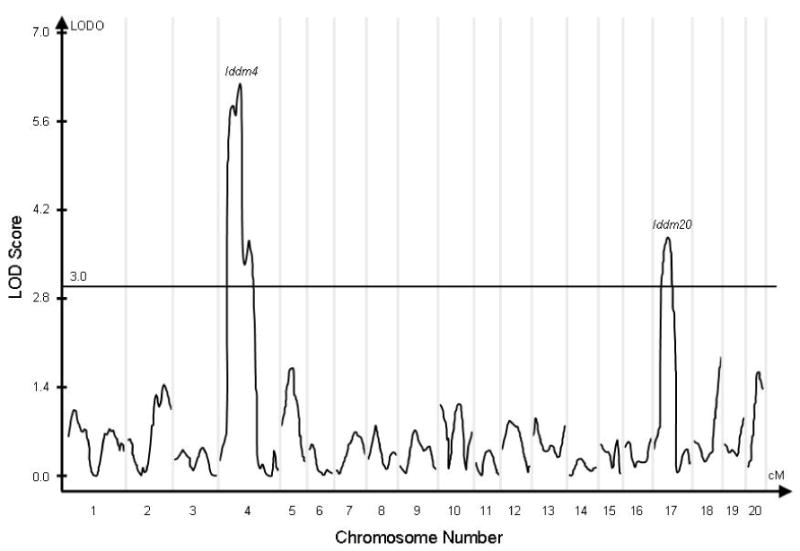
Composite interval analysis of linkage to the diabetes phenotype in 190 (WF × BBDR)F2 rats. Significant markers were first chosen using a linear regression model with a forward/backward selection procedure in the SRmapqtl module of QTL Cartographer. Markers flanking the test interval were added to the regression model to control for the presence of linked QTL. LOD scores are displayed as peaks over the entire genome, with chromosomes 1 through 20 displayed on the X axis. The curves are discontinuous, and the vertical gray lines delimit each individual chromosome and its associated curve. The horizontal reference line identifies the standard LOD=3 cutoff. The only peaks that significantly exceed this standard are on chromosome 4 (Iddm4) and on chromosome 17 (Iddm20).
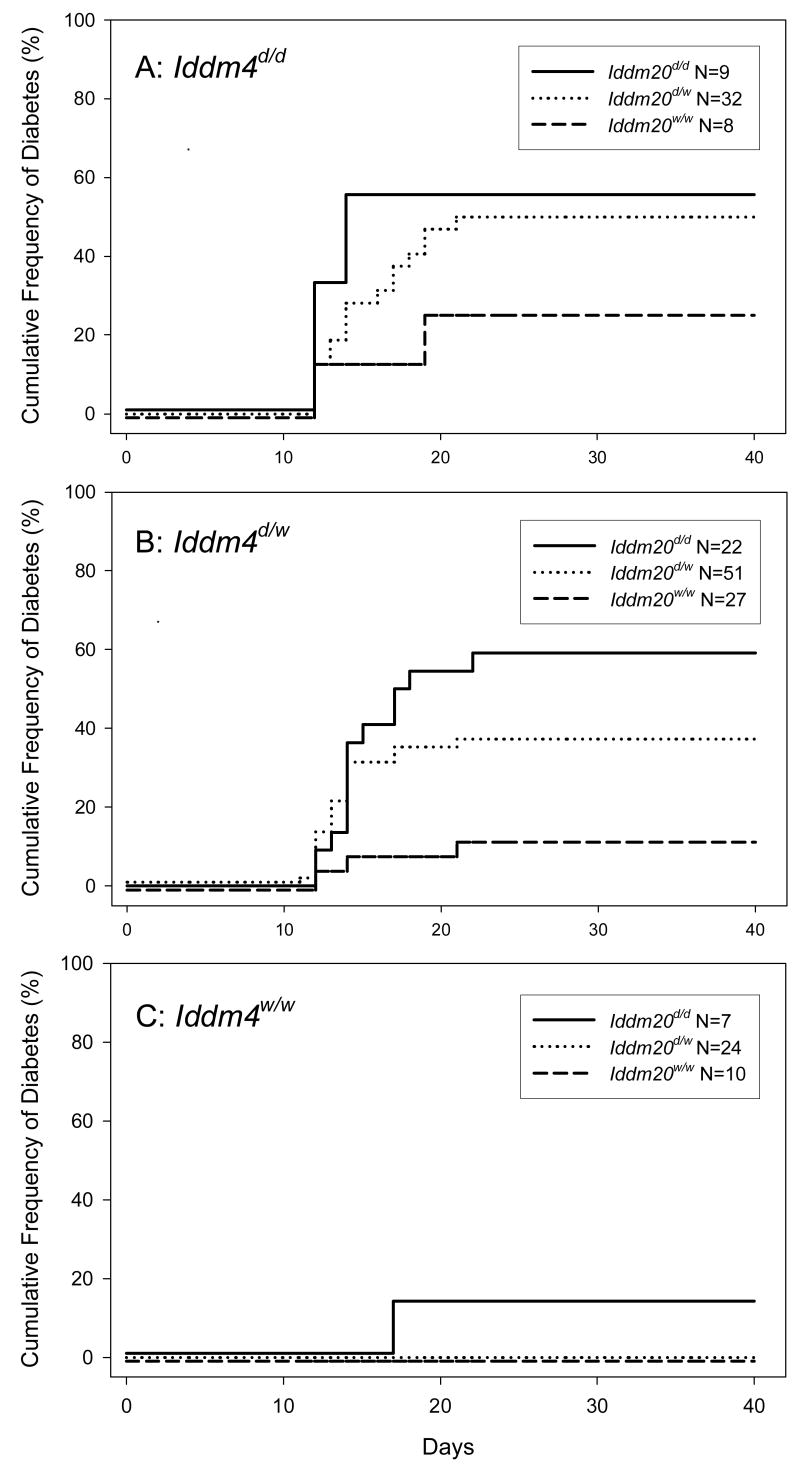
To determine the mode of inheritance and the interaction between these two loci, we analyzed the dataset using a life-table analysis. As shown in Figure 2, the highest likelihood of diabetes onset occurs in animals in which the BBDR-origin allele of Iddm4 is homozygous or heterozygous, and Iddm20 is homozygous.
Figure 2.
Kaplan Meier analysis of the cumulative frequency of autoimmune diabetes by inheritance of BBDR-origin diabetes susceptibility loci on chromosomes 4 and 17 in 190 (WF × BBDR)F2 rats. Time on the horizontal axis is in days after infection with KRV. Panel A presents data for F2 rats homozygous for the BBDR-origin allele of Iddm4; panel C presents data for F2 rats homozygous for the WF-origin allele of Iddm4; panel B present data for the heterozygotes. Within each panel, individual curves show data for subgroups expressing the indicated BBDR- and WF-origin alleles of Iddm20. The number of rats in each subgroup is indicated in the figure. Overall, 59 of 190 F2 animals (31%) became diabetic. Among the diabetic rats, 98% (58 of 59) expressed at least one BBDR-origin allele of Iddm4, and of these 91% (53 of 58) expressed at least one BBDR-origin allele of Iddm20. Among all rats homozygous for the WF-origin allele of Iddm20, 11% (5 of 45) became diabetic and all 5 expressed at least one BBDR-origin allele of Iddm4. Overall statistical analysis of the entire dataset for the effect of Iddm4 genotype stratified by Iddm20 genotype was statistically significant (log rank = 20.22, df=2, p<0.0001). Similarly, analysis for the effect of Iddm20 genotype stratified by Iddm4 genotype was statistically significant (log rank = 14.03, df=2, p<0.001). These data are shown in tabular form in Appendix, Table 3.
Candidate Gene Analysis
The genome-wide scan positioned the Iddm20 locus in a 1 LOD interval bounded by D17Rat61 (at 19.7 Mb) and D17Rat115 (at 27.7 Mb, http://genome.ucsc.edu). To identify candidate genes within this interval, we constructed a preliminary map using the databases at UCSC and Ensembl (http://genome.ucsc.edu/; http://www.ensembl.org/). The genes in the Iddm20 region have their human orthologues on human chromosome 5, 6, and 9 and on mouse chromosome 13. Of interest is a confirmed mouse diabetes QTL (Idd14) in this homologous region (14,15). Candidate genes in the Iddm20 interval are listed in Appendix Table 4
Histology
Histologic analysis of islets revealed nearly complete concordance of insulitis scores with diabetes phenotype. Among the 59 diabetic F2 animals, the mean insulitis score was 3.7 with 49 scored 4+ or end stage insulitis. In contrast, among 127 non-diabetic rats with technically satisfactory specimens, the mean insulitis score was 0.3 with 111 (87%) being entirely normal and 8 of the remaining 16 exhibiting only 1+ insulitis. Exocrine pancreatitis was absent.
Discussion
These data establish linkage of rat genotype to a form of environmental perturbation—infection—that is potentially important in the pathogenesis of autoimmunity. They confirm that Iddm4 is an exceptionally strong non-MHC determinant of susceptibility to autoimmune diabetes in the rat (6,9-11). In previous studies of Iddm4, diabetes was induced by chronic treatment with poly I:C plus Treg depletion. The present data now extend the role of Iddm4 in diabetes pathogenesis to virus-induced disease expression. They also illuminate the complexity of environmental interaction with genetic susceptibility. The diabetogenic potential of Iddm4 is readily discernable in congenic rats treated with poly I:C and Treg depletion, but is far less apparent in animals treated with KRV plus poly I:C unless additional BBDR genes are present. We have discovered at least one of these genes, designated Iddm20, on chromosome 17.
The Iddm20 interval (Appendix, Table 3) contains at least one gene of particular interest: Syk. This gene is involved in TCR-dependent signaling pathways and interacts with Cblb (16). This could be important because Cblb is a known rat diabetes susceptibility gene (17). Loss of function mutations in Cblb lead to activation of autoreactive diabetogenic T cells in the absence of full costimulation (17).
Iddm20, like Iddm4, appears to act as a genetic dominant with incomplete penetrance. There is clear disease-promoting activity in the Iddm20 heterozygote. In the poly I:C plus Treg system, ∼70% of both WF.Iddm4d/w heterozygotes and WF.Iddm4d/d homozygotes become diabetic (10). In the KRV plus poly I:C model, homozygosity for diabetogenic alleles at the Iddm20 locus increases the penetrance of diabetes in Iddm4d/d rats from 25% (in Iddm20w/w animals) to 56%, and it increases penetrance in Iddm4d/w rats from 11% to 59%. We therefore regard Iddm20 as a modifier of the Iddm4 locus.
The mechanisms by which poly I:C and KRV infection act to induce diabetes in genetically susceptible rats are not yet known. We speculate that Iddm4, Iddm20, or both define a strain-specific response of BBDR rats to KRV infection. KRV is known to infect lymphocytes in the pancreatic lymph nodes (but not the islets) of BBDR rats (18). More recent studies have revealed that KRV also causes a decrease in splenic CD4+CD25+ Treg cells in both BBDR and normal WF rats (8). In adult LEW rats, KRV-UMass infection is associated with several potentially important effects on both CD4+ and CD8+ T cell populations (19), but whether allelic variations in Iddm4 or Iddm20 regulate those responses is not yet known. By itself, KRV infection typically induces diabetes in about 30-40% of BBDR/Wor rats (8). Pre-treatment poly I:C, at a dose that is itself incapable of inducing disease, dramatically increases the penetrance of diabetes (8). The mechanism of this synergy is not clear, but is likely to relate to innate immunity because poly I:C is a ligand of TLR-3 and a potent inducer of type I interferon production by various cells (20) and IL-1 production by monocytes (21). It also activates NK (22) and B cells (23). In rats, it has been shown that interferon production in response to poly I:C varies substantially in different inbred strains (24), an effect that is presumably genetically determined. It will be of interest to determine if Iddm4 and/or Iddm20 is a determinant of the magnitude of the immune response to poly I:C.
Acknowledgments
We thank Dr. Michael Appel for scoring histology specimens, Michael Bates and Deborah Mullen for technical assistance, and Dennis Guberski for logistic support. Supported in part by grants DK49106 (DLG, JPM, EPB), DK 36024 (DLG), DK25306 (JPM), and Center Grant DK32520 from the National Institutes of Health. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Appendix.
Appendix: Figure 3.
Polymorphic markers used in this study. DNA samples and microsatellite markers. Genomic DNA was prepared using one of two protocols. Snap-frozen livers were ground on dry ice and the dispersed tissue was treated with Proteinase K in the presence of 10% sarkosyl and 0.5M EDTA (pH 8.0). DNA was purified from these digests by phenol-chloroform extraction and dialysis against Tris EDTA (0.01M Tris/0.001M EDTA, pH 7.4). Alternatively, genomic DNA was extracted from rat tail snips using the QIAamp Tissue kit (Qiagen, Stanford, CA) according to the manufacturer's instructions. Most microsatellite primers used in this study are available from Research Genetics, Inc. (Huntsville, AL). The general map location of these microsatellites was taken from our own segregating backcrosses, and from maps published by the Rat Genome Database (http://www.rgd.mcw.edu) and by Dr. R. Wilder and Dr. E. Remmers (www.nih.gov/niams/scientific/ratgbase/index.htm). Additional primer pairs were developed from unique sequences flanking short sequence repeats discovered by inspection of repeat regions in the Iddm4 and ART2 intervals (UCSC genome database). Primers found to be polymorphic between parental strains were used. The position of markers on the genetic map was established by inspection of the data set and conventional calculation methods to establish meiotic map distances, which are expressed in megabases (Mb) according to the rat genome sequence, June 2003 build (http://genome.ucsu.edu). Chr: chromosome.
Appendix Table 3. Frequency of Diabetes as a Function of Iddm4 and Iddm20 Genotype.
As documented by the low frequency of diabetes in the shaded cells, diabetes susceptibility in KRV-infected rats requires Iddm20, Iddm4, or both. “d” homozygous for the BBDR-origin allele; “”w” homozygous for the WF-origin alleles; “h” heterozygous.
| Iddm4→ | d | h | w | d | h | w |
|---|---|---|---|---|---|---|
| Iddm20↓ | sick | sick | sick | well | well | well |
| d | 5 | 13 | 1 | 4 | 9 | 6 |
| h | 16 | 19 | 0 | 16 | 32 | 24 |
| w | 2 | 3 | 0 | 6 | 24 | 10 |
Appendix Table 4. Candidate Genes in the Iddm20 Interval.
Genes in the Iddm20 Interval Abbreviations: idd14-NON is the interval determined in the NOD × NON cross (14) and idd14B6 is the interval determined in the NOD × B6 crosses and congenic (15). The rat Iddm20 supported interval ± 1 LOD is shown in gray shading. Chr: chromosome; bp: base pairs
| Rat Chr | Rat Chr Start (bp) | Description | Mouse Chr | Mouse Chr Start (bp) | Mouse Idd14 region | Rat Iddm20 |
|---|---|---|---|---|---|---|
| 17 | 29713540 | PAK/PLC-interacting protein 1 | 13 | 40443433 | idd14-NON | |
| 17 | 28451133 | Zinc finger protein 40 (Alpha A-crystallin-binding 1) | 13 | 41502581 | Idd14-NON | |
| 17 | 28306727 | Endothelin-1 precursor (ET-1). | 13 | 41748723 | Idd14-NON | |
| 17 | 27138814 | NAD-dependent deacetylase sirtuin 5 | 13 | 42822790 | Idd14-NON | Iddm20 |
| 17 | 27046741 | RAN BP9; B cell antigen receptor Ig beta associated p1 | 13 | 42855480 | Idd14-NON | Iddm20 |
| 17 | 26709913 | CD83 antigen. | 13 | 43237577 | Idd14-NON | Iddm20 |
| 17 | Jumonji protein | 13 | 44287373 | Idd14-NON | Iddm20 | |
| 17 | Adenylyl cyclase-associated protein 2 (CAP 2). | 13 | 46018959 | Idd14-NON | Iddm20 | |
| 17 | 23821596 | Kinesin-like protein KIF13A. | 13 | 46244160 | Idd14-NON | Iddm20 |
| 17 | 23728906 | Malin | 13 | 46507913 | Idd14-NON | Iddm20 |
| 17 | 23701239 | Thiopurine S-methyltransferase | 13 | 46519516 | Idd14-NON | Iddm20 |
| 17 | 23634062 | DEK oncogene (DNA binding). | 13 | 46579123 | Idd14-NON | Iddm20 |
| 17 | RNP particle component (Fragment). | 13 | 47794113 | Idd14-NON | Iddm20 | |
| 17 | 21994289 | Homeobox protein BarH-like 1. | 13 | 48158986 | idd14-NON | Iddm20 |
| 17 | 21806043 | PHD finger protein 2 (GRC5). | 13 | 48298466 | idd14B6 | Iddm20 |
| 17 | 21410701 | Ninjurin 1 (Nerve injury-induced protein 1). | 13 | 48685927 | idd14B6 | Iddm20 |
| 17 | 21015178 | Osteomodulin precursor (Osteoadherin) (OSAD) | 13 | 49088364 | idd14B6 | Iddm20 |
| 17 | 19659967 | Sphingosine 1-phosphate receptor Edg-3 | 13 | 50434260 | idd14B6 | Iddm20 |
| 17 | 19520647 | SHC transforming protein 3 (SH2 domain protein C3) | 13 | 50457472 | idd14B6 | Iddm20 |
| 17 | 19423770 | Cyclin-dependent kinases regulatory subunit 2 (CKS-2). | 13 | 50672083 | idd14B6 | Iddm20 |
| 17 | 19352632 | Semaphorin 4D precursor | 13 | 50728006 | idd14B6 | Iddm20 |
| 17 | 19230896 | GADD45 gamma (Cytokine responsive protein) | 13 | 50873436 | idd14B6 | Iddm20 |
| 17 | 18443785 | Tyrosine-protein kinase SYK (Spleen) | 13 | 51630629 | idd14B6 | Iddm20 |
| 17 | 18106038 | nuclear factor, interleukin 3, regulated. | 13 | 52003799 | idd14B6 | |
| 17 | 17736908 | Tyrosine-protein kinase ROR2 precursor 2. | 13 | 52145865 | idd14B6 | |
| 17 | 17243262 | Homeobox protein MSX-2 (Hox-8.1). | 13 | 52506491 | idd14B6 | |
| 17 | 16655985 | D(1A) dopamine receptor. | 13 | 53096324 | idd14B6 | |
| 17 | 16497967 | Histamine H2 receptor (H2R) | 13 | 53258774 | idd14B6 | |
| 17 | 16094098 | ADP-ribosylation factor-like 10A | 13 | 53665747 | idd14B6 | |
| 17 | 15595893 | Retinoid × receptor interacting protein 110. | 13 | 54118171 | idd14B6 | |
| 17 | 15512144 | Fibroblast growth factor receptor 4 precursor | 13 | 54244110 | idd14B6 | |
| 17 | 15353521 | Ras-related protein Rab-24 (Rab-16). | 13 | 54409106 | idd14B6 | |
| 17 | 15350379 | P×19-like protein. | 13 | 54409802 | idd14B6 | |
| 17 | 15346165 | Max dimerization protein 3. | 13 | 54414783 | idd14B6 | |
| 17 | 15313303 | Vesicular integral-membrane protein VIP36 precursor | 13 | 54432976 | idd14B6 | |
| 17 | 15222027 | Regulator of G-protein signaling 14 (RGS14) | 13 | 54459599 | idd14B6 | |
| 17 | 15261698 | Profilin III. | 13 | 54504680 | idd14B6 | |
| 17 | 15251796 | coagulation factor XII (Hageman factor) | 13 | 54507774 | idd14B6 | |
| 17 | 15194740 | Drebrin (Developmentally regulated brain protein). | 13 | 54563349 | idd14B6 | |
| 17 | 15147111 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 41 | 13 | 54627408 | idd14B6 | |
| 17 | 15033873 | Calcium-signal modulating cyclophilin ligand (CAML). | 13 | 54719996 | idd14B6 | |
| 17 | 14834123 | Pituitary homeobox 1 (Pt×1). | 13 | 54922627 | idd14B6 | |
| 17 | 14286947 | Small inducible cytokine B14 precursor (CXCL14) | 13 | 55389282 | idd14B6 | |
| 17 | 14068757 | Interleukin-9 precursor (IL-9) (T-cell growth factor P40) | 13 | 55582311 | idd14B6 | |
| 17 | 13995183 | Leukocyte cell-derived chemotaxin 2 precursor | 13 | 55645800 | idd14B6 | |
| 17 | 13905503 | transforming growth factor, beta induced 68kd | 13 | 55712518 | idd14B6 | |
| 17 | 13814377 | Mothers against decapentaplegic homolog 5 (SMAD5] | 13 | 55808625 | idd14B6 | |
| 17 | 13661202 | Short transient receptor potential channel 7 (TrpC7) | 13 | 55879544 | idd14B6 |
References
- 1.Hawa MI, Beyan H, Buckley LR, Leslie RDG. Impact of genetic and non-genetic factors in type 1 diabetes. Am J Med Genet. 2002;115:8–17. doi: 10.1002/ajmg.10339. [DOI] [PubMed] [Google Scholar]
- 2.Åkerblom HK, Vaarala O, Hyöty H, Ilonen J, Knip M. Environmental factors in the etiology of type 1 diabetes. Am J Med Genet. 2002;115:18–29. doi: 10.1002/ajmg.10340. [DOI] [PubMed] [Google Scholar]
- 3.Anjos S, Polychronakos C. Mechanisms of genetic susceptibility to type I diabetes: beyond HLA. Mol Genet Metab. 2004;81:187–195. doi: 10.1016/j.ymgme.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 4.Yoon JW, Jun HS. Role of viruses in the pathogenesis of type 1 diabees mellitus. In: LeRoith D, Taylor SI, Olefsky JM, editors. Diabetes mellitus. A fundamental and clinical text. Philadelphia: Lippincott Williams & Wilkins; 2004. pp. 575–590. [Google Scholar]
- 5.Mordes JP, Bortell R, Blankenhorn EP, Rossini AA, Greiner DL. Rat models of type 1 diabetes. Genetics, environment, and autoimmunity. ILAR Journal. 2004;45:278–291. doi: 10.1093/ilar.45.3.278. [DOI] [PubMed] [Google Scholar]
- 6.Martin AM, Blankenhorn EP, Maxson MN, Zhao M, Leif J, Mordes JP, Greiner DL. Non-major histocompatibility complex-linked diabetes susceptibility loci on chromosomes 4 and 13 in a backcross of the DP BB/Wor rat to the WF rat. Diabetes. 1999;48:50–58. doi: 10.2337/diabetes.48.1.50. [DOI] [PubMed] [Google Scholar]
- 7.Guberski DL, Thomas VA, Shek WR, Like AA, Handler ES, Rossini AA, Wallace JE, Welsh RM. Induction of type 1 diabetes by Kilham's rat virus in diabetes resistant BB/Wor rats. Science. 1991;254:1010–1013. doi: 10.1126/science.1658938. [DOI] [PubMed] [Google Scholar]
- 8.Zipris D, Hillebrands JL, Welsh RM, Rozing J, Xie JX, Mordes JP, Greiner DL, Rossini AA. Infections that induce autoimmune diabetes in BBDR rats modulate CD4+CD25+ T cell populations. J Immunol. 2003;170:3592–3602. doi: 10.4049/jimmunol.170.7.3592. [DOI] [PubMed] [Google Scholar]
- 9.Martin AM, Maxson MN, Leif J, Mordes JP, Greiner DL, Blankenhorn EP. Diabetes-prone and diabetes-resistant BB rats share a common major diabetes susceptibility locus, iddm4: Additional evidence for a “universal autoimmunity locus” on rat chromosome 4. Diabetes. 1999;48:2138–2144. doi: 10.2337/diabetes.48.11.2138. [DOI] [PubMed] [Google Scholar]
- 10.Mordes JP, Leif J, Novak S, DeScipio C, Greiner DL, Blankenhorn EP. The iddm4 locus segregates with diabetes susceptibility in congenic WF.iddm4 rats. Diabetes. 2002;51:3254–3262. doi: 10.2337/diabetes.51.11.3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hornum L, DeScipio C, Markholst H, Troutman SA, Novak S, Leif J, Greiner D, Mordes JP, Blankenhorn EP. Comparative mapping of rat Iddm4 to segments on HSA7 and MMU6. Mamm Genome. 2004;15:53–61. doi: 10.1007/s00335-003-3023-z. [DOI] [PubMed] [Google Scholar]
- 12.Butterfield RJ, Blankenhorn EP, Roper RJ, Zachary JF, Doerge RW, Teuscher C. Identification of genetic loci controlling the characteristics and severity of brain and spinal cord lesions in experimental allergic encephalomyelitis. Am J Pathol. 2000;157:637–645. doi: 10.1016/S0002-9440(10)64574-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellerman KE, Richards CA, Guberski DL, Shek WR, Like AA. Kilham rat virus triggers T-cell-dependent autoimmune diabetes in multiple strains of rat. Diabetes. 1996;45:557–562. doi: 10.2337/diab.45.5.557. [DOI] [PubMed] [Google Scholar]
- 14.Brodnicki TC, Quirk F, Morahan G. A susceptibility allele from a non-diabetes-prone mouse strain accelerates diabetes in NOD congenic mice. Diabetes. 2003;52:218–222. doi: 10.2337/diabetes.52.1.218. [DOI] [PubMed] [Google Scholar]
- 15.McAleer MA, Reifsnyder P, Palmer SM, Prochazka M, Love JM, Copeman JB, Powell EE, Rodrigues NR, Prins JB, Serreze DV, DeLarato NH, Wicker LS, Peterson LB, Schork NJ, Todd JA, Leiter EH. Crosses of NOD mice with the related NON strain - A polygenic model for IDDM. Diabetes. 1995;44:1186–1195. doi: 10.2337/diab.44.10.1186. [DOI] [PubMed] [Google Scholar]
- 16.Elly C, Witte S, Zhang Z, Rosnet O, Lipkowitz S, Altman A, Liu YC. Tyrosine phosphorylation and complex formation of Cbl-b upon T cell receptor stimulation. Oncogene. 1999;18:1147–1156. doi: 10.1038/sj.onc.1202411. [DOI] [PubMed] [Google Scholar]
- 17.Yokoi N, Komeda K, Wang HY, Yano H, Kitada K, Saitoh Y, Seino Y, Yasuda K, Serikawa T, Seino S. Cblb is a major susceptibility gene for rat type 1 diabetes mellitus. Nature Genet. 2002;31:391–394. doi: 10.1038/ng927. [DOI] [PubMed] [Google Scholar]
- 18.Brown DW, Welsh RM, Like AA. Infection of peripancreatic lymph nodes but not islets precedes Kilham rat virus-induced diabetes in BB/Wor rats. J Virol. 1993;67:5873–5878. doi: 10.1128/jvi.67.10.5873-5878.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKisic MD, Paturzo FX, Gaertner DJ, Jacoby RO, Smith AL. A nonlethal rat parvovirus infection suppresses rat T lymphocyte effector functions. J Immunol. 1995;155:3979–3986. [PubMed] [Google Scholar]
- 20.DeClercq E. Interferon induction by polynucleotides, modified polynucleotides, and polycarboxylates. Methods Enzymol. 1981;78:227–235. doi: 10.1016/0076-6879(81)78122-9. [DOI] [PubMed] [Google Scholar]
- 21.Akiyama Y, Stevenson GW, Schlick E, Matsushima K, Miller PJ. Differential ability of human blood monocyte subsets to release various cytokines. J Leukocyte Biol. 1985;37:519–530. doi: 10.1002/jlb.37.5.519. [DOI] [PubMed] [Google Scholar]
- 22.Fresa KL, Korngold R, Murasko DM. Induction of natural killer cell activity of thoracic duct lymphocytes by polyinosinic-polycytidylic acid (poly(I:C)) or interferon. Cell Immunol. 1985;91:336–343. doi: 10.1016/0008-8749(85)90231-x. [DOI] [PubMed] [Google Scholar]
- 23.Turner W, Chan SP, Chirigos MA. Stimulation of humoral and cellular antibody formation in mice by poly I:C. Proc Soc Exp Biol Med. 1970;133:334–338. doi: 10.3181/00379727-133-34469. [DOI] [PubMed] [Google Scholar]
- 24.Davis CT, Blankenhorn EP, Murasko DM. Genetic variation in the ability of several strains of rats to produce interferon in response to polyriboinosinic-polyribocytodilic acid. Infect Immun. 1984;43:580–583. doi: 10.1128/iai.43.2.580-583.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]