Summary
The vertebrate Fgf8 gene produces multiple protein isoforms by alternative splicing. Two evolutionarily conserved spliceforms, Fgf8a and Fgf8b, exhibit distinct bioactivities, with Fgf8b having a more potent inductive activity due to higher affinity for Fgf receptors. To investigate the in vivo requirement for Fgf8b, we created a splice site mutation abolishing Fgf8b expression in mice. Analysis of this mutant has uncovered a novel function of Fgf8 signaling before the onset of gastrulation. We show that the loss of Fgf8b disrupts the induction of the Brachyury gene in the pregastrular embryo, and in addition, disrupts the proper alignment of the anterior-posterior axis with the shape of the embryo and the uterine axes at E.6.5. Importantly, Fgf8-null embryos display the same phenotype as Fgf8b-deficient embryos at E.6.5, demonstrating that signaling by Fgf8b is specifically required for development of the pregastrular embryo. By contrast, during gastrulation, Fgf8a can partially compensate for the loss of Fgf8b in mesoderm specification. We show that an increased level of Fgf8a, which leads to Fgf4 expression in the primitive streak, can also promote mesoderm migration in the absence of Fgf8b. Therefore, different Fgf signals may have distinct requirements for the morphogenesis and gene regulation before and during gastrulation. Importantly, our findings implicate Fgf8 in the morphogenetic process that establishes the defined relationship between the axes of the embryo and the uterus at the beginning of gastrulation, a perplexing phenomenon discovered two decades ago.
Keywords: Fgf8, signaling, alternative splicing, anterior-posterior axis, embryo, uterus, remodeling
Introduction
Gastrulation is a morphogenetic process that leads to production of three different germ layers and establishes the embryonic body plan (Tam and Behringer, 1997). During gastrulation, epiblast cells traverse the primitive streak at the posterior end of the embryo and undergo an epithelial-to-mesenchymal transition. The newly generated mesoderm migrates away from distinct positions of the primitive streak to form different mesodermal lineages. When gastrulation starts at embryonic day 6.5 (E6.5), the mouse embryo exhibits a morphologically explicit anterior-posterior (AP) polarity. Interestingly, the AP axis tends to be perpendicular to the longitudinal axis (from the oviduct to the cervix) of the uterine horn (Smith, 1985). The mechanism and biological significance for the defined relationship between the embryonic and uterine axes is unclear. Molecules governing the orientation of embryonic AP axis with respect to the uterus have not yet been identified.
Recent studies have identified a series of patterning events in the mouse embryo before E6.5 (Rossant and Tam, 2004). Unexpectedly, the molecular markers that are characteristic for the anterior and posterior poles of the embryo are initially expressed at the opposite ends of the short transverse axis of the embryo between E5.75 and E6.0, when the embryo exhibits an ellipsoidal shape in the transverse plane (Mesnard et al., 2004; Perea-Gomez et al., 2004). Furthermore, the emerging AP axis does not relate with the uterine axis (Mesnard et al., 2004). After E5.75, the AP axis gradually shifts and eventually becomes parallel to the long axis of the embryo (Mesnard et al., 2004; Perea-Gomez et al., 2004). The shift of the AP axis was shown to be mainly caused by tissue remodeling, converting the short axis to long (Mesnard et al., 2004; Perea-Gomez et al., 2004). Concomitant with the remodeling, both the AP axis and the long axis of the embryo become perpendicular to the longitudinal axis of the uterine horn at E6.5 (Mesnard et al., 2004). Therefore, the embryo remodeling may be crucial for the final alignment of the AP axis with the long axis of the embryo and the uterus. Currently, little is known about the molecular mechanism underlying the morphogenetic remodeling.
Studies in chick and Xenopus embyryos have demonstrated that fibroblast growth factor (Fgf) signaling plays important roles before and during gastrulation, including the induction of the primitive streak and the mesoderm, and the control of mesoderm migration (Bottcher and Niehrs, 2005). In the mouse embryo, Fgf8 is the only Fgf that is known to play an essential role during gastrulation. Fgf8 is expressed in the epiblast and visceral endoderm at E5.75, and subsequently in the emerging primitive streak at E6.5 (Crossley and Martin, 1995). In the absence of Fgf8, or a component essential for Fgf8 signaling, the mesoderm is formed but fails to migrate away from the primitive streak at E7.5 (Ciruna and Rossant, 2001; Ciruna et al., 1997; Deng et al., 1997; Garcia-Garcia and Anderson, 2003; Sun et al., 1999; Yamaguchi et al., 1994). Therefore, Fgf8 signaling is essential for mesoderm migration during mouse gastrulation. However, whether Fgf8 plays any role before gastrulation remains unanswered.
The vertebrate Fgf8 gene produces multiple protein isoforms by alternative splicing (Crossley and Martin, 1995; Fletcher et al., 2006; Gemel et al., 1996; MacArthur et al., 1995; Sato et al., 2001). Two evolutionarily conserved isoforms, Fgf8a and Fgf8b, exhibit distinct bioactivities. When they are ectopically expressed in the developing brain, Fgf8a promotes cell proliferation in the midbrain, whereas Fgf8b transforms the midbrain into a cerebellum (Lee et al., 1997; Liu et al., 2003; Liu et al., 1999; Sato et al., 2001). In Xenopus, Fgf8b is the predominant Fgf8 spliceform involved in mesoderm induction, whereas Fgf8a appears to be involved in posterior neural development (Fletcher et al., 2006). The more potent inductive activity of Fgf8b is attributed to its higher affinity than Fgf8a for FGF receptors (Olsen et al., 2006). To investigate the in vivo function of Fgf8b, we mutated the alternative splice site of the Fgf8 gene, thereby abolishing Fgf8b expression in mice. Our analysis of this mutant has uncovered a novel function of Fgf8 signaling before the initiation of gastrulation. We show that there are different requirements for Fgf8b in embryo remodeling before gastrulation, in mesoderm specification, and mesoderm migration during gastrulation.
Materials and Methods
Generation of mouse mutants deficient for Fgf8b
By gene targeting, we changed the intron/exon junction sequence of Fgf8 exon 1D from taaagGTA to catatGTA, abolishing the 5′ alternative splice acceptor, while the downstream alternative splice acceptor in exon 1D remains intact (Fig. 1A and D). The mutation produced a new restriction endonuclease site for NdeI. A selectable marker, neo was placed 400 bp downstream of exon 1D. We also produced a mutation, designated as Fgf8neo that contains only the neo cassette at the same intronic region as Fgf8Δb-neo (Fig. 1D). Targeted ES cells were identified by a combination of Southern blot analysis, PCR and restriction analysis (Fig. 1B and C), and used to generate germline chimeras. The neo cassette, which is flanked with two loxP sites was subsequently removed by breeding Fgf8+/Δb-neo or Fgf8+/neo mice to CMV-Cre transgenic mice, which express Cre broadly (Li et al., 2002).
Figure 1.
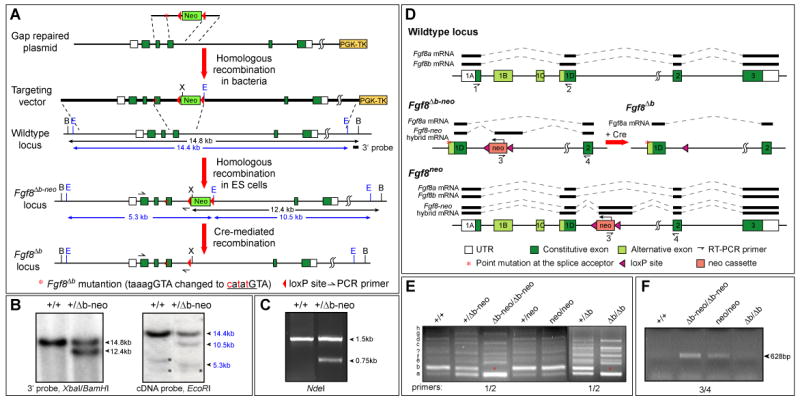
Generation of splice site mutation to abolish Fgf8b expression. (A) Schematic representation of mutations in the Fgf8 locus. The change of sequences shown in red at the junction of intron (small letters) and exon (capital letters) mutates the 5′ alternative splice acceptor and creates an NdeI site (underlined). (B) Southern blot analysis to identify targeted ES cell clones. Asterisks indicate non-specific signals. (C) PCR and restriction digestion analysis to verify the point mutation. (D) Schematic representation of Fgf8, Fgf8Δb-neo, Fgf8Δb and Fgf8neo loci, and alterations in RNA splicing due to the point mutation and neo insertion. The Fgf8-neo hybrid transcript results from a cryptic splice donor and acceptor in the neo and the intronic region 360 bp downstream to the neo, respectively. (E) RT-PCR analysis of different Fgf8 splice variants in E7.5 embryos of indicated genotypes using primers 1 and 2 (shown in E). Fgf8 splice variants (a-h and unknown) are marked to the left. Note that Fgf8b (asterisk) is missing in Fgf8Δb-neo/Δb-neo and Fgf8Δb/Δb embryos, while the neo insertion has no effect on the alternative splicing of the first four exons. (F) RT-PCR assay using primers 3 and 4 (indicated in D) reveals that the insertion of neo results in production of Fgf8-neo hybrid transcripts. Abbreviations: B, BamHI; E, EcoRI; X, XbaI.
Mouse breeding and genotyping
Mutant mouse strains were maintained in the CD1 (Charles River Laboratory) background. Noon of the day on which the vaginal plug was detected was designated as E0.5 in staging of embryos. Embryos at E5.5 and E5.75 were staged using morphological landmarks (Rivera-Perez et al., 2003) and EGFP fluorescence derived from a Hex-EGFP transgene, which is expressed in the visceral endoderm at the distal at E5.5 and the proximal anterior at E5.75 (Rodriguez et al., 2001). After primitive streak formation, embryos were staged according to Downs and Davies (Downs and Davies, 1993).
Genotyping was carried out by PCR analysis. Primers Fgf8-GT-f (CAGAGGGTTCAGAGGAGAGG) and Fgf8-GT-r1 (CCCGGAGTCTAACTTGCAGG) were used to produce 198 bp PCR products from the WT allele and 290 bp from the Fgf8Δb allele, respectively. Primers Fgf8-GT-r1 and Neo-pro-R (CGGTGGATGTGGAATGTGTGC) were used to produce 300 bp PCR products from the Fgf8Δb-neo or Fgf8neo allele.
Histological analysis
Embryos and uteri were recovered in PBS and fixed immediately in 4% paraformaldehyde at 4°C overnight. Serial transverse sections of the uterus were made at 7 μm across the mesometrium-antimesometrium axis. Wholemount or section RNA in situ hybridization was performed as described previously (Li and Joyner, 2001). Detection of Hex-EGFP was performed by immunohistochemistry (rabbit anti-GFP IgG fraction, 1:1,000 dilution, Invitrogen, A11122). Measurements of embryonic dimensions and axis angles were performed on digital images with ImageJ software. Student's t-Test and two-way ANOVA test were carried out with Microsoft Excel.
Results
Generation of mouse mutations abolishing Fgf8b
As shown previously (Crossley and Martin, 1995; MacArthur et al., 1995), alternative splicing of the first four exons of the mouse Fgf8 gene produces at least eight Fgf8 splice variants (a-h), with Fgf8b being the predominant one (Fig. 1D and E). To determine the in vivo function of Fgf8b, we mutated the 5′ alternative splice acceptor of Fgf8 exon 1D, designated as Fgf8Δb-neo. The neo selectable marker located in the Fgf8 intron was subsequently removed to produce the Fgf8Δb allele (Fig. 1A and D). RT-PCR analyses revealed that only Fgf8a, c, e and g splice variants, which utilize the remaining alternative splice acceptor in exon 1D, are expressed, at elevated levels, in Fgf8Δb-neo/Δb-neo and Fgf8Δb/Δb embryos at E7.5, whereas Fgf8b, d, f and h are absent (Fig. 1E). These results demonstrate that both Fgf8Δb-neo and Fgf8Δb mutations abolish Fgf8b and three minor b-type splice variants.
Fgf8Δb-neo/Δb-neo embryos display more severe defects than Fgf8Δb/Δb embryos
Fgf8Δb-neo/Δb-neo embryos exhibit severe abnormalities at E7.5. In all Fgf8Δb-neo/Δb-neo embryos, a mass of cells with the morphology and molecular characters of the nascent mesoderm was detected in the posterior region, bulging into the amniotic cavity (Fig. 2C-D, 4A, 4D, and 5). Furthermore, few mesodermal cells were evident outside of the primitive streak in the embryonic region of Fgf8Δb-neo/Δb-neo embryos (Fig. 2D). In the majority of Fgf8Δb-neo/Δb-neo embryos (23 out of 25, 92%, embryonic mesoderm-derived structures were completely absent at E8.5, whereas the allantois and the mesodermal components of the amnion and yolk sac, which are derived from the extraembryonic mesoderm were observed (Fig. 2F). In a few Fgf8Δb-neo/Δb-neo mutants (2/25, 8%), the heart mesoderm, the somites and the headfold were evident but severely malformed (data not shown). The phenotypes of Fgf8Δb-neo/Δb-neo embryos are remarkably similar to those described for Fgf8-null (Fgf8-/-) embryos at the morphological and histological level (Sun et al., 1999). These observations suggest that, similar to Fgf8-/- mutants, the embryonic mesoderm fails to migrate away from the primitive streak in Fgf8Δb-neo/Δb-neo embryos.
Figure 2.
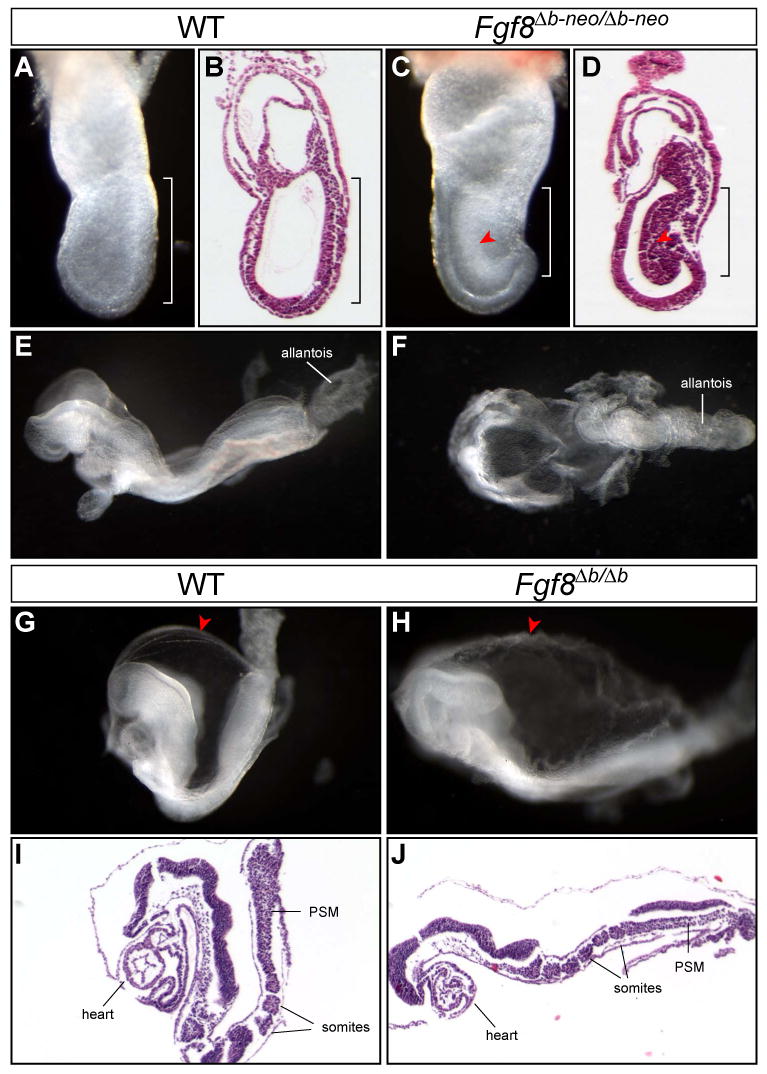
Fgf8Δb-neo/Δb-neo embryos have more severe defects than Fgf8Δb/Δb embryos. (A-D) Morphology and histology of WT and Fgf8Δb-neo/Δb-neo embryos at E7.5. (B) and (D) are H&E analysis of sagittal sections of the embryo shown in (A) and (B) respectively. Note the mesodermal cells (arrowhead) accumulated at the primitive streak (bracket) in Fgf8Δb-neo/Δb-neo embryo. (E and F) Lateral view of WT embryo (C) and dorsal view of Fgf8Δb-neo/Δb-neo embryo (D) at E8.5. (G-J) Morphology and histology of WT and Fgf8Δb/Δb embryos at E8.5. Arrowhead indicates the rough appearance of the amniotic membrane of Fgf8Δb/Δb embryos, comparing to the control. (I-J) H&E analysis of sagittal sections of the embryo in G and H, respectively. PSM, presomitic mesoderm.
Figure 4.
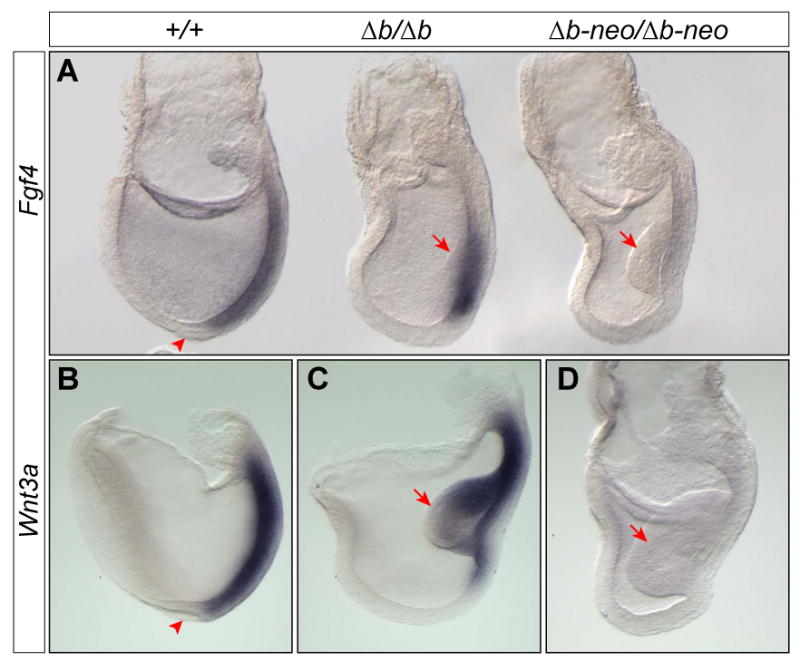
Fgf4 and Wnt3a are expressed in the primitive streak of Fgf8Δb/Δb, but not in Fgf8Δb-neo/Δb-neo embryos at E7.5. (A) Expression of Fgf4 in embryos of indicated genotypes. (B-D) Expression of Wnt3a in WT (B), Fgf8Δb/Δb (C) and Fgf8Δb-neo/Δb-neo (D) embryos at E7.75. Note that the morphologically distinct node (arrowhead) is absent in Fgf8Δb/Δb and Fgf8Δb-neo/Δb-neo embryos. Arrow marks the mass of mesodermal cells, which is variable and tends to be smaller in size than that found in Fgf8Δb-neo/Δb-neo embryos, at the primitive streak of Fgf8Δb/Δb embryos at E7.5.
Figure 5.
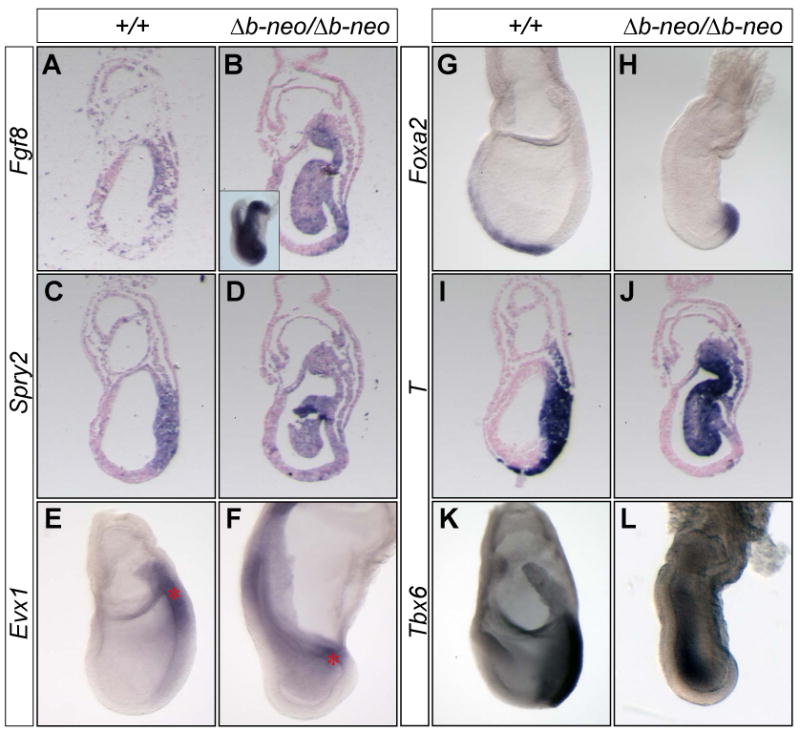
The regionalization of the primitive streak and the specification of the mesoderm are partially maintained in Fgf8Δb-neo/Δb-neo embryos. (A-L) In situ hybridization analysis of markers (indicated to the left) characteristic for Fgf8 signaling and mesoderm in WT and Fgf8Δb-neo/Δb-neo embryos at E7.5. (A-D and I-J) Adjacent sagittal sections of WT (A, C and I) and Fgf8Δb-neo/Δb-neo embryos (B, D and J). Inset in B shows wholemount analysis of Fgf8 expression. Note that axial mesoderm marked by T and Foxa2 expression is absent anterior to the streak of Fgf8Δb-neo/Δb-neo embryos, indicating a lack of mesoderm migration.
Significantly, removing the neo cassette led to relatively normal gastrulation in Fgf8Δb/Δb embryos. At E8.5, the somites and cardio-mesoderm were clearly discernable in most Fgf8Δb/Δb embryos (40/51, 78.4%, Fig. 2H and J), although some mutants (11/51, 21.6%) had more severe gastrulation defects similar to those found in Fgf8Δb-neo/Δb-neo mutants (data not shown). The amniotic membrane of all Fgf8Δb/Δb embryos appears rough and fluffy, probably due to abnormal development of the mesodermal component of the membrane (Fig. 2H and J). Analysis of region-specific markers demonstrated that the AP patterning of the neural plate of Fgf8Δb/Δb embryos is largely normal (Fig. 3). At E9.5, Fgf8Δb/Δb embryos are significantly smaller in size than their WT littermates and manifest multiple morphological defects, including open neural tube, malformed branchial arches and the heart tubes (data not shown). Therefore, Fgf8b is essential for proper developmental progression after E8.5. Collectively, our data demonstrate that the defect of mesoderm migration in Fgf8Δb-neo/Δb-neo embryos can be largely rescued by removing the neo cassette.
Figure 3.
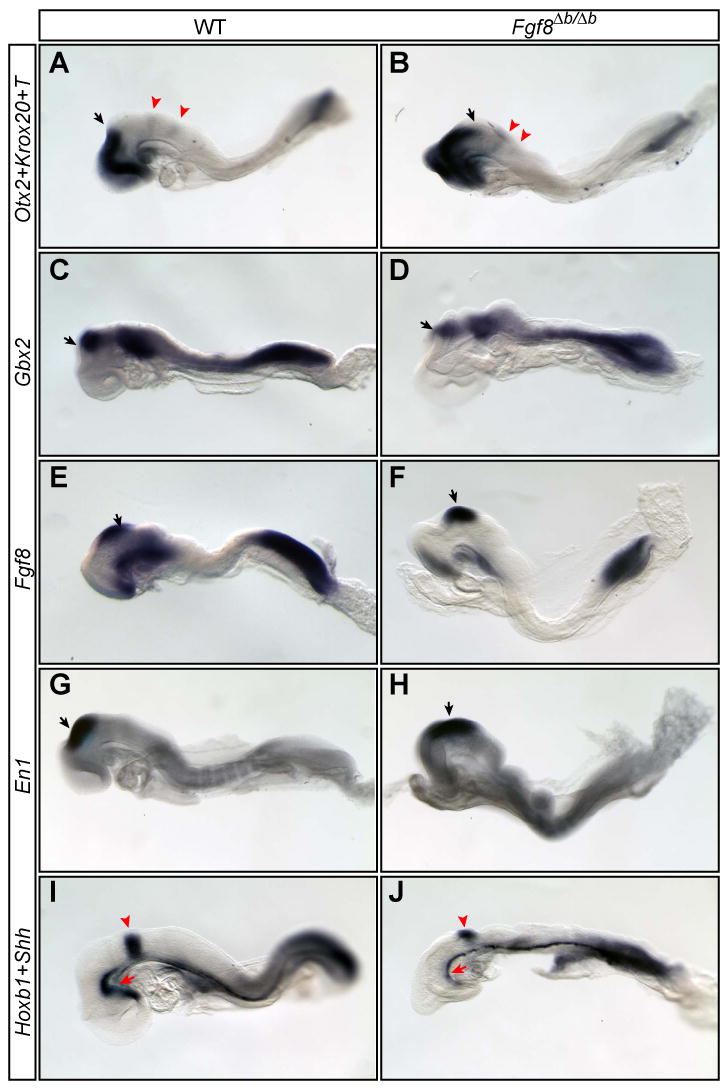
The pattern formation of the neural plate of Fgf8Δb/Δb embryos is largely normal. (A-J) Expression of region-specific markers as indicated to the left in the anterior neural plate of WT and Fgf8Δb/Δb embryos. Otx2, telencephalon and mesencephalon; Gbx2, rhombomere 1 (r1); Fgf8, r1; En1, mesencephalon and r1; Shh, head process and axial mesoderm. Black arrows mark mid/hindbrain junction. Arrowhead in (A-B and I-J) demarcates Krox20 expression in r3 and r5 (A-B) and Hoxb1 expression in r4 (I-J). Note that the head process (red arrow) appears abnormal in the mutants.
Fgf4 and Wnt3a are expressed in the primitive streak of WT and Fgf8Δb/Δb, but not in Fgf8Δb-neo/Δb-neo embryos
The more severe phenotype of Fgf8Δb-neo/Δb-neo than Fgf8Δb/Δb embryos prompted us to compare and contrast the underlying molecular defects that might reveal unique developmental contributions of Fgf8a and Fgf8b. We speculated that the presence of the neo cassette may interfere with expression of Fgf8 (Fgf8a, c, e and g only) from the Fgf8Δb-neo locus. Indeed, a homozygous mutation for Fgf8neo that contains an identical neo insertion to Fgf8Δb-neo (see Materials and Methods) results in defects similar to a homozygous mutation for an Fgf8 hypomorphic allele (Meyers et al., 1998), and the defects of Fgf8neo/neo mutants are completely rescued by removing the neo (data not shown). These data demonstrate that the neo insertion impairs Fgf8 expression, while the loxP sequence that is remained after neo removal has no effect on Fgf8 function (Fig. 1A and D). A similar neo insertion has been shown to cause aberrant splicing due to cryptic splice sites in the neo cassette (Meyers et al., 1998). Indeed, our RT-PCR and sequencing analyses demonstrated that both Fgf8 and Fgf8-neo hybrid transcripts are produced from Fgf8Δb-neo and Fgf8neo alleles, whereas only Fgf8 transcripts are generated from Fgf8Δb and Fgf8Δneo alleles (Fig. 1F and data not shown). The Fgf8-neo hybrid transcript contains stop codons in all three potential reading frames upstream of the coding sequences for the conserved FGF domain, and thus cannot produce functional Fgf8 proteins. Collectively, our genetic and molecular analyses strongly suggest that the presence of neo reduces functional Fgf8a expression from the Fgf8Δb-neo allele, and thus Fgf8Δb/Δb embryos produce a higher level of functional Fgf8a than that in Fgf8Δb-neo/Δb-neo embryos. Furthermore, our data indicate that an increase of Fgf8a expression can partially compensate for the loss of Fgf8b in promoting mesoderm migration.
We next sought to investigate the molecular basis for the rescued mesoderm migration in Fgf8Δb/Δb embryos. Fgf4 is co-expressed with Fgf8 in the primitive streak (Niswander and Martin, 1992), and Fgf4 expression is lost in Fgf8-/- mutants at E7.5 (Sun et al., 1999). Fgf4 and Fgf8 are known to play redundant roles in limb development (Boulet et al., 2004; Sun et al., 2002). In WT embryos at E7.5, Fgf4 is uniformly expressed in the primitive streak from the base of allantois to cells immediately posterior to the node (Fig. 4A). In Fgf8Δb/Δb embryos at E7.5, Fgf4 expression was detected in an increasing gradient from the anterior to the posterior of the primitive streak (4/4, Fig. 4A). By contrast, Fgf4 expression was absent (4/6), or greatly reduced in Fgf8Δb-neo/Δb-neo mutants (2/6, Fig. 4A).
The interplay between Wnt and Fgf signaling has been implicated in controlling mesoderm migration (Ciruna and Rossant, 2001). To investigate whether Wnt signaling is affected in Fgf8Δb-neo/Δb-neo mutants, we analyzed expression of Wnt3 and Wnt3a, which are expressed in the primitive streak at E7.5 (Takada et al., 1994). Wnt3 expression was indistinguishable between Fgf8Δb/Δb and Fgf8Δb-neo/Δb-neo mutants (data not shown). By contrast, Wnt3a transcripts were detected in the primitive streak of WT and Fgf8Δb/Δb (5/5), but not in Fgf8Δb-neo/Δb-neo mutants (3/3, Fig. 4B-D).
In summary, Fgf4 and Wnt3a are expressed in the primitive streak of Fgf8Δb/Δb, but not in Fgf8Δb-neo/Δb-neo embryos. The restoration of Fgf4 and Wnt3a, resulting from an elevated Fgf8a expression, may contribute to the rescue of mesoderm migration in Fgf8Δb/Δb embryos.
The remaining Fgf8a can promote mesoderm specification and regionalization of the primitive streak in Fgf8Δb-neo/Δb-neo embryos
Our RT-PCR analyses showed that the transcription of Fgf8 is independent of Fgf8b proteins (Fig. 1E). In situ hybridization analysis further demonstrated that there was robust Fgf8 expression in the mesodermal cells in the posterior of Fgf8Δb-neo/Δb-neo embryos (Fig. 5A). To determine whether the remaining Fgf8a isoform proteins elicit Fgf8 signaling in Fgf8Δb-neo/Δb-neo embryos, we analyzed expression of Spry2, which is a feedback inhibitor of Fgf signaling. Spry2 is expressed in the primitive streak at E7.5 and Spry2 expression is lost in the absence of Fgf8 signaling (Garcia-Garcia and Anderson, 2003; Minowada et al., 1999). Significantly, Spry2 expression was detected in the primitive steak of Fgf8Δb-neo/Δb-neo embryos at E7.5 (3/3, Fig. 5D). Therefore, the remaining Fgf8a isoforms activate Fgf8 signaling to a limited degree in the primitive streak of Fgf8Δb-neo/Δb-neo embryos.
We next sought to examine whether the residual Fgf8 signaling promotes mesoderm specification in Fgf8Δb-neo/Δb-neo embryos by analyzing expression of mesodermal markers at E7.5. Evx1 is expressed in the proximal part of the primitive streak (Fig. 5E), whereas Foxa2 (also known as HNF3β) is expressed in the distal end of the streak and the emerging axial mesoderm (Fig. 5G)(Ang et al., 1993; Dush and Martin, 1992; Sasaki and Hogan, 1993). Similar to that found in WT embryos, transcripts of Evx1 (n=5) and Foxa2 (n=3) were detected in the proximal and distal regions, respectively, of the primitive streak in Fgf8Δb-neo/Δb-neo embryos (Fig. 5F and H). These data suggest that the regionalization of the primitive streak in Fgf8Δb-neo/Δb-neo embryos is largely maintained.
T is expressed in the nascent mesoderm and in the axial mesoderm, while expression of Tbx6 demarcates the lineage of the paraxial mesoderm (Fig. 5I and K) (Chapman et al., 1996; Wilkinson et al., 1990). Prior studies have shown a great reduction of T and loss of expression of Tbx6 in embryos lacking Fgf8 signaling at E7.5 (Ciruna and Rossant, 2001; Garcia-Garcia and Anderson, 2003; Sun et al., 1999). By contrast, robust expression of T (n=9) and Tbx6 (n=3) was detected in the posterior region of Fgf8Δb-neo/Δb-neo embryos at E7.5 (Fig. 5J and L). These observations demonstrate that Fgf8a can partially compensate for the loss of Fgf8b in promoting the developmental program for mesoderm specification in Fgf8Δb-neo/Δb-neo embryos.
Loss of Fgf8b leads to abnormal orientation of the AP axis relative to the embryo shape at E6.5
Given the remarkably normal development of many Fgf8Δb/Δb embryos at E8.5, it was somewhat surprising that all the mutant embryos displayed significant abnormalities at E7.5, including a lack of morphologically distinct node structure and an accumulation of mesodermal cells at the primitive streak (Fig. 4A and C, n=41). These observations suggest that Fgf8b may play an essential role before E7.5. To test this hypothesis, we examined expression of the molecular markers that are characteristic for the anterior and posterior poles of the embryos at E6.5. At the prestreak and early streak stages, Fgf8 and Nodal are normally expressed in the posterior region of the mouse embryo, whereas Cer1 is expressed in the anterior visceral endoderm (AVE) (Supplementary Fig. 1SA, C and E). Transcripts of Fgf8 and Nodal were detected in the posterior side, whereas Cer1 expression was found in the AVE of Fgf8Δb/Δb embryos at E6.5 (Supplementary Fig. 1B, D and F). However, the expression domains of Fgf8, Nodal and Cer1 with respect to the shape of the embryo were found strikingly different between WT and Fgf8Δb/Δb embryos. As described previously (Mesnard et al., 2004; Perea-Gomez et al., 2004), Fgf8 and Nodal are expressed at one end of the long transverse axis of WT embryos, whereas Cer1 expressed in the opposite end. In contrast, Fgf8/Nodal and Cer1 expressing cells were found at the opposing ends of the short transverse axis of Fgf8Δb/Δb embryos (Supplementary Fig. S1). We next performed in situ hybridization with an RNA probe that recognizes both Lefty1 and Lefty2, which are respectively expressed in the AVE and the emerging primitive streak (Fig. 6A). We detected signals in the AVE, presumably Lefty1 expression, at one end of the short transverse axis in Fgf8Δb/Δb embryos at E6.5, whereas Lefty2 transcripts were largely absent (n=8, Fig. 6B). We also analyzed expression of T, which is a posterior marker of the pregastrular embryo. As described previously (Perea-Gomez et al., 2004; Rivera-Perez and Magnuson, 2005), T is expressed in the proximal-posterior epiblast and a radial ring of cells in the distal-most extraembryonic ectoderm between E6.25 and E6.5 (Fig. 6D). Interestingly, we found that T expression was absent in the distal extraembryonic ectoderm, while T transcripts were barely detected in the posterior epiblast of Fgf8Δb/Δb (3/3) embryos at E6.5 (Fig. 6E). Taken together, our data demonstrate that in the absence of Fgf8b, the AP axis aligns with the short, rather than the long, transverse axis of the embryo at E6.5. Furthermore, Fgf8b is required for the normal induction of T and Lefty2.
Figure 6.
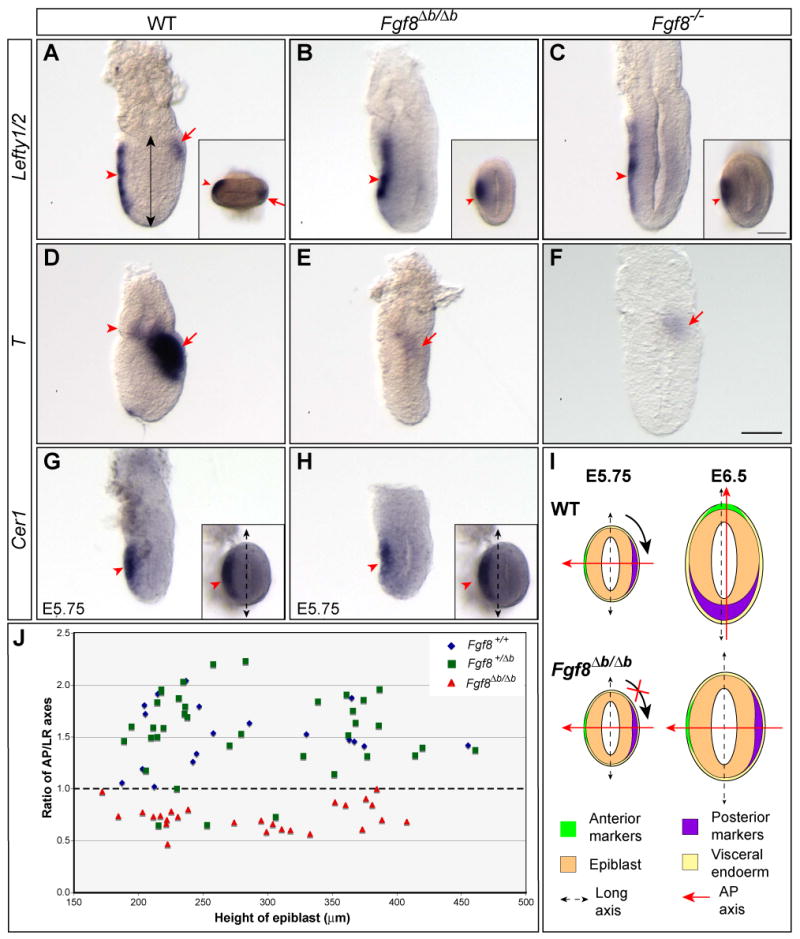
Fgf8b is required for normal induction of Lefty2 and T, and proper alignment between the AP axis and the shape of the embryo. (A-C) In situ hybridization with a RNA probe detecting both Lefty1 and Lefty2 in E6.5 embryos of indicated genotypes. Expression of Lefty1 in the AVE (marked by arrowhead) is unaffected in Fgf8Δb/Δb or Fgf8-/- embryos, whereas Lefty2 expression in the emerging primitive streak (arrow) is missing in the mutants. Insets show distal views of the embryos in A and C. (D-F) Analysis of T expression in E6.5 embryos of indicated genotype. Arrowhead marks T expression domain in the distal extraembryonic ectoderm, while arrow marks T expression in the posterior epiblast. (G-H) Expression of Cer1 in the AVE of WT (G) and Fgf8Δb/Δb embryos (H) at E5.75. Insets show distal views of the embryo. Dashed double-headed arrow marks the long axis of the embryo. (I) Schematic summary of the AP polarity with respect to the shape of WT and Fgf8Δb/Δb embryo at E5.75 and E6.5. Note that the shift of AP axis fails to occur in Fgf8Δb/Δb embryo. (J) Distribution of the ratio of AP and LR dimensions relating to the height of epiblast (indicated by double arrow in A) between E6.0 to E6.75 from intercrosses of Fgf8+/Δb mutants. Each dot represents one embryo. Scale bar in F that is equivalent to 50 μm is applicable to A-H, including insets in G and H. A different scale bar (50 μm) is used for the insets in A-C.
To determine whether the mutant phenotypes of Fgf8Δb/Δb embryos at E6.5 result from a developmental retardation, we measured the dimensions of Fgf8Δb/Δb and control embryos after in situ hybridization analysis with the above markers. The ratio of AP versus left-right (LR) dimension is significantly greater than 1 in control (Fgf8+/+ and Fgf8+/Δb) embryos (1.52± 0.38, n=59), but smaller than 1 in Fgf8Δb/Δb embryos (0.73±0.14, n=26), demonstrating that the loss of Fgf8b leads to abnormal alignment of the AP axis with the shape of the embryo (Fig. 6J). Importantly, no significant difference (F=0.208, two-way ANOVA test) was found in the height of epiblast (measured along the proximodistal axis of the egg cylinder) among Fgf8+/+ (270.42±82.91μm, n=20), Fgf8+/Δb (284.35±80.20μm, n=39) and Fgf8Δb/Δb (283.19±78.22μm, n=26) (Fig. 6J). Therefore, loss of Fgf8b does not cause a gross delay in development.
To ascertain whether the mutant phenotype of Fgf8Δb/Δb embryos at E6.5 is specific to the loss of Fgf8b, we re-examined the phenotype of Fgf8-/- mutants (Meyers et al., 1998). Fgf8-/- embryos display identical defects in the loss of Lefty2 (3/3) and T (5/5), and abnormal orientation of the AP axis as those observed in Fgf8Δb/Δb embryos at E6.5 (Fig. 6C and F). These results demonstrate that the Fgf8b proteins are essential for Fgf8 activity before E6.5.
Fgf8b is not required for the establishment of the AP polarity at E5.75
The abnormalities of Fgf8Δb/Δb and Fgf8-/- embryos at E6.5 prompted us to analyze expression patterns of Fgf8 in pregastrular embryos in greater details. At E5.5, diffuse signal of Fgf8 was detected in the epiblast (data not shown). By E5.75, Fgf8 transcripts were found in the AVE and throughout the epiblast (supplementary Fig. S2A). Between E5.75 and E6.0, Fgf8 expression is progressively confined to the proximal and subsequently to the proximal-posterior epiblast, while the expression in the AVE is down-regulated (supplementary Fig. S2B).
We next examine whether the specific expression of Fgf8 in the AVE may play a role in positioning the emerging AVE with reference to the shape of the embryo. As shown previously (Mesnard et al., 2004; Perea-Gomez et al., 2004), Lefty1 and Cer1 are expressed at one end of the short axis of WT embryos at E5.75 (Fig. 6G and data not shown). In Fgf8Δb/Δb embryos, the expression of Lefty1 and Cer1 was indistinguishable from that in WT (Fig. 6H and data not shown). Therefore, proper positioning of the AVE precursors along the short axis of the embryo at E5.75 does not depend on Fgf8b.
Loss of Fgf8b affects the orientation of the AP axis, but not the long axis, of the embryo relative to the longitudinal axis of the uterine horn
The AP axis and the long axis of the embryo are normally parallel to each other, and both axes tend to be perpendicular to the longitudinal axis of the uterine horn at E6.5 (Mesnard et al., 2004). As the AP axis becomes proximally perpendicular to the long axis of Fgf8Δb/Δb embryos, we sought to determine if loss of Fgf8b affects the orientation of the AP axis, the long axis of the embryo, or both with respect to the uterine horn. To do so, we examined the relative position of anterior pole of the embryo, the AVE cells, with respect to the long axes of the embryo and the uterus on cross sections of E6.5 embryos within the uterus. The Fgf8+/Δb males used for heterozygous intercrosses were homozygous for the Hex-EGFP transgene (Rodriguez et al., 2001), so that the AVE was marked by EGFP. In roughly three-quarters (73.0%) of the embryos (groups I, 27/37), the center of EGFP expression domain was close to one end of the long axis of the embryo (Fig. 7B and C). In the rest of the embryos [group II, 10/37 (27.0%)], the center of EGFP expression domain was near one end of the short axis (Fig. 7B and D). Remarkably, the long axis of the embryo in both groups tended to be perpendicular to the longitudinal axis of the uterus. The angles between the long axes of the embryo and the uterus are not significantly different (p=0.33, Student's t-Test) between group I (72.6±17.7, n=27) and group II (76.4±5.9, n=10) (Fig. 7E). As a result, the AP axis of group I embryos tends to be perpendicular to the longitudinal axis of the uterus, whereas the AP axis tends to be parallel to the longitudinal uterine axis of group II embryos (Fig. 7B-D). Based on the abnormal position of the AVE cells relative to the shape of the embryo, we suggest that the embryos of group II represent Fgf8Δb/Δb, whereas the embryos of group I represent WT (Fgf8+/+ and Fgf8+/Δb). Indeed, the percentage of embryos of group II (27%) is close to the expected Mendelian ratio (25%) for Fgf8Δb/Δb embryos.
Figure 7.
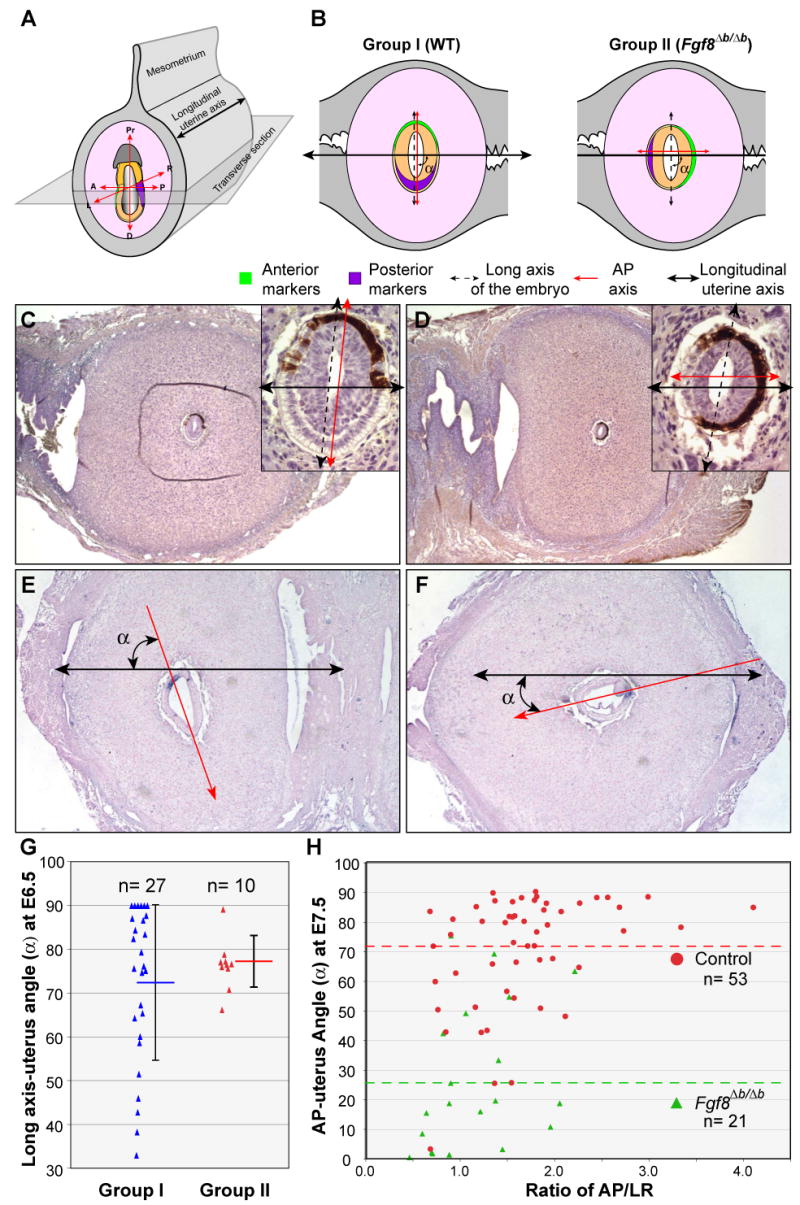
Loss of Fgf8b alters the orientation of AP axis, but not the long axis of the embryo, with respect to the longitudinal axis of the uterine horn. (A) Schema of orientation of the embryo within the uterus and the relative position of the sectioning plane. (B) Schematic representation of relationships between the axes of the embryo and uterus found in two groups (I and II) of embryos at E6.5 from intercrosses of Fgf8+/Δb parents. (C and D) Immunohistochemistrical analysis of EGFP, which marks the AVE (brown labeling) on transverse sections of E6.5 embryos within the uterus. Insets show embryos at higher magnification. (E-F) In situ hybridization of T transcripts on cross sections of WT and Fgf8Δb/Δb embryos at E7.5 within the uterus. Red arrow represents AP axis, black double arrow marks the long uterine axis. Note the abnormal bulge of cells (arrowhead) at the primitive steak of Fgf8Δb/Δb embryos in F. (G) Distribution of the angles (α) between the long axes of the embryo and the uterus at E6.5 for groups II and I. Each dot represents one embryo. The average angles (horizontal bar) and standard deviations (vertical bars) are indicated. (H) Distribution of the angles (α) between the AP axes and the uterus at E7.5 for control and Fgf8Δb/Δb embryos relating to the ration of AP/LR dimension. The green and red dashed lines indicate the average angle for control and Fgf8Δb/Δb embryos, respectively.
We next examined cross sections of E7.5 embryos within the uterus from the intercross of Fgf8+/Δb parents. At E7.5, Fgf8Δb/Δb embryos can be readily identified by the abnormal histology, while the orientation of the AP axis can be identified by the position of the primitive streak at the posterior end of the embryo based on histology and expression of T. In agreement with our analysis of Fgf8Δb/Δb embryos at E6.5, the AP axis of Fgf8Δb/Δb embryos tends to be parallel with the long uterine axis, whereas the AP axis of WT embryos is largely perpendicular to the long uterine axis (Fig. 7E-F and H). The average angle between the AP axis and the long uterine axis of control embryos (70.88±19.16, n=53) is significantly different from that of Fgf8Δb/Δb embryos (25.40±24.24, n=21, p=1.2×10-8). Taken together, these observations demonstrate that the loss of Fgf8b results in an almost 90 degree rotation of the AP axis relative to the longitudinal axis of the uterus. Nevertheless, the long axis of Fgf8b-deficient embryos is correctly in register with the uterine axis, suggesting that positioning of the embryo within the uterine lumen is dependent on the shape, rather than the AP axis of the embryo.
Discussion
Fgf8b is the major splice variant of the Fgf8 gene, and Fgf8b proteins have more potent activity than Fgf8a. In this study, by knocking out Fgf8b in the mouse embryo, we have uncovered distinct requirements for Fgf8b before and during gastrulation. We show that before the onset of gastrulation, Fgf8b is essential for Fgf8 activities in the induction of T and positioning the AP axis relative to the shape of the embryo and the uterine axis. Moreover, Fgf8b is required for the normal induction of Lefty2 in the emerging streak. During gastrulation, Fgf8a can partially compensate for the loss of Fgf8b. Our molecular and genetic studies suggest that a higher level of Fgf8a is required for promoting mesoderm migration than that for mesoderm specification.
Fgf8 is essential for embryo remodeling before gastrulation
Recent studies have demonstrated that the orientation of the AP axis shifts relative to the shape of the embryo between E5.75 to E6.5 due to morphogenetic remodeling of the epiblast (Mesnard et al., 2004; Perea-Gomez et al., 2004). Our studies have revealed a key function of Fgf8 in this morphogenetic process. In Fgf8Δb/Δb embryos at E6.5, the AP markers are expressed at the opposite ends of the short axis, rather than the long axis of the embryo. We have ruled out that loss of Fgf8b leads to a developmental arrest at the earlier stages, when the AP axis is parallel with the short axis. We further show that the AP axis is correctly aligned with the short axis of Fgf8Δb/Δb embryos before embryo remodeling occurs. Finally, we show that Fgf8Δb/Δb and Fgf8-/- embryos have the same defect in the orientation of the AP axis at E6.5 (Fig. 6A-F). Taken together, our results demonstrate that Fgf8b is responsible for Fgf8 signaling in mediating the morphogenetic remodeling of the embryo between E5.75 and E6.5 (see Fig. 6I).
The uterus influences positioning of pregastrular embryos according to the shape but not the AP axis of the embryo
It was discovered almost two decades ago that the AP axis of the mouse embryo tends to be perpendicular to the longitudinal uterine axis at E6.5 (Smith, 1985). This has led to the speculations that the uterus may influence formation of the AP axis, or that the positional cues for the prospective AP axis with reference to the uterus may be secured at the time of implantation (Rossant and Tam, 2004). Here, we show that in the absence of Fgf8b, the AP axis of the embryo becomes parallel, rather than perpendicular, to the longitudinal uterine axis at E6.5 and E7.5. This demonstrates that the uterus does not have an overriding role in orienting the AP axis, and argues against the hypothesis that the prospective AP positioning cues are fixed at the time of implantation.
What could be the mechanism for the defined relationship between the AP axis and the long uterine axis at the onset of gastrulation? Interestingly, the orientation of the long axis of the embryo with respect to the uterine axes is unaffected in the absence of Fgf8b (Fig. 7B). These findings have two important implications. One is that the uterus imposes positioning of the embryo according to its shape but not its AP axis. The second is that embryo remodeling is not necessary for the biased positioning of the embryo within the uterine lumen. We, therefore, propose that the predictable relationship between the AP axis and the uterine axes at E6.5 is a coincidental outcome of two independent and concurrent events. Between E5.75 and E6.5, the embryo undergoes remodeling leading to alignment of the AP axis with the long axis. We suggest that this morphogenetic process is driven by cues intrinsic to the embryo and is dependent on Fgf8 signaling. The fact that embryos cultured in vitro undergo similar shape changes is entirely consistent with our deduction (Mesnard et al., 2004; Perea-Gomez et al., 2004). Externally, interactions between the embryo and uterus lead to biased positioning of the embryo based on the shape of the embryo within the uterine lumen, probably due to physical constraints imposed on the developing embryo.
Does the abnormal alignment of the AP axis with the longitudinal uterine axis attribute to the abnormal development observed in Fgf8Δb/Δb embryos? We found that about half of the Fgf8Δb/Δb embryos (11/21, 52.4%) significantly extend along the AP axis at E7.5, with their AP:LR ratio being greater than 1 (Fig. 7H). However, the angle between the AP axis and the uterine axis does not correlate the AP:LR ratio, or the severity of the phenotype at the histological level (Fig. 7H and data not shown). Furthermore, a significant number of Fgf8Δb/Δb embryos are remarkably normal in the morphology at E8.5, although most mutants display an abnormal orientation with respect to the long uterine axis at E6.5. Our data, therefore, suggest that the alignment of the AP axis with the longitudinal uterine axis at the onset of mouse gastrulation is unlikely to play a critical role in subsequent development.
Fgf8b is involved in the initiation of T expression
Experiments in different model organisms have demonstrated that T and other T-box genes are the downstream target as well as the immediate mediators of Fgf signaling in mesoderm induction and patterning (Bottcher and Niehrs, 2005). Analysis of Fgf8 spliceforms in Xenopus demonstrates that Fgf8b, but not Fgf8a, has robust activity in inducing Xbra, the homologue of T (Fletcher et al., 2006). We show that in the absence of Fgf8b or Fgf8, T expression is lost in the extraembryonic ectoderm and greatly reduced in the proximal-posterior epiblast at E6.5. The lack of T expression in Fgf8Δb/Δb embryos is unlikely caused by a gross delay in development. The evidence for this assertion is that Fgf8Δb/Δb and WT embryos are comparable in size, and Nodal, Wnt3 and Fgf8 are normally induced in the posterior Fgf8Δb/Δb embryos at E6.5 (Supplementary Fig. S1 and data not shown). Our results thus demonstrate that there is an evolutionarily conserved requirement for Fgf8 signaling in the normal induction of T in the epiblast. Interestingly, T is eventually expressed in the primitive streak of Fgf8b-deficient embryos (Fig 5J), and in a reduced level in Fgf8-null embryos between E6.5 and E7.5 (Sun et al., 1999). These findings indicate that Fgf8 is not essential for T expression in the primitive streak of gastrulas.
Different levels of Fgf8a expression govern mesoderm specification and migration
Mesoderm specification and morphogenetic movement of mesodermal cells are two highly coordinated processes during vertebrate gastrulation. Distinct molecular pathways have been implicated in mesoderm specification and mesoderm migration (Carver et al., 2001; Nutt et al., 2001; Sivak et al., 2005; Zohn et al., 2006). Two Fgf inhibitors, Sprouty and Spred, which respectively inhibit MAPK and Ca2+/PLCδ signaling pathways, have been suggested to switch Fgf signal interpretation to coordinate mesoderm specification and migration in Xenopus embryos (Sivak et al., 2005). Are different Fgf signals involved in the differential control of specification and migration of the mesoderm? Our results show that in the absence of the Fgf8b, different levels of Fgf8a are required for promoting mesoderm specification and mesoderm migration. In Fgf8Δb-neo/Δb-neo embryos, the residual Fgf8a isoforms can compensate for the loss of Fgf8b in mesoderm specification, but not mesoderm migration (Figs 2, 4 and 5). Remarkably, the mesoderm migration defects of Fgf8Δb-neo/Δb-neo embryos are largely rescued by removing the neo cassette. We did not attempt to directly compare Fgf8a expression between Fgf8Δb-neo/Δb-neo and Fgf8Δb/Δb embryos due to complications arising from the variability of the mutant phenotype. However, we provide genetic and molecular evidence that the presence of the neo cassette impairs expression of Fgf8 (Fig. 1). The simplest interpretation of our results is that the Fgf8Δb/Δb embryo produces higher levels of Fgf8a than those in the Fgf8Δb-neo/Δb-neo embryo, thereby leading to normal gastrulation in the absence of Fgf8b. A critical yet unsolved question is how a higher level of Fgf8a can promote mesoderm migration. We show that Fgf4 is expressed in the primitive streak of Fgf8Δb/Δb embryos, but not in Fgf8Δb-neo/Δb-neo mutants at E7.5 (Fig. 4A). Clearly, the increased Fgf8a expression is sufficient for the induction of Fgf4, which may in turn activate a different signaling pathway controlling the mesoderm migration.
Supplementary Material
Supplementary Figure S1. Loss of Fgf8b alters the expression domains of Nodal, Cer1 and Fgf8 relative to the shape of the embryo. Expression of Nodal (A-B), Cer1 (C-D), and Fgf8 (E-F) in WT and Fgf8Δb/Δb embryos at E6.5. Images to the left show the lateral view (anterior to the right), images to the right show the distal view (C-D) or posterior view (A-B and E-F) of the same embryo. Asterisk marks the posterior pole of the epiblast.
Supplementary Figure S2. Dynamic changes of Fgf8 expression in pregastrula embryos. Expression of Fgf8 in embryos at E5.75 (A) an E6.25 (B). Arrowhead and arrow mark the AVE and posterior epiblast, respectively.
Acknowledgments
We are grateful to Drs. R. Behringer, A. Das, J. Rivera-Perez and W. Shawlot for discussion and critical reading of the manuscript. We thank Dr. Jianhui Guo for technical assistance, and the gene targeting and transgenic facility in UConn Health Center for the generation of germline chimeras. We thank Drs. A. Joyner and G. Martin for providing Fgf8 null mutants, and Drs. J. Rivera-Perez and T. Rodriguez for providing Hex-EGFP transgenic mice. We also thank Drs. R. Behringer, D. Chapman, A. Joyner, G. Martin, A. McMahon, V. Papaioannou, J. Rossant, W. Shawlot, and M. Wakamiya for in situ probes. This work was supported by a grant from the NIH (HD050474) to J. Y. L.
References
- Ang SL, Wierda A, Wong D, Stevens KA, Cascio S, Rossant J, Zaret KS. The formation and maintenance of the definitive endoderm lineage in the mouse: involvement of HNF3/forkhead proteins. Development. 1993;119:1301–15. doi: 10.1242/dev.119.4.1301. [DOI] [PubMed] [Google Scholar]
- Bottcher RT, Niehrs C. Fibroblast growth factor signaling during early vertebrate development. Endocr Rev. 2005;26:63–77. doi: 10.1210/er.2003-0040. [DOI] [PubMed] [Google Scholar]
- Boulet AM, Moon AM, Arenkiel BR, Capecchi MR. The roles of Fgf4 and Fgf8 in limb bud initiation and outgrowth. Dev Biol. 2004;273:361–72. doi: 10.1016/j.ydbio.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Carver EA, Jiang R, Lan Y, Oram KF, Gridley T. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol Cell Biol. 2001;21:8184–8. doi: 10.1128/MCB.21.23.8184-8188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman DL, Agulnik I, Hancock S, Silver LM, Papaioannou VE. Tbx6, a mouse T-Box gene implicated in paraxial mesoderm formation at gastrulation. Dev Biol. 1996;180:534–42. doi: 10.1006/dbio.1996.0326. [DOI] [PubMed] [Google Scholar]
- Ciruna B, Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev Cell. 2001;1:37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- Ciruna BG, Schwartz L, Harpal K, Yamaguchi TP, Rossant J. Chimeric analysis of fibroblast growth factor receptor-1 (Fgfr1) function: a role for FGFR1 in morphogenetic movement through the primitive streak. Development. 1997;124:2829–41. doi: 10.1242/dev.124.14.2829. [DOI] [PubMed] [Google Scholar]
- Crossley PH, Martin GR. The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development. 1995;121:439–51. doi: 10.1242/dev.121.2.439. [DOI] [PubMed] [Google Scholar]
- Deng C, Bedford M, Li C, Xu X, Yang X, Dunmore J, Leder P. Fibroblast growth factor receptor-1 (FGFR-1) is essential for normal neural tube and limb development. Dev Biol. 1997;185:42–54. doi: 10.1006/dbio.1997.8553. [DOI] [PubMed] [Google Scholar]
- Downs KM, Davies T. Staging of gastrulating mouse embryos by morphological landmarks in the dissecting microscope. Development. 1993;118:1255–66. doi: 10.1242/dev.118.4.1255. [DOI] [PubMed] [Google Scholar]
- Dush MK, Martin GR. Analysis of mouse Evx genes: Evx-1 displays graded expression in the primitive streak. Dev Biol. 1992;151:273–87. doi: 10.1016/0012-1606(92)90232-6. [DOI] [PubMed] [Google Scholar]
- Fletcher RB, Baker JC, Harland RM. FGF8 spliceforms mediate early mesoderm and posterior neural tissue formation in Xenopus. Development. 2006;133:1703–14. doi: 10.1242/dev.02342. [DOI] [PubMed] [Google Scholar]
- Garcia-Garcia MJ, Anderson KV. Essential role of glycosaminoglycans in Fgf signaling during mouse gastrulation. Cell. 2003;114:727–37. doi: 10.1016/s0092-8674(03)00715-3. [DOI] [PubMed] [Google Scholar]
- Gemel J, Gorry M, Ehrlich GD, MacArthur CA. Structure and sequence of human FGF8. Genomics. 1996;35:253–7. doi: 10.1006/geno.1996.0349. [DOI] [PubMed] [Google Scholar]
- Lee SM, Danielian PS, Fritzsch B, McMahon AP. Evidence that FGF8 signalling from the midbrain-hindbrain junction regulates growth and polarity in the developing midbrain. Development. 1997;124:959–69. doi: 10.1242/dev.124.5.959. [DOI] [PubMed] [Google Scholar]
- Li JY, Joyner AL. Otx2 and Gbx2 are required for refinement and not induction of mid-hindbrain gene expression. Development. 2001;128:4979–91. doi: 10.1242/dev.128.24.4979. [DOI] [PubMed] [Google Scholar]
- Li JY, Lao Z, Joyner AL. Changing requirements for Gbx2 in development of the cerebellum and maintenance of the mid/hindbrain organizer. Neuron. 2002;36:31–43. doi: 10.1016/s0896-6273(02)00935-2. [DOI] [PubMed] [Google Scholar]
- Liu A, Li JY, Bromleigh C, Lao Z, Niswander LA, Joyner AL. FGF17b and FGF18 have different midbrain regulatory properties from FGF8b or activated FGF receptors. Development. 2003;130:6175–85. doi: 10.1242/dev.00845. [DOI] [PubMed] [Google Scholar]
- Liu A, Losos K, Joyner AL. FGF8 can activate Gbx2 and transform regions of the rostral mouse brain into a hindbrain fate. Development. 1999;126:4827–38. doi: 10.1242/dev.126.21.4827. [DOI] [PubMed] [Google Scholar]
- MacArthur CA, Lawshe A, Xu J, Santos-Ocampo S, Heikinheimo M, Chellaiah AT, Ornitz DM. FGF-8 isoforms activate receptor splice forms that are expressed in mesenchymal regions of mouse development. Development. 1995;121:3603–13. doi: 10.1242/dev.121.11.3603. [DOI] [PubMed] [Google Scholar]
- Mesnard D, Filipe M, Belo JA, Zernicka-Goetz M. The anterior-posterior axis emerges respecting the morphology of the mouse embryo that changes and aligns with the uterus before gastrulation. Curr Biol. 2004;14:184–96. doi: 10.1016/j.cub.2004.01.026. [DOI] [PubMed] [Google Scholar]
- Meyers EN, Lewandoski M, Martin GR. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat Genet. 1998;18:136–41. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- Minowada G, Jarvis LA, Chi CL, Neubuser A, Sun X, Hacohen N, Krasnow MA, Martin GR. Vertebrate Sprouty genes are induced by FGF signaling and can cause chondrodysplasia when overexpressed. Development. 1999;126:4465–75. doi: 10.1242/dev.126.20.4465. [DOI] [PubMed] [Google Scholar]
- Niswander L, Martin GR. Fgf-4 expression during gastrulation, myogenesis, limb and tooth development in the mouse. Development. 1992;114:755–68. doi: 10.1242/dev.114.3.755. [DOI] [PubMed] [Google Scholar]
- Nutt SL, Dingwell KS, Holt CE, Amaya E. Xenopus Sprouty2 inhibits FGF-mediated gastrulation movements but does not affect mesoderm induction and patterning. Genes Dev. 2001;15:1152–66. doi: 10.1101/gad.191301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen SK, Li JY, Bromleigh C, Eliseenkova AV, Ibrahimi OA, Lao Z, Zhang F, Linhardt RJ, Joyner AL, Mohammadi M. Structural basis by which alternative splicing modulates the organizer activity of FGF8 in the brain. Genes Dev. 2006;20:185–198. doi: 10.1101/gad.1365406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea-Gomez A, Camus A, Moreau A, Grieve K, Moneron G, Dubois A, Cibert C, Collignon J. Initiation of gastrulation in the mouse embryo is preceded by an apparent shift in the orientation of the anterior-posterior axis. Curr Biol. 2004;14:197–207. doi: 10.1016/j.cub.2004.01.030. [DOI] [PubMed] [Google Scholar]
- Rivera-Perez JA, Mager J, Magnuson T. Dynamic morphogenetic events characterize the mouse visceral endoderm. Dev Biol. 2003;261:470–87. doi: 10.1016/s0012-1606(03)00302-6. [DOI] [PubMed] [Google Scholar]
- Rivera-Perez JA, Magnuson T. Primitive streak formation in mice is preceded by localized activation of Brachyury and Wnt3. Dev Biol. 2005;288:363–71. doi: 10.1016/j.ydbio.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Rodriguez TA, Casey ES, Harland RM, Smith JC, Beddington RS. Distinct enhancer elements control Hex expression during gastrulation and early organogenesis. Dev Biol. 2001;234:304–16. doi: 10.1006/dbio.2001.0265. [DOI] [PubMed] [Google Scholar]
- Rossant J, Tam PP. Emerging asymmetry and embryonic patterning in early mouse development. Dev Cell. 2004;7:155–64. doi: 10.1016/j.devcel.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Hogan BL. Differential expression of multiple fork head related genes during gastrulation and axial pattern formation in the mouse embryo. Development. 1993;118:47–59. doi: 10.1242/dev.118.1.47. [DOI] [PubMed] [Google Scholar]
- Sato T, Araki I, Nakamura H. Inductive signal and tissue responsiveness defining the tectum and the cerebellum. Development. 2001;128:2461–9. doi: 10.1242/dev.128.13.2461. [DOI] [PubMed] [Google Scholar]
- Sivak JM, Petersen LF, Amaya E. FGF signal interpretation is directed by Sprouty and Spred proteins during mesoderm formation. Dev Cell. 2005;8:689–701. doi: 10.1016/j.devcel.2005.02.011. [DOI] [PubMed] [Google Scholar]
- Smith LJ. Embryonic axis orientation in the mouse and its correlation with blastocyst relationships to the uterus. II. Relationships from 4 1/4 to 9 1/2 days. J Embryol Exp Morphol. 1985;89:15–35. [PubMed] [Google Scholar]
- Sun X, Mariani FV, Martin GR. Functions of FGF signalling from the apical ectodermal ridge in limb development. Nature. 2002;418:501–8. doi: 10.1038/nature00902. [DOI] [PubMed] [Google Scholar]
- Sun X, Meyers EN, Lewandoski M, Martin GR. Targeted disruption of Fgf8 causes failure of cell migration in the gastrulating mouse embryo. Genes Dev. 1999;13:1834–46. doi: 10.1101/gad.13.14.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S, Stark KL, Shea MJ, Vassileva G, McMahon JA, McMahon AP. Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes Dev. 1994;8:174–89. doi: 10.1101/gad.8.2.174. [DOI] [PubMed] [Google Scholar]
- Tam PP, Behringer RR. Mouse gastrulation: the formation of a mammalian body plan. Mech Dev. 1997;68:3–25. doi: 10.1016/s0925-4773(97)00123-8. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG, Bhatt S, Herrmann BG. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature. 1990;343:657–9. doi: 10.1038/343657a0. [DOI] [PubMed] [Google Scholar]
- Yamaguchi TP, Harpal K, Henkemeyer M, Rossant J. fgfr-1 is required for embryonic growth and mesodermal patterning during mouse gastrulation. Genes Dev. 1994;8:3032–44. doi: 10.1101/gad.8.24.3032. [DOI] [PubMed] [Google Scholar]
- Zohn IE, Li Y, Skolnik EY, Anderson KV, Han J, Niswander L. p38 and a p38-interacting protein are critical for downregulation of E-cadherin during mouse gastrulation. Cell. 2006;125:957–69. doi: 10.1016/j.cell.2006.03.048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Loss of Fgf8b alters the expression domains of Nodal, Cer1 and Fgf8 relative to the shape of the embryo. Expression of Nodal (A-B), Cer1 (C-D), and Fgf8 (E-F) in WT and Fgf8Δb/Δb embryos at E6.5. Images to the left show the lateral view (anterior to the right), images to the right show the distal view (C-D) or posterior view (A-B and E-F) of the same embryo. Asterisk marks the posterior pole of the epiblast.
Supplementary Figure S2. Dynamic changes of Fgf8 expression in pregastrula embryos. Expression of Fgf8 in embryos at E5.75 (A) an E6.25 (B). Arrowhead and arrow mark the AVE and posterior epiblast, respectively.


