Abstract
The evolutionarily conserved kelch-repeat protein muskelin was identified as an intracellular mediator of cell spreading. We discovered that its morphological activity is controlled by association with RanBP9/RanBPM, a protein involved in transmembrane signaling and a conserved intracellular protein complex. By subcellular fractionation, endogenous muskelin is present in both the nucleus and the cytosol. Muskelin subcellular localization is coregulated by its C terminus, which provides a cytoplasmic restraint and also controls the interaction of muskelin with RanBP9, and its atypical lissencephaly-1 homology motif, which has a nuclear localization activity which is regulated by the status of the C terminus. Transient or stable short interfering RNA–based knockdown of muskelin resulted in protrusive cell morphologies with enlarged cell perimeters. Morphology was specifically restored by complementary DNAs encoding forms of muskelin with full activity of the C terminus for cytoplasmic localization and RanBP9 binding. Knockdown of RanBP9 resulted in equivalent morphological alterations. These novel findings identify a role for muskelin–RanBP9 complex in pathways that integrate cell morphology regulation and nucleocytoplasmic communication.
Introduction
In eukaryotic cells, communication between the cytoplasm and nucleus is of central biological importance for the integration of cytoplasmic signaling, cytoskeletal organization, and the dynamic regulation of nuclear gene expression (Schwoebel and Moore, 2000; Xu and Massague, 2004; Davis et al., 2007). For proteins too large to diffuse through the nuclear pore complex, the regulation of nuclear localization by karyopherins and the Ran gradient are extensively understood (Mattaj and Englmeier, 1998; Pemberton and Paschal, 2005). However, the functional significance and molecular mechanisms by which proteins that lack classical nuclear localization sequences are trafficked remain poorly studied. The scale of this gap in knowledge is illustrated by the finding that 43% of nuclear-located proteins of Saccharomyces cerevisiae do not have classical nuclear import motifs (Lange et al., 2007). Furthermore, this figure does not include cytoplasmic-located proteins that enter the nucleus in response to specific cues. In this paper, we have identified a novel process involving the cytoplasmic protein muskelin that connects the regulation of cell morphology with nucleocytoplasmic communication.
Muskelin is an intracellular multidomain protein that was identified in an expression cloning screen for proteins functionally involved in cell spreading responses to the extracellular glycoprotein thrombospondin-1 (TSP-1; Adams et al., 1998). Muskelin is a member of the kelch-repeat superfamily, which includes numerous proteins with roles in actin binding or the regulation of ubiquitin-mediated protein degradation (Bork and Doolittle, 1994; Adams et al., 2000; Kobayashi and Yamamoto, 2005). The muskelin transcript is expressed throughout life in multiple tissues, including skeletal and smooth muscle, the central nervous system, and lung, and is up-regulated in several pathological conditions, including periodontitic neutrophils, ischemic vascular smooth muscle cells, and hyperosmotic intervertebral discs (Adams et al., 1998; Adams and Zhang, 1999; Kubota et al., 2001; Dhodda et al., 2004; Boyd et al., 2005; Ledee et al., 2005; Prag et al., 2007; Tagnaouti et al., 2007). The domain architecture of muskelin includes an amino-terminal discoidin-like domain (DD), a central α-helical region with lissencephaly-1 (LIS1) homology (LisH) and C-terminal to LisH (CTLH) motifs, six kelch repeats that form a β propeller structure, and a unique carboxyl-terminal region (Adams et al., 1998; Adams, 2002; Prag et al., 2004; Li et al., 2004).
Muskelin is unique within the kelch-repeat family: in addition to its unique domain architecture it has a unique phylogenetic distribution, being conserved in the animal, fungal, and protozoal kingdoms (Prag and Adams, 2003; Prag et al., 2004). In contrast, TSPs are restricted to the animal kingdom. Thus, it is likely that muskelin has additional TSP-independent cellular roles. Although cytoplasmically located and linked functionally to cytoskeletal organization in several cell types, muskelin does not specifically colocalize with F-actin or microtubules and is not a direct binding partner for monomeric or polymerized actin or tubulin (Adams et al., 1998; Prag et al., 2004). These findings motivated us to establish how muskelin participates in both TSP-dependent and -independent pathways.
Muskelin binding proteins reported from yeast two-hybrid screens include EP3α prostaglandin receptor, RanBP9/RanBPM, and the p39 activator of cyclin-dependent kinase 5 (Hasegawa et al., 2000; Umeda et al., 2003; Ledee et al., 2005). However, the tissue-restricted expression patterns of EP3α receptor and p39 are not congruent with a general significance for muskelin function in the many tissues where it is expressed. Additional complexity has been suggested by reports that muskelin and RanBP9 are components of a protein complex of unknown function that includes several nucleocytoplasmic proteins (Umeda et al., 2003; Kobayashi et al., 2007). In this paper, we establish that RanBP9 binding is needed for activity of muskelin in cell spreading and is integrated with mechanisms that regulate the distribution of muskelin between cytoplasm and nucleus. These data advance concepts of mechanisms of regulatory communication between the cytoplasm and the nucleus.
Results
Endogenous muskelin includes a nuclear pool
Muskelin is present in cytoplasm, yet a possible nuclear component was suggested by studies of GFP-tagged muskelin, and RanBP9 is present in both cytoplasm and the nucleus (Prag et al., 2004; Denti et al., 2004). We undertook rigorous subcellular fractionation of C2C12 skeletal myoblasts that have high levels of muskelin (Adams et al., 1998). Full-length muskelin was present in cytosol and the high-salt nuclear extract that is enriched for nonstructural nuclear proteins such as Ran (Fig. 1 A). The generality of full-length nuclear muskelin was established by its detection in human aortic smooth muscle cells and human SW1222 carcinoma cells (unpublished data). Recently, nuclear muskelin was reported in hippocampal neurons by immunofluorescence (Tagnaouti et al., 2007).
Figure 1.
Muskelin is a nucleocytoplasmic protein. (A) Western blots of subcellular fractions of C2C12 cells, equalized for protein load, were probed with antibodies to muskelin and also with GAPDH, Ran, and lamin B1 as quality control markers for the fractionation. Cy, cytosol; M, membranes; NE, nuclear extract; NP, nuclear pellet. Results are representative of three experiments. (B and C) The panel of muskelin domain proteins. (B) Schematic of muskelin domain deletions. (C) Representative expression levels of GFP-tagged muskelin domain deletion proteins in COS-7 cells. An overexposure to reveal the poorly expressed GFP-MKDDLH is shown on the right. All molecular mass markers are shown in kilodaltons.
Muskelin localization in cells is balanced by distinct domains
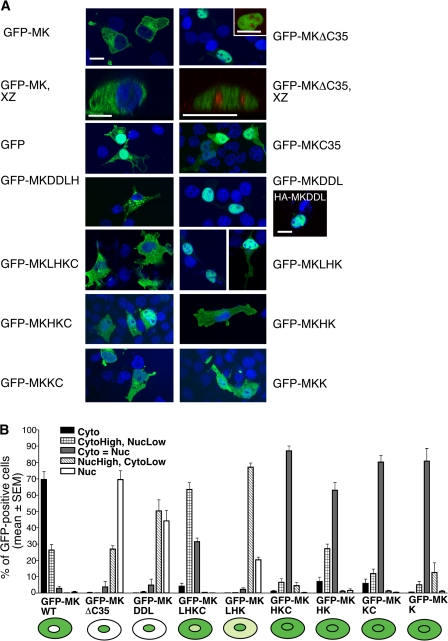
Toward defining the mechanisms underlying the multiple subcellular localizations of muskelin, we analyzed how muskelin domains contribute to its distribution. A panel of GFP-tagged mouse muskelin domain deletion mutants were developed according to the domain boundaries assigned from bioinformatic analyses (Fig. 1 B; Prag et al., 2004). With the exception of GFP-MKDDLH, the proteins were expressed at similar levels (Fig. 1 C). The subcellular localization of these proteins was examined in COS-7 epithelial cells (muskelin negative), C2C12 skeletal myoblastic cells, and mouse embryo fibroblasts (both muskelin positive), with identical results for all cells. Only results from COS-7 cells are depicted in figures.
GFP-MK was cytoplasmic in comparison to GFP alone and had the same distribution as endogenous muskelin (Fig. 2 A, xy and xz images of GFP-MK; Adams et al., 1998). We reported previously that GFP-MKDD (45.5 kD) is below the size threshold for diffusion through nuclear pores (Prag et al., 2004). The C-terminal domain alone, GFP-MKC35 (31.2 kD), is also below this threshold and also localized in the nucleus and cytoplasm indistinguishably from GFP (Fig. 2 A). Thus, neither the DD nor the C35 region contains active nuclear localization or export motifs. However, deletion of the C-terminal 35 aa (GFP-MKΔC35, predicted molecular mass 108.3 kD) resulted in a predominantly nuclear localization in 70% of the transfected cells. GFP-MKΔC35 was present in the nucleoplasm with exclusion from nucleoli, as established by costaining cells for the nucleolar component fibrillarin and examining confocal images in the xy and xz planes (Fig. 2 A). Because this large protein is above the molecular mass limit for diffusion through the nuclear pore complex, this result suggested the possible existence of nuclear localization or trafficking motifs within MKΔC35.
Figure 2.
Muskelin domain deletion proteins have distinct subcellular localizations. (A) Localization of GFP-MK and the panel of muskelin domain deletions, as shown in Fig. 1 B, visualized in fixed COS-7 cells by laser scanning confocal microscopy. Merged images show GFP proteins (green) and DAPI-stained nuclei (blue). For HA-DDL, anti-HA staining is green. Images are in the xy plane unless otherwise indicated. Exclusion of nuclear-located MKΔC35 from nucleoli is demonstrated in the inset and xz panels as merged images of GFP-MKΔC35 (green) and anti-fibrillarin (red) staining. Bars, 10 μm. (B) Quantification of the subcellular localizations of GFP-tagged muskelin domain deletion proteins by a five-point scoring scheme. The major localization of each protein is also shown schematically. All localizations were scored blind by two independent observers. Each column represents the mean from three to five independent experiments. Error bars indicate SEM. At least 1,000 cells were scored for each construct.
A five-point scoring scheme was used to quantify the localization of the different domain deletion proteins (Fig. 2 B). GFP-MKDD and GFP-MKC35 were excluded from this scoring because of their diffusion properties. GFP-MKDDLH (57.3 kD) was excluded because of its poor expression (Fig. 2 B). In the few cells where MKDDLH was detectable, it formed large cytoplasmic puncta (Fig. 2 A). Analysis of the other domain proteins revealed that GFP-MKDDL, although only 4 kD larger than GFP-MKDD, localized strongly to the nucleus with exclusion from nucleoli (Fig. 2 and not depicted). GFP-MKLHKC (93 kD) was mainly cytoplasmic, whereas GFP-MKLHK (89.4 kD) had pronounced nuclear localization (compare micrographs in Fig. 2 A). Other domain combinations from the C-terminal region of muskelin were the following: GFP-MKHKC (88.3 kD); GFP-MKHK (84.9 kD); GFP-MKKC (83.9 kD), and the kelch-repeats alone, GFP-MKK (78.8 kD), all distributed uniformly between the nucleus and the cytoplasm (Fig. 2). Additional experiments with influenza HA epitope-tagged MKDDL (HA-MKDDL; 24.7 kD) confirmed that the strong nuclear localization of MK-DDL was independent of GFP (Fig. 2 A).
These results revealed the following unexpected novel attributes of muskelin: First, its cytoplasmic localization depends on a cytoplasmic restraint, apparently provided by the C35 region; second, this restraint is only active in proteins that also contain the LisH motif (compare the major changes in distribution between WT and ΔC35 or LHKC and LHK, with the very similar distributions of HKC and HK or KC and K; Fig. 2); third, in the absence of C35 and the presence of the LisH motif, the major localization is in the nucleus; and fourth, any combination of the HKC domains partitions between nucleus and cytoplasm.
Muskelin does not undergo chromosome maintenance region 1 (Crm1)–mediated nuclear export
The propensity of GFP-MKΔC35 to localize to the nucleus could theoretically result either from deletion of a nuclear export sequence (NES) located in C35 or the dysregulated activity of a NES or NLS elsewhere in the protein. Because GFP-MKC35 had the same distribution as GFP (Fig. 2 A), our experiments did not support the presence of an NES in C35. An active NES would cause GFP to accumulate in the cytoplasm (Itahana et al., 2006). To examine whether a Crm1-dependent NES is present elsewhere within muskelin, GFP-MK–expressing cells were treated with leptomycin B to inhibit Crm1-mediated nuclear export (Fukuda et al., 1997; Wolff et al., 1997). After treatments from 30 min to 18 h, GFP-MK did not accumulate in the nucleus (Fig. 3, shown for 18-h treatment). Under the same conditions, cyclin B, which undergoes leptomycin B–sensitive nuclear export (Yang et al., 1998), became concentrated in the nucleus (Fig. 3 B). Thus, Crm1-mediated nuclear export of muskelin was not apparent.
Figure 3.
Muskelin does not undergo Crm1-mediated nuclear export. (A) Localization of GFP-MK in COS-7 cells treated with solvent (Con) or 10 μg/ml leptomycin B for 18 h. Bar, 10 μm. (B) Quantification of the cellular localizations of GFP-MK or cyclin B1 in cells treated with solvent or 10 μg/ml leptomycin B for 18 h. Only the distribution of cyclin B1 was significantly altered (*, P < 0.001). Each column represents the mean from three independent experiments. Error bars indicate SEM. At least 100 cells were scored in each experiment.
The LisH motif of muskelin has nuclear localization activity
From all the data in the previous sections, we hypothesized that the LisH motif of muskelin (LisHMK) has a major role in regulating the nuclear localization of muskelin. The LisH motif was identified originally as a short α-helical region at the amino terminus of LIS1 that is present in many other proteins (Emes and Ponting, 2001). In LIS1, the motif contributes to homodimerization (Cahana et al., 2001; Kim et al., 2004). Although all muskelins contain a central region with predicted α-helical secondary structure, not all contain motifs identified by the simple modular architecture research tool (SMART) database as LisH motifs (Prag et al., 2004). Thus, the relationship with LIS1 might not be the optimal way to consider the significance of this region of muskelin. We made an unbiased evaluation of the LisHMK sequence by the Sequence Logo method, a graphical method that displays the information content of a set of aligned sequences more comprehensively than a standard consensus sequence (Shaner et al., 2003). The Sequence Logo of LisH from 12 muskelins from chordates, insects, and fungi revealed strong conservation of multiple residues including arginines at positions 3 and 9, clustered basic residues at positions 13–16, and certain flanking acidic and neutral residues (Fig. 4 A; top). These conserved charged residues distinguish LisHMK from the general Sequence Logo for a LisH motif from these eukaryotes (Fig. 4 A, bottom).
Figure 4.
The LisH motif of muskelin has nuclear localization activity. (A) Sequence Logos for the consensus LisH motif of muskelin (LisHMK), derived by alignment of LisH motifs from 12 muskelin sequences from vertebrates, insects, and fungi, and the general consensus for a LisH motif, derived by alignment of 120 LisH motifs from vertebrates, insects, and fungi in the SMART database. Amino acids are shown in single letter code. The total height at each position is a measure of the strength of conservation at that position. Letter heights display the relative conservation of individual amino acids at that position. (B) Vinculin reporter constructs prepared without or with C-terminal fusion of GFP or LisHMK were expressed in COS-7 cells. Immunoblots of 5 and 20 μg of each whole cell extract demonstrate equivalent expression of vinculin, vinculin-LisHMK, and vinculin-LisHAA and higher expression of vinculin-GFP. Arrows indicate the ectopically expressed vinculins and asterisks indicate the endogenous vinculin. (C) LisHMK mediates nuclear localization of vinculin. Confocal xy or xz images of anti-vinculin–stained COS-7 cells expressing the indicated constructs. Bars, 10 μm. (D) Quantification of subcellular localizations of vinculin reporter proteins. Each column represents the mean from three independent experiments. Error bars indicate SEM. At least 500 cells were scored for each construct. (E) Immunoblot demonstrates equivalent expression of GFP-MK and GFP-MKΔC35, without or with point mutation of LisHMK, in COS-7 cells. Molecular mass markers are shown in kilodaltons. (F) Merged confocal images of representative localizations of the indicated GFP-tagged proteins (green) and DAPI-stained nuclei (blue). Bar, 10 μm. (G) Quantification of subcellular localizations. Each column represents the mean from three to five independent experiments. Error bars indicate SEM. At least 500 cells were scored for each construct.
The LisHMK consensus is distinct from a classical NLS (Mattaj and Englmeier, 1998). To test whether LisHMK confers nuclear import on a heterologous protein, we engineered a chimera of LisHMK with vinculin, a 1,068-aa 117-kD cytoplasmic protein that associates with F-actin and focal adhesions (Zamir and Geiger, 2001). Chimerization of vinculin with GFP (adding 265 aa, resulting in a predicted molecular mass of 143.9 kD; Fig. 4 B) did not alter the normal localization of vinculin in cytoplasm, membrane ruffles, and adhesion sites (Fig. 4, C and D [quantification]). In contrast, the addition of LisHMK (37 aa, resulting in a predicted molecular mass of 121.5 kD) did not change the expression level of the protein (Fig. 4 B) yet caused dramatic relocalization to the nucleus (Fig. 4, C and D [quantification]). To establish the role of the LisHMK basic cluster in this relocalization, we prepared a double alanine point mutation of positions 13 and 14 (Fig. 4 A, Sequence Logo). Vinculin-LisHAA had a majority cytoplasmic localization (Fig. 4, C and D [quantification]). These experiments establish that LisHMK can independently import a heterologous protein into the nucleus and that an intact basic cluster is necessary for this activity.
To establish the functional role of LisHMK within muskelin itself, we generated the same alanine point mutations in the context of GFP-MKΔC35 or GFP-MK. Both proteins were expressed at the same level as their control counterparts (Fig. 4 E) yet, in comparison to the mainly nuclear location of GFP-MKΔC35, GFP-MKΔC35/LisHAA had a dramatically altered uniform distribution between the nucleus and cytoplasm (Fig. 4, F and G [quantification]). This result was consistent with decreased nuclear entry of GFP-MKΔC35/LisHAA, as seen with the vinculin-LisHAA chimera. Counterintuitively, the point mutations in the context of full-length muskelin (GFP-MK/LisHAA) markedly increased the percentage of cells with equivalent cytoplasmic and nuclear muskelin compared with GFP-MK (Fig. 4, F and G [quantification]). This unexpected result pointed to the existence of additional intramolecular determinants that regulate muskelin nuclear localization.
The C35 region regulates the nucleocytoplasmic distribution of muskelin and RanBP9 binding
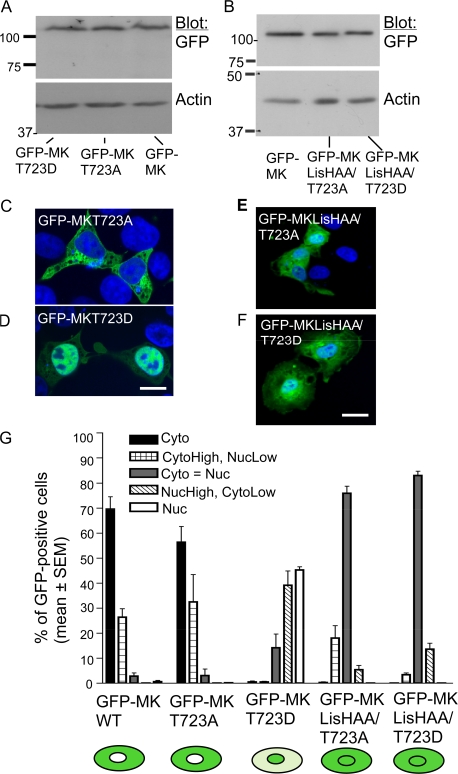
On the basis of our results and that C35 acts as a cytoplasmic restraint for full-length muskelin (Fig. 2), we hypothesized that LisHMK and C35 cocontrol the localization of full-length muskelin. To explore this concept, we analyzed the sequence of C35 for possible functional motifs. C35 from vertebrate muskelin contains a conserved consensus motif for phosphorylation by proline-directed serine/threonine kinases, T723PPK726 (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200801133/DC1; Brown et al., 1999). This is supported by the presence of a conserved consensus cyclin binding motif within the DD (Fig. S1; Brown et al., 1999). We addressed the putative functional significance of the TPPK motif through point mutations. GFP-MKT723A and GFP-MKT723D were expressed equivalently to GFP-MK (Fig. 5 A). Whereas GFP-MKT723A had the same cytoplasmic localization as GFP-MK, GFP-MKT723D was mostly nuclear (Fig. 5, C, D, and G). The redistribution of GFP-MKT723D was less than that of GFP-MKΔC35 (45% of GFP-MKT725D cells had nuclear muskelin compared with 70% of GFP-MKΔC35 cells; Fig. 2 B). Identical results were obtained in COS-7, C2C12, and A549 cells (Fig. 5, C and D; shown for COS-7 only). Proteins mutated in both the TPPK motif and LisHMK were also expressed at the same level as GFP-MK (Fig. 5 B). Both GFP-MKLisHAA/T723A and GFP-MKLisHAA/T723D were predominantly uniformly distributed between the cytoplasm and nucleus (Fig. 5, E–G). However, >10% of GFP-MKLisHAA/T723D–expressing cells had elevated nuclear muskelin unlike either GFP-MKLisHAA or GFP-MKLisHAA/T723A cells. 18% of GFP-MKLisHAA/T723A–expressing cells had elevated cytoplasmic muskelin (Fig. 5 G). Thus, whereas functional impairment of LisHMK strongly impacts the localization of GFP-MKΔC35, the LisH mutation counteracts but does not completely abolish the distinct localizations of the TPPK mutants.
Figure 5.
Role of the C-terminal TPPK motif in coregulating muskelin localization. (A and B) Immunoblots demonstrate equivalent expression of GFP-MK and the indicated point mutant proteins. Molecular mass markers are shown in kilodaltons. (C–F) Merged confocal images showing localization of GFP-tagged muskelin point mutants (green) in COS-7 cells costained with DAPI (blue). Bars, 10 μm. (G) Quantification of subcellular localizations. Each column represents the mean from two to three independent experiments. Error bars indicate SEM. At least 500 cells were scored for each construct. The localization of each protein is also shown schematically.
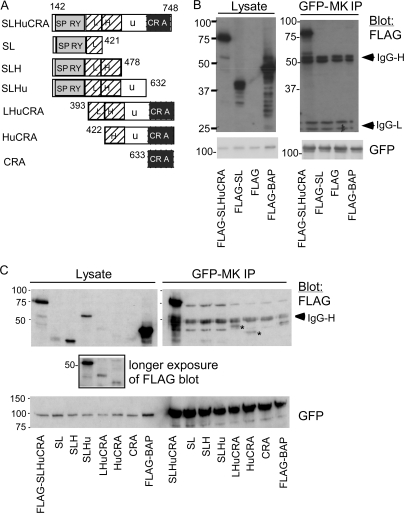
To identify a molecular basis for these activities, we performed a yeast two-hybrid screen of a human skeletal muscle cDNA library for muskelin binding proteins. Of several initial candidates, only one was confirmed for specific interaction with the muskelin bait under stringent conditions. This clone encoded aa 142–748 of RanBP9 (NP_005484). Muskelin has also been identified as a yeast two-hybrid binding partner of RanBP9 (Umeda et al., 2003). RanBP9 is a multidomain intracellular protein (Fig. 6 A) implicated as a scaffold of certain tyrosine kinase and adhesion receptors and nuclear signaling proteins (Wang et al., 2002; Rao et al., 2002; Denti et al., 2004; Cheng et al., 2005; Brunkhorst et al., 2005; Hafizi et al., 2005). Endogenous RanBP9 was principally located in cytoplasm and on membranes, with lower levels in the nuclear fractions (Fig. 6 B; Denti et al., 2004). We confirmed the specific physical association of endogenous RanBP9 with muskelin in mammalian cells by coimmunoprecipitation (Fig. 6 C). RanBP9 binding was mediated by the LHKC domains and not by the DD (Fig. 6 C). Further mapping within the LHKC domains demonstrated that the minimal fragment to mediate strong association of endogenous RanBP9 was GFP-MKHKC (Fig. 6 D). A lower level of binding occurred with the GPF-MKLHK protein that was equivalently immunoprecipitated (Fig. 6 D). Minor interaction was detected with the KC domains. There was no coimmunoprecipitation with HK, the kelch repeats, or C35 region alone. The status of C35 was important for association of RanBP9 with KC because no RanBP9 was detected in immunoprecipitates of an equivalently expressed mutant, GFP-MKKCT723D (Fig. 6 D).
Figure 6.
Role of the C35 region in regulating RanBP9 binding by muskelin. (A) Schematic diagram of RanBP9 domain architecture and the domains included in the yeast two-hybrid clone. (B) Localization of RanBP9 in subcellular fractions of C2C12 cells. Cy, cytosol; M, membranes; NE, nuclear extract; NP, nuclear pellet. (C) The domains sufficient for coimmunoprecipitation of endogenous RanBP9 with GFP-MK are included in the LHKC fragment. (D) Analysis of the minimal domain requirements within the LHKC domains for coimmunoprecipitation of endogenous RanBP9. (E and F) Coimmunoprecipitation of endogenous RanBP9 by GFP-MK is regulated by the status of muskelin C35 region. (F) RanBP9 signal intensities normalized relative to GFP-MK immunoprecipitation are given below the blot. All molecular mass markers are shown in kilodaltons. Results shown are representative of three experiments.
We investigated the role of muskelin C35 region in more detail, in the context of full-length muskelin. The association of RanBP9 with GFP-MKT723A was indistinguishable from that of GFP-MK, whereas association with GFP-MKT723D was reduced (Fig. 6 E). Deletion of the C35 region completely abolished binding (Fig. 6 F). GFP-MKLisHAA and GFP-MKLisHAA/T723A both associated strongly with RanBP9, whereas interaction with GFP-MKLisHAA/T723D was decreased (Fig. 6 F). Notably, these results demonstrated different RanBP9 binding capacities of muskelin mutants with similar majority localization in the cytoplasm (GFP-MKLisHAA/T723A vs. GFP-MKLisHAA/T723D), and also similar binding capacities by proteins with different distributions between cytoplasm and nucleus (GFP-MK vs. GFP-MKLisHAA). Thus, RanBP9 binding activity does not simply reflect cytoplasmic localization of muskelin but correlates precisely with the activity status of C35.
Multiple domains of RanBP9 are needed for efficient association with muskelin
We next developed a set of FLAG-tagged domain deletions of RanBP9[142–748] (Fig. 7 A) to map regions important for the association with muskelin. The SPRY (Sp1a and ryanodine receptor) domain of RanBP9 interacts with the cytoplasmic domains of several transmembrane receptors (Wang et al., 2002; Cheng et al., 2005; Hafizi et al., 2005); however, FLAG-RanBP9SL (containing the SPRY and LisH domains; Fig. 7 A) was not sufficient for interaction with muskelin (Fig. 7 B). Indeed, FLAG-RanBP9[142–748] was the only protein within the set to associate strongly with GFP-MK (Fig. 7 C). Interpretation of results with other domains was complicated by the low expression level of many of the domain deletions (Fig. 7 C and Fig. S2 [longer exposure], available at http://www.jcb.org/cgi/content/full/jcb.200801133/DC1), likely indicative of deficiencies in protein folding and/or stability. It was not possible to express a FLAG-RanBP9uCRA deletion protein (not depicted). The C-terminal fragments FLAG-RanBP9LHuCRA and FLAG-RabBP9HuCRA were very weakly expressed, yet specific coimmunoprecipitation with GFP-MK was detectable (Fig. 7 C). Overall, we conclude that multiple domains of RanBP9 are necessary for full muskelin-binding activity.
Figure 7.
Multiple RanBP9 domains are needed for association with muskelin. (A) Schematic diagram of the FLAG-tagged RanBP9 domains prepared for these experiments. SPRY, Sp1a and ryanodine receptor domain; L, LisH motif; H, CTLH motif; u, unique region; CRA, CT11/RanBP9 domain. (B) Specific coimmunoprecipitation of FLAG-RanBP9[142–748] with GFP-MK. (C) Analysis of RanBP9 domain requirements for coimmunoprecipitation with GFP-MK. The boxed inset is a longer exposure of three lysate lanes demonstrating the poorly expressed LHuCRA and HuCRA proteins. Asterisks indicate observed weak specific coimmunoprecipitations. All molecular mass markers are shown in kilodaltons. Results shown are representative of three experiments.
Morphological activity of muskelin depends on an active cytoplasmic restraint and RanBP9 binding
To understand how these newly defined attributes of muskelin and RanBP9 underpin the activity of muskelin in cell spreading and cytoskeletal organization, we prepared cells depleted of muskelin by two methods of siRNA-mediated transcriptional silencing. Pilot cyclohexamide treatment experiments indicated a protein half-life of less than 16 h (unpublished data). Effective silencing in A549 lung epithelial cells, which have strong expression of muskelin (Adams and Zhang, 1999) and RanBP9 (Fig. S3 C, available at http://www.jcb.org/cgi/content/full/jcb.200801133/DC1), was achieved by stable expression of short hairpin RNAs (shRNAs) targeting human muskelin. 62 clones were established with two different shRNAs, and 3 clones with the largest reductions in muskelin protein (50–65% reduction) representing both shRNAs (Fig. S3 A) were used for detailed analyses. Muskelin expression was silenced in C2C12 mouse skeletal myoblasts by an independent methodology: transfection with a chemical siRNA duplex targeting a distinct point on the mouse transcript from the shRNAs to human muskelin. In C2C12 cells, muskelin protein was depleted by 50–60% after 96 h compared with cells treated with control siRNA duplex (Fig. S3 B).
In agreement with previously reported effects of muskelin on adhesion, A549 muskelin-depleted clones were reduced in attachment to the TSP-1 C terminus (Fig. 8 A, shown for clones 2 and 3). The cells had altered spreading behavior on fibronectin (FN), with significantly enlarged cell perimeters caused by increased ruffling and protrusive edges (Fig. 8 B). These changes were muskelin-dependent because normal cell morphology was specifically restored by expression of GFP-MK (i.e., mouse MK which is not targeted by the shRNA) in multiple muskelin-depleted A549 clones (Fig. 8 B). The phenotype was confirmed in muskelin siRNA-treated C2C12 cells, which had similar specific increases in cell perimeter length and protrusions in FN-adherent cells (Fig. 8, C and D).
Figure 8.
The morphological activity of muskelin depends on cytoplasmic localization and RanBP9 binding. (A–D) siRNA-based depletion of muskelin leads to morphological alterations. (A) Reduced attachment of A549 cells with stable expression of muskelin-shRNA clone 2 (cl2) and clone 3 (cl3) cells to the TSP-1 C-terminal protein T35-7GCT. Adhesion assays were performed for 1 h under serum-free conditions as previously described (Kvansakul et al., 2004). (B and C) Cell perimeter measurements for control and muskelin-depleted A549 (B) or C2C12 (C) cells adherent on 50 nM FN for 1 h. (B) A549 lines were also compared after 48 h expression of GFP or GFP-MK. Only GFP-MK restored normal morphology. In each graph, each column represents the mean of three independent experiments. Error bars indicate SEM. *, P ≤ 0.0001. (D) Representative F-actin organization in control and muskelin-depleted C2C12 cells under the same experimental conditions as in C. Bar, 10 μm. (E) Activity of muskelin mutants in restoring the morphology of muskelin-depleted A549 clone 3 cells. GFP, GFP-MK, or the indicated mutants were expressed in clone 3 cells for 48 h, the cells adhered to 50 nM FN for 1 h, and cell perimeter lengths of GFP-positive cells were measured. Each column represents the mean of three independent experiments. Error bars indicate SEM. *, P ≤ 0.00001. At least 500 cells were scored per condition. (F) RanBP9 knockdown results in similar morphological changes in FN-adherent A549 cells. Where indicated, cells were treated with 0.75 μg/ml doxycycline for 36 h. Each column represents the mean from >50 cells. Error bars indicate SEM. *, P < 0.0001. Insets show representative merged cell images of F-actin (green), DAPI (blue), and, where induced, shRNA transcript expression (red; RFP). Bar, 10 μm.
To identify the molecular basis for these phenotypic effects, we tested the functional consequences of restoring muskelin-depleted A549 clone 3 cells with cDNAs encoding mutant (mouse) muskelins. GFP-MK/LisHAA and GFP-MKT723A had similar highly significant activities to GFP-MK in restoring normal cell morphology, whereas GFP-MKT723D and GFP-MKΔC35 were inactive (Fig. 8 E). Similarly, GFP-MKLisHAA/T723A was active, whereas GFP-MKLisHAA/T723D had reduced activity (Fig. 8 E). We also compared the activities of the N- and C-terminal regions. GFP-LHKC was active, whereas GFP-DDL was not. Identical results were obtained with A549 MK-depleted clone 2 cells (unpublished data). Thus, morphological activity depended on the status of C35 and not on LisHMK.
In view that the association of muskelin with RanBP9 also depends on C35 region, we examined whether depletion of RanBP9 resulted in similar phenotypic alterations. Because it did not prove possible to generate viable long-term RanBP9 knockdown cells (unpublished data), inducible transient knockdown of RanBP9 was used. In comparison to A549 control cells, or cells induced for an irrelevant shRNA, RanBP9-depleted A549 cells adherent on FN had enlarged cell perimeters, caused by protrusive morphologies, and altered F-actin distributions particularly at cell edges (Fig. 8 F). This phenotype matched that of muskelin-depleted A549 cells. We conclude that the morphological activity of muskelin depends on the cytoplasmic pool of muskelin and a physiological level of RanBP9 binding.
Discussion
These novel data advance concepts of the mechanisms by which regulation of cell morphology is integrated with nucleocytoplasmic signaling. The majority of muskelin appears cytoplasmic by immunofluorescence in multiple mammalian cells (Adams et al., 1998; Hasegawa et al., 2000; Prag et al., 2004; Ledee et al., 2005; Kobayashi et al., 2007), yet rigorous subcellular fractionation of endogenous muskelin revealed a nuclear pool. Analysis of the localizations of muskelin domain deletions defined that the distribution of the full-length protein depends on an intricate molecular balance, in which the extreme C terminus provides a cytoplasmic restraint and the LisH motif contains a nuclear localization activity that is largely cryptic under standard conditions. We identify that muskelin interacts with RanBP9 via its HKC domains and that the status of C35 is important in regulating this interaction. We establish that depletion of either muskelin or RanBP9 has an impact on cell morphology and that the morphological activity of muskelin depends on its C-terminal region, which mediates cytoplasmic localization and RanBP9 binding.
In other proteins with LisH motifs, which include both cytoplasmic- and nuclear-located proteins, the LisH motifs are typically uncharged (Emes and Ponting, 2001). Several have homodimerization activity that is important for protein function (Kim et al., 2004; Mikolajka et al., 2006). Interactions with cytoplasmic dynein or microtubules have been reported and the LisH motif of p220NPAT is required for activation of histone transcription (Cahana et al., 2001; Wei et al., 2003). A small number of proteins in mammalian genomes, including both muskelin and RanBP9, contain contiguous LisH and CTLH motifs. The functional significance of the combined motif is unclear. The presence of highly conserved charged residues within the muskelin LisH-like motif is a unique feature that we demonstrate for the first time here to have functional activity in regulating the nuclear localization of muskelin or a heterologous protein, vinculin. BLASTP searches of GenBank with the muskelin LisH-like motif identify only the LisH motifs from other muskelins. We suggest it is appropriate to redesignate the muskelin LisH-like motif as a nuclear localization motif.
Despite the potent activity of LisHMK in mediating nuclear localization of a heterologous protein, vinculin, under standard cell culture conditions only a small fraction of endogenous muskelin is nuclear located. In the vinculin chimera, the C-terminal–located LisHMK is very likely constitutively active. The experiments with muskelin mutants revealed that strong nuclear localization of muskelin depends on either deletion of the C terminus or inclusion of a phosphomimetic mutant within the C terminus. Thus, the major cytoplasmic localization of wild-type muskelin likely reflects the dominance of C35-dependent phosphoregulated binding interactions with cytoplasmic proteins over the intrinsic nuclear localization activity of LisHMK, with the net result that LisHMK activity appears cryptic. These counterbalancing activities remain functional in the LHKC deletion mutant. In addition, we previously identified that the amino-terminal DD and the C-terminal half, comprising the kelch-repeats and C35, bind in trans (Prag et al., 2004). Analogous to signal-activated cytoskeletal regulatory proteins (Pufall and Graves, 2002), molecular self-association might facilitate the autoinhibition of LisHMK by C35.
Muskelin exists in cells as multiple isoelectric variants, some of which result from phosphorylation by protein kinase C (Prag et al., 2007). Our data on the key role of the C35 region and the status of T723 for the subcellular localization of muskelin implicate phosphorylation by a proline-directed kinase as a physiological mechanism to relieve autoinhibition of LisHMK activity. Muskelin was reported to physically associate with the p39 activator of cdk5 (Ledee et al., 2005); however, we did not detect association of muskelin with p39, cdk5, or cyclins for cdk1 or cdk2 by coimmunoprecipitation (unpublished data). Instead, the presence of both phosphorylation and cyclin binding motifs in muskelin (Fig. S1) implicates a transient substrate interaction with active cdks. Further research will be needed to definitively identify the proline-directed kinases that mediate muskelin phosphorylation at residue T723. We speculate that dynamic phosphorylation and dephosphorylation might be linked to specific cell cycle stages and/or in response to specific environmental cues.
Another major conceptual outcome of this research is the identification of a novel functional role for the muskelin–RanBP9 complex. RanBP9 is an evolutionarily conserved nucleocytoplasmic protein implicated as a scaffold for receptors that network with the Erk1/2 pathway (Wang et al., 2002; Umeda et al., 2003; Cheng et al., 2005). An emerging consensus places RanBP9 as a multifunctional protein that interacts with the cytoplasmic domains of cell surface receptors including c-Met, integrin β subunit, and L1 adhesion protein. Interactions with nuclear proteins are also indicated (Rao et al., 2002; Wang et al., 2002; Denti et al., 2004; Brunkhorst et al., 2005; Cheng et al., 2005; Togashi et al., 2006; Kobayashi et al., 2007). Biochemically, RanBP9 is a component of a large protein complex of unknown function (Umeda et al., 2003; Kobayashi et al., 2007). In this complex, RanBP9 or its S. cerevisiae homologue, VID30p, associates with several proteins that are conserved from plants to animals. Current data suggest that the complex could be both cytoplasmic and nuclear (Pitre et al., 2006; Kobayashi et al., 2007). Muskelin is not encoded in plants or the S. cerevisiae genome (Prag et al., 2004), yet associates with the mammalian RanBP9 complex (Kobayashi et al., 2007). We propose that muskelin confers distinct functional capacities on the conserved complex in which RanBP9 participates.
Our data establish that the status of muskelin C terminus and, thereby, RanBP9 binding are essential for the morphological activity of muskelin. In general, the β propeller structure formed by repeated kelch motifs presents a large domain with multiple faces for binding interactions. There are precedents for intramolecular interactions with C-terminal sequences, and N-terminal sequences may also contribute to correct folding of the kelch-repeat β propeller (Prag and Adams, 2003; Li et al., 2004; Padmanabhan et al., 2006). For muskelin, C35 is not sufficient to bind RanBP9, yet its status profoundly regulates the physical association with RanBP9, which depends on the HKC domains. We propose that the status of C35 controls RanBP9 binding by the muskelin β propeller and that this is a central mechanism for regulating muskelin localization and, thereby, its morphological activity in cells. We have demonstrated that knockdown of either muskelin or RanBP9 leads to equivalent morphological alterations in which adherent cells adopt protrusive phenotypes. Receptor-bound RanBP9 might concentrate muskelin in proximity to submembranous sites of Erk signal activation and active morphological remodelling. For example, elevated RanBP9 levels alter cell migration behavior (Wang et al., 2002; Zou et al., 2003).
Our data also establish that nuclear-located forms of muskelin are impaired for morphological activity. Thus, physiological mechanisms to activate nuclear localization of muskelin can be seen as mechanisms that provide modulation of ECM-dependent cell adhesion and spreading. It is well established that conversion between a protrusive or contractile phenotype has a major impact on cell behavior (Cukierman et al., 2002). It will be of future interest to establish the functions of nuclear muskelin. The presence of muskelin in the salt-extractable nuclear fraction, but not the nuclear matrix, is suggestive of a regulatory rather than a structural role. One possibility is that nuclear muskelin functions via the RanBP9 complex. RanBP9 is also present in the salt-extractable nuclear fraction and is implicated in transcriptional regulation (Rao et al., 2002; Wang et al., 2002; Denti et al., 2004). In initial support, our coimmunoprecipitation data demonstrate that partially nuclear-located muskelin mutants associate efficiently with RanBP9. In conclusion, our data establish the muskelin–RanBP9 complex as a novel player in pathways of nucleocytoplasmic signaling and open new directions for understanding how nucleocytoplasmic communication and morphological processes are integrated by cells.
Materials and methods
Cell lines and materials
Green monkey kidney COS-7 cells and A549 human lung epithelial cells were grown in DME containing 10% FCS. C2C12 mouse skeletal myoblasts were grown in DME containing 20% FCS. Leptomycin B and phosphatase inhibitor cocktails 1 and 2 were obtained from Sigma-Aldrich. Rabbit polyclonal antibodies to muskelin were as previously described (Adams et al., 1998). In addition, a rabbit polyclonal antibody was raised against a MAP peptide corresponding to residues 37–51 of mouse muskelin according to standard procedures. Purified immunoglobulin was produced by passing the sera over protein G–Sepharose CL (GE Healthcare), with extensive washing and elution in 0.2 M glycine, pH 2.8. Mouse monoclonal antibodies to β-actin (clone AC-15), fibrillarin (clone 38F3), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were obtained from Abcam; to Ran (Clone 20) from BD Biosciences; to FLAG epitope (clone M2) and vinculin (clone VIN11.5) from Sigma-Aldrich; to GFP (clones 7.1 and 13.1 mixture) from Roche; and to HA tag (clone 262k) and cyclin B1 from Cell Signaling Technology. Goat anti–lamin B1 antibody was obtained from Santa Cruz Biotechnology, Inc. Goat anti-RanBP9 was obtained from Abcam. Mouse monoclonal to RanBP9 (Denti et al., 2004) was a gift from E. Bianchi (Institut Pasteur, Paris, France). Rhodamine-phalloidin was obtained from Sigma-Aldrich. TSP-1 C-terminal protein was prepared as previously described (Kvansakul et al., 2004). Human plasma FN was obtained from EMD.
Expression constructs
Mammalian expression plasmids encoding enhanced GFP-tagged mouse muskelin (GFP-MK); GFP-tagged DD (aa 1–159; GFP-MKDD), and GFP-tagged kelch repeats and C terminus (aa 243-735; GFP-MKKC) were as previously described (Prag et al., 2004). Additional deletion mutants (Fig. 1 B) were prepared in GFP-tagged form by PCR of the selected region from mouse muskelin cDNA with the forward oligonucleotide primer containing an EcoR1 site and the reverse primer containing a KpnI site, according to standard molecular biology procedures (Sambrook and Russel, 2001). The oligonucleotide primers used are listed in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200801133/DC1) and were synthesized by Sigma-Aldrich. Digested PCR products were ligated into the EcoRI and KpnI sites of pEGFP-C2 (Clontech Laboratories, Inc.) with QuickLigase (New England Biolabs, Inc.). HA-tagged MK-DDL was subcloned into pcDNA3.1. Gallus gallus vinculin cDNA (gift of Robert Kypta, Imperial College, London, England, UK) was subcloned into pcDNA3.1 V5 His (Invitrogen) by TOPO cloning, with introduction of a HpaI site at the 3′ end of the ORF. The LisH-encoding region of mouse muskelin (aa 172–204) was amplified from cDNA with PCR primers introducing a Hpa I site at the 5′ end and a XbaI site at the 3′ end, respectively, digested with HpaI and XbaI and ligated in frame at the 3′ end of the vinculin cDNA. cDNAs encoding domains of human RanBP9 were subcloned into p3xFLAG (Sigma-Aldrich), using the primer pairs listed in Table S1. Point mutations were prepared with QuikChange II site-directed mutagenesis kit (Stratagene) and GFP-MK or GFP-MKΔC35 as templates. All constructs were verified by DNA sequencing across the 5′ and 3′ junctions with vector-specific primers and by resequencing the complete cDNAs using internal primers. Automated DNA sequencing was performed by Cleveland Clinic Foundation Molecular Biotechnology and Genomics cores.
Bioinformatics analysis
Muskelin protein sequences were as previously identified (Adams et al., 1998; Adams and Zhang, 1999; Adams, 2002; Prag et al., 2004) or were identified by BLAST searches of GenBank and the database of expressed sequence tags with mouse MK as the query. Sequence Logo analysis was performed at WebLogo (http://weblogo.berkeley.edu; Shaner et al., 2003; Crooks et al., 2004). LisH motifs were collected from SMART (Schultz et al., 1998). Phosphorylation motifs were analyzed through Prosite (European Bioinformatics Institute) and Scansite (Massachussetts Institute of Technology).
Fluorescence microscopy
Transient transfection of COS-7 cells was performed as previously described (Prag et al., 2004). Cell-ECM adhesion of C2C12 or A549 cells was performed as previously described (Anilkumar et al., 2003; Kvansakul et al., 2004). All cells were fixed in 2% PFA. For immunofluorescent staining with antibodies to FLAG-tag, HA-tag, or vinculin, fixed cells were permeabilized in TBS containing 0.5% Triton X-100. For staining with antibody to fibrillarin, fixed cells were permeabilized for 1 min in ice-cold methanol. After incubation with primary antibody and appropriate FITC- or TRITC-conjugated secondary antibodies (MP Biomedicals or Sigma-Aldrich) all samples were prepared in DAPI mounting medium (Vector Laboratories). Epifluorescence microscopy was performed with an inverted microscope (DMIRE2; Leica) equipped with electronically controlled shutters, filter wheel, and focus. Images were captured under a 63× objective with a camera controller (C4742-95; Hamamatsu Photonics) run by Openlab software (version 3.1.3; Improvision). Cell perimeters were measured from calibrated images in Openlab. Confocal xy or xz images were taken at room temperature under a HCX PL APO 63× 1.4 NA oil immersion objective lens (zoom 1 to zoom 3) of a laser scanning confocal microscope (TCS-SP/SP-AOSB; Leica) using confocal image acquisition software version 2.5 (Leica). Fluorochromes used were DAPI, FITC, eGFP, TRITC, mRFP and Cy-5. The imaging medium was Vectashield with DAPI (Vector Laboratories).
Coimmunoprecipitation
106 COS-7 cells were transfected with 3 μg of the relevant pEGFP or p3XFLAG plasmids for 40 h and then lysed in 1% IGEPAL in TBS on ice. Lysates were precleared on protein G–agarose (Invitrogen) for 1 h and then incubated with 2 μg of GFP or FLAG antibody preimmobilized on protein G–agarose beads for 2 h at 4°C. Beads were extensively washed, boiled in SDS-PAGE buffer, and associated proteins analyzed by immunoblotting with appropriate antibodies.
Subcellular fractionation
Adherent cells in p90 dishes were washed twice in ice-cold PBS and swollen in 500 μl of ice-cold hypotonic buffer 1 (10 mM Tris-HCl, pH 7.4, containing 1 mM MgCl2, protease Complete cocktail [Roche], and phosphatase inhibitor cocktail 1 and 2) for 15 min. Cells were scraped from the dishes, Dounce-homogenized with 50 strokes, and centrifuged at 2,000 rpm for 10 min at 4°C to separate nuclei from cytosol. Nuclei were washed twice in 100 μl of buffer 1 and incubated in 100 μl of 25% glycerol, 20 mM Hepes, pH 7.9, 0.4 M NaCl, 1 mM EDTA, pH 8.0, 1 mM EGTA, 1 mM DTT, and 25 mM β-glycerophosphate containing protease and phosphatase inhibitor cocktails. Samples were rocked at 4°C for 20 min and centrifuged at 14,000 g for 20 min. This fraction was collected as nuclear extract. The remaining nuclear matrix pellet was washed twice in buffer 1 and solubilized in SDS-PAGE sample buffer. Meanwhile, the cytosolic fraction was centrifuged at 100,000 g for 1 h at 4°C to pellet the membrane fraction, which was solubilized in SDS-PAGE sample buffer. All fractions were brought to equal volumes and protein content was determined for the cytosol and nuclear extract fractions. Equalized aliquots (20 μg) were analyzed on 10% SDS-PAGE gels under reducing conditions and transferred to PVDF membranes. Blots were probed with antibodies to GAPDH, Ran, and lamin B1 for quality control of the fractionation, with anti-actin for normalization of loading, and with antibodies to muskelin or RanBP9, followed by appropriate secondary antibodies and development by enhanced chemiluminescence.
siRNA-mediated depletion of muskelin or RanBP9
Transient expression of siRNA duplexes was used to deplete muskelin in mouse C2C12 cells. To identify an active siRNA, COS-7 cells were transiently transfected with GFP or GFP-muskelin and, 24 h later, retransfected using 4 μl of 50 μM SMARTpool siRNA (Thermo Fisher Scientific) and 4 μl Oligofectamine (Invitrogen). Cells were lysed in SDS-PAGE sample buffer after 48 or 72 h and analyzed for GFP by immunoblotting, with normalization against β-actin. The SMARTpool siRNA produced an ∼45% decrease in muskelin protein at 72 h. Individual siRNA duplexes from the pool were tested for silencing by the same method, and the most active duplex (sense strand sequence 5′CAAAGGCGAUGGAGAAGAUUU), producing ∼70% depletion of GFP-MK, was chosen for depletion of endogenous mouse muskelin. C2C12 mouse skeletal myoblasts were plated at 25,000 cells per 37-mm well and transfected twice at 24-h intervals with 2 μl of 50 μM of the muskelin siRNA duplex or FITC-labeled scrambled control siRNA duplex and 10 μl of Lipofectamine 2000 reagent (Invitrogen) in 500 μl DME, according to manufacturer's instructions. Cells were lysed in SDS-PAGE sample buffer 48, 72, or 96 h later and analyzed for muskelin by immunoblotting. All blots were normalised against β-actin.
For stable depletion of human muskelin, A549 human lung carcinoma cells were stably transfected with human muskelin shRNAs, sh2038 (vector 1), or sh2024 (vector 2) in pSHAG-MAGIC2 vector or control shRNA vector (Thermo Fisher Scientific; Paddison et al., 2004), using Polyfect transfection reagent (QIAGEN) for 40 h, followed by selection with 7.5 μg/ml puromycin. Puromycin-resistant clones were analyzed for muskelin protein level by immunoblotting, with normalization against β-actin. Quantitation was performed using Image J version 1.38 (National Institutes of Health). For transient depletion of RanBP9, A549 cells were transfected with pTRIPZ plasmid encoding for RanBP9 (#68195) or nontargeting shRNA (#4743; Thermo Fisher Scientific), followed by selection and regrowth in 1.5 μg/ml puromycin for 10 d and then TET-ON induction of shRNA for 24–48 h with 0.75 μg/ml doxycycline. In this plasmid, RFP is induced as part of the same transcript as the shRNA. Induced and control cells were adhered to 50 nM FN for 1h, fixed, stained with FITC-phalloidin, and examined by laser confocal microscopy and morphometric scoring.
Yeast two-hybrid screen
A yeast two-hybrid library screen was performed with a human adult skeletal muscle cDNA library in the pACT2 vector, pretransformed into S. cerevisiae strain Y187 (Clontech Laboratories, Inc.). The library was screened for muskelin-interacting clones by mating for 20 h at 30°C with S. cerevisiae strain AH108 pretransformed with muskelin bait plasmid (pAS2-1/MK). This bait had no independent transcriptional activation activity or effect on mating efficiency. The library contained 3 × 106 independent clones. The mating mixture was plated on synthetic dropout −His/−Leu/−Trp minimal media, according to the library manufacturer's protocol, for 8 d at 30°C. The mating efficiency was 13.1% and a total of 2.85 × 106 library clones were screened. 54 colonies were recovered and replated under more stringent selection (SD minimal media −Ade/−His/−Leu/−Trp containing 40 μg/ml X-α-Gal (Clontech Laboratories, Inc.) to check for expression of the MEL1 reporter and eliminate false positives. 41 blue colonies were obtained and restreaked on SD −Leu/−Trp/+X-α-Gal and SD –Ade/−His/−Leu/−Trp +X-α-Gal plates to check for maintenance of phenotype. Plasmid DNA was then isolated from single yeast colonies by the alkaline lysis method, transformed into chemically competent Escherichia coli XL1Blue (Stratagene), and DNA minipreps prepared from multiple single colonies by QIAprep spin miniprep method (QIAGEN). To identify E. coli clones that contained pACT2 library clones, DNAs were digested with HaeIII and clones producing restriction digest patterns similar to that of the pACT2 vector were chosen for DNA sequencing. DNAs were also transformed into S. cerevisiae strain Y187 and tested for independent transcriptional activity and interaction with the muskelin bait by mating and plating on SD –Ade/−His/−Leu/−Trp +X-α-Gal. Clones passing these criteria were screened for specificity of interaction with muskelin by testing against lamin or syndecan-1 baits and pAS2-1 vector. Only one category of clone, encoding RanBP9 aa 142–748, was confirmed for specific interaction with muskelin bait by all these criteria.
Online supplemental material
Fig. S1 shows that vertebrate muskelin proteins contain a candidate conserved proline-directed kinase phosphorylation motif (A) and a cyclin-binding motif (B). Fig. S2 shows longer exposure of the FLAG blot samples shown in Fig. 7 C to demonstrate the weak expression of some RanBP9 domain deletions. Fig. S3 shows muskelin depletion by stable shRNA expression in A549 cells or transient expression of siRNAs in C2C12 cells. Table S1 show the oligonucleotide primers used to prepare muskelin or RanBP9 domain or point mutant constructs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200801133/DC1.
Supplementary Material
Acknowledgments
We thank Alan Tartakoff and Gregory Matera for conversations; Jason Bazil, Christine Baran, and Kaianna Austermiller for participation during research rotations; David Loftis, Naa Amponsah, and Raymond Monk for technical assistance; Elisabetta Bianchi for the gift of RanBP9 antibody; Robert Kypta for the gift of vinculin cDNA; Yosuke Hashimoto for advice on siRNA design; and The Cleveland Clinic Foundation Flow Cytometry, Imaging, Molecular Biotechnology, and Genomics cores for scientific support.
This research was supported in London by The Wellcome Trust (038284 to J.C. Adams). Research in Cleveland was supported by The Cleveland Clinic and by The National Institutes of Health (GM068073).
M. Valiyaveettil, A.A. Bentley, and P. Gursahaney contributed equally to this paper.
M. Valiyaveettil's present address is Novo Nordisk Research United States, North Brunswick, NJ 08902.
S. Prag's present address is Institute of Molecular Medicine, University of Lisbon, Lisbon, Portugal.
Abbreviations used in this paper: crm1, chromosome maintenance region 1; CTLH, C-terminal to LisH; DD, discoidin-like domain; FN, fibronectin; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; LIS1, lissencephaly-1; LisH, LIS1 homology; NES, nuclear export sequence; shRNA, short hairpin RNA; SMART, simple modular architecture research tool; TSP-1, thrombospondin-1.
References
- Adams, J.C. 2002. Characterization of a Drosophila melanogaster orthologue of muskelin. Gene. 297:69–78. [DOI] [PubMed] [Google Scholar]
- Adams, J.C., and L. Zhang. 1999. cDNA cloning of human muskelin and localisation of the muskelin (MKLN1) gene to human chromosome 7q32 and mouse chromosome 6 B1/B2 by physical mapping and FISH. Cytogenet. Cell Genet. 87:19–21. [DOI] [PubMed] [Google Scholar]
- Adams, J.C., B. Seed, and J. Lawler. 1998. Muskelin, a novel intracellular mediator of cell adhesive and cytoskeletal responses to thrombospondin-1. EMBO J. 17:4964–4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, J., R. Kelso, and L. Cooley. 2000. The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol. 10:17–24. [DOI] [PubMed] [Google Scholar]
- Anilkumar, N., M. Parsons, R. Monk, T. Ng, and J.C. Adams. 2003. Interaction of fascin and protein kinase Calpha: a novel intersection in cell adhesion and motility. EMBO J. 22:5390–5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork, P., and R.F. Doolittle. 1994. Drosophila kelch motif is derived from a common enzyme fold. J. Mol. Biol. 236:1277–1282. [DOI] [PubMed] [Google Scholar]
- Boyd, L.M., W.J. Richardson, J. Chen, V.B. Kraus, A. Tewari, and L.A. Setton. 2005. Osmolarity regulates gene expression in intervertebral disc cells determined by gene array and real-time quantitative RT-PCR. Ann. Biomed. Eng. 33:1071–1077. [DOI] [PubMed] [Google Scholar]
- Brown, N.R., M.E. Noble, J.A. Endicott, and L.N. Johnson. 1999. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat. Cell Biol. 1:438–443. [DOI] [PubMed] [Google Scholar]
- Brunkhorst, A., M. Karlen, J. Shi, M. Mikolajczyk, M.A. Nelson, M. Metsis, and O. Hermanson. 2005. A specific role for the TFIID subunit TAF4 and RanBPM in neural progenitor differentiation. Mol. Cell. Neurosci. 29:250–258. [DOI] [PubMed] [Google Scholar]
- Cahana, A., T. Escamez, R.S. Nowakowski, N.L. Hayes, M. Giacobini, A. von Holst, O. Shmueli, T. Sapir, S.K. McConnell, W. Wurst, et al. 2001. Targeted mutagenesis of Lis1 disrupts cortical development and LIS1 homodimerization. Proc. Natl. Acad. Sci. USA. 98:6429–6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, L., S. Lemmon, and V. Lemmon. 2005. RanBPM is an L1-interacting protein that regulates L1-mediated mitogen-activated protein kinase activation. J. Neurochem. 94:1102–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks, G.E., G. Hon, J.M. Chandonia, and S.E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukierman, E., R. Pankov, and K.M. Yamada. 2002. Cell interactions with three-dimensional matrices. Curr. Opin. Cell Biol. 14:633–639. [DOI] [PubMed] [Google Scholar]
- Davis, J.R., M. Kakar, and C.S. Lim. 2007. Controlling protein compartmentalization to overcome disease. Pharm. Res. 24:17–27. [DOI] [PubMed] [Google Scholar]
- Denti, S., A. Sirri, A. Cheli, L. Rogge, G. Innamorati, S. Putignano, M. Fabbri, R. Pardi, and E. Bianchi. 2004. RanBPM is a phosphoprotein that associates with the plasma membrane and interacts with the integrin LFA-1. J. Biol. Chem. 279:13027–13034. [DOI] [PubMed] [Google Scholar]
- Dhodda, V.K., K.A. Sailor, K.K. Bowen, and R. Vemuganti. 2004. Putative endogenous mediators of preconditioning-induced ischemic tolerance in rat brain identified by genomic and proteomic analysis. J. Neurochem. 89:73–89. [DOI] [PubMed] [Google Scholar]
- Emes, R.D., and C.P. Ponting. 2001. A new sequence motif linking lissencephaly, Treacher Collins and oral-facial-digital type 1 syndromes, microtubule dynamics and cell migration. Hum. Mol. Genet. 10:2813–2820. [DOI] [PubMed] [Google Scholar]
- Fukuda, M., S. Asano, T. Nakamura, M. Adachi, M. Yoshida, M. Yanagida, and E. Nishida. 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 390:308–311. [DOI] [PubMed] [Google Scholar]
- Hafizi, S., A. Gustafsson, J. Stenhoff, and B. Dahlback. 2005. The Ran binding protein RanBPM interacts with Axl and Sky receptor tyrosine kinases. Int. J. Biochem. Cell Biol. 37:2344–2356. [DOI] [PubMed] [Google Scholar]
- Hasegawa, H., H. Katoh, H. Fujita, K. Mori, and M. Negishi. 2000. Receptor isoform-specific interaction of prostaglandin EP3 receptor with muskelin. Biochem. Biophys. Res. Commun. 276:350–354. [DOI] [PubMed] [Google Scholar]
- Itahana, Y., E.T. Yeh, and Y. Zhang. 2006. Nucleocytoplasmic shuttling modulates activity and ubiquitination-dependent turnover of SUMO-specific protease 2. Mol. Cell. Biol. 26:4675–4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M.H., D.R. Cooper, A. Oleksy, Y. Devedjiev, U. Derewenda, O. Reiner, and J. Otlewski. 2004. The structure of the N-terminal domain of the product of the lissencephaly gene Lis1 and its functional implications. Structure. 12:987–998. [DOI] [PubMed] [Google Scholar]
- Kobayashi, M., and M. Yamamoto. 2005. Molecular mechanisms activating the Nrf2-Keap1 pathway of antioxidant gene regulation. Antioxid. Redox Signal. 7:385–394. [DOI] [PubMed] [Google Scholar]
- Kobayashi, N., J. Yang, A. Ueda, T. Suzuki, K. Tomaru, M. Takeno, K. Okuda, and Y. Ishigatsubo. 2007. RanBPM, muskelin, p48EMLP, p44CTLH and ARMC8α and ARMC8β are components of the CTLH complex. Gene. 396:236–247. [DOI] [PubMed] [Google Scholar]
- Kubota, T., T. Morozumi, K. Shimizu, N. Sugita, T. Kobayashi, and H. Yoshie. 2001. Differential gene expression in neutrophils from patients with generalized aggressive periodontitis. J. Periodontal Res. 36:390–397. [DOI] [PubMed] [Google Scholar]
- Kvansakul, M., J.C. Adams, and E. Hohenester. 2004. Structure of a thrombospondin C-terminal fragment reveals a novel calcium core in the type 3 repeats. EMBO J. 23:1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange, A., R.E. Mills, C.J. Lang, M. Stewart, S.E. Devine, and A.H. Corbett. 2007. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J. Biol. Chem. 282:5101–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledee, D.R., C.Y. Gao, R. Seth, R.N. Fariss, B.K. Tripathi, and P.S. Zelenka. 2005. A specific interaction between muskelin and the cyclin-dependent kinase 5 activator p39 promotes peripheral localization of muskelin. J. Biol. Chem. 280:21376–21383. [DOI] [PubMed] [Google Scholar]
- Li, X., D. Zhang, M. Hannink, and L.J. Beamer. 2004. Crystal structure of the kelch domain of human Keap1. J. Biol. Chem. 279:54750–54758. [DOI] [PubMed] [Google Scholar]
- Mattaj, I.W., and L. Englmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67:265–306. [DOI] [PubMed] [Google Scholar]
- Mikolajka, A., X. Yan, G.M. Popowicz, P. Smialowski, E.A. Nigg, and T.A. Holak. 2006. Structure of the N-terminal domain of the FOP (FGFR1OP) protein and implications for its dimerization and centrosomal localization. J. Mol. Biol. 359:863–875. [DOI] [PubMed] [Google Scholar]
- Paddison, P.J., J.M. Silva, D.S. Conklin, M. Schlabach, M. Li, S. Aruleba, V. Balija, A. O'Shaughnessy, L. Gnoj, K. Scobie, et al. 2004. A resource for large-scale RNA-interference-based screens in mammals. Nature. 428:427–431. [DOI] [PubMed] [Google Scholar]
- Padmanabhan, B., K.I. Tong, T. Ohta, Y. Nakamura, M. Scharlock, M. Ohtsuji, M.I. Kang, A. Kobayashi, S. Yokoyama, and M. Yamamoto. 2006. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell. 21:689–700. [DOI] [PubMed] [Google Scholar]
- Pemberton, L.F., and B.M. Paschal. 2005. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 6:187–198. [DOI] [PubMed] [Google Scholar]
- Pitre, S., F. Dehne, A. Chan, J. Cheetham, A. Duong, A. Emili, M. Gebbia, J. Greenblatt, M. Jessulat, N. Krogan, et al. 2006. PIPE: a protein-protein interaction prediction engine based on the re-occurring short polypeptide sequences between known interacting protein pairs. BMC Bioinformatics. 7:365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prag, S., and J.C. Adams. 2003. Molecular phylogeny of the kelch-repeat superfamily reveals an expansion of BTB/kelch proteins in animals. BMC Bioinformatics. 4:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prag, S., G.D.M. Collett, and J.C. Adams. 2004. Molecular analysis of muskelin identifies a conserved discoidin-like domain that contributes to protein self-association. Biochem. J. 381:547–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prag, S., A. De Arcangelis, E. Georges-Labouesse, and J.C. Adams. 2007. Regulation of post-translational modifications of muskelin by protein kinase C. Int. J. Biochem. Cell Biol. 39:366–378. [DOI] [PubMed] [Google Scholar]
- Pufall, M.A., and B.J. Graves. 2002. Autoinhibitory domains: modular effectors of cellular regulation. Annu. Rev. Cell Dev. Biol. 18:421–462. [DOI] [PubMed] [Google Scholar]
- Rao, M.A., H. Cheng, A.N. Quayle, H. Nishitani, C.C. Nelson, and P.S. Rennie. 2002. RanBPM, a nuclear protein that interacts with and regulates transcriptional activity of androgen receptor and glucocorticoid receptor. J. Biol. Chem. 277:48020–48027. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and D.W. Russel. 2001. Molecular Cloning, A Laboratory Manual. Third Edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. 745 pp.
- Schultz, J., F. Milpetz, P. Bork, and C.P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA. 95:5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwoebel, E.D., and M.S. Moore. 2000. The control of gene expression by regulated nuclear transport. Essays Biochem. 36:105–113. [DOI] [PubMed] [Google Scholar]
- Shaner, M.C., I.M. Blair, and T.D. Schneider. 2003. Sequence logos: a powerful yet simple tool. Version 3.03 of hawaii.txt http://www.lecb.ncifcrf.gov/∼toms/paper/ (accessed March, 2007).
- Tagnaouti, N., S. Loebrich, F. Heisler, Y. Pechmann, S. Fehr, A. De Arcangelis, E. Georges-Labouesse, J.C. Adams, and M. Kneussel. 2007. Neuronal expression of muskelin in the rodent central nervous system. BMC Neurosci. 8:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togashi, H., E.F. Schmidt, and S.M. Strittmatter. 2006. RanBPM contributes to Semaphorin3A signaling through plexin-A receptors. J. Neurosci. 26:4961–4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda, M., H. Nishitani, and T. Nishimoto. 2003. A novel nuclear protein, Twa1, and muskelin comprise a complex with RanBPM. Gene. 303:47–54. [DOI] [PubMed] [Google Scholar]
- Wang, D., Z. Li, E.M. Messing, and G. Wu. 2002. Activation of Ras/Erk pathway by a novel MET-interacting protein RanBPM. J. Biol. Chem. 277:36216–36222. [DOI] [PubMed] [Google Scholar]
- Wei, Y., J. Jin, and J.W. Harper. 2003. The cyclin E/Cdk2 substrate and Cajal body component p220NPAT activates histone transcription through a novel LisH-like domain. Mol. Cell. Biol. 23:3669–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff, B., J.J. Sanglier, and Y. Wang. 1997. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol. 4:139–147. [DOI] [PubMed] [Google Scholar]
- Xu, L., and J. Massague. 2004. Nucleocytoplasmic shuttling of signal transducers. Nat. Rev. Mol. Cell Biol. 5:209–219. [DOI] [PubMed] [Google Scholar]
- Yang, J., E.S. Bardes, J.D. Moore, J. Brennan, M.A. Powers, and S. Kombluth. 1998. Control of cyclin B1 localization through regulated binding of the nuclear export factor CRM1. Genes Dev. 12:2131–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir, E., and B. Geiger. 2001. Molecular complexity and dynamics of cell-matrix adhesions. J. Cell Sci. 114:3583–3590. [DOI] [PubMed] [Google Scholar]
- Zou, Y., S. Lim, K. Lee, X. Deng, and E. Friedman. 2003. Serine/threonine kinase Mirk/Dyrk1B is an inhibitor of epithelial cell migration and is negatively regulated by the Met adaptor Ran-binding protein M. J. Biol. Chem. 278:49573–49581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.