Abstract
Acquired mutations in the juxtamembrane region of MPL (W515K or W515L), the receptor for thrombopoietin, have been described in patients with primary myelofibrosis or essential thrombocythemia, which are chronic myeloproliferative disorders. We have developed a real-time polymerase chain reaction assay for the detection and quantification of MPL mutations that is based on locked nucleic acid fluorescent probes. Mutational analysis was performed using DNA from granulocytes. Reference curves were obtained using cloned fragments of MPL containing either the wild-type or mutated sequence; the predicted sensitivity level was at least 0.1% mutant allele in a wild-type background. None of the 60 control subjects presented with a MPLW515L/K mutation. Of 217 patients with myelofibrosis, 19 (8.7%) harbored the MPLW515 mutation, 10 (52.6%) with the W515L allele. In one case, both the W515L and W515K alleles were detected by real-time polymerase chain reaction. By comparing results obtained with conventional sequencing, no erroneous genotype attribution using real-time polymerase chain reaction was found, whereas one patient considered wild type according to sequence analysis actually harbored a low W515L allele burden. This is a simple, sensitive, and cost-effective procedure for large-scale screening of the MPLW515L/K mutation in patients suspected to have a myeloproliferative disorder. It can also provide a quantitative estimate of mutant allele burden that might be useful for both patient prognosis and monitoring response to therapy.
The Philadelphia chromosome-negative chronic myeloproliferative disorders include polycythemia vera (PV), essential thrombocythemia (ET), and primary myelofibrosis (PMF).1,2 Their molecular pathogeneses remained poorly characterized until the description of an acquired point mutation in exon 14 of JAK2 (V617F), which involves the substitution of a key valine for a phenylalanine residue in the JH2 pseudokinase domain.3,4,5,6 The frequency of the JAK2V617F mutation among patients with PV is more than 95%, whereas approximately half of those who are V617F-negative harbor different abnormalities located in JAK2 exon 12.7 On the other hand, approximately 60% of patients with ET or PMF possess the JAK2V617F mutant. In 25 to 30% of patients with either PV or PMF, the JAK2V617F mutation is harbored in a homozygous state due to mitotic recombination involving the short arm of chromosome 9.3,4,5,6,8 Homozygous patients are rare in ET and account for only 2 to 4%.9 The burden of the V617F allele influences disease phenotype9,10 and is associated with a greater risk of thrombosis in both ET9 and PV11 as well as with disease progression in PMF.12 This variable mutation frequency of patients with different clinical phenotypes,13 together with the demonstration that at least certain wild-type JAK2 patients with ET14,15 or PMF (unpublished data) present clonal hematopoiesis using assays for the X chromosome inactivation pattern (X-CIP),16 prompted a search for additional mutations in other genes involved in the JAK2/STAT pathway. Recently, two novel, recurrent, molecular abnormalities (W515L and W515K) were identified in MPL, the gene encoding the receptor for thrombopoietin.17 Thrombopoietin, the main humoral regulator of thrombopoiesis in humans,18 has been implicated in the pathogenesis of myelofibrosis in murine models.19,20 The 515 tryptophan residue is located in a unique amphipathic domain of MPL in the transmembrane-cytoplasmic hinge region that prevents spontaneous activation of the receptor.21 In fact, the tryptophan to leucine (W>L) substitution confers factor-independent growth to Ba/F3 cells that is associated with constitutive activation of downstream signaling pathways.17 Expression of MPLW515L in a murine bone marrow transplantation model resulted in an acute myeloproliferative disorder phenotype that recapitulated several aspects of human myelofibrosis.17 In humans, the MPL mutation occurs in multipotent hematopoietic stem cells, myeloid cells, purified B and T lymphocytes, and natural killer cells.22,23,24
The MPLW515L/K mutation has been detected in 5 to 7% of patients with PMF and in 1% of those with ET17,25,26; it has not been described in PV or myelodysplastic syndromes, including refractory anemia with ringed sideroblasts and thrombocytosis,27 or in acute myeloid leukemia.25 The coexistence of the MPLW515L/K and JAK2V617F mutations in myelofibrosis has also been reported.25,26,28 In a study of 217 patients with PMF, we found that this mutation was associated with more severe anemia and greater transfusion support.26 Therefore, a search for MPLW515 mutations in either PMF or ET may have both diagnostic and prognostic implications; indeed, the discovery of any of these molecular abnormalities establishes the presence of a clonal myeloproliferation, representing a major diagnostic criterion in the recent proposal for revision to the World Health Organization classification of myeloid neoplasms.2
Previously published assay methods for MPLW515L/K mutations have been represented by direct sequencing and melting curve analysis; these methods have a low sensitivity, unable to detect fewer than 10% and 3% mutant cells in a wild-type background, respectively, and are not suitable for high-throughput screening. Therefore, the aim of this study was to develop a novel real-time polymerase chain reaction (PCR) assay that could be used in a large series of patients and would be sensitive enough to detect a low MPL mutant allele burden. The latter point might be relevant to novel drugs that target members of the constitutively activated JAK/STAT pathway, as they are expected to enter the therapeutic scenario soon.29 This novel real-time PCR assay was validated by retrospectively analyzing a population of 217 patients with myelofibrosis who had been previously genotyped using direct sequencing.26
Materials and Methods
Patients
The characteristics of 217 patients with myelofibrosis analyzed in this study were previously described in detail26; 150 patients had PMF, 30 had prefibrotic myelofibrosis,1 and 37 had postpolycythemic/post-thrombocythemic myelofibrosis. Sixty healthy blood donors were used as controls. Furthermore, we evaluated 50 patients with PV who harbor the JAK2V617F mutation and were diagnosed according to the proposal for revision to the World Health Organization criteria.2 All patients and controls gave their consent for sample donation and informed consent for the study.
Sample Preparation
Peripheral blood granulocytes were separated by differential centrifugation over a Ficoll-Ipaque gradient. Briefly, 20 ml of whole blood were collected into EDTA-containing polypropylene tubes and processed within 4 hours. Blood was diluted 1:1 with Ca2+/Mg2+-free phosphate-buffered saline, carefully layered over 15 ml of Ficoll-Ipaque in a 50-ml tube, and centrifuged at 800 × g at room temperature for 20 minutes. Both the upper fraction and the Ficoll layer were carefully removed, while the lower fraction was collected and transferred to a fresh tube. Lysis of red blood cells was performed by the addition of a 10X volume of 1X BD PharmLyse solution (Becton Dickinson BD, San Jose, CA), centrifugation of the tube after a 15-minute incubation at room temperature, and removal of the supernatant. This step was repeated twice. After two washes in Ca2+/Mg2+-free phosphate-buffered saline, the dry granulocyte pellet was stored at −20°C until processed. Cytosmears of these cellular preparations routinely showed a granulocyte content greater than 95%. DNA was extracted using the QIAmp DNA blood kit (Qiagen, GmbH, Hilden, Germany) and quantified using the NanoDrop technology (ND-1000 spectrophotometer; NanoDrop Technology, Wilmington, DE). All samples used for real-time PCR had 260:280 ratios greater than 1.8.
Real-Time PCR Assay Development
MPL maps to human chromosome 1p33–34.2. GenBank sequence AL139289 was used for the design of primers and probes according to the Primer 3 software at http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi; accessed January 2007 and the MELT-CALC (version 2.0) software at http://www.meltcalc.de; accessed January 2007. The Tm of the locked nucleic acid (LNA) probes was verified using Exiqon Tm prediction (http://lna-tm.com; accessed March 2007). Sequences of primers and probes are provided in Table 1. This assay makes use of a single set of primers, with the advantage that amplification of mutant and wild-type alleles has the same efficiencies, while specificities are obtained with different probes that were modified according to the LNA chemistry (Sigma-Proligo, Paris, France).
Table 1.
Sequences of Primers and Probes Used for Real-Time PCR Assay of the MPLW515L or W515K Mutation
| Real-time PCR primers/probes | Sequence |
|---|---|
| MPL forward primer | 5′-agcctggatctccttggtgac-3′ |
| MPL reverse primer | 5′-accgccagtctcctgcct-3′ |
| MPL wild-type probe | 5′-ctgctg+Aggt+Ggc+Agtttc-3′ |
| MPL 515W>L probe | 5′-ctgc+Tgagg+T+Tgcag+T+Ttc-3′ |
| MPL 515W>K probe | 5′-tgc+Tgctgagg+A+Ag cagtttcc-3′ |
LNA bases are indicated by a capital letter with a plus sign before it. Probes were labeled with 5-carboxyfluorescein at their 5′ termini while Black Hole Quencher-1 was attached to the 3′ termini. The underlined nucleotides correspond to the 515 codon; the wild-type sequence is TGG, the W515L sequence is TTG, and the W515K sequence is AAG.
Real-time PCR was performed using both the ABI Prism 7000 platform and the StepOne real-time PCR system (Applied Biosystems, Foster City, CA) with similar results. Forty nanograms of granulocyte DNA were used in each real-time PCR assay. Three different real-time reactions were set up in triplicate (one each for W515L, W515K, and wild-type control) for each DNA sample. A 20-μl reaction contained 1X TaqMan universal PCR Master Mix (Applied Biosystems), 300 nmol/L each primer, and 200 nmol/L each LNA-modified probe. Control wells without template (NTC) were included in each assay. Amplification and detection were performed under the following conditions: initial hold at 50°C for 2 minutes, hold at 95°C for 10 minutes followed by 55 cycles at 95°C for 15 seconds and 66°C or 62°C for 1 minute for the case of MPL unmutated and W515L probe or W515K probe, respectively. The fluorescent signal intensities were recorded and analyzed during PCR amplification using the SDS software (Applied Biosystems). The mean ΔCT of triplicate determinations (CTMPLW515L/K − CTMPLwild-type) was calculated, and the percentage of mutant alleles in the sample was obtained by comparison with a reference curve of serial dilutions of mutant plasmid mixtures in wild-type plasmid DNA. Both positive and negative controls were included in each assay.
Preparation of Cloned MPL Fragments
To prepare a reference curve for the quantification of the mutant alleles, a 277-bp fragment of MPL from healthy subjects (wild-type sequence) or from patients harboring either the W515L or W515K mutation was subcloned into the pCR2.1 TOPO vector using the TOPO TA cloning kit (Invitrogen, Groningen, The Netherlands). The sequences of the forward and reverse primers for PCR were 5′-TGGGCCGAAGTCTGACCCTTT-3′ and 5′-AGAGGTGACGTGCAGGAAGTGGCGAAGC-3′, respectively. Recombinant plasmids were isolated and bi-directional sequencing was performed to validate the sequences before plasmid amplification. A dilution of the stock plasmid solution was prepared and preliminarily tested according to the standardization principles described in the Final Report on Plasmid Standards for Real Time PCR and GM Enforcement Testing developed by Department for Environment, Food, and Rural Affairs of the government of the United Kingdom at http://www.g-inspectorate.gov.uk/documents/PLASMID.pdf; accessed July 2007.
Comparison of Conventional Sequencing and Real-Time PCR Assay
DNA samples from 217 patients with myelofibrosis were genotyped for the MPLW5151L/K mutation using conventional sequencing. MPL mutation sequencing was performed by amplifying a 248-bp region of MPL exon 10 using the following primers: forward, 5′-TAGCCTGGATCTCCTTGGTG-3′ and reverse, 5′-AGAGGTGACGTGCAGGAAGT-3′. PCR products were subjected to bidirectional sequencing analysis using an ABI PRISM 3730DNA Analyzer (Applied Biosystems, Foster City, CA) as described.26
Results
In preliminary experiments we attempted to conduct a real-time PCR assay for the MPLW515L/K allele using conventional 5′-5-carboxyfluorescein- and 3′-Black Hole Quencher-1-labeled probes, with lengths varying from 16 to 28 bp; these probes were differentially spaced along the mutation site. However, we failed to obtain satisfactory results due to a high background generated by nonspecific binding of the probes for mutant alleles to the wild-type sequence (not shown in detail). Therefore, we subsequently explored the use of LNA-modified probes because of their anticipated greater specificity of hybridization and thermal stability. Both the primers and LNA probes, listed in Table 1, were chosen for their optimal performance among other sequences and were used in all of the experiments described herein. These probes were derived from a 28-bp probe originally designed for conventional PCR, progressively shortened by the introduction of LNA residues at various positions so that the final calculated Tms of the individual probes were similar.30 The Tms of the probes selected from a panel of five different sets that were experimentally tested in real-time PCR assays were 73°C, 74°C, and 72°C for the wild-type, W515L, and W515K probes, respectively. The probe corresponding to codon 515 had two LNA nucleotides for the mutant sequences and one LNA base in the wild-type sequence (underlined in Table 1). In our original assay design, we performed single real-time PCRs for each of these probes, all labeled with 5-carboxyfluorescein; however, subsequent experiments showed that comparable results could be obtained by using the 5-carboxyfluorescein-labeled wild-type probe with one of the 5-VIC-labeled mutant probes in the same reaction tube. Results presented in this manuscript were obtained using the single-probe assay.
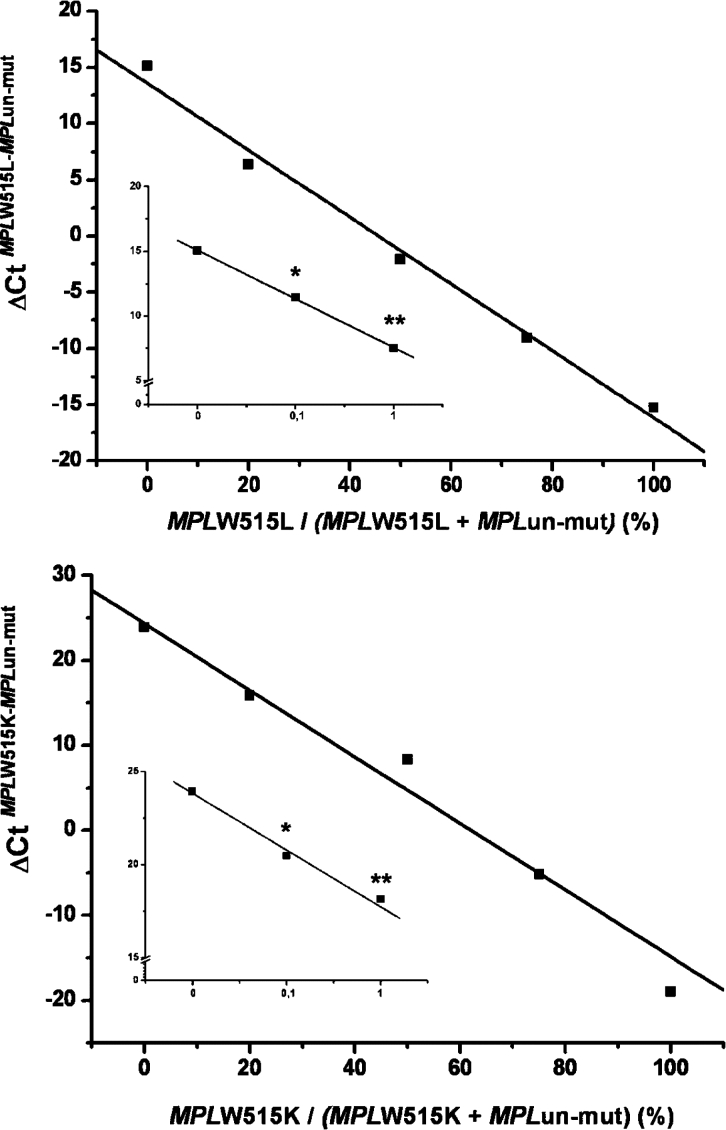
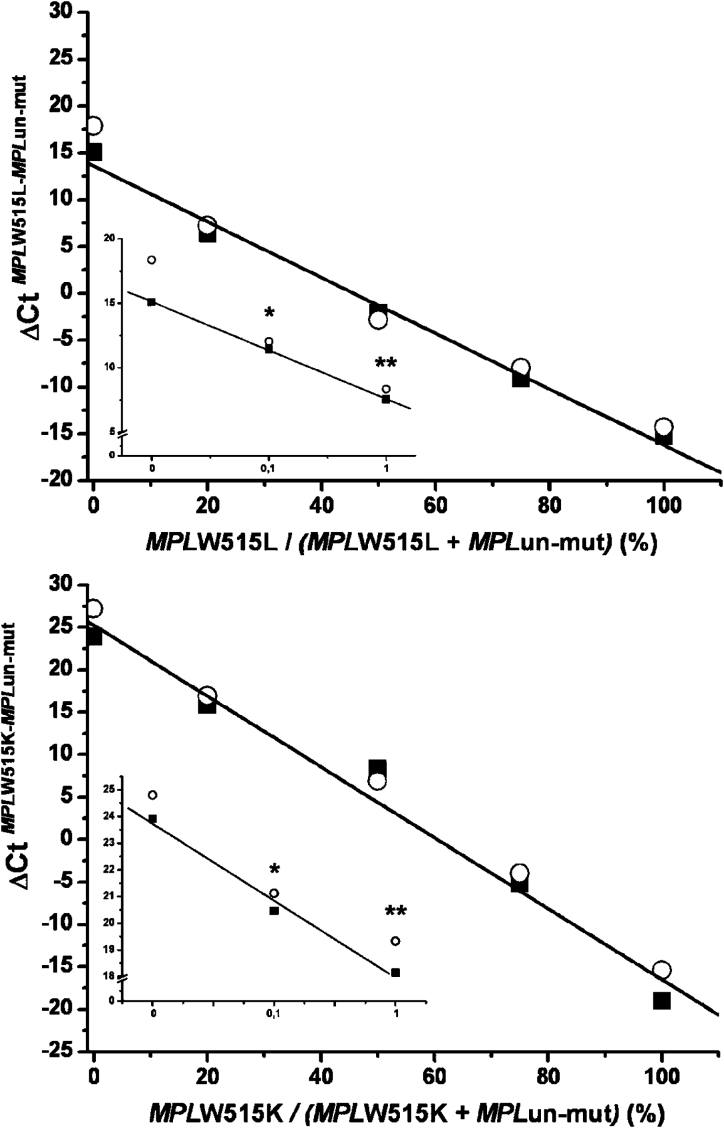
Standard curves were constructed for the wild-type and mutated sequences using serial dilutions of cloned MPL fragments in the range of 1 to 106 copies; amplification plots and corresponding regression lines are presented in Figure 1. As suggested by the slope of the curves, the efficiency of real-time PCR was almost absolute. Next, we performed progressive dilutions of each mutant plasmid allele, from 0.1% to 100%, in a solution of wild-type plasmid allele as in nonhomogeneous DNA samples used for diagnostic purposes. Figure 2 shows assay performance for the W515L and W515K alleles, which reproducibly allowed for the detection of at least 0.1% mutated allele in a wild-type background. This sensitivity level could be attained using 40 ng of genomic DNA from diagnostic samples, based on a 3.7 pg of DNA content per human haploid cell. To ascertain whether a similar level of sensitivity could be obtained in clinical samples, dilution experiments were also performed using 100% of mutated W515K or W515L patient DNA in an wild-type control DNA sample. As shown in Figure 3, the assay performance in genomic DNA samples was superimposable on the standard reference curve generated with plasmid DNA, suggesting that that 0.1% mutant allele could be reproducibly detected in granulocyte DNA samples.
Figure 1.
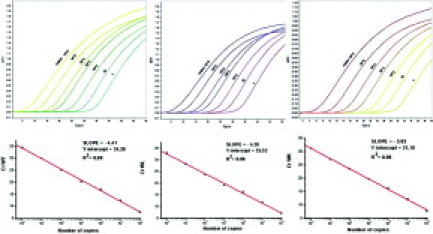
Amplification plots of wild-type and mutated MPLW515L and W515K alleles in the real-time PCR assay. Variations in threshold cycle reflected different input copy numbers of the respective cloned plasmids in the range of 1 to 106 copies. Linear regression analysis between the input number of copies and the CT value is presented in the lower part of the figure.
Figure 2.
Performance of the real-time PCR assay for quantification of the MPLW515L (upper panel) or W515K (lower panel) allele; curves were obtained by the progressive dilution of each mutated plasmid DNA in wild-type plasmid DNA. The SD of each experimental point was less than 4% in three individual real-time PCR assays and was not reported. The inset in each plot shows the sensitivity of the assay in the 0 to 1% mutant allele range and represents the mean (±SD) of five different preparations. *P < 0.01; **P < 0.001 compared with the 0 point.
Figure 3.
Comparative performance of the real-time PCR assay for quantification of the MPLW515L (upper panel) or W515K (lower panel) allele using plasmid DNA (circles) or patient DNA (gDNA; squares); curves were obtained by the progressive dilution of either each mutated plasmid DNA in wild-type plasmid DNA or a 100% mutated W515L or W515K patient in wild-type control DNA. The insert in each plot shows the sensitivity of the assay in the 0 to 1% mutant allele range and represents the mean of four different preparations. The SD of each experimental point was less than 3% and was not reported. *P < 0.01; **P < 0.001 compared with the 0 point.
The intra-assay variation coefficient was calculated using DNA samples from 10 control subjects and four patients with either the W515L or W515K mutation and assayed in triplicate in the same plate. The variation in the calculated percentage of mutant allele was less than 2% and 1% for the W515L and W5151K allele, respectively, while no control well was positive. The interassay variation test was performed using DNA samples from two patients, each with a 100% W515L or W515K allele, either undiluted or diluted in normal DNA to 10 and 50% of the mutant allele. These reference samples were analyzed in 10 different runs over a 3-month period, and the variation in the percentage of mutant allele was less than 2.5% for both mutated alleles. Therefore, this high reproducibility and the comparable performance of the reference curves obtained with plasmid or patient DNA (Figure 3) suggests that the assay does not require a standard curve for each run, but that the percentage of mutated allele in clinical samples may be obtained directly by the ΔCT calculation.
Under these experimental conditions, we evaluated sixty healthy blood donors, and none was found to have MPL mutant cells. Similarly, none of the 50 patients with PV had the MPLW515 mutation. We analyzed a population of 217 patients with myelofibrosis, both primary and secondary forms, who were previously genotyped using conventional bi-directional sequencing; this series has been already reported.26 According to sequencing results, nine patients presented with the W515L allele, eight patients had the W5151K allele, and one patient had both the W515L and W515K mutant alleles. Unfortunately, we were unable to collect fresh cells from this subject to evaluate whether these two mutations were either harbored simultaneously by a single cell or involved two different clonogenic progenitors. All patient results were confirmed using the novel real-time PCR assay. However, an additional patient who had been considered wild type using the sequencing approach was actually found to harbor low levels of the W515L allele (1.5%) by real-time PCR, and was accordingly rediagnosed as an MPL mutant. The mean calculated burden of MPL mutant alleles using real-time PCR was 51 ± 15% (range, 1.5–100%), not statistically different from that calculated using peak area integration of the chromatogram (55 ± 20%; range, 19–100%).
Discussion
Currently, genotyping of MPL mutations is performed using conventional bidirectional sequencing, a method that has a detection limit of 10 to 15%, is time consuming, and is relatively expensive; thus, it is not suitable for wide population screening. A melting curve technique, with a sensitivity of 3 to 5%, has also been described.25 We have developed a novel method for the detection and quantification of MPLW515L/K mutations that is based on LNA probes for real-time PCR. LNA probes have increased thermal stability and hybridization specificity due to their higher Tm, making them especially suitable for allelic discrimination.31 Indeed, LNA chemistry was key to the development of our assay in which a number of conventional TaqMan probes tested initially displayed very poor specificity. Using the optimized conditions described here, we were able to attain a sensitivity level of at least 0.1% mutant allele in a wild-type background, a value that is at least 100-fold higher than conventional sequencing. This high sensitivity of the assay is of particular relevance in the setting of myeloproliferative disorders in which only a proportion of hematopoietic clones and their differentiated progeny might harbor MPL mutations. It is also conceivable that the true incidence of MPL mutations in myeloproliferative disorders has been underestimated in previous studies where direct sequencing or melting curves were used for genotyping.17,25 As a matter of fact, one patient in this series who had been considered wild-type MPL using sequencing26 harbored a low W515L allele burden by real-time PCR. Furthermore, in a large series of patients with ET, we found that the frequency of MPL mutations using the real-time PCR assay was sevenfold higher than originally reported.25,30
The studies described herein have been conducted on purified granulocytes; due to the retrospective design of the study, we do not have stored samples of whole blood or bone marrow to perform comparative analyses among these different cellular sources. However, even in the case of JAK2V617F genotyping, it is still unknown whether fewer homogeneous cellular samples compared with granulocytes might be equally informative.33 In our opinion, purified blood granulocytes remain the most appropriate DNA source for genotyping in myeloproliferative disorders.36
In summary, we have developed a sensitive and convenient method for the detection and quantification of mutant alleles at MPL codon 515 based on real-time PCR in the presence of LNA-modified probes. This methodology is expected to facilitate the screening of MPL mutations in large patient series, helping to molecularly classify patients with ET or PMF and possibly provide a prognosis.26 In addition, this approach might be useful for measuring changes in mutant allele burdens in patients who will receive, in the near future, novel targeted drugs against MPL mutant cells and/or components of the JAK/STAT pathway, as well as in assisting disease monitoring after hematopoietic stem cell transplantation in myelofibrosis as described for the JAK2V617F mutation.35 However, assessment of the role of the MPLW515 mutant allele-burden quantification as a biomarker of disease or minimal residual disease will require prospective studies.
Acknowledgements
We thank all of our colleagues who referred patients for genotyping, and we thank patients and healthy donors for their contributions to the study.
Footnotes
Supported by Associazione Italiana per la Ricerca sul Cancro, Milano; grant COFIN 2006067001-003 from Ministero Italiano della Università e della Riccrea (to A.M.V.); and funds from the Istituto Toscano Tumori. A.P. was the recipient of a fellowship from Associazione Italiana per le Leucemie, Firenze.
References
- 1.Vardiman JW, Harris NL, Brunning RD. The World Health Organization (WHO) classification of the myeloid neoplasms. Blood. 2002;100:2292–2302. doi: 10.1182/blood-2002-04-1199. [DOI] [PubMed] [Google Scholar]
- 2.Tefferi A, Thiele J, Orazi A, Kvasnicka HM, Barbui T, Hanson CA, Barosi G, Verstovsek S, Birgegard G, Mesa R, Reilly JT, Gisslinger H, Vannucchi AM, Cervantes F, Finazzi G, Hoffman R, Gilliland DG, Bloomfield CD, Vardiman JW. Proposals and rationale for revision of the World Health Organization diagnostic criteria for polycythemia vera, essential thrombocythemia, and primary myelofibrosis: recommendations from an ad hoc international expert panel. Blood. 2007;110:1092–1097. doi: 10.1182/blood-2007-04-083501. [DOI] [PubMed] [Google Scholar]
- 3.James C, Ugo V, Le Couedic JP, Staerk J, Delhommeau F, Lacout C, Garcon L, Raslova H, Berger R, Bennaceur-Griscelli A, Villeval JL, Constantinescu SN, Casadevall N, Vainchenker W. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 4.Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, Vassiliou GS, Bench AJ, Boyd EM, Curtin N, Scott MA, Erber WN, Green AR. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 5.Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ, Boggon TJ, Wlodarska I, Clark JJ, Moore S, Adelsperger J, Koo S, Lee JC, Gabriel S, Mercher T, D'Andrea A, Frohling S, Dohner K, Marynen P, Vandenberghe P, Mesa RA, Tefferi A, Griffin JD, Eck MJ, Sellers WR, Meyerson M, Golub TR, Lee SJ, Gilliland DG. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 6.Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, Tichelli A, Cazzola M, Skoda RC. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 7.Scott LM, Tong W, Levine RL, Scott MA, Beer PA, Stratton MR, Futreal PA, Erber WN, McMullin MF, Harrison CN, Warren AJ, Gilliland DG, Lodish HF, Green AR. JAK2 exon 12 mutations in polycythemia vera and idiopathic erythrocytosis. N Engl J Med. 2007;356:459–468. doi: 10.1056/NEJMoa065202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kralovics R, Guan Y, Prchal JT. Acquired uniparental disomy of chromosome 9p is a frequent stem cell defect in polycythemia vera. Exp Hematol. 2002;30:229–236. doi: 10.1016/s0301-472x(01)00789-5. [DOI] [PubMed] [Google Scholar]
- 9.Vannucchi AM, Antonioli E, Guglielmelli P, Rambaldi A, Barosi G, Marchioli R, Marfisi RM, Finazzi G, Guerini V, Fabris F, Randi ML, De Stefano V, Caberlon S, Tafuri A, Ruggeri M, Specchia G, Liso V, Rossi E, Pogliani E, Gugliotta L, Bosi A, Barbui T. Clinical profile of homozygous JAK2V617F mutation in patients with polycythemia vera or essential thrombocythemia. Blood. 2007;110:840–846. doi: 10.1182/blood-2006-12-064287. [DOI] [PubMed] [Google Scholar]
- 10.Tefferi A, Lasho TL, Schwager SM, Strand JS, Elliott M, Mesa R, Li CY, Wadleigh M, Lee SJ, Gilliland DG. The clinical phenotype of wild-type, heterozygous, and homozygous JAK2V617F in polycythemia vera. Cancer. 2006;106:631–635. doi: 10.1002/cncr.21645. [DOI] [PubMed] [Google Scholar]
- 11.Vannucchi AM, Antonioli E, Guglielmelli P, Longo G, Pancrazzi A, Ponziani V, Bogani C, Ferrini PR, Rambaldi A, Guerini V, Bosi A, Barbui T. Prospective identification of high-risk polycythemia vera patients based on JAK2(V617F) allele burden. Leukemia. 2007;21:1952–1959. doi: 10.1038/sj.leu.2404854. [DOI] [PubMed] [Google Scholar]
- 12.Barosi G, Bergamaschi G, Marchetti M, Vannucchi AM, Guglielmelli P, Antonioli E, Massa M, Rosti V, Campanelli R, Villani L, Viarengo G, Gattoni E, Gerli G, Specchia G, Tinelli C, Rambaldi A, Barbui T. JAK2 V617F mutational status predicts progression to large splenomegaly and leukemic transformation in primary myelofibrosis. Blood. 2007;110:4030–4036. doi: 10.1182/blood-2007-07-099184. [DOI] [PubMed] [Google Scholar]
- 13.Nussenzveig RH, Swierczek SI, Jelinek J, Gaikwad A, Liu E, Verstovsek S, Prchal JF, Prchal JT. Polycythemia vera is not initiated by JAK2V617F mutation. Exp Hematol. 2007;35:32–38. doi: 10.1016/j.exphem.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 14.Antonioli E, Guglielmelli P, Pancrazzi A, Bogani C, Verrucci M, Ponziani V, Longo G, Bosi A, Vannucchi AM. Clinical implications of the JAK2 V617F mutation in essential thrombocythemia. Leukemia. 2005;19:1847–1849. doi: 10.1038/sj.leu.2403902. [DOI] [PubMed] [Google Scholar]
- 15.Gale RE, Allen AJ, Nash MJ, Linch DC. Long-term serial analysis of X-chromosome inactivation patterns and JAK2 V617F mutant levels in patients with essential thrombocythemia show that minor mutant-positive clones can remain stable for many years. Blood. 2007;109:1241–1243. doi: 10.1182/blood-2006-06-029769. [DOI] [PubMed] [Google Scholar]
- 16.Chen GL, Prchal JT. X-linked clonality testing: interpretation and limitations. Blood. 2007;110:1411–1419. doi: 10.1182/blood-2006-09-018655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pikman Y, Lee BH, Mercher T, McDowell E, Ebert BL, Gozo M, Cuker A, Wernig G, Moore S, Galinsky I, DeAngelo DJ, Clark JJ, Lee SJ, Golub TR, Wadleigh M, Gilliland DG, Levine RL. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med. 2006;3:e270. doi: 10.1371/journal.pmed.0030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaushansky K. The molecular mechanisms that control thrombopoiesis. J Clin Invest. 2005;115:3339–3347. doi: 10.1172/JCI26674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Villeval JL, Cohen-Solal K, Tulliez M, Giraudier S, Guichard J, Burstein SA, Cramer EM, Vainchenker W, Wendling F. High thrombopoietin production by hematopoietic cells induces a fatal myeloproliferative syndrome in mice. Blood. 1997;90:4369–4383. [PubMed] [Google Scholar]
- 20.Yan XQ, Lacey D, Hill D, Chen Y, Fletcher F, Hawley RG, McNiece IK. A model of myelofibrosis and osteosclerosis in mice induced by overexpressing thrombopoietin (mpl ligand): reversal of disease by bone marrow transplantation. Blood. 1996;88:402–409. [PubMed] [Google Scholar]
- 21.Staerk J, Lacout C, Sato T, Smith SO, Vainchenker W, Constantinescu SN. An amphipathic motif at the transmembrane-cytoplasmic junction prevents autonomous activation of the thrombopoietin receptor. Blood. 2006;107:1864–1871. doi: 10.1182/blood-2005-06-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu WY, Zhao Y, Ishii T, Sozer S, Shi J, Zhang W, Bruno E, Hoffman R, Xu M. Haematopoietic cell lineage distribution of MPLW515L/K mutations in patients with idiopathic myelofibrosis. Br J Haematol. 2007;137:378–379. doi: 10.1111/j.1365-2141.2007.06572.x. [DOI] [PubMed] [Google Scholar]
- 23.Pardanani A, Lasho TL, Finke C, Markovic SN, Tefferi A. Demonstration of MPLW515K, but not JAK2V617F, in in vitro expanded CD4+ T lymphocytes. Leukemia. 2007;21:2206–2207. doi: 10.1038/sj.leu.2404749. [DOI] [PubMed] [Google Scholar]
- 24.Chaligne R, James C, Tonetti C, Besancenot R, Le Couedic JP, Fava F, Mazurier F, Godin I, Maloum K, Larbret F, Lecluse Y, Vainchenker W, Giraudier S. Evidence for MPL W515L/K mutations in hematopoietic stem cells in primitive myelofibrosis. Blood. 2007;110:3735–3743. doi: 10.1182/blood-2007-05-089003. [DOI] [PubMed] [Google Scholar]
- 25.Pardanani AD, Levine RL, Lasho T, Pikman Y, Mesa RA, Wadleigh M, Steensma DP, Elliott MA, Wolanskyj AP, Hogan WJ, McClure RF, Litzow MR, Gilliland DG, Tefferi A. MPL515 mutations in myeloproliferative and other myeloid disorders: a study of 1182 patients. Blood. 2006;108:3472–3476. doi: 10.1182/blood-2006-04-018879. [DOI] [PubMed] [Google Scholar]
- 26.Guglielmelli P, Pancrazzi A, Bergamaschi G, Rosti V, Villani L, Antonioli E, Bosi A, Barosi G, Vannucchi AM. Anaemia characterises patients with myelofibrosis harbouring Mpl mutation. Br J Haematol. 2007;137:244–247. doi: 10.1111/j.1365-2141.2007.06565.x. [DOI] [PubMed] [Google Scholar]
- 27.Steensma DP, Caudill JS, Pardanani A, McClure RF, Lasho TL, Tefferi A. MPL W515 and JAK2 V617 mutation analysis in patients with refractory anemia with ringed sideroblasts and an elevated platelet count. Haematologica. 2006;91:ECR57. [PubMed] [Google Scholar]
- 28.Lasho TL, Pardanani A, McClure RF, Mesa RA, Levine RL, Gilliland DG, Tefferi A. Concurrent MPL515 and JAK2V617F mutations in myelofibrosis: chronology of clonal emergence and changes in mutant allele burden over time. Br J Haematol. 2006;135:683–687. doi: 10.1111/j.1365-2141.2006.06348.x. [DOI] [PubMed] [Google Scholar]
- 29.Pardanani A. JAK2 inhibitor therapy in myeloproliferative disorders: rationale, preclinical studies and ongoing clinical trials. Leukemia. 2008;22:23–30. doi: 10.1038/sj.leu.2404948. [DOI] [PubMed] [Google Scholar]
- 30.Latorra D, Arar K, Hurley JM. Design considerations and effects of LNA in PCR primers. Mol Cell Probes. 2003;17:253–259. doi: 10.1016/s0890-8508(03)00062-8. [DOI] [PubMed] [Google Scholar]
- 31.Latorra D, Campbell K, Wolter A, Hurley JM. Enhanced allele-specific PCR discrimination in SNP genotyping using 3′ locked nucleic acid (LNA) primers. Hum Mutat. 2003;22:79–85. doi: 10.1002/humu.10228. [DOI] [PubMed] [Google Scholar]
- 33.Hermouet S, Dobo I, Lippert E, Boursier MC, Ergand L, Perrault-Hu F, Pineau D. Comparison of whole blood vs purified blood granulocytes for the detection and quantitation of JAK2(V617F) Leukemia. 2007;21:1128–1130. doi: 10.1038/sj.leu.2404588. [DOI] [PubMed] [Google Scholar]
- 35.Kroger N, Badbaran A, Holler E, Hahn J, Kobbe G, Bornhauser M, Reiter A, Zabelina T, Zander AR, Fehse B. Monitoring of the JAK2–V617F mutation by highly sensitive quantitative real-time PCR after allogeneic stem cell transplantation in patients with myelofibrosis. Blood. 2007;109:1316–1321. doi: 10.1182/blood-2006-08-039909. [DOI] [PubMed] [Google Scholar]
Uncited references
- 32.Vannucchi AM, Antonioli E, Guglielmelli P, Pancrazzi A, Guerini V, Barosi G, Ruggeri M, Specchia G, LoCoco F, Delfini F, Villani L, Fiotto S, Ammatuna E, Alterini R, Carrai V, Capaccioli G, Di Lolo S, Liso V, Rambaldi A, Bosi A, Barbui T. Characteristics And Clinical Correlates of Mpl 515W>L/K Mutation In Essential Thrombocythemia. BLOOD. 2008 doi: 10.1182/blood-2008-01-135897. June 2, [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 34.Vannucchi AM, Antonioli E, Guglielmelli P, Pardanani A, Tefferi A. Clinical correlates of JAK2V617F presence or alelle burden in myeloproliferative neoplasms: a critical reappraisal. Leukemia. 2008;22:1299–1307. doi: 10.1038/leu.2008.113. [DOI] [PubMed] [Google Scholar]