Abstract
To test the significance of cyclic Notch activity for somite formation in mice, we analyzed embryos expressing activated Notch (NICD) throughout the presomitic mesoderm (PSM). Embryos expressing NICD formed up to 18 somites. Expression in the PSM of Hes7, Lfng, and Spry2 was no longer cyclic, whereas Axin2 was expressed dynamically. NICD expression led to caudalization of somites, and loss of Notch activity to their rostralization. Thus, segmentation and anterior–posterior somite patterning can be uncoupled, differential Notch signaling is not required to form segment borders, and Notch is unlikely to be the pacemaker of the segmentation clock.
Keywords: Notch signaling, segmentation, somite patterning, somitogenesis
Somitogenesis subdivides the paraxial mesoderm of vertebrate embryos into a series of homologous subunits, the somites. Somites form sequentially at the anterior end of the presomitic mesoderm (PSM). A molecular oscillator referred to as “segmentation clock” directs cyclic expression of genes in the PSM and is coupled with the progression of somitogenesis. Expression of cyclic genes is coordinated such that one wave of expression passes through the PSM during the formation of one somite (Palmeirim et al. 1997; McGrew et al. 1998; Jiang et al. 2000; Jouve et al. 2000; Aulehla et al. 2003). Microarray studies of the mouse PSM transcriptome showed in part mutually exclusive activation of the Notch, FGF, and Wnt pathways during each cycle, suggesting coordinated regulation of these three pathways (Dequeant et al. 2006). There is evidence that Wnt activity in the PSM acts upstream of Notch (Aulehla et al. 2003; Hofmann et al. 2004). However, cyclic Lfng expression was maintained in embryos with constitutive Wnt activity in the PSM (Aulehla et al. 2007). Similarly, the epistatic relationship of FGF and Wnt signaling is not clear, as studies in mice suggested both that FGF signaling acts upstream of Notch and Wnt (Wahl et al. 2007) and that Wnt is upstream of Notch and FGF (Dunty et al. 2008). Thus, it is still unclear how the activities of the Notch, FGF, and Wnt pathways are coordinated, and how these pathways contribute to segment border formation and somite patterning in mouse embryos.
Thus far, the most comprehensively studied genes displaying cyclic expression encode components of the Notch pathway. In mice, these are lunatic fringe (Lfng) and the bHLH genes Hes1, Hes7, and Hey2 (McGrew et al. 1998; Jouve et al. 2000; Leimeister et al. 2000; Bessho et al. 2001). Notch1 activity itself oscillates in the posterior PSM and appears to be arrested in the anterior PSM through Mesp2-induced repression by Lfng (Morimoto et al. 2005), a glycosyltransferase that modulates the receptiveness of Notch to various ligands (Hicks et al. 2000). Lfng expression is regulated by Notch activity (Morales et al. 2002), and Lfng was proposed to negatively regulate Notch1 activity in the PSM and to establish a negative feedback loop that drives cyclic Notch activity (Dale et al. 2003; Morimoto et al. 2005). Similarly, Hes7 oscillations are generated by a negative feedback loop of instable Hes7 protein periodically repressing Hes7 transcription (Bessho et al. 2001, 2003; Hirata et al. 2004). Mice homozygous for null alleles of Lfng and Hes7, respectively, display defects in somite compartmentalization, and somites are irregular in form and size (Zhang and Gridley 1998; Bessho et al. 2001).
Studies in different vertebrate species addressing Notch function during somitogenesis have led to various views as to the role of Notch in this process. It has been proposed that the segmentation clock regulates the periodic activation of Notch (Pourquie 1999; Serth et al. 2003), its signaling is required for the synchronization of the clock in neighboring cells (Jiang et al. 2000; Ozbudak and Lewis 2008), and the Notch pathway is part of the oscillator (Holley et al. 2002; Morales et al. 2002; Dale et al. 2003; Morimoto et al. 2005; Mara et al. 2007). In mouse embryos, extensive genetic studies led to the view that in the anterior PSM, Mesp2-induced down-regulation of Notch activity leads to somite boundaries where cells with activated Notch1 abut cells with repressed Notch1 activity, that the generation of somite polarity and borders is coupled, and Notch is part of the core of the segmentation clock (Takahashi et al. 2003; Morimoto et al. 2005; Saga 2007). This contrasts with recent findings in zebrafish that indicated a role for Notch signaling merely in the posterior PSM in the synchronization of cells (Ozbudak and Lewis 2008). To further study the role of Notch activity in somite formation and patterning in mice, we analyzed embryos expressing activated Notch (NICD) throughout the PSM. Our results argue against a role for Notch as a pacemaker of the clock, show that the confrontation of domains with and without Notch activity is not a prerequisite for border formation, and demonstrate that segmentation and anterior–posterior somite patterning can be uncoupled.
Results and Discussion
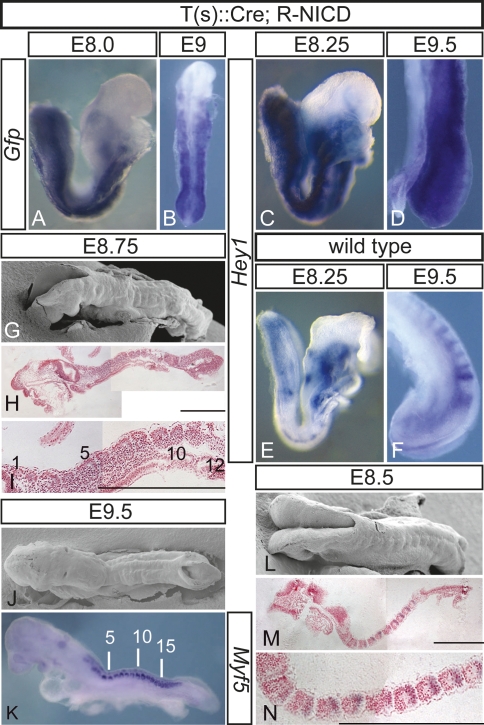
To activate Notch signaling throughout the PSM, we generated transgenic mice expressing Cre in the primitive streak under the control of regulatory elements of the brachyury gene (Stott et al. 1993), and crossed these mice to ROSANICD mice that allow for conditional expression of the constitutively active intracellular domain of Notch1 (NICD) by Cre-mediated excision of a stop cassette (Murtaugh et al. 2003). Cre activity resulted in reporter gene expression throughout mesodermal tissues (Supplemental Fig. 1), and in embryos carrying the ROSANICD allele and the Cre transgene (hereafter referred to as T-NICD embryos), NICD-Gfp fusion transcripts were detected throughout the PSM and in somites (Fig. 1A,B). The Notch target Hey1 was strongly up-regulated throughout the paraxial mesoderm of T-NICD embryos (Fig. 1, cf. C,D and E,F) demonstrating activation of the Notch pathway. T-NICD embryos did not complete turning, became severely retarded and distorted after embryonic day 9.5 (E9.5) with impaired elongation of the embryonic axis, and died around E10.5 with swollen pericardial sacs (data not shown), most likely due to cardiovascular defects. However, despite these defects, T-NICD embryos at E9.5 (n = 22) had formed on average 13 (range from 9 to 18) (Supplemental Fig. 2A) irregularly shaped somites of variable size (mean 70.35 μm, range 38–111 μm; n = 60) (Supplemental Fig. 2B) with clearly discernable segment borders (Fig. 1G–K), indicating that the presence of activated transgenic Notch (exoNICD) throughout the PSM did not prevent border formation.
Figure 1.
Segmentation in embryos with constitutive Notch activity. (A–F) In situ hybridization of embryos showing expression of the Nicd-Gfp fusion transcript throughout the paraxial mesoderm of T(s)∷Cre; R-NICD embryos (A,B) and up-regulation of Hey1 in the PSM and the somites (C,D) compared with wild-type embryos (E,F). (G,J,L) Scanning electron microscopic pictures illustrating somites in T(s)∷Cre; R-NICD (G,J) and wild-type (L) embryos. (H,I,M,N) Hemalaun-stained plastic sections of somites in T(s)∷Cre; R-NICD (H,I) and wild-type (M,N) embryos. (K) Segmented paraxial mesoderm in T(s)∷Cre; R-NICD embryos visualized by Myf5 expression. Developmental stages are indicated on top. Bars: H,I,M,N, 500 μm.
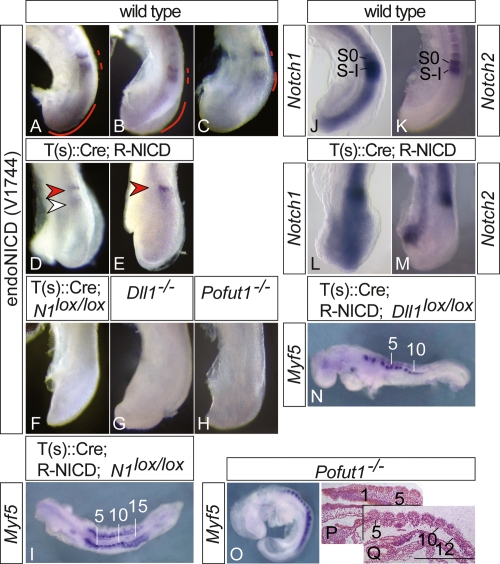
To address how activation of endogenous Notch1 is affected by exoNICD, we analyzed the presence of endogenous NICD using an antibody that specifically recognizes the cleaved form of Notch1 (but not the transgene-encoded NICD that lacks the V1744 epitope). In wild-type embryos, activated Notch1 (for clarity, from hereon referred to as endoNICD) was found in the posterior PSM in variable patterns, and in one or two bands in the anterior PSM (red lines in Fig. 2A–C), reflecting cyclic Notch activity. In contrast, T-NICD embryos (n = 21) showed one narrow stripe of endoNICD at the anterior end of the PSM (red arrowheads in Fig. 2D,E) that resembled the anterior endoNICD stripe of wild-type embryos, although levels appeared lower. In addition, a subset of T-NICD embryos (n = 13) had an additional weaker, fuzzy endoNICD stripe posterior to the anterior stripe (Fig. 2D, white arrowhead). In the posterior PSM region of T-NICD embryos, endoNICD was severely down-regulated (Fig. 2D,E), indicating that activation of Notch1 was disrupted in the T-NICD embryos.
Figure 2.
Endogenous Notch1 activity in wild-type and mutant embryos. (A–H) Immunohistochemical detection of activated endogenous Notch1 in the PSM. In contrast to wild-type embryos (A–C), endogenous Notch1 activity is down-regulated in the posterior but is found in one or two stripes in the anterior PSM (arrowheads in D,E) of T(s)∷Cre; R-NICD embryos. (F) Specificity of antibody is demonstrated by the lack of staining in T(s)∷Cre; N1loxp/loxP embryos. Loss of Dll1 (G) or Pofut1 (H) function completely abolishes Notch1 activity. (I,N,O) Segmented paraxial mesoderm in T(s)∷Cre; R-NICD; N1loxp/loxP (I), T(s)∷Cre; R-NICD; Dll1loxp/loxP (N), and Pofut−/− (O) embryos visualized by Myf5 expression. (J–M) Overlapping expression of Notch1 (J,L) and Notch2 (K,M) in the anterior PSM of wild-type (J,K) and T(s)∷Cre; R-NICD (L,M) embryos. (P,Q) Sections of plastic-embedded Pofut−/− embryos. Sections are 50 μm apart from the same serially sectioned embryo. Bar: Q, 500 μm.
The presence of a stripe(s) of endoNICD in the anterior PSM could generate regions where cells with higher levels of activated Notch (i.e., endoNICD plus exoNICD) abut cells with lower Notch activity (i.e., exoNICD only), which could underlie the generation of borders despite expression of exoNICD throughout the PSM. To test whether differential (endogenous) Notch1 activity in the anterior PSM might be responsible for border formation in T-NICD embryos, we simultaneously removed Notch1 (Supplemental Fig. 3A,B) from and expressed exoNICD in the paraxial mesoderm by combining a floxed Notch1 (Nicolas et al. 2003) and the ROSANICD allele. Embryos with this allele combination also formed ∼15 somites (Fig. 2I). In the anterior PSM of wild-type and T-NICD embryos, Notch2 expression overlaps with Notch1 expression in S0 and S-I (Fig. 2J–M). Loss of both Notch activities disrupts somitogenesis more severely than a mutation in either gene (Huppert et al. 2005), indicating some functional redundancy, and opening the possibility that Notch2 activity might contribute to somite border formation in T-NICD embryos. To test this possibility, we simultaneously removed Delta1 from (Supplemental Fig. 3C,D) and expressed exoNICD in the paraxial mesoderm by combining a floxed Delta1 (Hozumi et al. 2004) and the ROSANICD allele. Dll1 is the critical Notch ligand in the PSM (Fig. 2G). Its loss affects Notch target gene expression throughout the PSM (del Barco Barrantes et al. 1999; Jouve et al. 2000; Morales et al. 2002; Morimoto et al. 2005) and leads to somite patterning defects much more severe than loss of Notch1 (Conlon et al. 1995) and similar to the complete loss of Notch activity (Fig. 4, cf. E,F and G,H, below). Thus, removal of Dll1 activity should effectively eliminate endogenous Notch activation in the PSM. Embryos lacking Dll1 and expressing exoNICD were virtually identical to T-NICD embryos with respect to border formation (Fig. 2N), further supporting that segment borders form despite constitutive NICD expression in the paraxial mesoderm of T-NICD embryos. To address whether Notch activation in general is required for segment border formation, we analyzed embryos mutant for Pofut1, which encodes O-FucT-1, an enzyme essential for O-fucosylation of Notch EGF repeats and Notch activity (Fig. 2H). Loss of O-Fuct-1 activity abrogates all Notch activity and causes embryonic lethality at E9.5 (Fig. 2H; Shi and Stanley 2003), but mutant E9.25 embryos (n = 8) had on average 11 (range from 9 to 16) clearly discernable somites (Fig. 2O–Q). The apparently more severe segmentation defects in embryos without Notch activity compared with T-NICD embryos might be explained by the overall requirement for Notch during early embryonic development and the earlier arrest and demise of mutant embryos. Collectively, these data indicate that somite borders can form in the absence of Notch activity or the expression of exoNICD throughout the PSM, and thus without the confrontation of domains with and without Notch activity.
Figure 4.
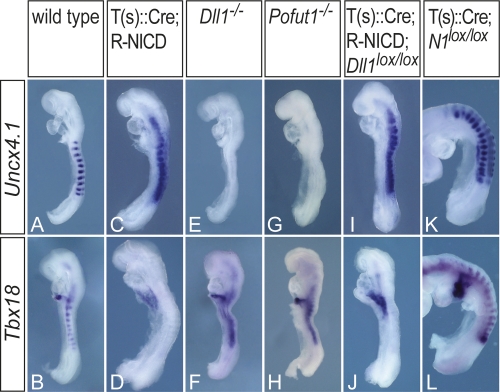
Disrupted somite polarity in transgenic embryos. Uncx4.1 (A,C,E,G,I,K) and Tbx18 (B,D,F,H,J,L) expression in wild-type (A,B), T(s)∷Cre; R-NICD (C,D), Dll1−/− (E,F), Pofut1−/− (G,H), T(s)∷Cre; R-NICD; Dlllox/lox (I,J), and T(s)∷Cre; N1loxp/loxP (K,L) embryos, indicating that constitutive Notch activity leads to caudalization of somites (C,D,I,J) and loss of all Notch activity to rostralization (E–H), whereas loss of Notch1 function alone has no significant impact on segment polarity (K,L).
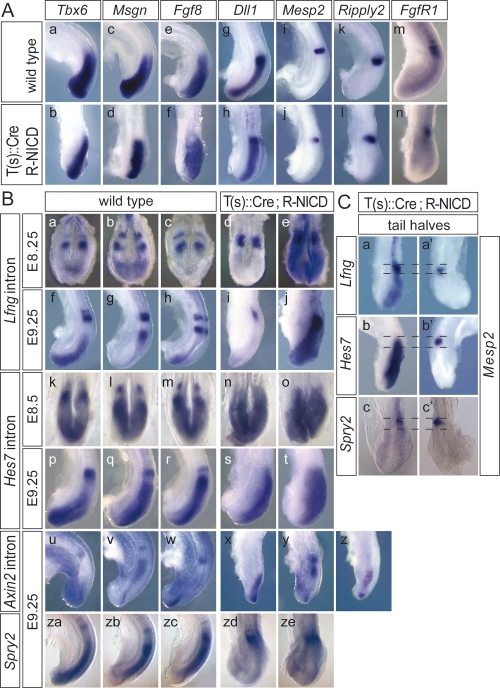
To address how exoNICD expression affects PSM patterning, we analyzed genes differentially expressed in the PSM. In T-NICD embryos, Tbx6, Msgn, and Fgf8 were expressed in apparently normal broad posterior domains (Fig. 3A, panels b,d,f). Likewise, Dll1 was expressed throughout the PSM with highest levels in the anterior region (Fig. 3A, panel h) similar to wild type. Mesp2, a gene essential for boundary formation (Saga et al. 1997), was expressed in T-NICD embryos (n = 18) in the anterior PSM in a single well defined stripe (Fig. 3A, panel j), whereas in wild-type embryos variable patterns of one or two stripes were found (data not shown; Takahashi et al. 2000). Since Mesp2 is synergistically activated in the anterior PSM by Tbx6 and Notch activity (Yasuhiko et al. 2006), and potentially repressed in the posterior PSM by FGF signaling (Delfini et al. 2005), it is possible that static Notch activity in T-NICD embryos disrupts dynamic Mesp2 activation but still allows for the stripe of Mesp2 expression, which appears to be sufficient to induce boundaries. Similarly, transcripts of Ripply2, a transcriptional target of Mesp2 (Morimoto et al. 2007), were found in one stripe in the anterior PSM (Fig. 3A, panel l), and the domain of high Fgfr1 expression in the anterior PSM was maintained (Fig. 3A, panel n). Collectively, these patterns suggested that the PSM was grossly patterned normal in the presence of constitutive Notch activity, but progression of somite maturation in the anterior PSM was affected. Importantly, oscillating expression of the Notch pathway genes Lfng and Hes7, two genes that were suggested to function as components of the segmentation clock (Bessho et al. 2003; Dale et al. 2003; Hirata et al. 2004; Morimoto et al. 2005), was not detected. In T-NICD embryos on E8.25 (n = 6) and 9.25 (n = 31), Lfng transcripts were found in a stripe in the anterior PSM at levels comparable to wild type (Fig. 3B, panels d,i), and throughout the posterior PSM at a significantly lower level that was detected only after prolonged color development (Fig. 3B, panels e,j). In contrast, Hes7 transcripts were detected uniformly throughout the PSM of E8.25 (n = 5) and E9.25 (n = 27) T-NICD embryos (Fig. 3, panels n,o,s,t), suggesting noncyclic or nonsynchronized expression. Expression tapered off in the anterior PSM in the region overlapping with Mesp2 and high levels of Lfng expression (Fig. 3C, panels b, b′).
Figure 3.
Patterning of the PSM in T(s)∷Cre; R-NICD embryos. (A) In situ hybridization of wild-type (panels a,c,e,g,i,k,m) and T(s)∷Cre; R-NICD (panels b,d,f,h,j,l,n) embryos with the probes indicated on top demonstrates grossly normal subdivision of the PSM of T(s)∷Cre; R-NICD embryos. (B) Cyclic expression of Lfng (panels a–c,f–h) and Hes7 (panels k–m,p–r) in wild-type and static expression throughout the PSM in T(s)∷Cre; R-NICD embryos (panels d,e,i,j,n,o,s,t). (Panels e,j) Extended color development revealed low levels of Lfng transcription throughout the posterior PSM. Cyclic expression of Axin2 both in wild-type (panels u–w) and T(s)∷Cre; R-NICD (panels x–z) embryos. Cyclic expression of Spry2 in wild-type (panels za–zc) is disrupted in T(s)∷Cre; R-NICD (panels zd,ze) embryos. (C) Tail halves of T(s)∷Cre; R-NICD hybridized with Mesp2 (panels a′,b′,c′) and Lfng (panel a), Hes7 (panel b), and Spry2 (panel c), indicating that in the anterior PSM the Mesp2 expression domain overlaps with the high Lfng and Spry2 expression and with the decreasing Hes7 expression domain. The dotted lines depict the overlapping domains.
Noncyclic expression of Hes7 and Lfng in T-NICD embryos is consistent with the transcriptional activation of both genes by Notch in the PSM (del Barco Barrantes et al. 1999; Morales et al. 2002; Bessho et al. 2003). Hes7 protein normally acts as a negative regulator of its own promoter (Bessho et al. 2003). Constitutive Hes7 transcription and therefore a presumably constant presence of Hes7 protein in the PSM could indicate that activation of Hes7 transcription by exoNICD overrides the repression by Hes7 protein. If that were the case, in wild-type embryos cyclic Notch inactivity might contribute to oscillating Hes7 transcription. In contrast, at the Lfng promoter, Hes7 protein appears to attenuate activation by exoNICD in the posterior PSM, leading to constant transcription but at lower levels than in wild type. Higher Lfng expression in the anterior PSM, where it overlaps with Hes7, could be due to the presence of Mesp2 (Fig. 3C, panels a, a′), which activates Lfng expression (Morimoto et al. 2005). Lfng was suggested to act as a negative regulator of Notch in the PSM (Morimoto et al. 2005). In the posterior PSM the low but constant Lfng activity in T-NICD embryos might be sufficient to repress endoNICD formation (Fig. 2D,E). However, high levels of Lfng in the anterior PSM should much more effectively repress endoNICD formation. Although the detected activation of Notch1 in the anterior PSM could reflect the significantly higher levels of Notch1 and Dll1 (Figs. 2L, 3A, panel h) in this region, our observations are much more compatible with Lfng promoting Notch activation by Dll1 as was shown in vitro (Hicks et al. 2000).
Since there are conflicting data concerning the epistatic relationship of Wnt, FGF, and Notch signaling in the PSM (Aulehla et al. 2003, 2007; Hofmann et al. 2004; Wahl et al. 2007; Dunty et al. 2008), we analyzed expression of Axin2 and Spry2, whose expression reflects Wnt and Fgf activity, respectively. Axin2 showed variable expression patterns in the PSM of T-NICD embryos (n = 19) (Fig. 3B, panels x–z). In contrast, Spry2, a negative regulator of FGF signaling, was expressed in a stripe in the anterior PSM but was severely down-regulated throughout the posterior PSM of T-NICD embryos (n = 28) (Fig. 3B [panels zd, ze], C [panel c]; Supplemental Fig. 4) similar to Lfng, suggesting that exoNICD impacts on FGF signaling. Therefore, we analyzed the expression of Snail, another FGF target. Snail expression (n = 8) appeared up-regulated in the PSM (Supplemental Fig. 5), suggesting that down-regulation of Spry2 in the posterior PSM of T-NICD embryos leads to overall increased FGF activity. This implies that down-regulation of Spry2 by exoNICD occurs independently from FGF signaling. Similarly, pharmacological blocking of FGF signaling appeared to block Lfng oscillations indirectly (Wahl et al. 2007). Thus, although Spry2 was repressed in the posterior PSM, we cannot rule out that FGF activity in T-NICD embryos might still be dynamic since cyclic Spry2 expression was observed in RBPjκ mutants (Dequeant et al. 2006). These uncertainties notwithstanding, our findings suggest that Notch signaling does not constitute the pacemaker of the segmentation clock in mice, and are consistent with previous observations that oscillations of Wnt activity are independent from Dll1 (Aulehla et al. 2003).
Notch signaling is essential for establishment of rostro-caudal somite polarity (Hrabe de Angelis et al. 1997; Takahashi et al. 2003). In T-NICD embryos, somites were completely caudalized (Fig. 4C,D) as indicated by the expression of Uncx4.1, a marker of the caudal compartment, throughout somites, and loss of Tbx18, which is normally expressed in rostral halves. Loss of Dll1 or Pofut1 function led to the opposite phenotype, i.e., loss of Uncx4.1 and expansion of Tbx18 expression throughout somites (Fig. 4E–H). The segment polarity defect in Dll1 mutants could completely be converted by T-NICD (Fig. 4I,J), whereas loss of Notch1 alone had no major effect on rostro-caudal patterning (Fig. 4K,L). Thus, under conditions of constant Notch activity or complete loss of Notch activity, somite polarity is not established, indicating that the generation of domains with and without Notch activity in the anterior PSM is essential for somite compartmentalization.
In summary, our data show that in mouse embryos, somite borders can form in the presence of constitutive Notch activity as well as without Notch activity. Thus, the confrontation of domains with and without active Notch is unlikely to underlie somite border formation in mice, implying that Notch-independent mechanisms operate during boundary formation. A recent study in zebrafish showed that also in teleost fish Notch signaling is not essential for somite boundary formation but synchronizes the segmentation clock (Ozbudak and Lewis 2008). Misalignment of borders and irregular size and shape of somites in mouse embryos with constitutive Notch activity, or without Notch activity, is consistent with similar Notch functions in mice. Thus, our results raise the possibility that in mouse embryos Notch signaling in the posterior PSM functions similar to zebrafish and might coordinate and synchronize cohorts of prospective somite cells. However, whereas Notch activity in the anterior PSM of zebrafish embryos appears to be dispensable for somite patterning (Ozbudak and Lewis 2008), our findings support that Notch activity in the anterior PSM of mice is essential for subdividing these cohorts into rostral and caudal compartments (Takahashi et al. 2000, 2003), and suggest that the generation of domains with and without active Notch underlies this process. Our results also show that segmentation does not depend on compartmentalization since borders can form in the absence of segment polarity. Furthermore, our findings and the observation that Notch signaling still cycles in the presence of constitutive Wnt activity in the PSM (Aulehla et al. 2007) suggest that both pathways can generate independent oscillations and act in parallel, whereas Notch and FGF activities might be linked.
Materials and methods
Mice and genotyping of transgenic mice
Regulatory elements of the brachyury gene directing gene expression in the primitive streak (Stott et al. 1993) were fused to a Cre cDNA, and T(s)∷Cre transgenic mice were generated by standard procedures. The other mouse lines have been described previously: ROSA-NICD (Murtaugh et al. 2003), floxed Notch1 (Nicolas et al. 2003), floxed Delta1 (Hozumi et al. 2004), and Pofut mutants (Shi and Stanley 2003). Genomic DNA isolated either from tails or yolk sacs of embryos was genotyped using the following primer combinations: Notch1LoxP, N1-5′Lox2 (GTAGACTGGAAGCTACTG) and N1-3′Lox2 (GCATGAAGTGGTC CAGGGTG); RosaNICD, NICDfor (AAAGTCGCTCTGAGTTGTTAT) and SA-rev (GAAAGACCGCGAAGAGTTTG) for the transgene and Wt-rev (TAAGCCTGCCCAGAAGACTC) for the wild-type locus; Dll12LoxP, Dll1-5′lox (GCATTTCTCACACACCTC) and Dll1-3′lox (GAGAGTACTTGATGGAGCAAG); Pofut, Pofut-for (AATGCCGTGC TGAGAGTAAAGG) and Pofut-rev (ACCCACAGGCTGTGCAGT CTTTG); T(s)∷Cre, T-for (AATCTTTGGGCTCCGCAGAG) and Cre-rev (ACGTTCACCGGCATCAACG); Dll1LacZ, Melta38 (ATCCCTGGG TCTTTGAAGAAG) and LacZ-rev (CAAATTCAGACGGCAAAC) for the mutant and Dll1-Wt-for (CTGCTGCGCGGTGGAGGGAGG) and Dll1-Wt-rev (GGAGTCGACACCCAGCACTGGCG) for the wild-type allele.
Whole-mount in situ hybridization
Whole-mount in situ hybridizations were performed following standard procedures with digoxygenin-labeled antisense riboprobes. All probes were cDNA probes except for Lfng, Hes7, and Axin2, for which probes derived from the respective first intron were used.
Histology and scanning electron microscopy
Embedding, sectioning of embryos, and scanning electron microscopy were performed by standard procedures.
Antibody staining
Embryos were dissected in PBS and fixed for 1 min with 50% Methanol/50% DMSO (Fluka), and treated with 15% H2O2 in PBS containing 50 mM NH4Cl for 15 min at 4°C. Primary (anti-cleaved Notch1, Cell Signaling), secondary (anti-rabbit biotinylated BA1000, Vector), and tertiary (Streptavidin-HRP, Perkin Elmer) antibodies were each diluted 1:100 in 1%Triton X-100, 10% FCS-PBS (TFP) and sequentially incubated o/n at 4°C with intermediate washes in TFP. Visualization was done using 4-chloro-1-naphtol as substrate.
Somite size measurement
Ten clearly discernable somites per wild-type and T-NICD embryo (n = 6, respectively) were measured in dorsal photomicrographs using the ImageJ software (National Institutes of Health, Bethesda, MD). Analysis was performed with Prism software (GraphPad).
Acknowledgments
We thank Julian Lewis and Katsuto Hozumi, Freddy Radtke, Doug Melton, and Pamela Stanley for providing Dll1loxP, N1loxP, ROSA-NICD, and Pofut1 mutant mice, respectively; Bernhard Herrmann for the T-promoter streak element; David Ish-Horowicz, Ryoichiro Kageyama, Yumiko Saga, Andreas Kispert, and Gerry Weinmaster for probes; and Anatoli Heiser for help with mouse husbandry and genotyping. This work was supported by a DFG grant to A.G.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.480408.
References
- Aulehla A., Wehrle C., Brand-Saberi B., Kemler R., Gossler A., Kanzler B., Herrmann B.G. Wnt3a plays a major role in the segmentation clock controlling somitogenesis. Dev. Cell. 2003;4:395–406. doi: 10.1016/s1534-5807(03)00055-8. [DOI] [PubMed] [Google Scholar]
- Aulehla A., Wiegraebe W., Baubet V., Wahl M.B., Deng C., Taketo M., Lewandoski M., Pourquie O. A β-catenin gradient links the clock and wavefront systems in mouse embryo segmentation. Nat. Cell Biol. 2007;10:186–193. doi: 10.1038/ncb1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho Y., Sakata R., Komatsu S., Shiota K., Yamada S., Kageyama R. Dynamic expression and essential functions of Hes7 in somite segmentation. Genes & Dev. 2001;15:2642–2647. doi: 10.1101/gad.930601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho Y., Hirata H., Masamizu Y., Kageyama R. Periodic repression by the bHLH factor Hes7 is an essential mechanism for the somite segmentation clock. Genes & Dev. 2003;17:1451–1456. doi: 10.1101/gad.1092303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon R.A., Reaume A.G., Rossant J. Notch1 is required for the coordinate segmentation of somites. Development. 1995;121:1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- Dale J.K., Maroto M., Dequeant M.L., Malapert P., McGrew M., Pourquie O. Periodic Notch inhibition by Lunatic Fringe underlies the chick segmentation clock. Nature. 2003;421:275–278. doi: 10.1038/nature01244. [DOI] [PubMed] [Google Scholar]
- del Barco Barrantes I., Elia A.J., Wünsch K., Hrabe De Angelis M., Mak T.W., Rossant R., Conlon R.A., Gossler A., de la Pompa J.-L. Interaction between L-fringe and Notch signalling in the regulation of boundary formation and posterior identity in the presomitic mesoderm of the mouse. Curr. Biol. 1999;9:470–480. doi: 10.1016/s0960-9822(99)80212-7. [DOI] [PubMed] [Google Scholar]
- Delfini M.C., Dubrulle J., Malapert P., Chal J., Pourquie O. Control of the segmentation process by graded MAPK/ERK activation in the chick embryo. Proc. Natl. Acad. Sci. 2005;102:11343–11348. doi: 10.1073/pnas.0502933102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dequeant M.L., Glynn E., Gaudenz K., Wahl M., Chen J., Mushegian A., Pourquie O. A complex oscillating network of signaling genes underlies the mouse segmentation clock. Science. 2006;314:1595–1598. doi: 10.1126/science.1133141. [DOI] [PubMed] [Google Scholar]
- Dunty W.C., Biris K.K., Chalamalasetty R.B., Taketo M.M., Lewandoski M., Yamaguchi T.P. Wnt3a/β-catenin signaling controls posterior body development by coordinating mesoderm formation and segmentation. Development. 2008;135:85–94. doi: 10.1242/dev.009266. [DOI] [PubMed] [Google Scholar]
- Hicks C., Johnston S.H., diSibio G., Collazo A., Vogt T.F., Weinmaster G. Fringe differentially modulates Jagged1 and Delta1 signalling through Notch1 and Notch2. Nat. Cell Biol. 2000;2:515–520. doi: 10.1038/35019553. [DOI] [PubMed] [Google Scholar]
- Hirata H., Bessho Y., Kokubu H., Masamizu Y., Yamada S., Lewis J., Kageyama R. Instability of Hes7 protein is crucial for the somite segmentation clock. Nat. Genet. 2004;36:750–754. doi: 10.1038/ng1372. [DOI] [PubMed] [Google Scholar]
- Hofmann M., Schuster-Gossler K., Watabe-Rudolph M., Aulehla A., Herrmann B.G., Gossler A. WNT signaling, in synergy with T/TBX6, controls Notch signaling by regulating Dll1 expression in the presomitic mesoderm of mouse embryos. Genes & Dev. 2004;18:2712–2717. doi: 10.1101/gad.1248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holley S.A., Julich D., Rauch G.J., Geisler R., Nusslein-Volhard C. her1 and the notch pathway function within the oscillator mechanism that regulates zebrafish somitogenesis. Development. 2002;129:1175–1183. doi: 10.1242/dev.129.5.1175. [DOI] [PubMed] [Google Scholar]
- Hozumi K., Negishi N., Suzuki D., Abe N., Sotomaru Y., Tamaoki N., Mailhos C., Ish-Horowicz D., Habu S., Owen M.J. Delta-like 1 is necessary for the generation of marginal zone B cells but not T cells in vivo. Nat. Immunol. 2004;5:638–644. doi: 10.1038/ni1075. [DOI] [PubMed] [Google Scholar]
- Hrabe de Angelis M., McIntyre J., Gossler A. Maintenance of somite borders in mice requires the Delta homologue DII1. Nature. 1997;386:717–721. doi: 10.1038/386717a0. [DOI] [PubMed] [Google Scholar]
- Huppert S.S., Ilagan M.X., De Strooper B., Kopan R. Analysis of Notch function in presomitic mesoderm suggests a γ-secretase-independent role for presenilins in somite differentiation. Dev. Cell. 2005;8:677–688. doi: 10.1016/j.devcel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Jiang Y.J., Aerne B.L., Smithers L., Haddon C., Ish-Horowicz D., Lewis J. Notch signalling and the synchronization of the somite segmentation clock. Nature. 2000;408:475–479. doi: 10.1038/35044091. [DOI] [PubMed] [Google Scholar]
- Jouve C., Palmeirim I., Henrique D., Beckers J., Gossler A., Ishhorowcz D., Pourquié O. Notch signaling is required for cyclic expression of the hairy-like gene HES1 in the presomitic mesoderm. Development. 2000;127:1421–1429. doi: 10.1242/dev.127.7.1421. [DOI] [PubMed] [Google Scholar]
- Leimeister C., Dale K., Fischer A., Klamt B., Hrabe de Angelis M., Radtke F., McGrew M.J., Pourquie O., Gessler M. Oscillating expression of c-Hey2 in the presomitic mesoderm suggests that the segmentation clock may use combinatorial signaling through multiple interacting bHLH factors. Dev. Biol. 2000;227:91–103. doi: 10.1006/dbio.2000.9884. [DOI] [PubMed] [Google Scholar]
- Mara A., Schroeder J., Chalouni C., Holley S.A. Priming, initiation and synchronization of the segmentation clock by deltaD and deltaC. Nat. Cell Biol. 2007;9:523–530. doi: 10.1038/ncb1578. [DOI] [PubMed] [Google Scholar]
- McGrew M.J., Dale J.K., Fraboulet S., Pourquie O. The lunatic Fringe gene is a target of the molecular clock linked to somite segmentation in avian embryos. Curr. Biol. 1998;8:979–982. doi: 10.1016/s0960-9822(98)70401-4. [DOI] [PubMed] [Google Scholar]
- Morales A.V., Yasuda Y., Ish-Horowicz D. Periodic lunatic fringe expression during segmentation is controlled by a cyclic transcriptional enhancer responsive to Notch signalling. Dev. Cell. 2002;3:63–74. doi: 10.1016/s1534-5807(02)00211-3. [DOI] [PubMed] [Google Scholar]
- Morimoto M., Takahashi Y., Endo M., Saga Y. The Mesp2 transcription factor establishes segmental borders by suppressing Notch activity. Nature. 2005;435:354–359. doi: 10.1038/nature03591. [DOI] [PubMed] [Google Scholar]
- Morimoto M., Sasaki N., Oginuma M., Kiso M., Igarashi K., Aizaki K., Kanno J., Saga Y. The negative regulation of Mesp2 by mouse Ripply2 is required to establish the rostro-caudal patterning within a somite. Development. 2007;134:1561–1569. doi: 10.1242/dev.000836. [DOI] [PubMed] [Google Scholar]
- Murtaugh L.C., Stanger B.Z., Kwan K.M., Melton D.A. Notch signaling controls multiple steps of pancreatic differentiation. Proc. Natl. Acad. Sci. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas M., Wolfer A., Raj K., Kummer J.A., Mill P., van Noort M., Hui C.C., Clevers H., Dotto G.P., Radtke F. Notch1 functions as a tumor suppressor in mouse skin. Nat. Genet. 2003;33:416–421. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- Ozbudak E.M., Lewis J. Notch signalling synchronizes the zebrafish segmentation clock but is not needed to create somite boundaries. PLoS Genet. 2008;4:e15. doi: 10.1371/journal.pgen.0040015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmeirim I., Henrique D., Ish-Horowicz D., Pourquie O. Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell. 1997;91:639–648. doi: 10.1016/s0092-8674(00)80451-1. [DOI] [PubMed] [Google Scholar]
- Pourquie O. Notch around the clock. Curr. Opin. Genet. Dev. 1999;9:559–565. doi: 10.1016/s0959-437x(99)00011-8. [DOI] [PubMed] [Google Scholar]
- Saga Y. Segmental border is defined by the key transcription factor Mesp2, by means of the suppression of Notch activity. Dev. Dyn. 2007;236:1450–1455. doi: 10.1002/dvdy.21143. [DOI] [PubMed] [Google Scholar]
- Saga Y., Hata N., Koseki H., Taketo M.M. Mesp2: A novel mouse gene expressed in the presegmented mesoderm and essential for segmentation initiation. Genes & Dev. 1997;11:1827–1839. doi: 10.1101/gad.11.14.1827. [DOI] [PubMed] [Google Scholar]
- Serth K., Schuster-Gossler K., Cordes R., Gossler A. Transcriptional oscillation of lunatic fringe is essential for somitogenesis. Genes & Dev. 2003;17:912–925. doi: 10.1101/gad.250603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S., Stanley P. Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc. Natl. Acad. Sci. 2003;100:5234–5239. doi: 10.1073/pnas.0831126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stott D., Kispert A., Herrmann B.G. Rescue of the tail defect of Brachyury mice. Genes & Dev. 1993;7:197–203. doi: 10.1101/gad.7.2.197. [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Koizumi K., Takagi A., Kitajima S., Inoue T., Koseki H., Saga Y. Mesp2 initiates somite segmentation through the Notch signalling pathway. Nat. Genet. 2000;25:390–396. doi: 10.1038/78062. [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Inoue T., Gossler A., Saga Y. Feedback loops comprising Dll1, Dll3 and Mesp2, and differential involvement of Psen1 are essential for rostrocaudal patterning of somites. Development. 2003;130:4259–4268. doi: 10.1242/dev.00629. [DOI] [PubMed] [Google Scholar]
- Wahl M.B., Deng C., Lewandoski M., Pourquie O. FGF signaling acts upstream of the NOTCH and WNT signaling pathways to control segmentation clock oscillations in mouse somitogenesis. Development. 2007;134:4033–4041. doi: 10.1242/dev.009167. [DOI] [PubMed] [Google Scholar]
- Yasuhiko Y., Haraguchi S., Kitajima S., Takahashi Y., Kanno J., Saga Y. Tbx6-mediated Notch signaling controls somite-specific Mesp2 expression. Proc. Natl. Acad. Sci. 2006;103:3651–3656. doi: 10.1073/pnas.0508238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Gridley T. Defects in somite formation in lunatic fringe-deficient mice. Nature. 1998;394:374–377. doi: 10.1038/28625. [DOI] [PubMed] [Google Scholar]