Abstract
The 274 tRNA genes in Saccharomyces cerevisiae are scattered throughout the linear maps of the 16 chromosomes, but the genes are clustered at the nucleolus when compacted in the nucleus. This clustering is dependent on intact nucleolar organization and contributes to tRNA gene-mediated (tgm) silencing of RNA polymerase II transcription near tRNA genes. After examination of the localization mechanism, we find that the chromosome-condensing complex, condensin, is involved in the clustering of tRNA genes. Conditionally defective mutations in all five subunits of condensin, which we confirm is bound to active tRNA genes in the yeast genome, lead to loss of both pol II transcriptional silencing near tRNA genes and nucleolar clustering of the genes. Furthermore, we show that condensin physically associates with a subcomplex of RNA polymerase III transcription factors on the tRNA genes. Clustering of tRNA genes by condensin appears to be a separate mechanism from their nucleolar localization, as microtubule disruption releases tRNA gene clusters from the nucleolus, but does not disperse the clusters. These observations suggest a widespread role for condensin in gene organization and packaging of the interphase yeast nucleus.
Keywords: tRNA gene, condensin, microtubules, nuclear organization, nucleolus
Numerous levels of preferential DNA positioning within nuclei have now been described (Heun et al. 2001b; Parada et al. 2003, 2004; Hurst et al. 2004; Misteli 2004; Taddei et al. 2004; Foster and Bridger 2005; Bartova and Kozubek 2006; Cremer et al. 2006; Thanbichler and Shapiro 2006). Examples include global effects like chromosome territories (Foster and Bridger 2005; Cremer et al. 2006) and transcription factories (Faro-Trindade and Cook 2006), and specific effects such as telomeres and centromeres at the yeast nuclear periphery (Heun et al. 2001a; Hediger et al. 2002), the large ribosomal DNA repeats at the nucleoli of all organisms (Shaw et al. 1995), and the 5S rDNA at or near the nucleoli of many organisms (Haeusler and Engelke 2006). Multiple genes alter their position when transcriptionally activated (Buchenau et al. 1997; Brown et al. 1999; Volpi et al. 2000; Kosak et al. 2002; Casolari et al. 2004, 2005; Harnicarova et al. 2006), and some genes are brought together with linearly distant enhancer elements (Liu and Garrard 2005; Spilianakis et al. 2005; Liu and Francke 2006; Su et al. 2006). In some cases, genes that are coregulated but not linearly connected are brought together in three dimensions, including β-globin genes that are found clustered when active (Brown et al. 2006), and the nucleolar clustering of most of the 274 tRNA genes in Saccharomyces cerevisiae, which are transcribed by RNA polymerase III (pol III) (Thompson et al. 2003; L. Wang et al. 2005). Although the mechanisms of spatial genome organization are not well defined, a few elements have been determined for specific cases. The long-range movement of at least one inducible gene requires actin/myosin (Chuang et al. 2006). The transcription-dependent tethering of some genes at the nuclear pore depends on interactions between the gene promoters and the basket of the nuclear pore complex (Schmid et al. 2006). Centromere clustering at the spindle pole body requires microtubules (Bystricky et al. 2004), as does homologous recombination, which requires spatial proximity of the two loci (Thrower et al. 2003).
The nucleolar localization of most tRNA genes in S. cerevisiae is a particularly pervasive example of genome-wide gene positioning. The spatial clustering was originally discovered because proximity to tRNA genes leads to transcriptional silencing of nearby RNA polymerase II (pol II) promoters, termed tRNA gene mediated (tgm) silencing (Kinsey and Sandmeyer 1991; Hull et al. 1994; Kendall et al. 2000). The colocalization of the tRNA genes may also contribute to the elevated rate of recombination between active tRNA genes in yeast (Pratt-Hyatt et al. 2006).
To identify requirements for nucleolar clustering of tRNA genes, we first examined the influence of the cell cycle and the involvement of microtubules on tRNA gene positions. We observed that tRNA genes associate with the nucleolus throughout division, and that microtubule depolymerization releases intact clusters of tRNA genes away from the nucleolus, suggesting that some separate mechanism was responsible for bringing the tRNA genes together.
To investigate the mechanism of tRNA gene clustering, the involvement of the protein complex condensin was examined. This highly conserved complex (Legagneux et al. 2004; Hirano 2005) is involved in the compaction of chromosomes, and the mechanism of compaction is generally thought to involve direct condensin–condensin interactions among condensin complexes bound to dispersed sites in the genome (Losada and Hirano 2005; Nasmyth and Haering 2005; Thanbichler and Shapiro 2006). Condensin complexes have five essential subunits: two core “structural maintenance of chromosomes” (SMC) subunits (Smc2 and Smc4), which are thought to be targeted to chromosomes by the three regulatory non-SMC subunits (Ycs4, Ycg1, and Brn1 in S. cerevisiae). Prior studies of yeast condensin have emphasized its role in the dramatic condensation of the nucleolar rDNA locus (Guacci et al. 1994; Freeman et al. 2000; Lavoie et al. 2000, 2002; Bhalla et al. 2002), and using chromatin immunoprecipitation (ChIP) with PCR and standard microarray chips, the complex has previously been found to bind to multiple preferential sites along every yeast chromosome (B.D. Wang et al. 2005). However, binding of individual condensin complexes to a short DNA elements such as tRNA genes might not be detected at the resolution used in the earlier microarray study. High-resolution, genome-wide ChIP results presented in an accompanying paper (D’Ambrosio et al. 2008) show that sharply localized condensin is preferentially bound at tRNA genes, prompting us to test whether condensin participated in clustering.
The role of condensin in structural organization of nucleolar DNA and its specific binding to tRNA genes prompted the examination of mutations in all five condensin subunits for effects on tRNA gene organization: first using tgm silencing as an indirect, functional evaluation for proper localization and then directly by DNA in situ fluorescence microscopy. The results show that the condensin complex is required for clustering of dispersed tRNA genes in yeast. We go on to show that condensin is stably associated with a complex of bound transcription factors on the tRNA genes, accounting for the specificity of the condensin interactions at tRNA gene sites and pol III transcription factor sites in the yeast genome.
Results
tRNA genes remain clustered at the nucleolus throughout the cell cycle
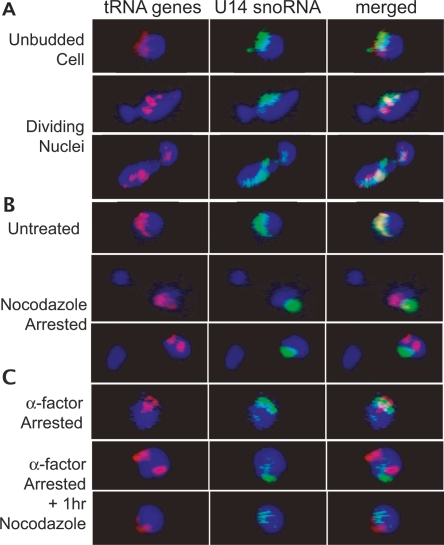
Clustering of most of the 274 yeast tRNA genes near the nucleolus affects the organization of much of the interphase genome, but previously little was known about the mechanism. As a first step, we asked whether the localization was dynamic during the cell cycle or whether tRNA gene clusters continually associated with the nucleolus, which remains intact throughout mitosis in S. cerevisiae (Loidl 2003). Staining of nucleolar proteins and ribosomal DNA (rDNA) throughout mitosis has indicated that this region is one of the last in the genome to divide (Fuchs and Loidl 2004; Machin et al. 2005). To examine the nucleolar association of tRNA genes during mitosis, extensive imaging was first undertaken in unsynchronized cell populations, using in situ hybridization to directly visualize a representative tRNALeu(CAA) gene family, one of several tRNA gene families originally tested (Thompson et al. 2003). Cells captured throughout mitosis, as determined by bud size in the DIC image, were imaged and analyzed. Representative cells undergoing nuclear division are shown in Figure 1A. In these cells and in cells from each visually distinct phase (additional images in Supplement Fig. S1), we observed nearly complete overlap of tRNA gene signal with a nucleolar marker. Similar results were obtained when cells were synchronized in G1 with the mating pheromone α factor or synchronized with α factor and then released for varying time periods, or synchronized in S phase with hydroxyurea (data not shown). Although it is possible that individual tRNA genes become transiently dissociated, since signal from individual genes is not detectable, the observation that tRNA genes continue to associate with a dividing nucleolus argues that the bulk of the tRNA genes remain clustered with the nucleolus during its partitioning late in nuclear division.
Figure 1.
In situ hybridization of unsynchronized and nocodazole-treated cells. In each panel, fluorescent oligonucleotide probes complementary to the U14 snoRNA (green) or 10 tRNALeu(CAA) genes (red) were used for hybridization. Blue represents DAPI staining of nucleoplasmic DNA. (A) Nuclei from unsynchronized cells show that the tRNA gene signal (red) consistently overlaps the nucleolar signal (green) prior to and throughout division of the nucleus. (B) Depolymerization of microtubules by arrest in nocodazole causes partially divided nuclei in which tRNA genes remain clustered, but clusters are divorced from the nucleolus. (C) This effect is not due to nocodozole blockage prior to nuclear division, as demonstrated by the release of tRNA gene clusters from the nucleolus by depolymerization of microtubules in cells arrested in G1 by α factor.
Nucleolar localization of tRNA gene clusters requires microtubules
One hypothesis for the mechanism of nucleolar tRNA gene clustering was that a structural element, such as microtubules, might be involved. Deletions and mutations of various microtubule motor and actin microfilament components did not affect the silencing of pol II transcription near tRNA genes, which requires nucleolar clustering of tRNA genes (L. Wang et al. 2005), but many of these genes are redundant or essential, so deletions that are viable might not be informative. To directly assess the involvement of microtubules in tRNA gene positioning, nocodazole was used at conditions known to cause microtubule disassembly (Jacobs et al. 1988), and tRNA genes were subsequently visualized as described (Thompson et al. 2003). The nocodazole treatment of these cells did not strongly inhibit tRNA expression or release tgm silencing (Supplement Fig. S2) but did result in a shift in the subnuclear positioning of the tRNA gene clusters. As shown in Figure 1B, cells arrested with nocodazole in late G2 phase of the cell cycle show tRNA genes that are gathered in one or two clusters but divorced from the nucleolus. The proportion of cells with a single nucleolar cluster of tRNA genes was greatly reduced from 76% in wild type, unsynchronized cells to 30% in nocodazole-arrested cells. A similar pattern was observed for cells treated with nocodazole for only 1 h (data not shown). This phenotype is distinct from one in which tRNA genes become dispersed throughout the nucleoplasm, as seen in some nucleolar mutants that release tgm silencing (L. Wang et al. 2005). To ascertain whether nonnucleolar tRNA gene clusters were due to nocodazole disruption of nuclear structure, and not as a result of arrest in G2/M, untreated cells in a large-budded state (similar to nocodazole-arrested cells) were evaluated. The proportion of cells with a single cluster of tRNA genes, where the cluster was nucleolar, was similar to the overall unsynchronized cell population (83%), indicating that cell cycle stage was not the cause of the divorce of tRNA gene clusters from the nucleolus. Furthermore, nocodazole depolymerization of microtubules in G1-arrested cells also loses nucleolar localization of tRNA gene clusters. In contrast, cells arrested in G1 with α factor alone show tRNA genes at the nucleolus (Fig. 1C). Thus, maintenance of tRNA gene positioning is dependent throughout the cell cycle on intact microtubules, although this requirement could be either direct or indirect. In either case, the persistent tRNA gene clustering in the absence of microtubules suggests tRNA gene clustering is caused by a separate clustering mechanism that is independent of overall nuclear organization.
Clustering of tRNA genes is condensin-dependent
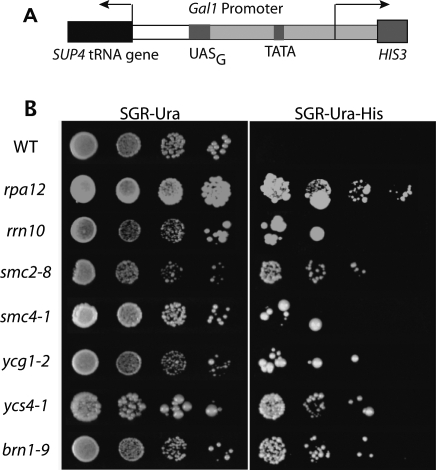
To identify potential genetic components of tRNA gene clustering, release of tgm silencing of pol II transcription was used as an initial screen. tgm silencing requires nucleolar clustering of tRNA genes (L. Wang et al. 2005). The condensin complex was chosen as one target in this screen because of its known involvement in chromosome organization, particularly at the nucleolar rDNA repeats, and the recent discovery of condensin occupancy at yeast tRNA genes (D’Ambrosio et al. 2008). Because the five condensin subunits are all essential, temperature-sensitive mutants in all five subunits were tested: smc2-8 (Freeman et al. 2000), smc4-1 (Freeman et al. 2000), ycg1-2 (Lavoie et al. 2002), ycs4-1 (Bhalla et al. 2002), and brn1-9 (Lavoie et al. 2002). Loss of tgm silencing from a reporter plasmid in mutant strains allows growth on media lacking histidine (Fig. 2A; Hull et al. 1994; Kendall et al. 2000; L. Wang et al. 2005). Plates were grown at 23°C, the permissible temperature for each strain (Freeman et al. 2000; Bhalla et al. 2002; Lavoie et al. 2002), and at 30°C, where the strains can still grow but the mutant proteins may be partially defective. The silencing phenotypes were the same at both temperatures, and 30°C data are shown. Growth of these mutants is arrested at 37°C.
Figure 2.
Condensin mutants release tgm silencing. Strains containing mutations in each condensin subunit were transformed with a plasmid containing a tgm silencing test construct (shown in A) (Hull et al. 1994; Kendall et al. 2000; L. Wang et al. 2005), where the SUP4 tRNA gene silences HIS3 expression in wild-type cells. Cells with defective tgm silencing are able to grow on media lacking histidine (SGR-Ura-His) (B). Growth at semi-permissive temperature for the temperature-sensitive mutants (30°C) is shown, although growth at 23°C gave similar results (data not shown). All five condensin mutants release tgm silencing to varying extents, similar to deletions of RNA polymerase I components Δrpa12 and Δrrn10, strains that are known to release tgm silencing (L. Wang et al. 2005).
Growth at 30°C is shown in Figure 2B. In contrast to wild-type yeast, each of the condensin mutants shows some growth in the absence of histidine, consistent with release of tgm silencing. As with all mutations previously shown to release tgm silencing (Kendall et al. 2000; L. Wang et al. 2005), a minority of cells are able to grow on selective media, and the extent varies between strains. For comparison, loss of silencing phenotype is shown for two previously characterized strains (rpa12 and rrn10) with deletions affecting ribosomal RNA gene transcription and nucleolar integrity and causing dispersal of the tRNA genes. Mutations affecting nucleolar function and compaction do not normally release tgm silencing and tRNA gene position, however (L. Wang et al. 2005) Many nucleolar defects were shown to not affect tgm silencing, including the toposisomerases implicated in ribosomal DNA repeat condensation (Supplemental Fig. S3; L. Wang et al. 2005). Surprisingly, each condensin mutant also demonstrates release of silencing at 23°C, where the defects are not sufficient to cause severe growth defects (data not shown). The variability in colony size seen in some mutants is of unknown origin but is reproducible at both growth temperatures, and restreaking various sized colonies continues to give mixed colony sizes. The release of tgm silencing in condensin mutants suggests its involvement in the behavior of tRNA genes, potentially their clustering and/or localization mechanisms.
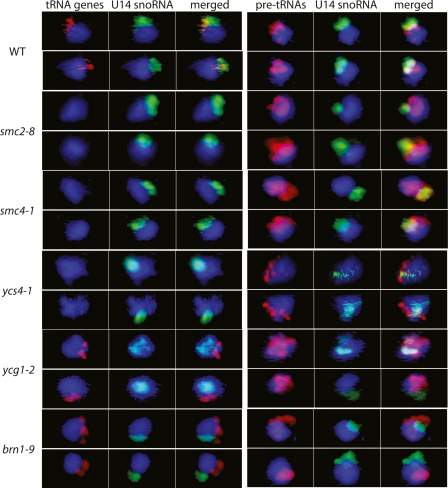
The subnuclear distribution of tRNA genes was analyzed by in situ hybridization in wild-type cells and in each mutant strain, where yeast were grown at 30°C prior to fixation and slide preparation. As noted previously, the permissive growth temperature for all five mutant strains is 23°C, and growth stops at 37°C. We used 30°C as an intermediate condition where the complex might be partially defective. In wild-type cells, tRNA genes are clustered at the nucleolus (Fig. 3; Thompson et al. 2003; L. Wang et al. 2005). However in all five mutant strains, tRNA gene clustering is either absent or defective. In smc2-8, smc4-1, and ycs4-1, the tRNA gene (red) signal is no longer detectable above the background, as previously observed for nucleolar disruptions that disperse the tRNA genes (L. Wang et al. 2005). In ycg1-2 and brn1-9, tRNA genes appear to still be at least partially clustered but divorced from the nucleolus. In ycg1-2, only 36% of all nuclei showed a single tRNA gene cluster (compared with 83% in wild type), and of those single-cluster nuclei, only 23% showed nucleolar localization (compared with 76% in wild type). Many more ycg1-2 nuclei showed two or more clusters (51%), and the clusters were nonnucleolar in 94% of these nuclei. A small number of ycg1-2 nuclei showed no clustering (13%). In brn1-9, 49% of nuclei showed a single tRNA gene cluster, where only 27% of these were nucleolar. 38% of brn1-9 nuclei showed two or more clusters, and clusters were nonnucleolar in 88% of these nuclei. Thirteen percent of brn1-9 cells showed no clusters. The different phenotypes observed may be due either to differences in mutation severity or to the different functions of the subunits. It can be concluded that defective mutations in any of the five condensin subunits can cause severe defects in tRNA gene positioning.
Figure 3.
Condensin mutants show loss of tRNA gene and pre-tRNA nucleolar localization. Fluorescence in situ hybridization to the tRNALeu genes and the early precursor tRNAs (pre-tRNALeu) in wild-type cells shows the genes and their transcripts clustered at the nucleolus, as described previously (Bertrand et al. 1998; Kendall et al. 2000; Thompson et al. 2003; L. Wang et al. 2005). However, mutations in condensin subunits change the patterns of tRNA gene clusters. In some cases, the tRNA genes disperse (smc2-8, smc4-1, ycs4-1) such that the fluorescent signal is no longer detectable above the background, and in other cases, they mislocalize outside of the nucleolus (ycg1-2, brn1-9). Pre-tRNA positions are also altered, in some cases localizing near the nuclear periphery (ycs4-1, brn1-9) and in others diffusely in the nucleoplasm (smc2-8, smc4-1, ycg1-2).
As confirmation of mislocalized pre-tRNA synthesis in strains where the genes are too dispersed to give focal signals, the locations of pre-tRNALeu(CAA) primary transcripts were also probed (Bertrand et al. 1998; L. Wang et al. 2005). Because there are hundreds of copies of this precursor RNA per nucleus, the fluorescent signal is detectable even when the corresponding tRNA genes are dispersed in the nucleoplasm (L. Wang et al. 2005). In wild-type cells, pre-tRNAs are concentrated at the nucleolus (Fig. 3), as expected (Bertrand et al. 1998; L. Wang et al. 2005). In each condensin mutant, pre-tRNAs are aberrantly localized. Instead of predominantly overlapping the nucleolar marker, pre-tRNAs in the mutants are either more dispersed throughout the nucleoplasm or localized mostly to the nuclear periphery. Representative cells are shown in Figure 3. The more diffuse nucleoplasmic staining seen in smc2-8, smc4-1, and ycg1-2 is similar to that seen in several RNA polymerase I mutants and rRNA gene transcription factor mutants that also release tgm silencing (L. Wang et al. 2005). The pre-tRNAs in ycs4-1 and brn1-9 appear concentrated at the nuclear periphery, a phenotype that has not previously been seen. The reason for this is unclear, but it is possible that residual function of these subunits at 30°C leads to partial, but aberrant, pre-tRNA localization. Tests of whether pre-tRNA synthesis and processing were substantially compromised by the condensin-deficient mislocalization did not show dramatic defects (Supplemental Fig. S4).
Condensin is bound preferentially to tRNA genes
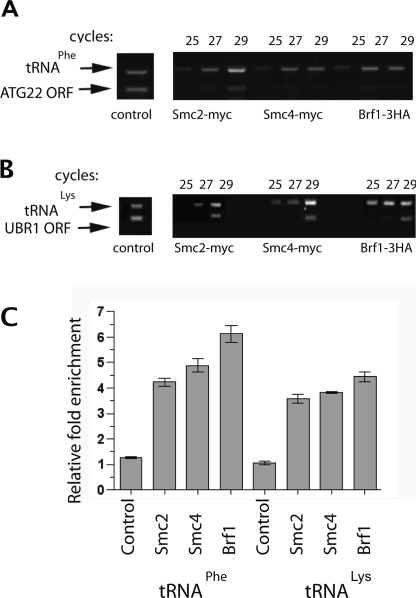
The involvement of condensin at tRNA gene loci suggests a possible direct interaction of the complex with tRNA genes, with subsequent clustering due to interactions between condensin complexes bound at different loci, as has been proposed for other forms of condensin-mediated chromosome condensation. Recent ChIP analysis with high-resolution oligonucleotide arrays shows a sharp peak of condensin bound to each tRNA gene in the yeast genome (D’Ambrosio et al. 2008). To independently verify the binding of condensin to tRNA genes, we tested condensin occupancy of two distinct tRNA genes relative to chromosomal locations devoid of tRNA genes. Chromatin associated with affinity-tagged Smc2p or Smc4p was used for semiquantitative PCR amplification of a tRNALys on chromosome VII or a tRNAPhe on chromosome XVI (Fig. 4). Two separate internal amplification controls were from pol II-transcribed genes (coding regions) with no nearby tRNA genes or TFIIIC-binding sites. Both Smc2p and Smc4p show specific binding to each of the representative tRNA genes, with enrichment similar to that of affinity-tagged Brf1p, a subunit of the pol III transcription factor TFIIIB, which continuously occupies activated tRNA genes.
Figure 4.
Condensin subunits Smc2 and Smc4 interact with tRNA genes. (A,B) ChIPs were performed using epitope-tagged versions of Smc2, Smc4, and Brf1. Twenty-five, 27, or 29 rounds of PCR are shown with primers that simultaneously amplify a tRNAPhe gene and the ATG22 ORF segment (A) or a tRNALys gene and a UBR1 ORF segment (B). Controls show representative signals rounds of PCR performed in parallel precipitations for which the primary antibodies were omitted (29–33 rounds to amplify background). Pulldowns using anti-myc primary antibody in untagged strains also did not enrich for tRNA genes (data not shown). (C) Ratios of relative signal intensity were quantified for each tRNA/ORF pair at subsaturating points (29 cycles in the examples shown). Error bars represent the standard deviation for four amplifications in two experiments.
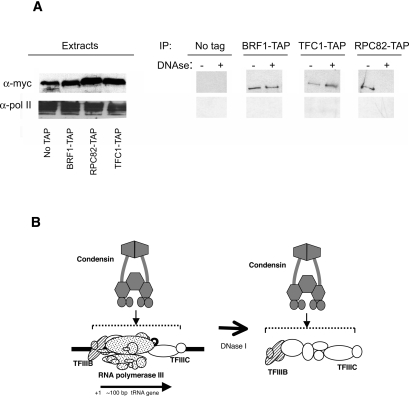
The preferential binding of condensin to tRNA genes strongly implies that condensin is interacting specifically with some component(s) of the pol III transcription complex on the genes, since the only conserved sequences of all tRNA genes are the internal promoters within the coding sequences (Paule and White 2000). tRNA genes are recognized by initial binding of the multisubunit TFIIIC to these internal promoters, followed by binding of TFIIIB upstream of the gene but in contact with TFIIIC, followed by pol III (Huibregtse and Engelke 1989; Kassavetis et al. 1989; Bartholomew et al. 1991). We tested for direct association of pol III components with condensin using coimmunoprecipitation from soluble cell extracts. TAP-tagged proteins from pol III (Rpc82), TFIIIB (Brf1), or TFIIIC (Tfc1) were isolated and blotted for a 13xMyc-tagged condensin subunit (Smc2 or Smc4, depending on viability of dual-tagged strains). We observed a low-level association with all three pol III components in cell lysates (Fig. 5), suggesting condensin was stably associated with the assembled tRNA gene complexes released by the lysis procedure, but not preferentially with any single soluble component, since these components do not form stable complexes away from the DNA. To test if these interactions were direct versus DNA-mediated, we performed the same immunoprecipitation in parallel after treating lysates with enough DNase I to completely digest any exposed DNA. This DNase treatment consistently removed the association of pol III with condensin but left the condensin signal in TFIIIB and TFIIIC pulldowns essentially unchanged (Fig. 5). This strongly suggests that there is a direct interaction between condensin and a stable TFIIIB/TFIIIC complex, and that maintenance of this complex does not require the presence of RNA polymerase III.
Figure 5.
Condensin subunits Smc2 and Smc4 interact with subunits of RNA pol III and its transcription factors. (A) Strains containing a Myc-tagged SMC subunit and either no TAP-tagged protein, or a TAP-tagged subunit of RNA pol III, TFIIIB, or TFIIIC were lysed, and TAP-containing complexes were affinity isolated and probed for Myc-tagged condensin. Whole extracts blotted with anti-Myc antibody to detect condensin subunits or anti-pol II (large subunit C-terminal domain) are shown on the left. A portion of the condensin found in the extracts coisolated with each of the pol III complexes, whereas little or none of the pol II control did. (Faint signals of pol II large subunit were reproducible in the TFC1-tagged strain pulldown.) Aggressive DNase I treatment removed pol III, as well as DNA, from the complexes while leaving condensin associated with TFIIIB and TFIIIC. (B) A model for the association of condensin with the known complex of pol III transcription factors TFIIIC and TFIIIB. Although the DNase resistance of the complex suggests a direct and stable protein–protein association, it does not rule out contact of condensin with the DNA as well.
Discussion
The spatial organization of genes within a eukaryotic nucleus is extraordinarily complex. Examples of this in yeast are the preference of telomeres (Hediger et al. 2002), centromeres (Heun et al. 2001a), and silenced chromatin (Maillet et al. 1996; Feuerbach et al. 2002) for the nuclear periphery, the localization of various highly expressed and inducible genes to nuclear pores (Brickner and Walter 2004; Casolari et al. 2004, 2005; Schmid et al. 2006), and the colocalization of ribosomal DNA and a majority of tRNA genes at the nucleolus. To accommodate each of these known levels of organization, there are likely to be multiple collaborating mechanisms. Two factors are shown here to affect the positions of tRNA genes: condensin-dependent clustering and separable association of clusters with the nucleolus. Because the tRNA genes are distributed throughout the linear maps of the 16 chromosomes, this phenomenon affects the condensation and spatial organization of much of the yeast genome.
The involvement of microtubules in nuclear genome organization has previously been observed in a variety of other studies, although the mechanistic details for these effects are not entirely clear. Microtubules were found to be responsible for the constraint on chromosome motion in the yeast nucleus (Marshall et al. 1997), and they are also required for centromere clustering at the spindle pole body (Bystricky et al. 2004). Our observation that intact microtubules are required for localizing tRNA genes is consistent with microtubule-dependent structures being required for the general positioning of chromosomes. It is possible that, in addition to passive foundational support, microtubules might be involved in the dynamic positioning of specific DNA regions. For example, homologous recombination, which requires two DNA loci to be spatially adjacent, is facilitated by microtubules in S. cerevisiae (Thrower et al. 2003). While preliminary investigations of microtubules interacting directly with pol III transcription units in yeast have proven inconclusive (R.A. Haeusler and D.R. Engelke, unpubl.), in human cell culture β-tubulin is one of the few nonsubunit proteins associated with highly purified RNA polymerase III (Hu et al. 2002, 2003), and it remains possible that microtubules have a direct effect on tRNA gene positioning.
The association of the condensin complex with tRNA gene complexes appears to be direct. Thus, although it is possible that condensin mutants decluster the tRNA genes through disorganization of nucleolar architecture, direct condensin participation in the clustering seems likely. Prior to this work, only a small number of interphase condensin functions were explored in various organisms (Hagstrom and Meyer 2003), including gene regulation and centromere organization. Of particular interest is the condensin-like dosage compensation complex in Caenorhabditis elegans, which is responsible for the sex-determining X-chromosome repression (Hagstrom and Meyer 2003). In S. cerevisiae, condensin’s role in the architecture of the rDNA locus during division has been documented (Freeman et al. 2000; Lavoie et al. 2000, 2002; Bhalla et al. 2002), but the complex might have other nonmitotic functions that are distinct from its role in chromosome segregation. For example, condensin has been implicated in the SIR2-mediated silencing at the rDNA and telomeres (Machin et al. 2004), and certain condensin mutations can also affect silencing at the mating-type loci (Bhalla et al. 2002), which is also SIR2-mediated. While silencing near tRNA genes (and their nucleolar localization) is mechanistically distinct from the other types of silencing and independent of SIR2 (L. Wang et al. 2005), it has been suggested previously that condensin may influence interphase chromosome arrangements in ways that impact gene regulation (Hagstrom and Meyer 2003). Our combined evidence that condensin is required for tRNA gene clustering and for tgm silencing suggests that the complex plays a very important role in the nuclear architecture and that this influences nuclear functions. The nucleolar clustering does not appear to be necessary for tRNA transcription and processing, since these condensin mutations did not dramatically affect tRNA precursor accumulation at both viable (though defective) and nonviable temperatures (Supplement Fig. S4).
As yet, the process by which condensin condenses chromosome structure is unknown, although several models have been suggested (Hagstrom and Meyer 2003; Hirano 2005; Huang et al. 2005; Stray et al. 2005). Because the Smc2/4 complex is large (capable of spanning up to 150 base pairs [bp] of DNA) (Hirano 2005), it is theoretically possible that each condensin molecule is associated with two or more regions of dsDNA. It is also possible that condensin molecules oligomerize, bringing together the chromatin regions to which they are bound. The condensin-dependent clustering of tRNA genes from multiple chromosomes argues for a model in which multiple DNA strands are brought together by multiple condensin molecules.
In order to bind tRNA genes, condensin appears to target one or more of the many proteins in the transcription factors TFIIIC or TFIIIB. The TFIIIC transcription factor, which initially recognizes the genes, has been identified as a likely binding partner (D’Ambrosio et al. 2008), since genome sites bound by TFIIIC but not other members of the transcription complex also bind condensin. Most models for SMC complex interactions with bacterial and eukaryotic chromosomes assume direct contacts with DNA, and yeast condensin SMC complexes are known to bind naked DNA avidly in a non-sequence-dependent fashion (Stray et al. 2005). Our observation of a DNase-resistant TFIIIB/TFIIIC/condensin complex indicates that condensin association with the complex is stable once the exposed DNA is removed, although it does not address whether condensin is also in contact with the DNA.
The interaction of condensin at sites that bind TFIIIC but do not stably interact with the other pol III components (D’Ambrosio et al. 2008) supports a model whereby condensin is brought to the tRNA gene through TFIIIC’s sequence-specific binding to the internal promoters. It remains possible that additional contacts might also be made with the RNA polymerase, since Smc4 has also been found to coisolate with Rpc40 (Ho et al. 2002). It is potentially interesting that Rpc40 is a subunit of both pol I and pol III, but not pol II. Thus condensin might be targeted to both tRNA genes and rDNA as a part of nucleolar organization through this interaction. However, the present data do not provide support for a stable interaction between polymerase subunits and condensin.
The condensin-dependent clustering of linearly scattered tRNA genes affects the spatial organization of most of the yeast genome, but the extent to which this phenomenon affects larger eukaryotic genomes has not yet been explored. Vertebrate genomes can be two orders of magnitude larger than yeast, yet the number of active tRNA genes is estimated to be only a few fold higher (Goodenbour and Pan 2006) and their positions in nuclei have not been determined. It would be surprising, though not impossible, for the metazoan transcribed tRNA genes to also be localized to nucleoli during interphase. It is perhaps also worth considering the extent to which the many nontranscribed, tRNA-like promoter elements are organized. Vertebrate genomes tend to have over a million copies of short interspersed nuclear elements (SINEs) (Okada 1991; Weiner 2002) that were originally derived from retrotransposition of either tRNA genes or 7SL RNA genes, which have tRNA-class pol III promoters. Although these elements are normally expressed only at very low levels, if at all, most have consensus TFIIIC-binding sites and cloned copies can be transcribed by RNA polymerase III in vitro. It is not currently known to what extent TFIIIC and other pol III components are bound to the elements in vivo, but imaging of SINE (human Alu element) positions in HeLa cells shows a punctate pattern (Kaplan et al. 1993) that is consistent with local clustering. If pol III components and condensin associate with some subset of the SINEs, it is interesting to speculate that they might serve as symbiotic, rather than selfish DNA elements, in that they provide condensation signals for the relatively large metazoan genomes.
Materials and methods
Yeast strains
For tgm silencing assays and in situ hybridizations, wild-type parent strains included BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 GAL4 GAL80) and W3031A (MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15). Mutant strains included 1bAS330 (MATa smc2-8 ade2 leu2 lys2 his3 trp1 ura3) and 1bAS342 (MATα smc4-1 ade2 leu2 lys2 his3 ura3) (Freeman et al. 2000), BLY07 (MATa ycg1-2:KAN ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2-801 ade2-101) (Lavoie et al. 2002), NBY481 (MATa ycs4-1 ura3-1∷ PGAL-MPS1∷TRP1 leu2-3,112 his3-11,15 trp1-1 ade2-1 can1-100 lys2Δ hmlΔ∷LEU2) (Bhalla et al. 2002), and BHY78 (MATa brn1-9:TRP1 ura3-52 leu2Δ1 trp1Δ63 his3∷natMX4), which was created by replacing HIS3 with the natMX4 nourseothricin resistance cassette in the strain CH2524 (Lavoie et al. 2002).
For ChIP assays, strains were derived from YPH500α (ΜΑΤα ade2-101 his3-Δ200 leu2Δ1 lys2-801 trp1-Δ63 ura3-52) (Sikorski and Hieter 1989) (gift of Olivier Lefebvre). Smc2p and Smc4p were C-terminally tagged with 13xMyc epitope by methods previously described (Longtine et al. 1998). Brf1p is C-terminally tagged with 3xHA in strain MW4034 (Harismendy et al. 2003) (derivative of YPH500α, gift of Olivier Lefebvre). For protein–protein interaction studies, the C-terminally 13xMyc epitope-tagged Smc2p was also incorporated into BRF1-TAP and RPC82-TAP strains. Smc4p was C-terminally tagged with 13xmyc in a Tcf1-TAP strain, since tagging Smc2p in this strain did not give healthy isolates. Tandem affinity strains were obtained from Open Biosystems (Ghaemmaghami et al. 2003).
Nocodazole treatment and cell cycle synchronization
BY4741 cultures were grown in SDC media to an OD600 of 0.2–0.4 prior to addition of drug or pheromone. Nocodazole (Sigma-Aldrich) was added to a final concentration of 15 μg/mL, and 30°C incubation continued for either 1 h or until >80% of cells appeared by microscopy to be arrested in a large-budded state, 3.5–4.5 h. Cells were subsequently fixed, and immunofluorescence with an anti-α-tubulin antibody confirmed the absence of microtubules in treated cells (Supplemental Fig. S5). DMSO added with the nocodazole, a potential source of nucleolar disruption, did not release tRNA gene clusters from the nucleolus (Supplemental Fig. S6). For α factor synchronization, α factor was added to 10 μg/mL and 30°C incubation continued until cells arrested in a G1 “shmoo” state, 2–2.5 h. Cells were fixed at this point, or for subsequent treatment, yeast were harvested, washed in prewarmed SDC, and resuspended in prewarmed SDC containing 15 μg/mL nocodazole for 1 h of additional incubation at 30°C prior to fixation.
tgm silencing growth assay
To test for release of tgm silencing, cells were transformed with pSUP4(-77)o (Hull et al. 1994; Kendall et al. 2000; L. Wang et al. 2005) and plated in serial dilutions on synthetic media containing galactose and raffinose (SGR) or dextrose (SD): SGR-ura, SGR-ura-his, SD-ura-his. There was no growth on SD-ura-his. Mutant cells were able to grow on SGR-ura-his at both 23°C and 30°C, although the percentage of cells that grew was variable between repeat experiments. This phenotype is typical for mutations that relieve tgm silencing (Kendall et al. 2000; L. Wang et al. 2005). Large and small colonies from mutant strains were restreaked onto SGR-ura-his and grown at 23°C, which resulted in growth of mixed sized colonies, regardless of original colony size.
In situ hybridization and microscopy
Yeast were grown in synthetic dextrose complete (SDC) media and fixed for 30 min by adding 3.6% paraformaldehyde directly to the growth media, to minimally disturb cellular and nuclear architecture (Bertrand et al. 1998; Thompson et al. 2003; L. Wang et al. 2005; Moir et al. 2006). After fixation, cells were treated as previously described (Long et al. 1995; Bertrand et al. 1998), except that spheroplasting was performed by treating with 0.38 μg/μL Zymolyase 20T (Seikagaku) for 45 min at 37°C. Oligonucleotide probes, labeled with Oregon Green 488 (Molecular Probes) or Cy3 (Amersham), to U14 snoRNA, tRNALeu(CAA), and pretRNALeu(CAA) were previously described, and in situ hybridization, microscopy, and deconvolution were performed as published (Bertrand et al. 1998; Thompson et al. 2003; L. Wang et al. 2005). After high-resolution imaging, cells that retained tRNA gene signal were scored: 216 untreated and 89 nocodazole-arrested cells, as well as 61 ycg1-2 and 45 brn1-9 mutant cells.
ChIP
ChIP procedures were adapted from Ren et al. (2000), with these exceptions: Fixation was performed with 1% paraformaldehyde for 30 min at room temperature and stopped with 400 mM glycine for 10 min. Cell pellets were resuspended in lysis buffer (50 mM HEPES-KOH at pH 7.5, 100 mM NaCl, 1 mM EDTA, 1% Triton, 0.1% deoxycholate, complete protease inhibitors [Roche]), and incubated with 0.3 μg/μL Zymolyase for 30 min at 30°C. Samples were lysed with glass beads in a Bead Beater (Biospec Products), sonicated, and spun out. Samples were incubated overnight with antibody against either Myc (9E10) or HA (Y11) (Santa Cruz Biotechnology). Myc-tagged samples were also incubated with 1μL of a rabbit anti-mouse IgG (Jackson ImmunoResearch) for compatibility with Protein A beads. Antibody-bound samples were added to prewashed, immobilized Protein A beads (RepliGen) and incubated for 3 h. Remaining procedures were as described (Ren et al. 2000).
PCR and quantitation
Precipitated DNA was used as template in PCR reactions cycled for 30 sec at 94°C, for 30 sec at 63°C, and for 60 sec at 72°C × 25, 27, 29, or 31 cycles. Primers used were as follows: for tRNALys(CUU) on chromosome VII, 5′-AGTATATTCCAAA CATGTGTTAATC-3′ and 5′-CCAGAGCACTTTTATGTGG GAC-3′; for tRNAPhe(GAA) on chromosome XVI, 5′-GACGCT TGGACCATTTATAAAGCAC-3′ and 5′-CCATAAGAGAAG GAGCAGTCAAGTTCA-3′; for UBR1, 5′-GCATCCATATTT GGAGAACATCCCATA-3′ and 5′-CGTAGCTTGATTTCCT TAGATTGGGTA-3′; and for ATG22, 5′-CAAAGTTTCGGTG GACTTCTAGTCAAA-3′ and 5′-GCTTTAAACCGAACGCA TTGAAGAAAA-3′. ATG22 and UBR1 genomic controls were selected because there was no pol III transcribed region within 5000 bp. EtBr-stained PCR products were imaged on a Syngene Bioimaging System with GeneSnap and quantitated with GeneTools software (Syngene). Typically immunoaffinity isolation of any gene from the sheared chromatin was less than 1% efficient. Specificity of pulldowns were assured by co-PCR UBR1 and ATG22 ORFs in the same reactions as the tRNA gene amplifications (below).
Condensin–TFIIIC/TFIIIB/Pol III interaction study
Cells were grown in YPD to an OD600 of 0.6–0.8, and pellets were stored at −80°C. Cells were thawed, lysed in buffer (175 mM NaCl, 20 mM Tris-HCl at pH 7.9, 0.5% NP-40, complete protease inhibitors [Roche]), subjected to mechanical bead disruption, and spun at 14,000 rpm for 20 min. Of each supernatant, half was DNase I-treated with 50 U of DNase I (Cooper Biomedical) per 3-mL extract. Elimination of DNA from extracts was verified by extracting and precipitating samples, then performing PCR using primers flanking the tRNALys locus. DNased samples produced no product after 40 cycles, whereas non-DNased samples produced product in less than 30 cycles.
Strains used in the pulldown were SMC2-myc (no TAP tag), SMC4-myc (no TAP tag), BRF1-TAP + SMC2-myc, RPC82-TAP + SMC2-Myc, and TFC1-TAP + SMC4-myc. Extracts were incubated with IgG Sepharose (GE Healthcare) for 2.5 h, then washed four times with lysis buffer and once with TEV cleavage buffer (50 mM Tris-HCl at pH 8.0, 0.5 mM EDTA, 1 mM DTT). Samples were eluted overnight with TEV protease, precipitated with trichloroacetic acid, and electrophoresed, and blots were probed in parallel with either a Myc monoclonal antibody (9E10) (Santa Cruz Biotechnologies) or monoclonal antibody directed against the C-terminal domain of the largest subunit of RNA pol II (Abcam 8WG16) as a control.
Acknowledgments
We thank Alexander Strunnikov, Brigitte Lavoie, Andrew Murray, and Olivier Lefebvre for yeast strains; Claudio D’Ambrosio and Frank Uhlmann for communicating results prior to publication; and Engelke laboratory members for helpful discussion. This work was funded by NIH grant GM061342 to D.R.E., a predoctoral fellowship from the University of Michigan Rackham Graduate School and NIH Genetics Predoctoral Training Grant T32 GM007544 to R.A.H., and NIH Cellular and Molecular Biology Training Grant T32 GM007315 to M.P-H..
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1675908.
References
- Bartholomew B., Kassavetis G.A., Geiduschek E.P. Two components of Saccharomyces cerevisiae transcription factor IIIB (TFIIIB) are stereospecifically located upstream of a tRNA gene and interact with the second-largest subunit of TFIIIC. Mol. Cell. Biol. 1991;11:5181–5189. doi: 10.1128/mcb.11.10.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartova E., Kozubek S. Nuclear architecture in the light of gene expression and cell differentiation studies. Biol. Cell. 2006;98:323–336. doi: 10.1042/BC20050099. [DOI] [PubMed] [Google Scholar]
- Bertrand E., Houser-Scott F., Kendall A., Singer R.H., Engelke D.R. Nucleolar localization of early tRNA processing. Genes & Dev. 1998;12:2463–2468. doi: 10.1101/gad.12.16.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla N., Biggins S., Murray A.W. Mutation of YCS4, a budding yeast condensin subunit, affects mitotic and nonmitotic chromosome behavior. Mol. Biol. Cell. 2002;13:632–645. doi: 10.1091/mbc.01-05-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner J.H., Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2:e342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K.E., Baxter J., Graf D., Merkenschlager M., Fisher A.G. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol. Cell. 1999;3:207–217. doi: 10.1016/s1097-2765(00)80311-1. [DOI] [PubMed] [Google Scholar]
- Brown J.M., Leach J., Reittie J.E., Atzberger A., Lee-Prudhoe J., Wood W.G., Higgs D.R., Iborra F.J., Buckle V.J. Coregulated human globin genes are frequently in spatial proximity when active. J. Cell Biol. 2006;172:177–187. doi: 10.1083/jcb.200507073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchenau P., Saumweber H., Arndt-Jovin D.J. The dynamic nuclear redistribution of an hnRNP K-homologous protein during Drosophila embryo development and heat shock. Flexibility of transcription sites in vivo. J. Cell Biol. 1997;137:291–303. doi: 10.1083/jcb.137.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystricky K., Heun P., Gehlen L., Langowski J., Gasser S.M. Long-range compaction and flexibility of interphase chromatin in budding yeast analyzed by high-resolution imaging techniques. Proc. Natl. Acad. Sci. 2004;101:16495–16500. doi: 10.1073/pnas.0402766101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari J.M., Brown C.R., Komili S., West J., Hieronymus H., Silver P.A. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- Casolari J.M., Brown C.R., Drubin D.A., Rando O.J., Silver P.A. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes & Dev. 2005;19:1188–1198. doi: 10.1101/gad.1307205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang C.H., Carpenter A.E., Fuchsova B., Johnson T., de Lanerolle P., Belmont A.S. Long-range directional movement of an interphase chromosome site. Curr. Biol. 2006;16:825–831. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- Cremer T., Cremer M., Dietzel S., Muller S., Solovei I., Fakan S. Chromosome territories—A functional nuclear landscape. Curr. Opin. Cell Biol. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- D’Ambrosio C., Schmidt C.K., Katou Y., Kelly G., Itoh T., Shirahige K., Uhlmann F. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes & Dev. 2008 doi: 10.1101/gad.1675708. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faro-Trindade I., Cook P.R. Transcription factories: Structures conserved during differentiation and evolution. Biochem. Soc. Trans. 2006;34:1133–1137. doi: 10.1042/BST0341133. [DOI] [PubMed] [Google Scholar]
- Feuerbach F., Galy V., Trelles-Sticken E., Fromont-Racine M., Jacquier A., Gilson E., Olivo-Marin J.C., Scherthan H., Nehrbass U. Nuclear architecture and spatial positioning help establish transcriptional states of telomeres in yeast. Nat. Cell Biol. 2002;4:214–221. doi: 10.1038/ncb756. [DOI] [PubMed] [Google Scholar]
- Foster H.A., Bridger J.M. The genome and the nucleus: a marriage made by evolution. Genome organisation and nuclear architecture. Chromosoma. 2005;114:212–229. doi: 10.1007/s00412-005-0016-6. [DOI] [PubMed] [Google Scholar]
- Freeman L., Aragon-Alcaide L., Strunnikov A. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J. Cell Biol. 2000;149:811–824. doi: 10.1083/jcb.149.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs J., Loidl J. Behaviour of nucleolus organizing regions (NORs) and nucleoli during mitotic and meiotic divisions in budding yeast. Chromosome Res. 2004;12:427–438. doi: 10.1023/B:CHRO.0000034726.05374.db. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O'Shea E.K., Weissman J.S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Goodenbour J.M., Pan T. Diversity of tRNA genes in eukaryotes. Nucleic Acids Res. 2006;34:6137–6146. doi: 10.1093/nar/gkl725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V., Hogan E., Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J. Cell Biol. 1994;125:517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler R.A., Engelke D.R. Spatial organization of transcription by RNA polymerase III. Nucleic Acids Res. 2006;34:4826–4836. doi: 10.1093/nar/gkl656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom K.A., Meyer B.J. Condensin and cohesin: More than chromosome compactor and glue. Nat. Rev. Genet. 2003;4:520–534. doi: 10.1038/nrg1110. [DOI] [PubMed] [Google Scholar]
- Harismendy O., Gendrel C.G., Soularue P., Gidrol X., Sentenac A., Werner M., Lefebvre O. Genome-wide location of yeast RNA polymerase III transcription machinery. EMBO J. 2003;22:4738–4747. doi: 10.1093/emboj/cdg466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harnicarova A., Kozubek S., Pachernik J., Krejci J., Bartova E. Distinct nuclear arrangement of active and inactive c-myc genes in control and differentiated colon carcinoma cells. Exp. Cell Res. 2006;312:4019–4035. doi: 10.1016/j.yexcr.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Hediger F., Neumann F.R., Van Houwe G., Dubrana K., Gasser S.M. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr. Biol. 2002;12:2076–2089. doi: 10.1016/s0960-9822(02)01338-6. [DOI] [PubMed] [Google Scholar]
- Heun P., Laroche T., Raghuraman M.K., Gasser S.M. The positioning and dynamics of origins of replication in the budding yeast nucleus. J. Cell Biol. 2001a;152:385–400. doi: 10.1083/jcb.152.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun P., Taddei A., Gasser S.M. From snapshots to moving pictures: New perspectives on nuclear organization. Trends Cell Biol. 2001b;11:519–525. doi: 10.1016/s0962-8924(01)02174-2. [DOI] [PubMed] [Google Scholar]
- Hirano T. Condensins: Organizing and segregating the genome. Curr. Biol. 2005;15:R265–R275. doi: 10.1016/j.cub.2005. 03.037. [DOI] [PubMed] [Google Scholar]
- Ho Y., Gruhler A., Heilbut A., Bader G.D., Moore L., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Hu P., Wu S., Sun Y., Yuan C.C., Kobayashi R., Myers M.P., Hernandez N. Characterization of human RNA polymerase III identifies orthologues for Saccharomyces cerevisiae RNA polymerase III subunits. Mol. Cell. Biol. 2002;22:8044–8055. doi: 10.1128/MCB.22.22.8044-8055.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P., Wu S., Hernandez N. A minimal RNA polymerase III transcription system from human cells reveals positive and negative regulatory roles for CK2. Mol. Cell. 2003;12:699–709. doi: 10.1016/j.molcel.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Huang C.E., Milutinovich M., Koshland D. Rings, bracelet or snaps: Fashionable alternatives for Smc complexes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005;360:537–542. doi: 10.1098/rstb.2004.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse J.M., Engelke D.R. Genomic footprinting of a yeast tRNA gene reveals stable complexes over the 5′-flanking region. Mol. Cell. Biol. 1989;9:3244–3252. doi: 10.1128/mcb.9.8.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull M.W., Erickson J., Johnston M., Engelke D.R. tRNA genes as transcriptional repressor elements. Mol. Cell. Biol. 1994;14:1266–1277. doi: 10.1128/mcb.14.2.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst L.D., Pal C., Lercher M.J. The evolutionary dynamics of eukaryotic gene order. Nat. Rev. Genet. 2004;5:299–310. doi: 10.1038/nrg1319. [DOI] [PubMed] [Google Scholar]
- Jacobs C.W., Adams A.E., Szaniszlo P.J., Pringle J.R. Functions of microtubules in the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 1988;107:1409–1426. doi: 10.1083/jcb.107.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F.S., Murray J., Sylvester J.E., Gonzalez I.L., O'Connor J.P., Doering J.L., Muenke M., Emanuel B.S., Zasloff M.A. The topographic organization of repetitive DNA in the human nucleolus. Genomics. 1993;15:123–132. doi: 10.1006/geno.1993.1020. [DOI] [PubMed] [Google Scholar]
- Kassavetis G.A., Riggs D.L., Negri R., Nguyen L.H., Geiduschek E.P. Transcription factor IIIB generates extended DNA interactions in RNA polymerase III transcription complexes on tRNA genes. Mol. Cell. Biol. 1989;9:2551–2566. doi: 10.1128/mcb.9.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall A., Hull M.W., Bertrand E., Good P.D., Singer R.H., Engelke D.R. A CBF5 mutation that disrupts nucleolar localization of early tRNA biosynthesis in yeast also suppresses tRNA gene-mediated transcriptional silencing. Proc. Natl. Acad. Sci. 2000;97:13108–13113. doi: 10.1073/pnas.240454997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsey P.T., Sandmeyer S.B. Adjacent pol II and pol III promoters: transcription of the yeast retrotransposon Ty3 and a target tRNA gene. Nucleic Acids Res. 1991;19:1317–1324. doi: 10.1093/nar/19.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak S.T., Skok J.A., Medina K.L., Riblet R., Le Beau M.M., Fisher A.G., Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- Lavoie B.D., Tuffo K.M., Oh S., Koshland D., Holm C. Mitotic chromosome condensation requires Brn1p, the yeast homologue of Barren. Mol. Biol. Cell. 2000;11:1293–1304. doi: 10.1091/mbc.11.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie B.D., Hogan E., Koshland D. In vivo dissection of the chromosome condensation machinery: Reversibility of condensation distinguishes contributions of condensin and cohesin. J. Cell Biol. 2002;156:805–815. doi: 10.1083/jcb.200109056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legagneux V., Cubizolles F., Watrin E. Multiple roles of condensins: A complex story. Biol. Cell. 2004;96:201–213. doi: 10.1016/j.biolcel.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Liu J., Francke U. Identification of cis-regulatory elements for MECP2 expression. Hum. Mol. Genet. 2006;15:1769–1782. doi: 10.1093/hmg/ddl099. [DOI] [PubMed] [Google Scholar]
- Liu Z., Garrard W.T. Long-range interactions between three transcriptional enhancers, active Vκ gene promoters, and a 3′ boundary sequence spanning 46 kilobases. Mol. Cell. Biol. 2005;25:3220–3231. doi: 10.1128/MCB.25.8.3220-3231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J. Chromosomes of the budding yeast Saccharomyces cerevisiae. Int. Rev. Cytol. 2003;222:141–196. doi: 10.1016/s0074-7696(02)22014-8. [DOI] [PubMed] [Google Scholar]
- Long R.M., Elliott D.J., Stutz F., Rosbash M., Singer R.H. Spatial consequences of defective processing of specific yeast mRNAs revealed by fluorescent in situ hybridization. RNA. 1995;1:1071–1078. [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Losada A., Hirano T. Dynamic molecular linkers of the genome: The first decade of SMC proteins. Genes & Dev. 2005;19:1269–1287. doi: 10.1101/gad.1320505. [DOI] [PubMed] [Google Scholar]
- Machin F., Paschos K., Jarmuz A., Torres-Rosell J., Pade C., Aragon L. Condensin regulates rDNA silencing by modulating nucleolar Sir2p. Curr. Biol. 2004;14:125–130. [PubMed] [Google Scholar]
- Machin F., Torres-Rosell J., Jarmuz A., Aragon L. Spindle-independent condensation-mediated segregation of yeast ribosomal DNA in late anaphase. J. Cell Biol. 2005;168:209–219. doi: 10.1083/jcb.200408087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet L., Boscheron C., Gotta M., Marcand S., Gilson E., Gasser S.M. Evidence for silencing compartments within the yeast nucleus: A role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes & Dev. 1996;10:1796–1811. doi: 10.1101/gad.10.14.1796. [DOI] [PubMed] [Google Scholar]
- Marshall W.F., Straight A., Marko J.F., Swedlow J., Dernburg A., Belmont A., Murray A.W., Agard D.A., Sedat J.W. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr. Biol. 1997;7:930–939. doi: 10.1016/s0960-9822(06)00412-x. [DOI] [PubMed] [Google Scholar]
- Misteli T. Spatial positioning: A new dimension in genome function. Cell. 2004;119:153–156. doi: 10.1016/j.cell.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Moir R.D., Lee J., Haeusler R.A., Desai N., Engelke D.R., Willis I.M. Protein kinase A regulates RNA polymerase III transcription through the nuclear localization of Maf1. Proc. Natl. Acad. Sci. 2006;103:15044–15049. doi: 10.1073/pnas.0607129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K., Haering C.H. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- Okada N. SINEs. Curr. Opin. Genet. Dev. 1991;1:498–504. doi: 10.1016/s0959-437x(05)80198-4. [DOI] [PubMed] [Google Scholar]
- Parada L.A., Roix J.J., Misteli T. An uncertainty principle in chromosome positioning. Trends Cell Biol. 2003;13:393–396. doi: 10.1016/s0962-8924(03)00149-1. [DOI] [PubMed] [Google Scholar]
- Parada L.A., Sotiriou S., Misteli T. Spatial genome organization. Exp. Cell Res. 2004;296:64–70. doi: 10.1016/j.yexcr.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Paule M.R., White R.J. Survey and summary: Transcription by RNA polymerases I and III. Nucleic Acids Res. 2000;28:1283–1298. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt-Hyatt M.J., Kapadia K.M., Wilson T.E., Engelke D.R. Increased recombination between active tRNA genes. DNA Cell Biol. 2006;25:359–364. doi: 10.1089/dna.2006.25.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B., Robert F., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Schmid M., Arib G., Laemmli C., Nishikawa J., Durussel T., Laemmli U.K. Nup-PI: The nucleopore-promoter interaction of genes in yeast. Mol. Cell. 2006;21:379–391. doi: 10.1016/j.molcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Shaw P.J., Highett M.I., Beven A.F., Jordan E.G. The nucleolar architecture of polymerase I transcription and processing. EMBO J. 1995;14:2896–2906. doi: 10.1002/j.1460-2075.1995.tb07289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilianakis C.G., Lalioti M.D., Town T., Lee G.R., Flavell R.A. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- Stray J.E., Crisona N.J., Belotserkovskii B.P., Lindsley J.E., Cozzarelli N.R. The Saccharomyces cerevisiae Smc2/4 condensin compacts DNA into (+) chiral structures without net supercoiling. J. Biol. Chem. 2005;280:34723–34734. doi: 10.1074/jbc.M506589200. [DOI] [PubMed] [Google Scholar]
- Su J.S., Tsai T.F., Chang H.M., Chao K.M., Su T.S., Tsai S.F. Distant HNF1 site as a master control for the human class I alcohol dehydrogenase gene expression. J. Biol. Chem. 2006;281:19809–19821. doi: 10.1074/jbc.M603638200. [DOI] [PubMed] [Google Scholar]
- Taddei A., Hediger F., Neumann F.R., Gasser S.M. The function of nuclear architecture: A genetic approach. Annu. Rev. Genet. 2004;38:305–345. doi: 10.1146/annurev.genet.37.110801.142705. [DOI] [PubMed] [Google Scholar]
- Thanbichler M., Shapiro L. Chromosome organization and segregation in bacteria. J. Struct. Biol. 2006;156:292–303. doi: 10.1016/j.jsb.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Thompson M., Haeusler R.A., Good P.D., Engelke D.R. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302:1399–1401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower D.A., Stemple J., Yeh E., Bloom K. Nuclear oscillations and nuclear filament formation accompany single-strand annealing repair of a dicentric chromosome in Saccharomyces cerevisiae. J. Cell Sci. 2003;116:561–569. doi: 10.1242/jcs.00251. [DOI] [PubMed] [Google Scholar]
- Volpi E.V., Chevret E., Jones T., Vatcheva R., Williamson J., Beck S., Campbell R.D., Goldsworthy M., Powis S.H., Ragoussis J., et al. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell Sci. 2000;113:1565–1576. doi: 10.1242/jcs.113.9.1565. [DOI] [PubMed] [Google Scholar]
- Wang B.D., Eyre D., Basrai M., Lichten M., Strunnikov A. Condensin binding at distinct and specific chromosomal sites in the Saccharomyces cerevisiae genome. Mol. Cell. Biol. 2005;25:7216–7225. doi: 10.1128/MCB.25.16.7216-7225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Haeusler R.A., Good P.D., Thompson M., Nagar S., Engelke D.R. Silencing near tRNA genes requires nucleolar localization. J. Biol. Chem. 2005;280:8637–8639. doi: 10.1074/jbc.C500017200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A.M. SINEs and LINEs: The art of biting the hand that feeds you. Curr. Opin. Cell Biol. 2002;14:343–350. doi: 10.1016/s0955-0674(02)00338-1. [DOI] [PubMed] [Google Scholar]