Abstract
Proteins unfold constantly in cells, especially under stress conditions. Degradation of denatured polypeptides by Lon and related ATP-dependent AAA+ proteases helps prevent toxic aggregates formation and other deleterious consequences, but how these destructive enzymatic machines distinguish between damaged and properly folded proteins is poorly understood. Here, we show that Escherichia coli Lon recognizes specific sequences—rich in aromatic residues—that are accessible in unfolded polypeptides but hidden in most native structures. Denatured polypeptides lacking such sequences are poor substrates. Lon also unfolds and degrades stably folded proteins with accessible recognition tags. Thus, protein architecture and the positioning of appropriate targeting sequences allow Lon degradation to be dependent or independent of the folding status of a protein. Our results suggest that Lon can recognize multiple signals in unfolded polypeptides synergistically, resulting in nanomolar binding and a mechanism for discriminating irreversibly damaged proteins from transiently unfolded elements of structure.
Keywords: Lon, protease, AAA+ proteins, quality control, proteolysis
Protein quality-control systems have evolved in all cells to provide protection from the harmful effects of protein unfolding. The key components of these systems include ATP-dependent proteases, chaperones, heat-shock proteins, and regulatory molecules (for review, see Sauer et al. 2004; Bukau et al. 2006). These networks assist in the folding of newly synthesized proteins, in the disaggregation and refolding of misfolded molecules, and in the degradation of irreparably damaged proteins. The mechanisms that allow the selective identification of unfolded or misfolded proteins by these enzymes are critical for proper function but, in most cases, are poorly understood. Recognition might depend on sequence-independent interactions with long stretches of the polypeptide backbone, which would be inaccessible in folded proteins. Alternatively, recognition could depend on interactions with specific amino acid sequences that are hidden in the native structure of a protein and become exposed only upon unfolding. For example, the DnaK and GroEL chaperones of Escherichia coli interact with short hydrophobic sequences that are typically exposed in misfolded proteins but buried in native structures (Rudiger et al. 1997; Chen and Sigler 1999).
To avoid unnecessary degradation of cellular proteins, substrate selection by quality-control proteases is tightly regulated. In AAA+ proteases, a complex architecture minimizes inadvertent interactions with proteins by steric hindrance but requires ATP hydrolysis to power degradation. The active proteolytic sites of these proteases are sequestered inside of a barrel-shaped structure that can only be reached after passing through a narrow axial pore. Protein substrates are recognized by a protease-associated AAA+ ATPase, unfolded if necessary, and translocated in an ATP-dependent process into the proteolytic chamber for degradation (Gottesman 2003; Baker and Sauer 2006).
Lon, a highly conserved member of the AAA+ superfamily, plays a pivotal role in protein quality control by degrading damaged proteins in diverse bacteria and in the organelles of eukaryotes (Shineberg and Zipser 1973; Kowit and Goldberg 1977; Gottesman and Zipser 1978; Chung and Goldberg 1981; Rotanova et al. 2004; Tsilibaris et al. 2006). For example, Lon is responsible for ∼50% of the turnover of proteins resulting from premature translational termination or from the incorporation of amino acid analogs in Escherichia coli (Kowit and Goldberg 1977). Clearly, Lon must be capable of recognizing the majority of E. coli proteins when damage precludes their proper folding. Although genetic experiments have identified determinants of Lon recognition for some substrates (Gonzalez et al. 1998; Johansson and Uhlin 1999; Ishii and Amano 2001; Shah and Wolf 2006), it is not clear whether Lon recognizes a native or unfolded form of these proteins, and a detailed understanding of the mechanism by which Lon recognizes diverse substrates has been elusive.
To probe the interaction of Lon with misfolded proteins, we sought a good nonnative substrate that would allow us to characterize determinants of recognition and degradation. Here, we show that an unstructured fragment of β-galactosidase is bound tightly and degraded rapidly by E. coli Lon, whereas full-length native β-galactosidase is resistant to degradation. Lon recognition of this fragment depends upon a specific sequence in the unfolded polypeptide. A cluster of aromatic side chains, which are buried in the native protein, is an important positive recognition determinant. The absence of small polar amino acids appears to be a secondary recognition determinant. We show that similar recognition sequences are present in other Lon substrates, that unfolded proteins or hydrophobic peptides per se need not be good Lon substrates, and that attaching a recognition tag to a stably folded protein results in efficient degradation by Lon. Our results provide a simple mechanism that accounts for the ability of Lon to degrade a wide variety of denatured proteins, explain why most native proteins are impervious to Lon, demonstrate that very stable native proteins can be unfolded and degraded by Lon, and reveal synergistic recognition of multiple signals in nonnative proteins.
Results
Degradation of a β-galactosidase fragment
It has been known for more than 30 years that nonsense fragments of β-galactosidase are degraded rapidly in a lon-dependent manner in E. coli (Lin and Zabin 1972; Shineberg and Zipser 1973; Kowit and Goldberg 1977; Miller and Zipser 1977). To determine if this behavior could be reproduced using purified components in vitro, we cloned, purified, and studied Lon degradation of an N-terminal fragment corresponding to residues 3–93 of β-galactosidase. Mass spectrometry showed that the 3–93 β-galactosidase fragment had a mass within 1 Da of the expected value (10,503 Da), ruling out the presence of post-translational modifications.
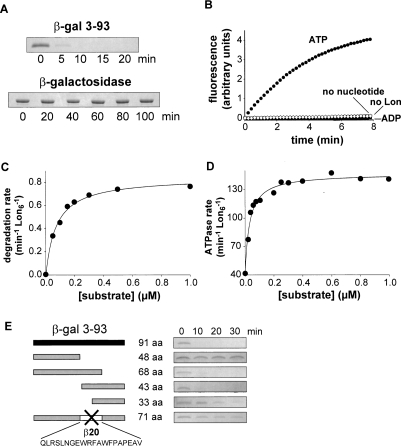
The purified 3–93 β-galactosidase fragment had a circular–dichroism spectrum characteristic of an unfolded protein and was degraded rapidly by proteases, including trypsin and chymotrypsin, which recognize simple sequences in denatured polypeptides (data not shown). As assayed by SDS-PAGE, purified Lon degraded the 3–93 fragment rapidly but did not degrade native β-galactosidase (Fig. 1A). To assess the nucleotide dependency of Lon degradation, we assayed increases in fluorescence upon proteolysis of a fluorescein-labeled variant of the 3–93 fragment. Robust Lon degradation of this fragment required ATP. Little, if any, degradation was observed in the presence of ADP or without nucleotide (Fig. 1B). To determine steady-state kinetic parameters, we measured Lon degradation rates for different concentrations of the 3–93 fragment labeled at its single cysteine with 14C-iodoacetic acid and fit the resulting data to the Henri-Michaelis-Menten equation (Fig. 1C; Table 1). At substrate saturation, the turnover number was 0.84 ± 0.02 molecules of substrate per enzyme per min. KM for degradation was 73 ± 7.3 nM, indicating that Lon binds this substrate tightly.
Figure 1.
Lon recognition and degradation of denatured β-galactosidase fragments. (A) Lon (150 nM hexamer) degraded the 3–93 fragment of β-galactosidase (5 μM) but not native β-galactosidase (5 μM) as assayed by SDS-PAGE. (B) Rapid degradation of fluorescein-labeled 3–93 fragment (5 μM) by Lon (150 nM hexamer) required ATP and was not observed with ADP, without nucleotide, or without Lon. (C) Steady-state rates of degradation of different concentrations of the 3–93 fragment by Lon (10 nM hexamer) were assayed by release of acid soluble 14C-labeled peptides. The solid curve is a fit to the Henri-Michaelis-Menten equation (R2 = 0.99). KM and Vmax are listed in Table 1. (D) Stimulation of Lon S679A (10 nM hexamer) ATP-hydrolysis rates by increasing concentrations of the 3–93 fragment. The curve is a fit (R2 = 0.98) to the hyperbolic binding isotherm (rate = basal + max ⋅ [S]/(Kapp + [S])) with Kapp = 32 ± 3 nM. (E) Deletion analysis of the 3–93 fragment. Truncated variants of the 3–93 fragment (5 μM) were assayed for degradation by Lon (150 nM hexamer) by SDS-PAGE.
Table 1.
Steady-state kinetic parameters for Lon degradation
The ATPase activity of Lon increases upon substrate binding (Waxman and Goldberg 1986). Titration of the 3–93 β-galactosidase fragment against a constant concentration of the proteolytically inactive Lon S679A mutant stimulated ATPase activity in a hyperbolic manner with an activation constant of 32 ± 3.3 nM (Fig. 1D). These results confirm that Lon binds tightly to the 3–93 fragment. Lon S679A was used for these studies to avoid substrate depletion and complications of product interactions during the long times required to obtain accurate rates of ATP hydrolysis at low concentrations of enzyme (10 nM), but this mutant has the same ATPase activity as the wild-type enzyme (Fischer and Glockshuber 1993; Starkova et al. 1998). The maximal rate of ATP hydrolysis by wild-type Lon was 147 ± 2.0 min−1 in the presence of saturating amounts of the 3–93 fragment. This value in combination with Vmax for degradation shows that Lon hydrolyzes about 175 ATP molecules in the time required to degrade a single 3–93 β-galactosidase fragment. Hence, the energetic cost of Lon degradation of the unfolded 3–93 substrate (≈2 ATPs per residue) is substantial.
Identification of a recognition element
To determine if specific amino acid sequences were required for recognition, we constructed and purified deletion mutants of the 3–93 fragment and assayed their degradation by Lon (Fig. 1E). Some truncations resulted in a dramatic decrease in the degradation rate. For example, Lon degraded a 48-residue N-terminal variant and a 33-residue C-terminal variant very slowly. In contrast, Lon rapidly degraded a variant containing the 68 N-terminal residues and a variant containing the C-terminal 43 residues of the 3–93 fragment. These results show that unfolded polypeptides are not necessarily good Lon substrates, and suggest that specific sequences between residues 49 and 68 of the 3–93 β-galactosidase fragment play important roles in Lon recognition. To test the importance of these residues directly, we deleted a 20-residue segment (QLRSLNGEWRFAWFPAPEAV; hereafter called β20) consisting of residues 49–68 and found that Lon degraded the resulting variant very slowly (Fig. 1E). We conclude that residues in the deleted segment play important roles in Lon recognition.
An autonomous degradation tag
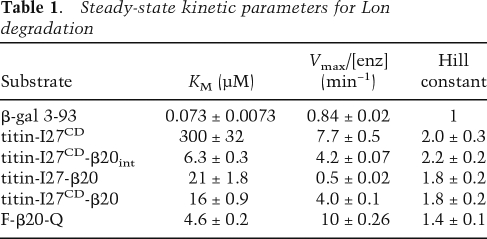
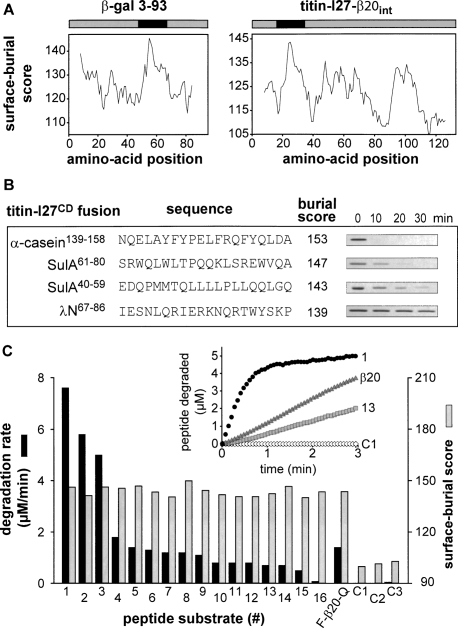
If the β20 sequence is sufficient for Lon recognition, then it should direct Lon degradation of other polypeptides. As a test of this hypothesis, we used a variant of the I27 domain of human titin (titin-I27CD) containing aspartic acids in place of both wild-type cysteines, which are normally buried in the native hydrophobic core. The titin-I27CD variant had a circular–dichroism spectrum characteristic of an unfolded protein (Fig. 2A) but was a poor Lon substrate with a KM of ≈300 μM (Fig. 2B,C).
Figure 2.
Properties and Lon degradation of substrates. (A) Circular–dichroism spectra (25°C) show that titin-I27 (40 μM) is natively folded and titin-I27CD (40 μM) is denatured. (B) SDS-PAGE assays of degradation of 5 μM titin-I27CD or β20-tagged variants by Lon (150 nM hexamer). The internal β20 tag was cloned between residues 17 and 18 of titin-I27CD. (C) Steady-state rates of Lon (100 nM hexamer) degradation of 35S-labeled titin-I27CD with or without an internal β20 insertion were assayed by release of acid-soluble peptides. The solid curves are fits (R2 = 0.99 for both curves) to the Hill equation (V = Vmax ⋅ [S]n/(KMn + [S]n); values for Vmax, KM, and the Hill constant are listed in Table 1. (D) Steady-state rates of degradation of 35S-labeled native titin-I27-β20 and denatured titin-I27CD-β20 by Lon (100 nM hexamer) were determined and fitted as described in C (R2 = 0.99 for both curves). Values of KM, Vmax, and the Hill constant are listed in Table 1. (Inset) As assayed by changes in circular–dichroism ellipticity at 228 nm, titin-I27 (closed symbols) and titin-I27-β20 (open symbols) had the same thermal stability. (E) SDS-PAGE assays of Lon degradation (150 nM hexamer) of the N-terminal domain of λ cI repressor (5 μM) with or without a β20 tag. (F) Degradation of GFP or GFP-β20 (3 μM each) by Lon (6 μM hexamer). Reactions were measured by decreases in GFP fluorescence.
Lon degradation was stimulated substantially when the β20 sequence was cloned at the N terminus, at the C terminus, or at an internal position of titin-I27CD (Fig. 2B). The titin-I27CD variant with the internal insertion was the best substrate. KM for Lon degradation of this variant was ∼6 μM (Fig. 2C), about 50-fold tighter than titin-I27CD alone. Thus, the β20 sequence enhances recognition of polypeptides that, by themselves, are intrinsically poor Lon substrates. Lon degradation of titin-I27CD, with or without a β20 insert, was positively cooperative with a Hill coefficient close to 2 (Fig. 2C). Similar behavior has been reported for other Lon substrates (Thomas-Wohlever and Lee 2002).
Native titin-I27 provides a stringent test of the ability of AAA+ proteases to denature stably folded substrates because it resists mechanical denaturation until very high forces are applied (Politou et al. 1995; Carrion-Vazquez et al. 1999; Kenniston et al. 2003; Burton et al. 2005). Thus, we appended the β20 sequence to the C terminus of native titin-I27 (titin-I27-β20) and assayed degradation by Lon (Fig. 2D). KM for Lon degradation of native titin-I27-β20 (21 ± 1.8) was slightly higher than that for denatured titin-I27CD-β20 (16 ± 0.9 μM), whereas Vmax was about eightfold higher for the denatured substrate (4.0 ± 0.1 min−1 versus 0.5 ± 0.02 min−1). Native titin-I27-β20 had the same thermal stability as unmodified titin-I27 (Fig. 2D, inset), establishing that addition of the unstructured β20 sequence did not destabilize the native portion of the fusion protein. Lon also degraded two additional folded substrates tagged with β20, the N-terminal domain of λ cI repressor and GFP, faster than their untagged counterparts (Fig. 2E,F).
We conclude that Lon has a powerful protein unfoldase activity and β20 functions as a Lon recognition tag when attached to a native protein. Because native titin-I27-β20 was degraded substantially more slowly than denatured titin-I27CM-β20 under conditions of substrate saturation, protein unfolding by Lon is likely to be the slow step in the steady-state degradation of the native substrate.
Peptide degradation
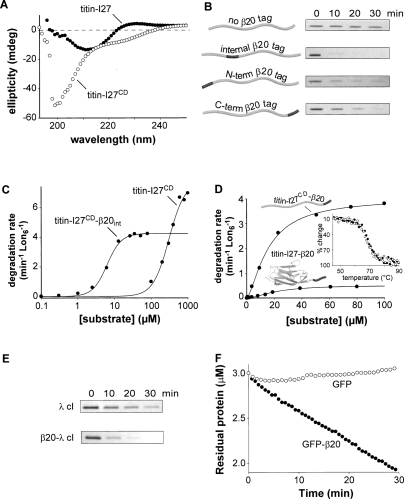
Does the β20 sequence contain all of the elements required for Lon recognition and degradation? To address this question, we synthesized a peptide consisting of this sequence flanked by a fluorophore-quencher pair (F-β20-Q) and tested for Lon cleavage by increases in fluorescence. In the presence of ATP, Lon degraded the F-β20-Q peptide with a KM of 4.6 ± 0.23 μM (Fig. 3A), establishing that amino acid sequences as short as 20 residues can be recognized efficiently by Lon. Indeed, the KM for Lon degradation of the β20 peptide is similar to those for degradation of α-casein (2.5 μM) and λ N protein (13 μM) (Lee and Berdis 2001).
Figure 3.
Degradation directed by peptide-tag sequences (A) Concentration dependence of the steady-state rate of F-β20-Q degradation by Lon (100 nM hexamer). The solid line is a fit (R2 = 0.99) to the Hill equation (V = Vmax ⋅ [S]n/(KMn + [S]n). KM, Vmax, and Hill constants are listed in Table 1. (B) Degradation of the F-β20-Q peptide (5 μM) by Lon (0.3 μM hexamer) was assayed by increased fluorescence in the presence of 2 mM ATP, ATPγS, AMPPNP, or without nucleotide. No significant degradation was observed using LonS679A (0.3 μM hexamer) and ATP. (C) Degradation of F-β20-Q, F-SulA150–169-Q, and F-SoxS1–21-Q (5 μM each) by Lon (0.3 μM hexamer). (D) Degradation of unfolded titin substrates by Lon (0.3 μM hexamer) or ClpXP (0.3 μM ClpX6, 0.9 μM ClpP14) was assayed by SDS-PAGE.
Lon degraded the F-β20-Q peptide more slowly when ATPγS was substituted for ATP, and displayed little if any activity with AMPPNP or ADP (Fig. 3B). These results suggest that efficient Lon degradation of F-β20-Q requires hydrolysis of ATP or ATPγS under our assay conditions, which include 100 mM KCl. It is known that AMPPNP, which is nonhydrolyzable, supports Lon degradation of some peptides in very low-salt buffer (Thomas-Wohlever and Lee 2002), and we confirmed that this was also the case for F-β20-Q (data not shown). However, Lon appears to aggregate in the absence of salt, and we worried that degradation observed under these nonphysiological conditions might not reflect the activity of native Lon hexamers.
Comparison with other Lon tags
Previous studies identified Lon targeting sequences in several substrates, including residues 15–29 of UmuD (Gonzalez et al. 1998), the C-terminal region of SulA (Ishii et al. 2000), the N-terminal 21 residues of SoxS (Shah and Wolf 2006), and the ssrA tag (Choy et al. 2007). For comparative purposes, we assayed the interaction of peptides containing these sequences or the β20 sequence with Lon. A peptide corresponding to residues 12–31 of UmuD stimulated Lon ATPase activity with an apparent affinity of 67 μM, but peptides corresponding to residues 150–169 of SulA, residues 1–21 of SoxS, and the ssrA tag showed no stimulation at concentrations up to 500 μM (Table 2). The β20 peptide half stimulated Lon at a concentration of 10.7 μM (Table 2). Hence, peptide sequences that appear to interact with Lon show a wide range of behaviors in their ability to bind Lon and/or to stimulate ATP hydrolysis.
Table 2.
%Peptide binding to Lon
Apparent affinities (Kapp) were determined by peptide-dependent stimulation of ATP hydrolysis by Lon (150 nM hexamer).
Because peptides could bind Lon without affecting ATPase activity, we assayed Lon degradation of the SoxS and SulA peptides flanked by fluorophore and quencher groups. The SoxS peptide was cleaved slowly, whereas the SulA peptide was cleaved at a rate similar to the β20 peptide (Fig. 3C). Recognition of the ssrA tag by ClpXP (another AAA+ protease) requires the α-carboxyl group (Kim et al. 2000), and thus we did not want to compromise potential Lon interactions by flanking the ssrA peptide with fluorophore and quencher groups. Instead, we assayed degradation of carboxymethylated (CM) titin-I27, which is unfolded (Kenniston et al. 2003), with or without an appended ssrA tag. Lon degraded titin-I27CM and titin-I27CM-ssrA at comparable rates that were much slower than degradation of titin-I27CM-β20 (Fig. 3D). For comparison, ClpXP degraded titin-I27CM-ssrA rapidly but did not degrade titin-I27CM or titin-I27CM-β20 (Fig. 3D). Thus, the ssrA tag is a relatively poor degradation tag for Lon, whereas the β20 tag is a good tag for Lon degradation but not for ClpXP degradation. Taken together, these results show that peptide targeting sequences display a wide range of Lon interactions. Some sequences (β20, UmuD11–31, and SulA150–169) are recognized reasonably well in the absence of additional binding determinants. Other sequences (SoxS1–21 and ssrA) do not seem to interact well with Lon, and substrates may require these and additional targeting determinants to ensure efficient Lon degradation.
Native burial as a Lon recognition determinant
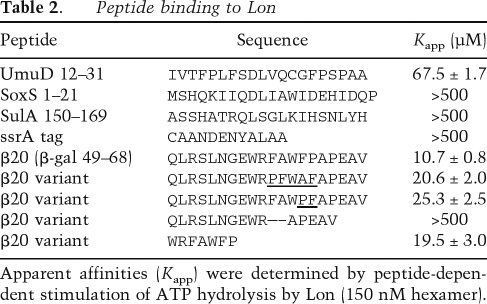
What makes the β20 tag such a good Lon recognition sequence? Strong hydrophobicity is one possibility, consistent with Lon’s ability to recognize a diverse group of unfolded proteins and the fact that aromatic and nonpolar residues comprise 60% of the β20 sequence. Moreover, other quality-control proteins have been shown to interact with hydrophobic sequences (Rudiger et al. 1997, 2001; Chen and Sigler 1999; Patzelt et al. 2001). Using most hydrophobicity scales, β20 was not the highest scoring sequence segment in either the β-galactosidase 3–93 fragment or in titin-I27 with the β20 insertion (data not shown). In contrast, when we used a scale based on average surface area buried in native proteins (Rose et al. 1985), a portion of the β20 sequence was the highest scoring region in both the 3–93 β-galactosidase fragment and in titin-I27 with the β20 insertion (Fig. 4A).
Figure 4.
Correlation of surface-burial scores with Lon degradation. (A) Surface-burial scores for β-gal 3–93 and titin-I27-β20int were calculated as described in Experimental Procedures. The β20 sequence scored highest in both substrates. (B) Sequences with the highest surface-burial scores from three Lon substrates were inserted by cloning between residues 17 and 18 of unfolded titin-I27CD and each fusion protein (5 μM) was assayed for degradation by Lon (150 nM hexamer) by SDS-PAGE. (C) Peptides corresponding to 15-residue β-galactosidase sequences with the highest surface-burial scores (numbered 1–16) were synthesized with flanking fluorophore and quencher groups. Each peptide (5 μM) was incubated with Lon (300 nM hexamer) and the rate of degradation was assayed by changes in fluorescence, as shown for representative peptides in the inset. C1–C3 are 15-residue control β-galactosidase peptides with low surface-burial scores. Degradation rates (dark bars) and surface-burial scores (light bars) are plotted for each peptide. Peptide sequences were RWQFNRQSGFLSQMW (1), YWQAFRQYP RLQGGF (2), FAKYWQAFRQYPRLQ (3), HYPNHPLWYTLC DRY (4), MWRMSGIFRDVSLLH (5), RWDLPLSDMYTPYVF (6), RWLPAMSERVTRMVQ (7), EYLFRHSDNELLHWM (8), YLEDQDMWRMSGIFR (9), LTEAKHQQQFFQFRL (10), LRA GENRLAVMVLRW (11), LLIRGVNRHEHHPLH (12), RMVQR DRNHPSVIIW (13), FRQYPRLQGGFVWDW (14), HQWRGD FQFNISRYS (15), FVWDWVDQSLIKYDE (16), GETQVASG TAPFGGE (C1), RPVQYEGGGADTTAT (C2), and GIGGDDS WSPSVSAE (C3).
To test the significance of this correlation, we scanned three Lon substrates (α-casein, SulA, and the λN-protein) for the highest scoring 15-residue sequences using the surface-burial algorithm. These sequences were then cloned into titin-I27CD with a few flanking residues, and the resulting proteins were assayed for degradation by Lon (Fig. 4B). The insert with the highest score (residues 139–158 of α-casein) was degraded most rapidly, the next highest scoring inserts (SulA residues 61–80 and 40–59, respectively) caused intermediate levels of degradation, and the insert with the lowest score (λN residues 67–86) stimulated little if any Lon degradation. Below, we show that surface-burial scores can generally identify peptide sequences that are degraded well by Lon but do not necessarily predict the relative rates of degradation. Nevertheless, propensity for surface burial can be used to rationalize Lon recognition of the β20 tag and to identify sequences from Lon substrates that serve as Lon recognition tags.
To test the apparent correlation between surface-burial scores and Lon degradation in greater detail, we scanned the sequence of full-length β-galactosidase and identified 15-residue segments with scores >140. Peptides corresponding to each of these sequences were synthesized with flanking fluorophore and quencher groups, and 16 of 25 were sufficiently soluble to allow assays of Lon degradation at a substrate concentration of 5 μM (Fig. 4C). Three peptides (#1, #2, and #3) were degraded considerably faster than the F-β20-Q peptide, and 10 peptides were degraded at rates within 50% of F-β20-Q. Only one high-scoring peptide (#16) showed much slower degradation. As controls, we synthesized three 15-residue β-galactosidase peptides with very low surface-burial scores. Two of these peptides were not degraded at a detectable rate by Lon (C1 and C2) and one (C3) was degraded >30-fold slower than the F-β20-Q reference peptide.
Taken together, these results strengthen the conclusion that sequences rich in residues with a high probability of being buried in native proteins are determinants of Lon recognition. It is important to note, however, that there was no strict correlation between peptide degradation rates and surface-burial scores among the high-scoring peptides. For example, peptides with almost identical scores were degraded at rates differing by >10-fold (Fig. 4C). Thus, other sequence features must also contribute to Lon recognition. For instance, Lon may recognize a sequence pattern in which the positions and/or identities of specific residues are important.
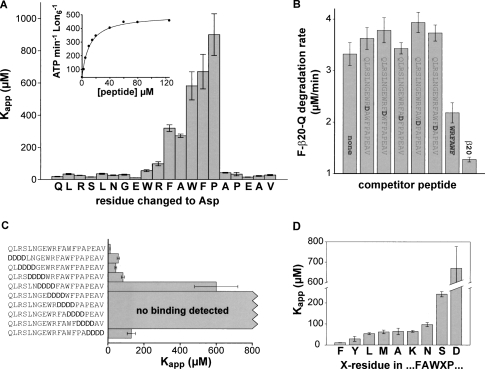
Mutational analysis of β20
To probe the importance of different residues in the β20 tag, we synthesized peptides in which each amino acid was individually mutated to aspartic acid, a highly polar residue with a low probability of burial in native structures. Each peptide was titrated against a fixed concentration of Lon, and apparent interaction constants (Kapp) were determined by fitting rates of ATP hydrolysis to a hyperbolic binding isotherm (see inset in Fig. 5A). For the wild-type β20 peptide, Kapp was 10.7 ± 0.80 μM (Table 2). Substitutions for the first ten and last five residues of the β20 peptide caused modest reductions in apparent affinity (Fig. 5A). In contrast, single aspartate substitutions in the FAWFP sequence of β20 reduced apparent affinity by a factor of 20 or more (Fig. 5A). We also assayed inhibition of Lon degradation of F-β20-Q by the unmodified β20 peptide or variants with aspartic acid substitutions in the FAWFP sequence (Fig. 5B). Competition was observed with the β20 peptide but not with the mutants, confirming the importance of the FAWFP sequence for tight binding to Lon. Consistently, deletion of the FAWFP sequence prevented detectable peptide binding to Lon (Table 2).
Figure 5.
Mutational analysis of β20-peptide recognition by Lon. (A) The apparent Lon affinities of β20-peptide variants, each containing a single aspartic acid substitution, were determined by assaying changes in the ATPase rate of Lon (150 nM hexamer) at a series of peptide concentrations (bars represent standard errors for the Kapp values). The inset shows the data and fitted affinity curve for the wild-type β20 peptide (Kapp = 10.7 ± 0.8 μM). R2 values for all fitted curves were >0.97. For weakly binding peptides, saturation was not reached and Κapp was calculated by assuming that Vmax was ≈500 ATP/min ⋅ Lon6. (B) Degradation of F-β20-Q (25 μM) by Lon (300 nM hexamer) was measured in the presence/absence of the designated competitor peptides (125 μΜ each). (R2 = 0.99 for both curves). (C) Values of Kapp for Lon binding were measured for β20-peptide variants in which a block of four consecutive residues was substituted with DDDD as described in A. R2 values for all fitted curves were >0.97. (D) Values of Kapp for Lon binding, measured as described in A, for β20-peptides with different residues at the X position in the FAWXP subsequence. Values for the F and D sequences are taken from A. R2 values for all fitted curves were >0.97.
A heptapeptide (WRFAWFP) within β20 stimulated the ATPase activity of Lon in a hyperbolic fashion with an apparent binding constant of 19.5 ± 3.0 μM (Table 2) and also inhibited Lon degradation of F-β20-Q (Fig. 5B). Because the affinity of Lon for this heptapeptide was within twofold of the full β20 peptide, other β20 residues appear to play only minor roles in Lon recognition. In another test of the importance of β20 residues, we substituted blocks of four residues with four aspartic acids (DDDD) and assayed Lon binding of the mutant peptides using the ATPase assay (Fig. 5C). As expected, binding was eliminated by DDDD substitutions that changed any part of the FAWFP core sequence. In addition, DDDD substitutions of flanking residues also weakened binding by factors ranging from eightfold to 50-fold, with the largest effect observed when the DDDD sequence was placed immediately before the FAWFP core.
To determine if the precise sequence of the FAWFP core was critical for Lon recognition, we performed several experiments. First, we reversed the positions of the last two residues (FAWPF) and also inverted the entire sequence (PFWAF). In both cases, Lon-binding affinity dropped <2.5-fold (Table 2), suggesting that the exact order of residues in the core affects recognition but is not a crucial determinant. Second, we substituted the phenylalanine at the fourth position of the core sequence (FAWFP) with alanine, asparagine, aspartic acid, leucine, lysine, methionine, serine, or tyrosine, and measured binding of the mutant peptides to Lon (Fig. 5D). The largest reductions in binding were observed for the aspartate (70-fold) and serine substitutions (20-fold), but even the most conservative substitutions (leucine and tyrosine) decreased binding roughly eightfold. Thus, the identity and positioning of side chains in the FAWFP core play roles in determining the strength of Lon recognition. Nevertheless, our results suggest that Lon recognition tolerates considerable variation, allowing a large number of different sequences to bind tightly enough to serve as degradation tags.
Discussion
Recognition of sequence signals in nonnative substrates
Our results show that Lon recognition is mediated by specific sequences in nonnative substrates. For example, a 20-residue sequence from β-galactosidase binds to Lon and targets other proteins for Lon degradation. Within this sequence, a cluster of five to seven relatively hydrophobic residues is both critical and sufficient for recognition. We found no evidence for models in which Lon recognizes unfolded peptides or proteins per se. Indeed, we find that unfolded peptides or polypeptides of comparable length can be degraded at vastly different rates by Lon, with some showing no detectable proteolysis. The simplest and most appealing model is that sequence signals in denatured polypeptides bind directly to a docking site in Lon. Several lines of evidence argue against an alternative model, in which these sequences drive aggregation, with aggregates then being the species recognized by Lon. First, the rate of Lon degradation of the 3–93 fragment of β-galactosidase changed as a simple hyperbolic function of substrate concentration, whereas a strong power dependence would be expected for an aggregation model. The concentration dependence of Lon ATPase stimulation by different substrates was also consistent with a 1:1 binding model. Lon degradation of several substrates did show Hill constants as high as 2. However, one of these proteins (titin-I27-β20) was highly soluble and chromatographed as a monomer in gel filtration experiments (data not shown), and thus the oligomeric species degraded by Lon is unlikely to be a higher-order aggregate.
In native β-galactosidase, the backbone and most side chains of the critical FAWFP segment of β20 are embedded in the three-dimensional structure. Hence, it is not surprising that Lon does not degrade folded β-galactosidase. In contrast, these residues would be accessible to Lon in unstructured N-terminal fragments, explaining why Lon degrades these molecules. When the β20 sequence was transferred to other proteins, it directed Lon degradation when present at the N terminus, the C terminus, or an internal position of an unfolded substrate. Although degradation tags are frequently found at protein termini, Lon can obviously recognize internal tags as has previously been shown for the related ClpXP, ClpAP, and FtsH proteases (Hoskins et al. 2002; Okuno et al. 2006). The fusion protein with the internal β20 tag was a slightly better Lon substrate than proteins with terminal tags for reasons that are not yet clear. The β20 sequence also directed Lon degradation when fused to natively folded proteins, showing that Lon is capable of unfolding stable protein structures. Finally, Lon degraded the isolated β20 peptide efficiently. Thus, an accessible β20 sequence acts as an autonomous tag that allows Lon to bind denatured or native substrates. In this regard, Lon is similar to other AAA+ proteases that recognize specific peptide signals in substrates (for review, see Sauer et al. 2004).
Recognition rules
The results presented here suggest that Lon recognition sequences are likely to be hidden in the hydrophobic cores of many proteins. These sequences must meet the dual constraints of mediating stable packing in the context of the native three-dimensional structure and binding Lon with reasonable affinity in the denatured state. Given the diversity of protein folds, it would be difficult to imagine this system functioning well if Lon recognition required one or a few closely related sequences. Indeed, our results show that significant variation is allowed in sequences that mediate Lon recognition. For example, we found that reversing the order of residues in the critical FAWFP segment of β20 caused only a twofold reduction in Lon affinity. Moreover, considerable variation in hydrophobic core packing is tolerated in protein folding (Lim and Sauer 1989; Baldwin and Matthews 1994). Thus, the ability of a broad set of “hydrophobic” sequences to be incorporated into native protein structures and to be recognized with reasonable affinity by Lon helps explain why such a diverse array of denatured proteins can be recognized and degraded by this protease.
Experiments based on mutagenesis and informatics established the general importance of hydrophobicity in Lon recognition. A hydrophobicity algorithm based on surface-area burial (Rose et al. 1985) allowed us to identify numerous peptide sequences in β-galactosidase and other substrates that serve as efficient Lon recognition signals. This scale, which did a good job of identifying peptide sequences that Lon degrades but a poor job of predicting relative degradation rates, ranks aromatic side chains most highly. Indeed, hydrophobic sequences that lacked aromatics were usually weak degradation tags, and the best Lon recognition tags had a cluster of hydrophobic residues, with at least two aromatics (preferentially phenylalanine and tryptophan). For example, the WRFAWFP heptamer, which contains four aromatic residues, was sufficient for Lon recognition. The aliphatic portions of lysine and arginine side chains apparently satisfy a need for nonpolar packing in Lon recognition, as these basic residues were less deleterious than small polar side chains in substitution experiments and were reasonably common in peptides that Lon recognized efficiently. In contrast, negatively charged residues were absent from the core regions of the best recognition tags and substitution of core and flanking residues with aspartic acid decreased Lon affinity. Thus, our studies reveal the basic principles of Lon recognition of misfolded proteins.
Lon recognition strategies
In addition to misfolded proteins, Lon degrades undamaged proteins. For example, Lon recognizes a degradation tag close to the N terminus of UmuD, which is similar to the β20 tag in containing several critical aromatic residues (Gonzalez et al. 1998). The UmuD tag is accesssible in the native protein, and thus Lon probably has to unfold UmuD to ensure complete degradation. Indeed, our results show that Lon has a robust unfolding activity. Some substrates, like the N protein of bacteriophage λ, appear to expose Lon recognition elements in a free but not bound state. This transcriptional anti-terminator lacks defined tertiary structure when it is not bound to RNA and is rapidly degraded by Lon (Legault et al. 1998).
Some AAA+ proteases recognize multiple classes of peptide signals (Flynn et al. 2003), and Lon appears to share this characteristic. For example, the C-terminal sequence of SulA is important for Lon degradation (Ishii et al. 2000; Ishii and Amano 2001), but there is little similarity between this recognition element and recognition tags like β20. Nevertheless, we found that Lon degraded a C-terminal SulA peptide at a rate comparable with the β20 peptide. Moreover, unlike β20, the SulA peptide did not stimulate Lon’s ATPase activity, suggesting that these tags interact with Lon in fundamentally different manners. Thus, Lon may recognize native and denatured proteins using different classes of sequence signals. We note that the extreme hydrophobicity of the β20 tag could easily create problems in using it as a degradation tag for native proteins in vivo. For example, when we fused β20 to the N or C terminus of GFP, a large portion of the resulting protein was insoluble in the cell.
Consistent with previous studies (Ebel et al. 1999), our results indicate that the Lon hexamer contains more than one recognition site for protein substrates. First, more than one site is required to explain the Hill constant of 2 observed for degradation of titin-I27 variants. Second, the 3–93 fragment of β-galactosidase bound Lon with an apparent affinity of 30–75 nM, whereas the β20 and WRFAWFP subfragments bound ∼100-fold more weakly. From a biological perspective, the ability of Lon hexamers to recognize multiple sequences in an unfolded polypeptide chain would allow individual interactions of modest affinity to be coupled to produce much tighter binding and thus to ensure efficient elimination of potentially deleterious nonnative substrates. Shah and Wolf (2006) identified several regions of SoxS, including the first 21 residues that appear to play roles in degradation by Lon. We found that the SoxS1–21 peptide was degraded relatively slowly by Lon, suggesting that additional determinants are likely to be important in efficient degradation of intact SoxS by Lon. Strikingly, some of the other SoxS sequences that influence degradation have high surface-burial scores and thus may resemble the β20 tag. For many AAA+ proteases, tighter binding is mediated by adaptor proteins that tether specific substrates to the protease (for review, see Baker and Sauer 2006). By using one region to tether themselves to Lon, many substrates may function as their own adaptors for degradation.
Lon is clearly well suited for specific recognition and degradation of a broad array of incomplete, damaged, and nonnative proteins. For example, unfolding of a single domain containing a strong Lon recognition signal should suffice for proteolysis, as Lon would be able to unfold and degrade any attached regions of native protein. Hence, unfolding of the N-terminal domain (residues 1–219) of β-galactosidase (1029 residues total) should produce an excellent substrate for complete Lon degradation, with a KM near or below 75 nM. Proteins also undergo transient local denaturation, however, and it would be wasteful if these events triggered efficient degradation. If just the β20 sequence of β-galactosidase became transiently exposed, for example, the probability of Lon recognition and degradation would be relatively small because the KM for Lon recognition would be much higher and refolding would occur rapidly. Hence, long-lived protein unfolding events that exposed multiple Lon recognition elements would result in efficient degradation, whereas transient events that exposed just a single element would be relatively benign.
Interplay with chaperones
Proteases and chaperones represent two sides of the same coin, acting in opposing pathways to clear unfolded proteins from the cell. From an energetic perspective, protein refolding is more cost effective than degradation and resynthesis. Thus, mechanisms should exist to ensure that denatured proteins have a good chance to fold or refold before they are degraded. The competition is probably biased toward refolding because chaperones and chaperonins are more abundant in the cell than Lon (Phillips et al. 1984; Lorimer 1995; Mogk et al. 1999). There also appear to be more recognition sites in denatured polypeptides for chaperones than for Lon. For example, DnaK and its cochaperone, DnaJ (Hsp40), bind hydrophobic sequences that are thought to occur roughly once every 40 residues in unfolded proteins (Rudiger et al. 1997, 2001; Patzelt et al. 2001), whereas potential Lon-binding sites with surface-burial scores >140 are less frequent. Although Lon, DnaKJ, and GroEL all recognize hydrophobic sequences, their recognition sites may not overlap significantly. For example, DnaK and GroEL prefer some peptide sequences (Rudiger et al. 1997; Chen and Sigler 1999), which our results suggest would be bound poorly by Lon. Moreover, we found that DnaKJE and GroEL did not compete with Lon for binding to the β20 peptide (data not shown). Nevertheless, it is straightforward to imagine how binding of GroEL or DnaK to a nonnative protein could block Lon recognition. At specific stages in their ATPase cycles, GroEL and DnaK release nonnative protein substrates (Bukau et al. 2006). If the released substrate is damaged and cannot refold, however, then rebinding by the chaperone/chaparonin versus binding by Lon will become a race. Even if the protease wins this race only infrequently, statistical considerations will eventually result in Lon binding and proteolytic removal of the damaged protein.
Materials and methods
Protein purification and modification
The Lon purification was based on the procedure of Goldberg et al. (1994). A 2-L culture of E. coli strain ER2566 (New England Biolabs) carrying plasmid pBAD33-lon (Christensen et al. 2004) was grown at 37°C in 2XYT medium supplemented with chloramphenicol (34 μg/mL). At an OD600 of 1.0, L-arabinose was added (0.2%, w/v) and the culture was grown for an additional 3 h before harvesting cells and freezing them at –80°C. After thawing cells in 25 mL of cold buffer A (100 mM potassium phosphate at pH 6.5, 1 mM DTT, 1 mM EDTA, 10% glycerol), the cell suspension was lysed by a French pressure cell press. The lysate was centrifuged (18,000g, 30 min), and the supernatant was decanted to a 50-mL tube containing 15 mL of P11-phosphocellulose (Whatman) prewashed with buffer A. More buffer was added to a final volume of 50 mL and the suspension was slowly shaken for 30 min at 4°C. After washing twice with buffer A and twice with a similar buffer containing 200 mM potassium phosphate (pH 6.5), the protein was eluted with 35 mL buffer containing 400 mM potassium phosphate (pH 6.5). The protein solution was filtered (0.22 μm), concentrated to 5 mL using an Amicon concentrator with a 100-kDa cutoff, and loaded on a 26/60 sephacryl-S300 gel filtration column (GE-Healthcare) preequilibrated with 50 mM HEPES (pH 7.5), 1 mM DTT, 1 mM EDTA, and 20% glycerol. Fractions containing Lon at purity >95% were concentrated as described above, aliquoted, and kept frozen at −80°C. Lon S679A was purified using the same procedure. Purified E. coli ClpX and ClpP were a gift form Mary Lee (MIT).
The 3–93 fragment of β-galactosidase and derivatives were expressed from a T7 promoter (pET.β-gal 3–93 in E. coli ER2566 cells) in 1-L cultures of 2XYT broth at 37°C. Following induction with 0.5 mM IPTG for 3 h, cells were harvested, resuspended in 25 mL of T25 (25 mM Tris-HCl at pH 8.0) plus 1 mM MgCl2 and lysozyme (0.02 mg/mL). Cells were lysed using a French pressure cell and the lysate was centrifuged (18,000g, 10 min). The β-galactosidase fragments were found in the insoluble fraction. The pellets were resuspended in 25 mL of T25 plus 0.02 mg/mL lysozyme. After 20 min at room temperature, the suspension was recentrifuged. To remove membrane proteins, the pellets were washed three times with a solution of 1 mg/mL deoxycholic acid plus 1 mM EDTA, and then washed once with T25. After solubilization of the pellets in T25 plus 6 M GuHCl, the solutions were centrifuged and supernatants were passed through a 0.22-μm filter. After this step, fragments were >95% pure. To exchange buffers, the protein was first diluted to 200 μM in T25, 2 M GuHCl, and 1 mM DTT. 2.5 mL of this solution was chromatographed on a PD-10 column (GE Healthcare) equilibrated with T25 plus 1 mM DTT. After this step, the protein solution was centrifuged (5000g, 10 min), 2.5 mL was passed through a second PD-10 column, and the protein was centrifuged again, filtered, and stored at 4°C.
For radiolabeling, a freshly prepared solution of 3–93 fragment in T25 plus 6 M GuHCl was filtered, carboxymethylated with 10 mM 14C-iodoacetic acid (GE Healthcare) for 2 h at room temperature, precipitated with 10% trichloroacetic acid, and washed twice with acetone. After drying, the protein was solubilized in T25, 6 M GuHCl, and 1 mM DTT and exchanged into buffer without denaturant as described for the unlabeled protein. Labeling of the 3–93 fragment labeled with 5-iodoacetamidofluorescein (Molecular Probes) was performed in the same manner, except the precipitation step was skipped and a final gel filtration step was performed using a 16/60 Superdex-75 (GE Healthcare) column in T25 plus 1 mM DTT.
Variants of His6-tagged titin-I27 were expressed from a T7 promoter in 1-L cultures. Harvested cells were resuspended in 25 mL of T25, 500 mM NaCl, 20 mM imidazole, and 0.02 mg/mL lysozyme (without NaCl for titin-I27-β20) and lysed either with a French pressure cell press or by sonication. For variants other than titin-I27CD fusions, the supernatant was mixed in a 50-mL tube with 2 mL Ni-NTA resin (Qiagen), prewashed with the same buffer, and incubated at 4°C for 15 min. The resin was washed three times with 20 mL of the same buffer, resuspended in 5 mL buffer, and transferred to a gravity column, and washed once with 10 mL of buffer. For titin-Ι27tβ20, an additional wash was carried out with 10 mL of T25, 20 mM imidazole, and 20% ethylene glycol, following a second wash with 10 mL of T25 plus 20 mM imidazole. Protein was eluted with 3 mL of T25, 500 mM NaCl, and 250 mM imidazole (without NaCl for titin-I27-β20). For titin-I27-β20, an additional gel filtration step (GE Healthcare 16/60 Superdex-75; T25) was carried out after passing the protein solution through a 0.22-μm filter. Following purification, titiin-I27 variants were stored at −70°C. For 35S-labeling, cells were grown in rich defined medium (TekNova), a 35S-methionine/35S-cysteine mixture (Perkin-Elmer) was introduced during expression, and purification through the Ni++-NTA step was performed. Carboxymethylation with 10 mM iodoacetic acid was carried out at room temperature for 2 h. After expression, titin-I27CD fusions were present in inclusion bodies and were purified as described for the 3–93 fragment of β-galactosidase. Following solubilization in 6 M GuHCl, the protein was bound to a Ni-NTA resin under denaturing conditions, washed with T25, eluted, and filtered (0.22 μM) prior to storage.
Tagged and untagged variants of the N-terminal domain of λ cI (residues 1–97) and GFP were expressed under T7-promoter control (see above), and were purified by Ni++-NTA affinity and gel filtration on a Superdex-S75 column in T25 buffer plus 300 mM NaCl.
Peptides
The 33-residue C-terminal peptide of the 3–93 fragment was obtained by tryptic digestion, followed by a mono-Q purification step. Other peptides were synthesized by the MIT Biopolymers Lab, purified by HPLC, and resolubilized in T25 or dimethylsulfoxide. For fluorophore-quencher labeling, para-aminobenzoic acid (PABA) was used as the fluorophore at the N terminus, and nitrotyrosine was used as a quencher at the penultimate C-terminal position followed by a single alanine. Concentrations of peptides containing nitrotyrosine were determined from absorbance at 381 nm (ε = 2200 M−1cm−1).
Degradation and ATPase assays
Degradation and ATPase assays were carried out at 37°C. Lon degradation buffer contained T25, 100 mM KCl, 10 mM MgCl2, 1 mM DTT, 2 mM ATP, 20 mM phosphoenolpyruvate, and 10 U/mL Pyruvate Kinase. Degradation rates were determined as described (Thomas-Wohlever and Lee 2002). For degradation of radioactive substrates, TCA precipitation was carried out as described (Gottesman et al. 1998). For ATPase measurements, NADH (1 mM) and LDH (10 U/mL) were added to the degradation buffer and assays were performed as described (Norby 1988; Lindsley 2001). For ClpXP degradation, the buffer contained 25 mM HEPES (pH 7.6), 100 mM KCl, 5 mM MgCl2, 1 mM DTT, 10% glycerol, 2 mM ATP, 20 mM phosphoenolpyruvate, and 10 U/mL Pyruvate Kinase.
Hydrophobicity calculations
Surface-burial scores were calculated at http://www.expasy.org/tools/protscale.html using the “average area buried” scale with a 15-residue window.
Acknowledgments
We thank L. Van Melderen for plasmid pBAD33-lon, C. Hayes for the titin-I27CD clone, and B. Cezairliyan, O. Kandror, M. Laub, A. Martin, J. Sohn, and S. Sundar for helpful discussions. This work was supported by NIH grant AI-16892.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1670908.
References
- Baker T.A., Sauer R.T. ATP-dependent proteases of bacteria: Recognition logic and operating principles. Trends Biochem. Sci. 2006;31:647–653. doi: 10.1016/j.tibs.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin E.P., Matthews B.W. Core-packing constraints, hydrophobicity and protein design. Curr. Opin. Biotechnol. 1994;5:396–402. doi: 10.1016/0958-1669(94)90048-5. [DOI] [PubMed] [Google Scholar]
- Bukau B., Weissman J., Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Burton R.E., Baker T.A., Sauer R.T. Nucleotide-dependent substrate recognition by the AAA+ HslUV protease. Nat. Struct. Mol. Biol. 2005;12:245–251. doi: 10.1038/nsmb898. [DOI] [PubMed] [Google Scholar]
- Carrion-Vazquez M., Oberhauser A.F., Fowler S.B., Marszalek P.E., Broedel S.E., Clarke J., Fernandez J.M. Mechanical and chemical unfolding of a single protein: A comparison. Proc. Natl. Acad. Sci. 1999;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Sigler P.B. The crystal structure of a GroEL/peptide complex: Plasticity as a basis for substrate diversity. Cell. 1999;99:757–768. doi: 10.1016/s0092-8674(00)81673-6. [DOI] [PubMed] [Google Scholar]
- Choy J.S., Aung L.L., Karzai A.W. Lon protease degrades transfer-messenger RNA-tagged proteins. J. Bacteriol. 2007;189:6564–6571. doi: 10.1128/JB.00860-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S.K., Maenhaut-Michel G., Mine N., Gottesman S., Gerdes K., Van Melderen L. Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: Involvement of the yefM–yoeB toxin–antitoxin system. Mol. Microbiol. 2004;51:1705–1717. doi: 10.1046/j.1365-2958.2003.03941.x. [DOI] [PubMed] [Google Scholar]
- Chung C.H., Goldberg A.L. The product of the lon (capR) gene in Escherichia coli is the ATP-dependent protease, protease La. Proc. Natl. Acad. Sci. 1981;78:4931–4935. doi: 10.1073/pnas.78.8.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel W., Skinner M.M., Dierksen K.P., Scott J.M., Trempy J.E. A conserved domain in Escherichia coli Lon protease is involved in substrate discriminator activity. J. Bacteriol. 1999;181:2236–2243. doi: 10.1128/jb.181.7.2236-2243.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer H., Glockshuber R. ATP hydrolysis is not stoichiometrically linked with proteolysis in the ATP-dependent protease La from Escherichia coli. J. Biol. Chem. 1993;268:22502–22507. [PubMed] [Google Scholar]
- Flynn J.M., Neher S.B., Kim Y.I., Sauer R.T., Baker T.A. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell. 2003;11:671–683. doi: 10.1016/s1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- Goldberg A.L., Moerschell R.P., Chung C.H., Maurizi M.R. ATP-dependent protease La (lon) from Escherichia coli. Methods Enzymol. 1994;244:350–375. doi: 10.1016/0076-6879(94)44027-1. [DOI] [PubMed] [Google Scholar]
- Gonzalez M., Frank E.G., Levine A.S., Woodgate R. Lon-mediated proteolysis of the Escherichia coli UmuD mutagenesis protein: In vitro degradation and identification of residues required for proteolysis. Genes & Dev. 1998;12:3889–3899. doi: 10.1101/gad.12.24.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S. Proteolysis in bacterial regulatory circuits. Annu. Rev. Cell Dev. Biol. 2003;19:565–587. doi: 10.1146/annurev.cellbio.19.110701.153228. [DOI] [PubMed] [Google Scholar]
- Gottesman S., Zipser D. Deg phenotype of Escherichia coli lon mutants. J. Bacteriol. 1978;133:844–851. doi: 10.1128/jb.133.2.844-851.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S., Roche E., Zhou Y., Sauer R.T. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes & Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins J.R., Yanagihara K., Mizuuchi K., Wickner S. ClpAP and ClpXP degrade proteins with tags located in the interior of the primary sequence. Proc. Natl. Acad. Sci. 2002;99:11037–11042. doi: 10.1073/pnas.172378899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii Y., Amano F. Regulation of SulA cleavage by Lon protease by the C-terminal amino acid of SulA, histidine. Biochem. J. 2001;358:473–480. doi: 10.1042/0264-6021:3580473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii Y., Sonezaki S., Iwasaki Y., Miyata Y., Akita K., Kato Y., Amano F. Regulatory role of C-terminal residues of SulA in its degradation by Lon protease in Escherichia coli. J. Biochem. 2000;127:837–844. doi: 10.1093/oxfordjournals.jbchem.a022677. [DOI] [PubMed] [Google Scholar]
- Johansson J., Uhlin B.E. Differential protease-mediated turnover of H-NS and StpA revealed by a mutation altering protein stability and stationary-phase survival of Escherichia coli. Proc. Natl. Acad. Sci. 1999;96:10776–10781. doi: 10.1073/pnas.96.19.10776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenniston J.A., Baker T.A., Fernandez J.M., Sauer R.T. Linkage between ATP consumption and mechanical unfolding during the protein processing reactions of an AAA+ degradation machine. Cell. 2003;114:511–520. doi: 10.1016/s0092-8674(03)00612-3. [DOI] [PubMed] [Google Scholar]
- Kim Y.I., Burton R.E., Burton B.M., Sauer R.T., Baker T.A. Dynamics of substrate denaturation and translocation by the ClpXP degradation machine. Mol. Cell. 2000;5:639–648. doi: 10.1016/s1097-2765(00)80243-9. [DOI] [PubMed] [Google Scholar]
- Kowit J.D., Goldberg A.L. Intermediate steps in the degradation of a specific abnormal protein in Escherichia coli. J. Biol. Chem. 1977;252:8350–8357. [PubMed] [Google Scholar]
- Lee I., Berdis A.J. Adenosine triphosphate-dependent degradation of a fluorescent λ N substrate mimic by Lon protease. Anal. Biochem. 2001;291:74–83. doi: 10.1006/abio.2001.4988. [DOI] [PubMed] [Google Scholar]
- Legault P., Li J., Mogridge J., Kay L.E., Greenblatt J. NMR structure of the bacteriophage lambda N peptide/boxB RNA complex: Recognition of a GNRA fold by an arginine-rich motif. Cell. 1998;93:289–299. doi: 10.1016/s0092-8674(00)81579-2. [DOI] [PubMed] [Google Scholar]
- Lim W.A., Sauer R.T. Alternative packing arrangements in the hydrophobic core of λ repressor. Nature. 1989;339:31–36. doi: 10.1038/339031a0. [DOI] [PubMed] [Google Scholar]
- Lin S., Zabin I. β-galactosidase. Rates of synthesis and degradation of incomplete chains. J. Biol. Chem. 1972;247:2205–2211. [PubMed] [Google Scholar]
- Lindsley J.E. Use of a real-time, coupled assay to measure the ATPase activity of DNA topoisomerase II. Methods Mol. Biol. 2001;95:57–64. doi: 10.1385/1-59259-057-8:57. [DOI] [PubMed] [Google Scholar]
- Lorimer G. A quantitative assessment of the role of the chaperonin proteins in protein folding in vivo. FASEB J. 1995;10:5–9. doi: 10.1096/fasebj.10.1.8566548. [DOI] [PubMed] [Google Scholar]
- Miller C.G., Zipser D. Degradation of Escherichia coli β-galactosidase fragments in protease-deficient mutants of Salmonella typhimurium. J. Bacteriol. 1977;130:347–353. doi: 10.1128/jb.130.1.347-353.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A., Tomoyasu T., Goloubinoff P., Rüdiger S., Röder D., Langen H., Bukau B. Identification of thermolabile Escherichia coli proteins: Prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 1999;18:6934–6949. doi: 10.1093/emboj/18.24.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norby J.G. Coupled assay of Na+,K+-ATPase activity. Methods Enzymol. 1988;156:116–119. doi: 10.1016/0076-6879(88)56014-7. [DOI] [PubMed] [Google Scholar]
- Okuno T., Yamanaka K., Ogura T. An AAA protease FtsH can initiate proteolysis from internal sites of a model substrate, apo-flavodoxin. Genes Cells. 2006;11:261–268. doi: 10.1111/j.1365-2443.2006.00940.x. [DOI] [PubMed] [Google Scholar]
- Patzelt H., Rudiger S., Brehmer D., Kramer G., Vorderwulbecke S., Schaffitzel E., Waitz A., Hesterkamp T., Dong L., Schneider-Mergener J., et al. Binding specificity of Escherichia coli trigger factor. Proc. Natl. Acad. Sci. 2001;98:14244–14249. doi: 10.1073/pnas.261432298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips T.A., VanBogelen R.A., Neidhardt F.C. Lon gene product of Escherichia coli is a heat-shock protein. J. Bacteriol. 1984;159:283–287. doi: 10.1128/jb.159.1.283-287.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politou A.S., Thomas D.J., Pastore A. The folding and stability of titin immunoglobulin-like modules, with implications for the mechanism of elasticity. Biophys. J. 1995;69:2601–2610. doi: 10.1016/S0006-3495(95)80131-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose G.D., Geselowitz A.R., Lesser G.J., Lee R.H., Zehfus M.H. Hydrophobicity of amino acid residues in globular proteins. Science. 1985;229:834–838. doi: 10.1126/science.4023714. [DOI] [PubMed] [Google Scholar]
- Rotanova T.V., Melnikov E.E., Khalatova A.G., Makhovskaya O.V., Botos I., Wlodawer A., Gustchina A. Classification of ATP-dependent proteases Lon and comparison of the active sites of their proteolytic domains. Eur. J. Biochem. 2004;271:4865–4871. doi: 10.1111/j.1432-1033.2004.04452.x. [DOI] [PubMed] [Google Scholar]
- Rudiger S., Germeroth L., Schneider-Mergener J., Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 1997;16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudiger S., Schneider-Mergener J., Bukau B. Its substrate specificity characterizes the DnaJ co-chaperone as a scanning factor for the DnaK chaperone. EMBO J. 2001;20:1042–1050. doi: 10.1093/emboj/20.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer R.T., Bolon D.N., Burton B.M., Burton R.E., Flynn J.M., Grant R.A., Hersch G.L., Joshi S.A., Kenniston J.A., Levchenko I., et al. Sculpting the proteome with AAA+ proteases and disassembly machines. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah I.M., Wolf R.E. Sequence requirements for Lon-dependent degradation of the Escherichia coli transcription activator SoxS: Identification of the SoxS residues critical to proteolysis and specific inhibition of in vitro degradation by a peptide comprised of the N-terminal 21 amino acid residues. J. Mol. Biol. 2006;357:718–731. doi: 10.1016/j.jmb.2005.12.088. [DOI] [PubMed] [Google Scholar]
- Shineberg B., Zipser D. The lon gene and degradation of beta-galactosidase nonsense fragments. J. Bacteriol. 1973;116:1469–1471. doi: 10.1128/jb.116.3.1469-1471.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkova N.N., Koroleva E.P., Rumsh L.D., Ginodman L.M., Rotanova T.V. Mutations in the proteolytic domain of Escherichia coli protease Lon impair the ATPase activity of the enzyme. FEBS Lett. 1998;422:218–220. doi: 10.1016/s0014-5793(98)00012-x. [DOI] [PubMed] [Google Scholar]
- Thomas-Wohlever J., Lee I. Kinetic characterization of the peptidase activity of Escherichia coli Lon reveals the mechanistic similarities in ATP-dependent hydrolysis of peptide and protein substrates. Biochemistry. 2002;41:9418–9425. doi: 10.1021/bi0255470. [DOI] [PubMed] [Google Scholar]
- Tsilibaris V., Maenhaut-Michel G., Van Melderen L. Biological roles of the Lon ATP-dependent protease. Res. Microbiol. 2006;157:701–713. doi: 10.1016/j.resmic.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Waxman L., Goldberg A.L. Selectivity of intracellular proteolysis: Protein substrates activate the ATP-dependent protease (La) Science. 1986;232:500–503. doi: 10.1126/science.2938257. [DOI] [PubMed] [Google Scholar]