Abstract
Multiple centrosomes in tumor cells create the potential for multipolar divisions that can lead to aneuploidy and cell death. Nevertheless, many cancer cells successfully divide because of mechanisms that suppress multipolar mitoses. A genome-wide RNAi screen in Drosophila S2 cells and a secondary analysis in cancer cells defined mechanisms that suppress multipolar mitoses. In addition to proteins that organize microtubules at the spindle poles, we identified novel roles for the spindle assembly checkpoint, cortical actin cytoskeleton, and cell adhesion. Using live cell imaging and fibronectin micropatterns, we found that interphase cell shape and adhesion pattern can determine the success of the subsequent mitosis in cells with extra centrosomes. These findings may identify cancer-selective therapeutic targets: HSET, a normally nonessential kinesin motor, was essential for the viability of certain extra centrosome-containing cancer cells. Thus, morphological features of cancer cells can be linked to unique genetic requirements for survival.
Keywords: Centrosomes, mitosis, actin, adhesion, cancer, cell cycle
Centrosomes play a crucial role in the equal segregation of chromosomes by contributing to bipolar spindle assembly during mitosis (Doxsey 2001). The tight control of centrosome duplication, limiting it to once per cell cycle, ensures that normal cells enter mitosis with two centrosomes or microtubule organizing centers (MTOCs). Failure to properly control centrosome number and function can lead to multipolar spindles, aneuploidy, disruption of cell polarity, and failure of asymmetric cell divisions (Heneen 1970; Nigg 2002).
Increased centrosome number, often termed centrosome amplification, is a common characteristic of solid and hematological cancers. Centrosome amplification correlates with aneuploidy and malignant behavior in tumor cell lines, mouse tumor models, and human tumors (Levine et al. 1991; Lingle et al. 1998; D’Assoro et al. 2002; Pihan et al. 2003; Giehl et al. 2005). Mutation or misregulation of a variety of tumor suppressors or oncogenes are correlated with centrosome amplification (Fukasawa 2007). Centrosome amplification can, in principle, arise from several types of cell division errors: centrosome overduplication, de novo synthesis of centrosomes, cell fusion, or cytokinesis failure (Boveri 1929; Nigg 2002; Ganem et al. 2007).
The role of supernumerary centrosomes in tumor biology is likely to be multifaceted. Whereas multiple centrosomes might facilitate tumorigenesis by promoting aneuploidy and/or disrupting cell polarity, they may also impose a fitness cost on the growth of mature cancers because of the potential for multipolar mitoses. To circumvent this problem, many cancer cells appear to have mechanisms that suppress multipolar mitoses, the best studied being clustering of supernumerary centrosomes into two groups enabling a bipolar mitosis (Ring et al. 1982; Brinkley 2001; Nigg 2002). Centrosome clustering in tumor cells is incompletely understood; however, it is expected to rely to a significant degree on microtubule-associated proteins (MAPs) and motors that organize the spindle poles (Karsenti and Vernos 2001; Nigg 2002). For example, recent work uncovered a requirement of cytoplasmic dynein, a minus end-directed microtubule (MT) motor, and NuMA, a spindle associated MAP, in centrosome clustering (Quintyne et al. 2005). The existence of mechanisms that suppress multipolar mitoses raises the possibility of a novel therapeutic strategy for cancer: Drugs that interfere with centrosome clustering mechanisms could be lethal to tumor cells containing multiple centrosomes, but potentially spare normal cells. Although several drugs, including Taxol, can promote multipolar mitosis, none are specific to cells with multiple centrosomes (Chen and Horwitz 2002; Rebacz et al. 2007).
To comprehensively define the genes required to suppress multipolar mitoses, we performed a genome-wide RNAi screen in near tetraploid Drosophila S2 cells. The classes of genes discovered by this screen enabled the identification of processes that suppress multipolar mitoses. The spindle assembly checkpoint (SAC) ensures adequate time for clustering of centrosomes prior to anaphase. Centrosome clustering is achieved by a combination of spindle-intrinsic MT-binding forces and actin-regulating forces at the cell cortex. The distribution of these cortical forces is determined by the interphase adhesion pattern. Thus, the tumor microenvironment can determine the success of mitosis in cells with extra centrosomes. Finally, we validate the idea that some tumors might be uniquely dependent on centrosome clustering for survival.
Results
RNAi screen for genes required for clustering supernumerary centrosomes
We used an RNAi screen to comprehensively define the molecular pathways required for clustering supernumerary centrosomes (Fig. 1). Of eight Drosophila cell lines characterized, the near-tetraploid S2 cells were most suitable for our screen because >50% of cells contain extra centrosomes that are efficiently clustered into two poles during mitoses (>95%) (Supplemental Figs. S1, S2).
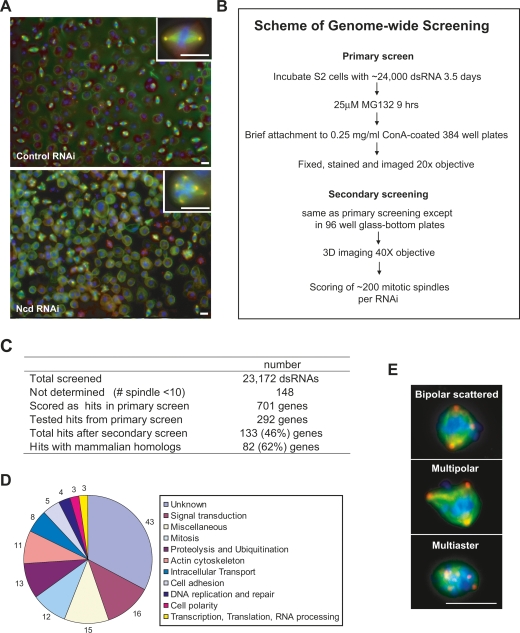
Figure 1.
Genome-wide screen for genes required for clustering supernumerary centrosomes in S2 cells. (A) Sample images from screen. Normal bipolar spindles in control EGFP RNAi (top panel) and multipolar spindles in Ncd RNAi (bottom panel) are shown. Cells were stained for MTs (α-tubulin, green), centrosomes (γ-tubulin, red), and DNA (blue). (Insets) Images at high magnification. (B) Scheme of genome-wide screen in S2 cells. Procedure for the primary and secondary screens. Addition of MG132, a proteasome inhibitor, at the end of the RNAi treatment was used to increase mitotic index. Immediate transfer of cells to Con-A was then used to facilitate cell attachment. (C) Table summarizing the screen results. (D) Gene Ontology (GO) annotations for the 133 genes from the screen. (E) Images showing classes of centrosome clustering defects in S2 cells. Cells were stained for MTs (α-tubulin, green), centrosomes (γ-tubulin, red), and DNA (blue). Bar, 10 μm.
We screened 23,172 dsRNAs targeting ∼99% of the Drosophila genome (∼14,000 genes) to identify genes whose knockdown leads to multipolar spindles (centrosome declustering) in S2 cells (Fig. 1A,B). S2 cells were exposed to dsRNA for 4 d and mitotic figures were enriched by treatment with the proteasome inhibitor MG132 during the last 9 h of the RNAi treatment (Supplemental Fig. S2). Cells were stained for DNA, MTs, and centrosomes, and images were acquired with a 20× objective, using a high-throughput automated microscope (Fig. 1B; see the Supplemental Material for detail).
By visual inspection of ∼96,000 images, we scored the percentage of multipolar spindles for each RNAi condition. Using a 95% confidence interval, the primary screen identified 701 candidates associated with a multipolar spindle phenotype (Fig. 1C; Supplemental Fig. S3). Two-hundred-ninety-two genes were selected as initial cohort for further study based on the strength of the phenotype, the existence of readily identifiable mammalian homologs, and few or no predicted off-target effects. Additionally, we eliminated most genes that were previously determined to be required for cytokinesis in Drosophila cells (Echard et al. 2004; Eggert et al. 2004) because spindle multipolarity can be a secondary effect of cytokinesis failure (Goshima et al. 2007). Of the 292 genes selected for the secondary screen, 133 were confirmed to have a bona fide role in centrosome clustering (Fig. 1C; Supplemental Table S1). Among the validated genes, 62% of the genes on our list (82 out of 133 genes) have mammalian homologs (Fig. 1C), and 33% of the genes (43) do not have a known function (Fig. 1D). Centrosome clustering can occur with varying efficiency. Knockdown caused a variety of centrosome clustering defects, which we classified into three groups: bipolar scattered (spindles with multiple centrosomes scattered around the spindle), multipolar spindles, and multiaster spindles (small and more compact multipolar spindles) (Fig. 1E).
We identified genes involved in a diverse range of cellular processes, suggesting unappreciated complexity in the mechanisms controlling organization of supernumerary centrosomes (Fig. 1D). We found a number of genes that promote the bundling of spindle MTs, including the minus end-directed kinesin Ncd (human HSET). The screen also unexpectedly identified genes involved in the SAC, actin regulation, cell polarity, and cell adhesion. Below we present experiments that define three overlapping mechanisms that suppress multipolar mitoses: a timing mechanism employing the SAC, intrinsic pole clustering mechanisms relying on MT regulators, and a novel mechanism requiring actin and cell adhesion.
The spindle assembly checkpoint (SAC) prevents multipolar mitoses
The SAC components Mad2, BubR1 (human Bub1), and CENP-Meta (human CENP-E) are required for centrosome clustering (Fig. 2A; Supplemental Table S1), suggesting a role for the SAC in this process. This requirement was even more evident in cells that were not treated with MG132 (Fig. 2A), indicating that the short treatment with MG132 employed in our screen partially masked the effect of SAC gene RNAi on spindle multipolarity. This finding was somewhat surprising, given previous work in PtK1 cells suggesting that the SAC is not activated by multipolar spindles or multiple centrosomes (Sluder et al. 1997).
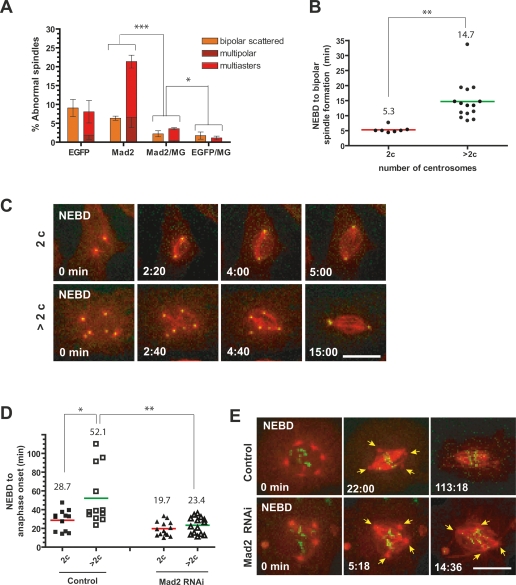
Figure 2.
The SAC prevents multipolar mitoses in S2 cells. (A) Mad2 requirement for centrosome clustering. Centrosome clustering defects were scored in S2 cells upon RNAi of EGFP, Mad2 alone, and EGFP or Mad2 plus 7 h of MG132 treatment. Graph shows the average of three independent experiments; mean ± SD. (*) P < 0.05; (***) P < 0.001, Student’s t-test. (B) Cell cycle delay associated with multipolar mitoses. Comparison of the time (minutes) taken from NEBD to bipolar spindle formation in S2 cells that have two centrosomes (2c) or extra centrosomes (>2c) in mitosis. Dots and bars indicate individual cells and the average, respectively, in minutes. (**) P < 0.002, Student’s t-test. (C) Examples of time-lapse images from mitotic S2 cells with two (top panels) or more than two (bottom panels; Supplemental Movie S1) centrosomes. Centrosomes and MTs are labeled by GFP-SAS-6 (green) and mCherry-α-tubulin (red). (D) Delay induced by extra centrosomes requires Mad2. Time taken from NEBD to anaphase onset was obtained from S2 cells expressing GFP-Cid and mCherry-α-tubulin upon RNAi of EGFP control or Mad2 (average times are shown with red and green bars from cells with two and more than two centrosomes, respectively. (*) P < 0.02; (**) P < 0.002, Student’s t-test. Anaphase delay is not due to cell undergoing mitotic slippage because it is observed in cells with normal number of kinetochores: data were only obtained from S2 cells containing 20–26 Cid dots per cell. (E) Examples of time lapse images (see Supplemental Movies S3, S4). (Bottom panels) Premature anaphase onset occurs before centrosome clustering in Mad2 depleted cells. Arrows indicate centrosomes. Bar, 10 μm.
Time-lapse imaging supported a role for the SAC in preventing multipolar mitoses. In S2 cells, where centrioles and MTs were labeled with GFP-SAS-6 and mCherry α-tubulin, there was a clear correlation between an increased number of centrosomes and a prolonged time required to form a bipolar spindle (2.7-fold) (Fig. 2B,C; Supplemental Table S3; Supplemental Movie S1). We therefore measured the interval between NEBD and anaphase onset (visualized with GFP-Cid, Drosophila CENP-A) comparing cells with two or more than two centrosomes. Relative to cells with two centrosomes, cells with multiple centrosomes exhibited a marked delay in anaphase onset (1.8-fold) (Fig. 2D; Supplemental Table S3; Supplemental Movies S2, S3). Moreover, the delay in anaphase onset was abolished by Mad2 RNAi, and cells entered anaphase with declustered centrosomes and misaligned kinetochores (Fig. 2D,E; Supplemental Table S3). Further suggesting SAC activation, multipolar spindles had a strong increase in the number of BubR1 foci relative to bipolar metaphase spindles (Supplemental Fig. S4). Finally, the requirement for the SAC to prevent multipolar mitoses can be partially suppressed by an artificial metaphase delay imposed by treatment with MG132 (Fig. 2A). This suggests that the SAC does not monitor multipolar mitosis per se, but rather that SAC activation, likely triggered by abnormal kinetochore attachment or tension, provides sufficient time for compensatory mechanisms to organize multiple centrosomes.
Spindle-intrinsic pole clustering forces prevent multipolar mitoses
Previous work in S2 cells has demonstrated a critical role for MT motors and MAPs in spindle pole focusing (Goshima et al. 2005; Morales-Mulia and Scholey 2005). We identified Ncd, a Kinesin-14 family member, as the strongest hit in the primary screen. Ncd is a minus end-directed motor that bundles MTs at the spindle poles (Karabay and Walker 1999). By GFP-SAS-6 labeling, we demonstrated that Ncd is required to cluster multiple centrosomes (Supplemental Movie S6). Drosophila dynein was not identified in our screen. This was expected because in S2 cells loss of dynein does not significantly induce multipolar mitoses, although it does compromise centrosome attachment and tight focusing of the spindle poles (Goshima et al. 2005). Further validating the screen, we confirmed the role of the MAP Asp in pole focusing (Wakefield et al. 2001; Morales-Mulia and Scholey 2005). Additionally, our screen identified several other factors that contribute to the intrinsic cohesion of spindle MTs (Supplemental Table S1). We identified the requirement of Bj1/RCC1 (RanGEF) for centrosome clustering, consistent with its role in preventing multipolar mitosis in mammalian cells (Chen et al. 2007). We also identified the ADP-ribosylation factors Tankyrase and CG15925, a putative human PARP-16 homolog. ADP-ribosylation by tankyrase is thought to contribute to spindle bipolarity by providing a static matrix that may anchor MT motors and other spindle proteins (Chang et al. 2005). A role for PARP-16 in mitosis has not been previously described.
Actin-dependent forces and spindle intrinsic forces cooperatively prevent multipolar mitoses
In addition to genes that likely contribute to the bundling and organization of spindle MTs, we unexpectedly identified genes involved in the organization and regulation of the actin cytoskeleton, such as the formin Form3/INF2 (Fig. 1D; Supplemental Table S1; Chhabra and Higgs 2006). Knockdown of most of these genes does not induce cytokinesis failure (Echard et al. 2004; Eggert et al. 2004). Further supporting a direct role for actin in preventing multipolar mitoses, a brief (2 h) treatment with actin inhibitors similarly induced multipolar mitoses in S2 cells (Fig. 3A). Live cell imaging of S2 cells revealed that actin is indeed required for the initial clustering of multiple centrosomes. Relative to controls (14.7 ± 6.4 min), there was a 1.5-fold delay in centrosome clustering in 13 of 15 LatA-treated cells (22.1 ± 12.3 min). The remaining two of 15 cells completely failed to cluster extra centrosomes (Fig. 3B; Supplemental Table S3; Supplemental Movie S5). The cell cycle delay induced by LatA treatment is likely due to activation of the SAC, as evidenced by prominent labeling of kinetochores with BubR1 in LatA-treated cells (Supplemental Fig. S4). The fact that LatA treatment was synergistic with either Ncd or Bj1/RCC1-depletion suggested that the actin cytoskeleton and intrinsic spindle forces cooperate to suppress spindle multipolarity (Fig. 3C).
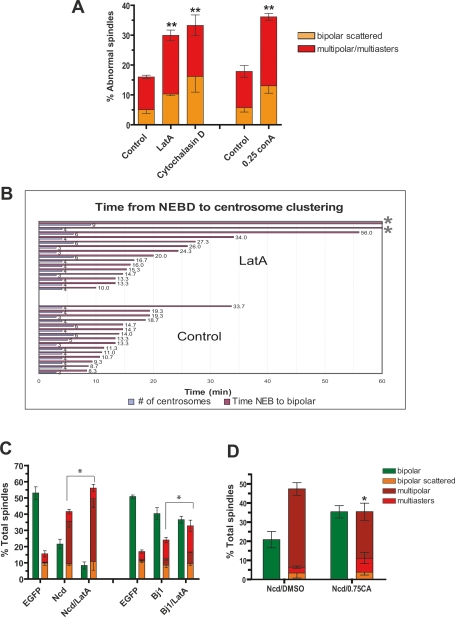
Figure 3.
Actin-dependent forces cooperate with spindle intrinsic forces to cluster supernumerary centrosomes. (A) Actin requirement for centrosome clustering in S2 cells. Cells were treated with Latrunculin (40 μM LatA), Cytochalasin D (20 μM), or Con-A (0.25 mg/mL) for 2 h and the percentage of centrosome clustering defects was determined. Graph shows the average of three independent experiments (mean ± SD, [**] P < 0.005, Student’s t-test). (B) Live cell imaging was used to measure the time from NEBD to centrosome clustering in S2 cells expressing GFP-SAS-6 and mCherry-α-tubulin in the presence or absence of LatA. There is a delay in centrosome clustering in LatA-treated cells (P < 0.02, Student’s t-test), and asterisks indicate the cells that failed to cluster centrosomes (Supplemental Movie S5). (C) Percentage of cells with centrosome clustering defects after RNAi of Ncd or Bj1 (RCC1) alone or in conjunction with LatA treatment (2 h). (D) Calyculin A (0.75 nM for 2 h) partially rescues the centrosome clustering defect in Ncd RNAi-treated cells. Graph shows the average of three independent experiments; mean ± SD. (*) P < 0.05, Student’s t-test. Bar, 10 μm.
Next, we determined if cortical contraction is required for centrosome clustering. Cells were exposed to soluble tetravalent lectin concanavalin A (Con-A), which cross-links the plasma membrane and thus globally blocks cortical contraction (Canman and Bement 1997). This treat-ment induced centrosome clustering defects (Fig. 3A). Furthermore, we found that enhancing myosin II-based contractility can suppress spindle multipolarity. Low concentrations of calyculin A (CA) inhibit the myosin light-chain phosphatase (MLCP) and promote myosin II activation without altering its distribution (Gupton and Waterman-Storer 2006). Wild-type S2 cells treated with CA had a modest decrease in centrosome clustering defects (15%–9%). Moreover, CA treatment partially rescued the centrosome clustering defect induced by Ncd RNAi (Fig. 3D). Thus, in cells with extra centrosomes, normal actin and actin-based contractility promotes bipolar spindle assembly.
Time-lapse imaging enabled the visualization of actin-dependent movement of centrosomes toward the cell cortex. Spinning disc microscopy was used to define the trajectory of centrosome movement in Ncd-depleted S2 cells. Centrosomes in Ncd-depleted cells exhibited a striking increase in mobility; both the speed and extent of movement was increased (Fig. 4A panel b vs. panel a; Supplemental Movie S6). Moreover, the bulk of the centrosome movement was directed away from the spindle and toward the cell cortex. By contrast, transient LatA treatment of cells reduced centrosome mobility (Fig. 4A, panel c; Supplemental Movie S5). Moreover, the dramatic mobility of centrosomes in Ncd-depleted cells was suppressed by exposure of cells to LatA (Fig. 4A, panel e; Supplemental Movie S7). Together, these results reveal actin-dependent forces on centrosomes that, if properly organized, might suppress spindle multipolarity.
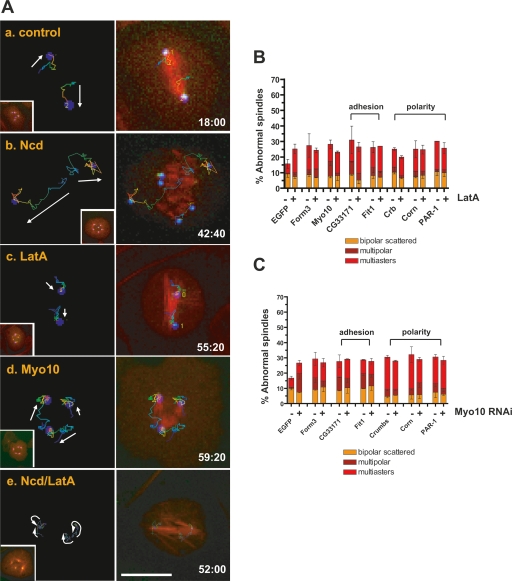
Figure 4.
Genes involved in cell adhesion and cell polarity affect spindle multipolarity via the actin cytoskeleton. (A) Visualization of the actin-dependent cortical forces on centrosomes. Centrosome tracking in S2 cells expressing GFP-SAS-6 and mCherry-α-tubulin (inset, t = 0 min, NEBD) treated with control RNAi (panel a), Ncd RNAi (panel b) (Supplemental Movie S6), LatA (panel c) (Supplemental Movie S5), Myo10A RNAi (panel d) (Supplemental Movie S8), and Ncd RNAi/LatA (panel e) (Supplemental Movie S7). Images were taken at the indicated time (minutes) after NEBD. Centrosome trajectories are shown from blue (start of path) to red (end of path). The direction of movements is indicated with white arrows. (B) Requirement of actin, cell polarity, and adhesion genes for clustering supernumerary centrosomes in S2 cells. Centrosome clustering defects after RNAi with or without LatA treatment (2 h). (C) Percentage of centrosome clustering defects after RNAi of genes involved in cell adhesion, polarity, or actin alone or in conjunction with Myo10A RNAi. Graph shows the average of three independent experiments (mean ± SD). LatA treatment alone or all single gene knockdowns (−LatA) induce centrosome clustering defects significantly above control RNAi levels. P < 0.05, Student’s t-test. Bar, 10 μm.
Our results also provided insight into the nature of the cortical force generators that regulate spindle multipolarity. We found that the MT + tip CLIP-190 and the myosin Myo10A are important for centrosome clustering (Supplemental Table S1). Drosophila Myo10A is a human Myo15 homolog that can bind MTs via a unique MyTH4-FERM domain. Myo10, a member of mammalian MyTH4-FERM-containing myosin, is known to be required for spindle positioning (Sousa and Cheney 2005; Toyoshima and Nishida 2007). RNAi of Myo10A, but not the other Drosophila MyTH4-FERM-containing myosin Myo7, induced a twofold increase in centrosome clustering defects (Fig. 4B) without cytokinesis failure (data not shown). Moreover, knockdown of Myo10A did not have an additive effect on spindle multipolarity if cells were concomitantly treated with LatA (Fig. 4B). Finally, centrosome tracking of Myo10A-depleted cells revealed a similar effect on centrosome movement as LatA treatment (Figs. 4A, panels c,d; Supplemental Movie S8); in cells depleted of Myo10, nondirected or greatly reduced movements of centrosomes were detected in contrast to the extensive cortically directed movement shown in Ncd-depleted cells (Fig. 4A, panels b,d). Together, our data suggest that multiple centrosomes are organized combinatorially by spindle-intrinsic forces and by actin-dependent cortical forces acting at least in part on astral MTs.
Cell shape, cell polarity, and adhesion effects on spindle multipolarity
Our screen identified a requirement for genes implicated in cell adhesion for centrosome clustering: Turtle, Echinoid, Cad96Ca, CG33171, and Fit1 (Supplemental Table S1). The Drosophila FERM domain containing protein Fit1 appears to have a highly conserved function in regulating cell–matrix adhesion in higher eukaryotes (Tu et al. 2003). The mammalian Fit1 homolog, Mig-2/human PLEKHC1, localizes to focal adhesions (FAs) and is important for integrin-mediated cell adhesion and modulation of cell shape by linking integrins to actin cytoskeleton (Tu et al. 2003). The uncharacterized CG33171 protein has homology with mammalian Col18A, previously implicated in the regulation of cell matrix adhesion (Dixelius et al. 2002). Turtle and Echinoid containing fibronectin (FN) type III domains are involved in cell–cell adhesion (Bodily et al. 2001; Wei et al. 2005). In addition we identified the posterior/lateral polarity gene PAR-1 (PAR-1/MARK/KIN1 family member) and the apical polarity genes Crumbs and Cornetto, which are important for astral MT function, asymmetric cell division and epithelial polarity (Bulgheresi et al. 2001; Tepass et al. 2001; Munro 2006). A number of these genes have been identified previously because of their requirement to maintain normal interphase cell shape and adhesion (Kiger et al. 2003; Bakal et al. 2007).
How might genes controlling interphase cell shape and adhesion affect spindle morphology? One appealing hypothesis is that these genes control the organization of cortical actin-based forces that act on astral MTs. Indeed, LatA treatment or Myo10 depletion showed no enhancement of spindle multipolarity when combined with depletion of CG33171, Fit1, Crumbs, Cornetto, or PAR-1 proteins (Fig. 4B,C), suggesting that these genes influence centrosome clustering via the actin cytoskeleton.
Conservation of the mechanisms to prevent multipolar mitoses
We determined if mammalian cancer cells use similar mechanisms to cluster multiple centrosomes to examine the relevance of our screen to cancer. Furthermore, techniques available for manipulating cell adhesion in mammalian cells enabled us to directly characterize how the adhesion pattern influences spindle organization. Although there is some variability in its efficiency, clustering of extra centrosomes is commonly observed in mammalian cells (Supplemental Table S4 describes the cells characterized below).
Transient actin disruption led to a significant increase in the frequency of multipolar spindles in cell lines that contained multiple centrosomes but not in cells with normal centrosome number (Fig. 5A; Supplemental Table S4). These multipolar spindles result from declustering of extra centrosomes and were not due to the splitting/fragmentation of centrioles (Fig. 5A). Actin presumably influences centrosome positioning via forces on astral MTs. Consistent with this idea, low-dose nocodazole treatment, selectively disassembling astral MTs, increased the frequency of multipolar spindles specifically in cells with extra centrosomes (Supplemental Fig. S5).
Figure 5.
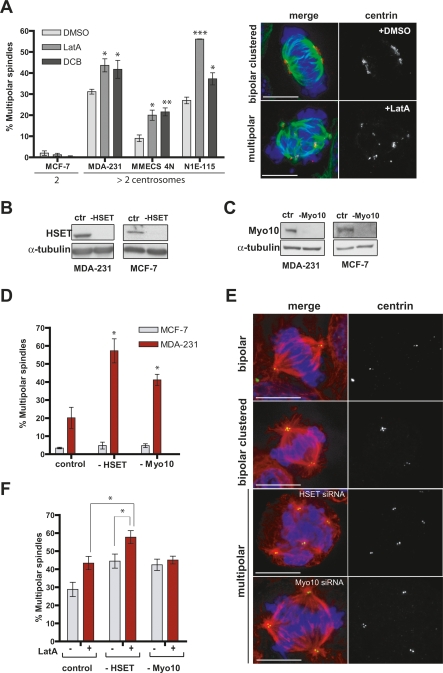
Mechanisms for centrosome clustering are conserved in mammalian cells. (A) Actin requirement for clustering centrosomes in cancer cells. Quantitation of multipolar spindles upon treatment with DMSO, LatA (5 μM) or DCB (10 μM) for 2 h in cancer cell lines. N1E-115 cells were stained for MTs (α-tubulin, green), centrioles (centrin2, red) and DNA (blue). (B,C) Western blots showing the depletion of HSET (B) or Myo10 (C) after 3 d of siRNA in MDA-231 and MCF-7 cells (ctr, control). (D) HSET and Myo10 requirement for centrosome clustering. Quantitation of mitotic cells with multipolar spindles upon treatment with HSET (−HSET) or Myo10 (−Myo10) siRNA for 3 d in MDA-231 and MCF-7. (E) Representative images from D (α-tubulin, red; centrin2, green; DNA, blue). (F) The percentage of mitotic MDA-231 cells with multipolar spindles upon treatment siRNA of HSET or Myo10, both with (+) and without (−) LatA. Note that the percentage of multipolar spindles is among cells with extra centrosomes in all graphics of this figure. Graph shows the average of three independent experiments; mean ± SD. (*) P < 0.05; (**) P < 0.005; (***) P < 0.001, Student’s t-test. Bar, 10 μm.
The parallel between Drosophila and mammalian cells extended to the genetic requirements for centrosome clustering. siRNA of the Ncd homolog HSET (a Kinesin-14 member) and Myo10 increased the frequency of multipolarity, specifically in cells harboring multiple centrosomes (Fig. 5B–E). As in S2 cells, Myo10-induced multipolarity is not a consequence of cytokinesis failure (data not shown). Finally, we determined whether the actin cytoskeleton influences centrosome organization in parallel with intrinsic spindle pole clustering forces. Disruption of both actin and HSET had a combinatorial effect. By contrast, LatA treatment did not increase the frequency of spindle multipolarity in cells where Myo10 was depleted (Fig. 5F). Thus, similar overlapping mechanisms prevent multipolar mitoses in mammalian cancer cells and Drosophila S2 cells.
Interphase cell shape, adhesion, and multipolar mitoses
Although cells round up in mitosis, they preserve a memory of their interphase shape by retaining actincontaining retraction fibers (RFs) linked to sites of strong cell–matrix adhesion (Mitchison 1992; Thery and Bornens 2006). The interphase adhesion pattern and the distribution of actin-containing RFs are known to strongly influence spindle orientation during mitosis (Thery et al. 2005). The finding from our screen that preventing multipolar mitoses requires both cell–matrix adhesion genes and actin regulators, suggested an appealing hypothesis: These gene products could act cooperatively to organize extra centrosomes by affecting the distribution and/or the composition of RFs and thus cortical force generators.
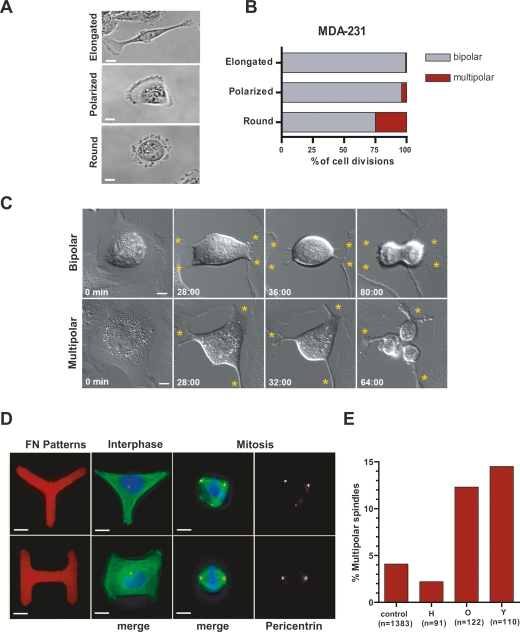
Several lines of evidence support this hypothesis. First, we used live-cell imaging to correlate interphase cell shape with the pattern of cell division during mitosis. MDA-231 (breast cancer cells containing extra centrosomes) that assumed an elongated or polarized shape in interphase, almost uniformly underwent bipolar divisions (Fig. 6A,B; Supplemental Fig. S6). By contrast, MDA-231 cells that assumed a round shape in interphase had an increased frequency of multipolar divisions. Note that in fixed MDA-231 cells all multipolar anaphases contain extra centrosomes (Supplemental Fig. S6). Sec ond, in tetraploid BSC-1 cells, whose thick RFs are readily visualized by DIC imaging, we noted a strong correlation between the positioning of the RFs and whether cells underwent bipolar (bipolar distribution of RFs) or multipolar division (isotrophic distribution of RFs) (Fig. 6C; Supplemental Movies S9, S10). Third, RFs accumulate specific proteins, such as the ERM protein ezrin, which are implicated in cortical heterogeneity and thus local force generation on astral MTs. Disruption of this cortical heterogeneity by the src kinase inhibitor PP2 (Thery et al. 2005), also induced multipolar spindles in MDA-231 but not in MCF-7 cells (Supplemental Fig. S7A). Fourth, to evaluate the role of cell–matrix adhesion for the efficiency of mitosis, we examined cells plated on different concentrations of FN to vary the strength of cell–matrix attachment. We observed that concentrations of FN that inhibit FA turnover (30 μg/mL) increased the frequency of multipolar spindles in MDA-231 cells but not in MCF-7 cells (Supplemental Fig. S7B). Moreover, this effect could be reversed by CA, which promotes FA turnover by increasing cortical contractility (Supplemental Fig. S7C; Gupton and Waterman-Storer 2006).
Figure 6.
Interphase adhesion pattern controls the bipolar/multipolar decision. (A) Bright-field images representing the different interphase cell shapes of MDA-231 cells: elongated, polarized, and round. (B) Elongated cell morphology is correlated with bipolar division. Quantitation of bipolar and multipolar divisions by live-cell imaging of MDA-231 cells with different interphase shapes (n = 279; 45% elongated, 46% polarized, 9% round). (C) RF position correlates with bipolar or multipolar divisions. Representative images from DIC movies of 4N BSC-1 cells from interphase (left) through mitosis (right). Yellow asterisks indicate RFs. Sixty-six binucleated cells were imaged in three independent experiments. (D) Interphase cell shape controls spindle morphology in cells with extra centrosomes. MDA-231 cells were plated on FN micropatterns (H, O, and Y) and stained for MTs (α-tubulin, green), centrosomes (pericentrin, red), and DNA (blue). (E) Percentage of cells with multipolar spindles in MDA-231 cells after plating on different patterns (H, O, and Y). Note that the percentage of multipolar spindles was quantified from all metaphase cells. Controls were obtained from cells that were attached to unconstrained FN micropatterns. Graph represents three independent experiments. Bar, 10 μm.
To directly test the role of the cell adhesion pattern and RF positioning in centrosome clustering, we used FN micropatterns to manipulate cell adhesion patterns (Thery et al. 2005). We observed that MDA-231 cells plated onto Y-shaped or O-shaped micropatterns caused a significant (three- to fourfold) increase in multipolar spindles compared with the controls (Fig. 6D,E). In contrast, plating cells on H-shaped micropattern suppressed the frequency of multipolar spindles relative to control cells (2%, half of the control). Thus, O and Y arrangements of adhesive contacts bias cells into multipolar mitoses, whereas bipolar arrangements of adhesive contacts (H-shape) promote bipolar mitoses. Our findings demonstrate that interphase cell adhesion pattern and cell shape can have a remarkable influence on the fidelity of mitosis, specifically in cancer cells containing extra centrosomes.
Disruption of centrosome clustering can selectively kill cancer cells
In principle, disruption of centrosome clustering could have a selective effect on the viability of cancer cells containing multiple centrosomes because most somatic cells have two centrosomes during mitosis. As a first step toward evaluating this potential therapeutic strategy we characterized the sensitivity of different cancer cell lines to knockdown of HSET.
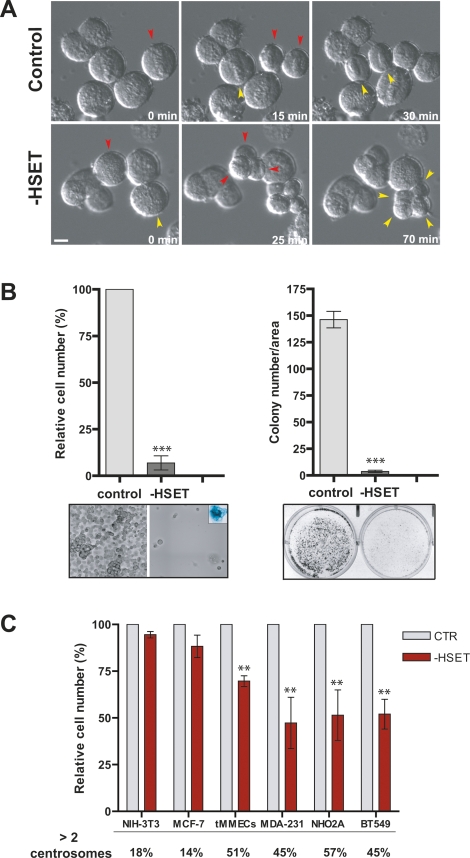
HSET is a particularly interesting therapeutic target because it is nonessential for cell division in normal cells, and kinesins are amenable to small molecule inhibition (Mayer et al. 1999; Mountain et al. 1999). We found that depletion of HSET by siRNA leads to an increase in multipolar spindles in human cancer cells containing multiple centrosomes (Fig. 5D). To determine the consequences of centrosome declustering, we monitored cell division in multiple cell lines that contain extra centrosomes using DIC microscopy (Fig. 7A; Supplemental Fig. S8). We found that depletion of human HSET confirmed with three independent siRNAs induced a dramatic increase in multipolar anaphases (88%) in N1E-115 cells where nearly 100% of cells contain extra centrosomes (Fig. 7A; Supplemental Movies S11, S12; Spiegelman et al. 1979). A similar result was obtained with MDA-231 cells where ∼50% of cells contain extra centrosomes (24% multipolar anaphases after HSET depletion) (Supplemental Fig. S8) and with tetraploid BJ and NIH-3T3 cells with extra centrosomes. By contrast, HSET knockdown had no effect on cell division in a variety of diploid control cells (Supplemental Fig. S8).
Figure 7.
Forced declustering of centrosomes kills cancer cells with extra-centrosomes. (A) Multipolar anaphases induced by HSET depletion in N1E-115 cells. Representative still images from DIC movies of N1E-115 cells after HSET (n = 435) or control (n = 265) siRNA. Colored arrowheads indicate mitotic cells immediately before and after the onset of anaphase. Bar, 10 μm. (B) Loss of cell viability and inhibition of colony formation by N1E-115 cells after 6 d of HSET siRNA. (Left panel) Relative cell number in control and HSET-depleted (−HSET) cells from three independent experiments after 6 d post-transfection. Bright-field images of control and HSET-depleted N1E-115 cells. (Inset) A surviving N1E-115 cell after HSET depletion stained for senescence-associated β-galactosidase activity (blue). (Right panel) Graph represents the average colony number from two independent experiments in four different areas (area, 10 mm2). The corresponding images are shown below. (C) HSET RNAi-induced cell death in various cancer cell lines in proportion to the fraction of cells with extra centrosomes. Relative cell number in control (CTR) and HSET-depleted (−HSET) cells after 6 d post-transfection. HSET can be efficiently depleted from the different cell lines as shown in Supplemental Figure S9. Percentage of cells with more than two centrosomes (>2c) is indicated below the graph. Graph shows the average of three independent experiments. All graphics represent mean ± SD. (**) P < 0.005; (***) P < 0.001, Student’s t-test.
Strikingly, depletion of HSET from N1E-115 cells for 6 d reduced cell viability by >90%. By DIC imaging, most cells appeared to undergo apoptosis, and many of the surviving cells were found to be senescent (Fig. 7B). HSET depletion also induced cell death in three other cancer cells lines (MDA-231, BT-549, and NHO2A) in rough proportion to the fraction of cells containing extra centrosomes (Fig. 7C; Supplemental Table S4). In contrast, the viability of cells that mostly possess two centrosomes (MCF-7 and NIH-3T3) was only slightly reduced in the absence of HSET (Fig. 7C) although HSET is efficiently knocked down by RNAi (Fig. 5B; Supplemental Fig. S9). Thus, centrosome declustering can induce cell death selectively in cells with supernumerary centrosomes.
Discussion
The association between multiple centrosomes and cancer has been known for nearly 100 years (Boveri 1929). Nevertheless, an understanding of how multiple centrosomes affect the division and survival of tumor cells remains poorly understood. Here we used a genome-wide RNAi approach to define the mechanisms by which cells suppress multipolar mitoses. Our experiments lead to four novel conclusions. First, we found that the minus end-directed motor HSET/Ncd is essential for clustering extra centrosomes. Second, we uncovered a previously unappreciated role for the SAC in suppressing spindle multipolarity. Third, we demonstrate that interphase adhesion pattern and actin organization can profoundly influence the success of mitosis if cells contain multiple centrosomes. Finally, our findings suggest a new therapeutic strategy: Blocking centrosome clustering and promoting multipolar mitoses may selectively induce death in tumors with a high proportion of cells containing multiple centrosomes.
Our findings illustrate the fact that centrosome clustering uses processes present in normal cells rather than being a unique feature of cancer cells. Consistent with this idea, several nontransformed cell types cluster extra centrosomes after cytokinesis failure (Supplemental Table S4; Sluder and Nordberg 2004). Furthermore, clustering of extra centrosomes in tumors can be inferred by comparing the frequency of extra centrioles with the frequency of bipolar mitoses (Supplemental Table S4; Levine et al. 1991; Lingle and Salisbury 1999). Most strikingly, flies containing extra centrosomes are viable because of robust centrosome clustering (Basto et al. 2008). Nevertheless, the efficiency of centrosome clustering varies among cancer cell lines; it remains an interesting possibility that, under selective pressure, the strength of these pathways is modulated in different tumor types.
The SAC and multipolar mitoses
A mitotic timing mechanism is important for suppressing multipolar mitoses. S2 cells with multiple centrosomes spend more time in mitosis than cells with two centrosomes. This delay is mediated by SAC activation. Therefore, SAC activation suppresses multipolar mitoses by providing additional time for centrosome clustering mechanisms to facilitate bipolar spindle assembly. Thus, our data demonstrate that multipolar mitoses can activate the SAC in S2 cells, and very recently a similar checkpoint-mediated mitotic delay was reported in mammalian tissue culture cells (Yang et al. 2008). Indeed, the SAC becomes essential in flies containing extra centrosomes (Basto et al. 2008).
Our findings contrast with a previous study reporting that multipolar mitoses do not activate the SAC in PtK1 cells (Sluder et al. 1997). Differences in cell type might explain the contrasting results. We also note that Mad2 RNAi abrogated the cell cycle delay when the spindle was grossly abnormal, but had little effect if extra centrosomes were scattered throughout a bipolar spindle. Thus, in agreement with the main conclusions of Sluder et al. (1997), the SAC is activated when spindles are defective enough to produce abnormal kinetochore attachment or tension, but the SAC does not appear to recognize abnormal spindles per se.
The influence of actin and matrix adhesion on the frequency of multipolar mitoses
Our screen identified a critical role for the actin cytoskeleton in suppressing multipolar mitoses in cells with extra centrosomes. The actin cytoskeleton could affect the distribution of centrosomes by at least three mechanisms: by controlling cell shape, by controlling the contractility and stiffness of the cell cortex, and by influencing the cell adhesion pattern and the organization and integrity of RFs. Our experiments support a key role for the interphase adhesion pattern and thus the distribution of RFs.
The effect of the adhesive geometry on centrosome clustering was most dramatically demonstrated in cells plated on FN micropatterns. Interphase cell adhesion appears to be translated into cortical forces pulling on spindle poles during mitosis. This is made possible by the accumulation of actin-related cues in specific cortical regions facing cell adhesion areas/RFs (Thery et al. 2005). Cortical cues activate force generators that pull astral MTs under tension and induce spindle pole movement (Grill et al. 2001). Based on theoretical considerations, numerical simulations, and experiments, it has been proposed recently that the spatial distribution of these cues induce torque on astral MTs, guiding spindle positioning (Thery et al. 2007). According to this view, spindle pole separation and spindle positioning depend on both pole–pole and pole–cortex mechanical interactions (Supplemental Fig. S10). Thus, in cells containing multiple centrosomes, the specific localization of cortical cues and the corresponding distribution of forces could either promote (O and Y patterns) or suppress (H pattern) multipolar mitoses (Fig. 7E). An important direction for future work will be to define how the various factors identified by our experiments (e.g., PAR-1, PI-3 kinase, Myo10, and myosin II) coordinately influence the forces exerted on centrosomes.
Multipolar mitoses and cancer therapeutics
Our experiments have potential translational implications: Inhibiting centrosome clustering in tumors with a high proportion of cells with supernumerary centrosomes may selectively kill tumor cells. Indeed, we found that HSET, which is nonessential for mitosis in normal cells, is essential for the survival of cancer cells with extra centrosomes. Thus, like the ploidy-specific lethal mutations we described previously (Storchova et al. 2006), HSET has important characteristics for an appealing chemotherapeutic target: It is not essential in normal cells, but becomes essential for the survival of certain tumor cells.
There is already evidence for cell context-specific requirements for HSET/Ncd. Although it is not required for pole organization in normal somatic cells, Ncd is essential for female meiosis in the fly where cells lack centrosomes (Endow and Komma 1998). Centrosomes impose a strong bundling force on the spindle poles that in somatic cells may override the requirement for HSET/Ncd. The force generated by HSET/Ncd is necessary in meiotic divisions where centrosomes are absent (Mountain et al. 1999). Thus, we speculate that the forces holding centrosomes together in cancer cells have much in common with the forces that bundle the minus ends of MTs in acentrosomal spindles (Karsenti and Vernos 2001; Nigg 2002). Although HSET would not necessarily be mutated, amplified, over- or underexpressed in tumors, it is interesting to note that there is a wide variation in HSET expression in tumors (http://cgap.nci.nih.gov/SAGE/AnatomicViewer). Perhaps in some tumors such as glioblastomas HSET expression is increased as an adaptive response to promote centrosome clustering. HSET therefore, illustrates a class of cancer-dependency factors that could only be identified by functional screens or directed experiments.
In addition to multiple centrosomes, our screen identified other features of cells that influence the extent to which cells require HSET/Ncd. The strength and organization of interphase adhesion sites strikingly influences whether cells with multiple centrosomes undergo bipolar or multipolar mitoses. These findings suggest an interesting mechanism by which the tumor microenvironment might influence genetic stability. N1E-115 neuroblastoma cells are a classic “small round blue-cell” tumor. These cells have low integrin β-1 expression and can attach to laminin but not FN in vitro (Sarner et al. 2000; Amendola et al. 2001). The round shape and inefficient substrate attachment of these cells thus may contribute to their dramatic requirement for HSET. Likewise, the results of our screen suggest that inhibition of HSET might be synergistic with a PI-3 kinase inhibitor, which is an important cancer therapeutic target (Ward et al. 2003). Finally, our data suggest that cancer cells with compromised SAC signaling might also be more dependent on HSET.
Despite the exciting development of oncogene-specific targeted therapies, at present these therapies are available for a small number of tumor types (Bild et al. 2006). Conventional cytotoxic chemotherapies remain the mainstay of clinical practice, and there is a strong need to develop more rational, personalized utilization of these drugs. Recently, there has been significant progress in using genetic information to individually tailor the use of DNA-damaging agents (O’Connor et al. 2007). Similarly, the work described here suggests ways in which morphological features of cells might be used to individualize the use of anti-mitotic drugs.
Materials and methods
RNAi screen and data analysis
Detailed descriptions of the RNAi screen and data analyses can be found in the Supplemental Material. S2 cells were treated with dsRNA for 3.5 d followed by 9 h of MG132. Cells were imaged with an automated microscope using a 20× air objective. In the secondary screen, high-resolution 3D images were taken with a Zeiss Axiovert microscope using a 40× air ELWD objective (Zeiss). Multipolar spindles were scored from ∼200 mitotic spindles per RNAi condition. The follow-up experiments after RNAi screen were repeated without MG132, and data presented in all figures are from unperturbed cycling cells, unless otherwise specified. Statistical analysis of the data is described in the Supplemental Material.
Cell culture and drug treatment
Cell lines, drug treatments, fluorescent protein constructs, and transfection conditions are described in the Supplemental Material.
Immunofluorescence microscopy and live cell imaging
Protocols for immunofluorescence, live-cell imaging, and antibodies used in this study are described in the Supplemental Material. Briefly, 3D time-lapse (4D) imaging was performed in S2 cells using a spinning-disk confocal microscopy (Yokogawa). Long-term DIC imaging of mammalian cells was performed on a Nikon TE2000E inverted microscope in a temperature- and CO2-controlled environment.
Microcontact printing
FN micropatterns were made on glass coverslips using a microcontact printing technique adapted from the work of Fink et al. (2007) with some important modifications to allow a better stamping of the FN patterns on the glass (Supplemental Material). MDA-231 cells were plated on micropatterns for 4–5 h prior to fixation.
Acknowledgments
We thank B. Mathey-Prevot and the HMS Drosophila RNAi Screening Center (DRSC) and Institute of Chemistry and Cell Biology (ICCB) Longwood for help with the screen; D. Glover, M. Bettencourt-Dias, J. Raff, R. Basto, G. Goshima, C. Sunkel, G. Rogers, S. Rogers, M. Bornens, S. Henikoff, C. Walczak, and S. Cai for reagents; R. Basto and J. Raff for communicating unpublished results; and M. Bettencourt-Dias, A. Kung, R. King, S. Rogers, and S. Yoshida for comments on the manuscript. This work was supported by a Susan Komen grant to M.K., by an FCT grant to S.A.G., by a fellow award from the LLS to N.J.G., and by NIH grant GM083299 and a DFCI/Novartis Drug Discovery Grant to D.P.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1700908.
References
- Amendola R., Martinez R., Negroni A., Venturelli D., Tanno B., Calabretta B., Raschella G. DR-nm23 expression affects neuroblastoma cell differentiation, integrin expression, and adhesion characteristics. Med. Pediatr. Oncol. 2001;36:93–96. doi: 10.1002/1096-911X(20010101)36:1<93::AID-MPO1021>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Bakal C., Aach J., Church G., Perrimon N. Quantitative morphological signatures define local signaling networks regulating cell morphology. Science. 2007;316:1753–1756. doi: 10.1126/science.1140324. [DOI] [PubMed] [Google Scholar]
- Basto R., Brunk K., Vinadogrova T., Peel N., Franz A., Khodjakov A., Raff J.W. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–1042. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bild A.H., Yao G., Chang J.T., Wang Q., Potti A., Chasse D., Joshi M.B., Harpole D., Lancaster J.M., Berchuck A., et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- Bodily K.D., Morrison C.M., Renden R.B., Broadie K. A novel member of the Ig superfamily, turtle, is a CNS-specific protein required for coordinated motor control. J. Neurosci. 2001;21:3113–3125. doi: 10.1523/JNEUROSCI.21-09-03113.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boveri T. The origin of malignant tumors. Williams and Wilkins; Baltimore, MD: 1929. [Google Scholar]
- Brinkley B.R. Managing the centrosome numbers game: From chaos to stability in cancer cell division. Trends Cell Biol. 2001;11:18–21. doi: 10.1016/s0962-8924(00)01872-9. [DOI] [PubMed] [Google Scholar]
- Bulgheresi S., Kleiner E., Knoblich J.A. Inscuteable-dependent apical localization of the microtubule-binding protein Cornetto suggests a role in asymmetric cell division. J. Cell Sci. 2001;114:3655–3662. doi: 10.1242/jcs.114.20.3655. [DOI] [PubMed] [Google Scholar]
- Canman J.C., Bement W.M. Microtubules suppress actomyosin-based cortical flow in Xenopus oocytes. J. Cell Sci. 1997;110:1907–1917. doi: 10.1242/jcs.110.16.1907. [DOI] [PubMed] [Google Scholar]
- Chang P., Coughlin M., Mitchison T.J. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat. Cell Biol. 2005;7:1133–1139. doi: 10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- Chen J.G., Horwitz S.B. Differential mitotic responses to microtubule-stabilizing and -destabilizing drugs. Cancer Res. 2002;62:1935–1938. [PubMed] [Google Scholar]
- Chen T., Muratore T.L., Schaner-Tooley C.E., Shabanowitz J., Hunt D.F., Macara I.G. N-terminal α-methylation of RCC1 is necessary for stable chromatin association and normal mitosis. Nat. Cell Biol. 2007;9:596–603. doi: 10.1038/ncb1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra E.S., Higgs H.N. INF2 Is a WASP homology 2 motif-containing formin that severs actin filaments and accelerates both polymerization and depolymerization. J. Biol. Chem. 2006;281:26754–26767. doi: 10.1074/jbc.M604666200. [DOI] [PubMed] [Google Scholar]
- D’Assoro A.B., Barrett S.L., Folk C., Negron V.C., Boeneman K., Busby R., Whitehead C., Stivala F., Lingle W.L., Salisbury J.L. Amplified centrosomes in breast cancer: A potential indicator of tumor aggressiveness. Breast Cancer Res. Treat. 2002;75:25–34. doi: 10.1023/a:1016550619925. [DOI] [PubMed] [Google Scholar]
- Dixelius J., Cross M., Matsumoto T., Sasaki T., Timpl R., Claesson-Welsh L. Endostatin regulates endothelial cell adhesion and cytoskeletal organization. Cancer Res. 2002;62:1944–1947. [PubMed] [Google Scholar]
- Doxsey S. Re-evaluating centrosome function. Nat. Rev. Mol. Cell Biol. 2001;2:688–698. doi: 10.1038/35089575. [DOI] [PubMed] [Google Scholar]
- Echard A., Hickson G.R., Foley E., O’Farrell P.H. Terminal cytokinesis events uncovered after an RNAi screen. Curr. Biol. 2004;14:1685–1693. doi: 10.1016/j.cub.2004.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert U.S., Kiger A.A., Richter C., Perlman Z.E., Perrimon N., Mitchison T.J., Field C.M. Parallel chemical genetic and genome-wide RNAi screens identify cytokinesis inhibitors and targets. PLoS Biol. 2004;2:e379. doi: 10.1371/journal.pbio. 0020379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow S.A., Komma D.J. Assembly and dynamics of an anastral:astral spindle: The meiosis II spindle of Drosophila oocytes. J. Cell Sci. 1998;111:2487–2495. doi: 10.1242/jcs.111.17.2487. [DOI] [PubMed] [Google Scholar]
- Fink J., Thery M., Azioune A., Dupont R., Chatelain F., Bornens M., Piel M. Comparative study and improvement of current cell micro-patterning techniques. Lab Chip. 2007;7:672–680. doi: 10.1039/b618545b. [DOI] [PubMed] [Google Scholar]
- Fukasawa K. Oncogenes and tumour suppressors take on centrosomes. Nat. Rev. Cancer. 2007;7:911–924. doi: 10.1038/nrc2249. [DOI] [PubMed] [Google Scholar]
- Ganem N.J., Storchova Z., Pellman D. Tetraploidy, aneuploidy and cancer. Curr. Opin. Genet. Dev. 2007;17:157–162. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Giehl M., Fabarius A., Frank O., Hochhaus A., Hafner M., Hehlmann R., Seifarth W. Centrosome aberrations in chronic myeloid leukemia correlate with stage of disease and chromosomal instability. Leukemia. 2005;19:1192–1197. doi: 10.1038/sj.leu.2403779. [DOI] [PubMed] [Google Scholar]
- Goshima G., Nedelec F., Vale R.D. Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J. Cell Biol. 2005;171:229–240. doi: 10.1083/jcb.200505107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Goodwin S.S., Zhang N., Scholey J.M., Vale R.D., Stuurman N. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill S.W., Gonczy P., Stelzer E.H., Hyman A.A. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegansembryo. Nature. 2001;409:630–633. doi: 10.1038/35054572. [DOI] [PubMed] [Google Scholar]
- Gupton S.L., Waterman-Storer C.M. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell. 2006;125:1361–1374. doi: 10.1016/j.cell.2006.05.029. [DOI] [PubMed] [Google Scholar]
- Heneen W.K. In situ analysis of normal and abnormal patterns of the mitotic apparatus in cultured rat-kangaroo cells. Chromosoma. 1970;29:88–117. doi: 10.1007/BF01183663. [DOI] [PubMed] [Google Scholar]
- Karabay A., Walker R.A. Identification of microtubule binding sites in the Ncd tail domain. Biochemistry. 1999;38:1838–1849. doi: 10.1021/bi981850i. [DOI] [PubMed] [Google Scholar]
- Karsenti E., Vernos I. The mitotic spindle: A self-made machine. Science. 2001;294:543–547. doi: 10.1126/science.1063488. [DOI] [PubMed] [Google Scholar]
- Kiger A.A., Baum B., Jones S., Jones M.R., Coulson A., Echeverri C., Perrimon N. A functional genomic analysis of cell morphology using RNA interference. J. Biol. 2003;2:27. doi: 10.1186/1475-4924-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine D.S., Sanchez C.A., Rabinovitch P.S., Reid B.J. Formation of the tetraploid intermediate is associated with the development of cells with more than four centrioles in the elastase-simian virus 40 tumor antigen transgenic mouse model of pancreatic cancer. Proc. Natl. Acad. Sci. 1991;88:6427–6431. doi: 10.1073/pnas.88.15.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle W.L., Salisbury J.L. Altered centrosome structure is associated with abnormal mitoses in human breast tumors. Am. J. Pathol. 1999;155:1941–1951. doi: 10.1016/S0002-9440(10)65513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle W.L., Lutz W.H., Ingle J.N., Maihle N.J., Salisbury J.L. Centrosome hypertrophy in human breast tumors: Implications for genomic stability and cell polarity. Proc. Natl. Acad. Sci. 1998;95:2950–2955. doi: 10.1073/pnas.95.6.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer T.U., Kapoor T.M., Haggarty S.J., King R.W., Schreiber S.L., Mitchison T.J. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- Mitchison T.J. Actin based motility on retraction fibers in mitotic PtK2 cells. Cell Motil. Cytoskeleton. 1992;22:135–151. doi: 10.1002/cm.970220207. [DOI] [PubMed] [Google Scholar]
- Morales-Mulia S., Scholey J.M. Spindle pole organization in Drosophila S2 cells by dynein, abnormal spindle protein (Asp), and KLP10A. Mol. Biol. Cell. 2005;16:3176–3186. doi: 10.1091/mbc.E04-12-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountain V., Simerly C., Howard L., Ando A., Schatten G., Compton D.A. The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J. Cell Biol. 1999;147:351–366. doi: 10.1083/jcb.147.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro E.M. PAR proteins and the cytoskeleton: A marriage of equals. Curr. Opin. Cell Biol. 2006;18:86–94. doi: 10.1016/j.ceb.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Nigg E.A. Centrosome aberrations: Cause or consequence of cancer progression? Nat. Rev. Cancer. 2002;2:815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- O’Connor M.J., Martin N.M., Smith G.C. Targeted cancer therapies based on the inhibition of DNA strand break repair. Oncogene. 2007;26:7816–7824. doi: 10.1038/sj.onc.1210879. [DOI] [PubMed] [Google Scholar]
- Pihan G.A., Wallace J., Zhou Y., Doxsey S.J. Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res. 2003;63:1398–1404. [PubMed] [Google Scholar]
- Quintyne N.J., Reing J.E., Hoffelder D.R., Gollin S.M., Saunders W.S. Spindle multipolarity is prevented by centrosomal clustering. Science. 2005;307:127–129. doi: 10.1126/science.1104905. [DOI] [PubMed] [Google Scholar]
- Rebacz B., Larsen T.O., Clausen M.H., Ronnest M.H., Loffler H., Ho A.D., Kramer A. Identification of griseofulvin as an inhibitor of centrosomal clustering in a phenotype-based screen. Cancer Res. 2007;67:6342–6350. doi: 10.1158/0008-5472.CAN-07-0663. [DOI] [PubMed] [Google Scholar]
- Ring D., Hubble R., Kirschner M. Mitosis in a cell with multiple centrioles. J. Cell Biol. 1982;94:549–556. doi: 10.1083/jcb.94.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarner S., Kozma R., Ahmed S., Lim L. Phosphatidylinositol 3-kinase, Cdc42, and Rac1 act downstream of Ras in integrin-dependent neurite outgrowth in N1E-115 neuroblastoma cells. Mol. Cell. Biol. 2000;20:158–172. doi: 10.1128/mcb.20.1.158-172.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluder G., Nordberg J.J. The good, the bad and the ugly: The practical consequences of centrosome amplification. Curr. Opin. Cell Biol. 2004;16:49–54. doi: 10.1016/j.ceb.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Sluder G., Thompson E.A., Miller F.J., Hayes J., Rieder C.L. The checkpoint control for anaphase onset does not monitor excess numbers of spindle poles or bipolar spindle symmetry. J. Cell Sci. 1997;110:421–429. doi: 10.1242/jcs.110.4.421. [DOI] [PubMed] [Google Scholar]
- Sousa A.D., Cheney R.E. Myosin-X: A molecular motor at the cell’s fingertips. Trends Cell Biol. 2005;15:533–539. doi: 10.1016/j.tcb.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Spiegelman B.M., Lopata M.A., Kirschner M.W. Aggregation of microtubule initiation sites preceding neurite outgrowth in mouse neuroblastoma cells. Cell. 1979;16:253–263. doi: 10.1016/0092-8674(79)90003-5. [DOI] [PubMed] [Google Scholar]
- Storchova Z., Breneman A., Cande J., Dunn J., Burbank K., O’Toole E., Pellman D. Genome-wide genetic analysis of polyploidy in yeast. Nature. 2006;443:541–547. doi: 10.1038/nature05178. [DOI] [PubMed] [Google Scholar]
- Tepass U., Tanentzapf G., Ward R., Fehon R. Epithelial cell polarity and cell junctions in Drosophila. Annu. Rev. Genet. 2001;35:747–784. doi: 10.1146/annurev.genet.35.102401.091415. [DOI] [PubMed] [Google Scholar]
- Thery M., Bornens M. Cell shape and cell division. Curr. Opin. Cell Biol. 2006;18:648–657. doi: 10.1016/j.ceb.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Thery M., Racine V., Pepin A., Piel M., Chen Y., Sibarita J.B., Bornens M. The extracellular matrix guides the orientation of the cell division axis. Nat. Cell Biol. 2005;7:947–953. doi: 10.1038/ncb1307. [DOI] [PubMed] [Google Scholar]
- Thery M., Jimenez-Dalmaroni A., Racine V., Bornens M., Julicher F. Experimental and theoretical study of mitotic spindle orientation. Nature. 2007;447:493–496. doi: 10.1038/nature05786. [DOI] [PubMed] [Google Scholar]
- Toyoshima F., Nishida E. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J. 2007;26:1487–1498. doi: 10.1038/sj.emboj.7601599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y., Wu S., Shi X., Chen K., Wu C. Migfilin and Mig-2 link focal adhesions to filamin and the actin cytoskeleton and function in cell shape modulation. Cell. 2003;113:37–47. doi: 10.1016/s0092-8674(03)00163-6. [DOI] [PubMed] [Google Scholar]
- Wakefield J.G., Bonaccorsi S., Gatti M. The Drosophila protein asp is involved in microtubule organization during spindle formation and cytokinesis. J. Cell Biol. 2001;153:637–648. doi: 10.1083/jcb.153.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S., Sotsios Y., Dowden J., Bruce I., Finan P. Therapeutic potential of phosphoinositide 3-kinase inhibitors. Chem. Biol. 2003;10:207–213. doi: 10.1016/s1074-5521(03)00048-6. [DOI] [PubMed] [Google Scholar]
- Wei S.Y., Escudero L.M., Yu F., Chang L.H., Chen L.Y., Ho Y.H., Lin C.M., Chou C.S., Chia W., Modolell J., et al. Echinoid is a component of adherens junctions that cooperates with DE-Cadherin to mediate cell adhesion. Dev. Cell. 2005;8:493–504. doi: 10.1016/j.devcel.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Yang Z., Loncarek J., Khodjakov A., Rieder C.L. Extra centrosomes and/or chromosomes prolong mitosis in human cells. Nat. Cell Biol. 2008;10:748–751. doi: 10.1038/ncb1738. [DOI] [PMC free article] [PubMed] [Google Scholar]