Abstract
In a forward genetic screen for chemotaxis mutants in Dictyostelium discoideum, we identified a loss-of-function mutation, designated tsunami, encoding a homolog of the Fused kinase. Cells lacking tsuA function could not effectively perform chemotaxis and were unable to become polarized or correctly orient pseudopods in chemotactic gradients. While tsuA− cells were able to couple receptor occupancy to phosphatidylinositol (3,4,5) trisphosphate (PIP3) production and actin polymerization, the PIP3 response was prolonged and basal F-actin levels were increased. Interestingly, TsuA localizes to the microtubule network and puncta mainly found at the cell periphery. Analysis of the gene uncovered a novel C-terminal domain that we designated the Tsunami Homology (TH) domain. Both the kinase domain and the TH domain are required to rescue the phenotypic defects of tsuA− cells. While kinase activity is not required for localization to microtubules, the TH domain is essential. Thus, localization of kinase activity to microtubules is critical for TsuA function. We propose that functions in association with the microtubule network may underlie the divergent roles of Fused kinase proteins in different organisms.
Keywords: Chemotaxis, Dictyostelium, Fused, polarization
Chemotaxis refers to the capacity of cells to move directionally in a gradient of an external signaling molecule. This process guides the coordinated cell movements that produce morphogenesis during development and the migration of immune cells around the body and contributes to the pathology of cancer metastasis, asthma, and rheumatoid arthritis (Ridley et al. 2003). The mechanisms of chemotaxis are very similar in evolutionarily diverse organisms (Bagorda et al. 2006).
Powerful genetic and cell biological approaches available in the model organism Dictyostelium discoideum have led to many conceptual advances in our understanding of directed cell migration (Franca-Koh et al. 2006; Willard and Devreotes 2006). In particular, the parallels between this simple amoebae and leukocytes are remarkable. In these two cell types, chemotaxis is mediated by G-protein-coupled receptors and can conceptually be divided into three interrelated processes: membrane extension, directional sensing, and polarization (Iglesias and Devreotes 2008). Membrane extensions are protrusions driven by actin polymerization that mediate cell motility. In the absence of a chemoattractant gradient, membrane extensions are formed randomly along the cell perimeter. In a gradient, however, the process of directional sensing generates intracellular asymmetries that serve to focus signaling events and membrane extensions in the direction of the gradient. Together, membrane extensions coupled to directional sensing are sufficient to mediate chemotaxis. The third process, polarization, refers to the elongation of cell morphology and stable asymmetric localization of internal components and serves to increase the efficiency and speed of chemotaxis (Devreotes and Janetopoulos 2003). Chemotaxing neutrophils and starved D. discoideum amoebae are highly polarized. In both cell types, the poles can be distinguished morphologically, and the front is more sensitive to chemoattractants than the back. In contrast, vegetative D. discoideum cells sensing bacterial metabolites do not become elongated and are equally responsive to chemoattractants around the cell perimeter. This correlates with reduced speed and directionality compared with starved cells performing chemotaxis.
Recent studies have begun to elucidate some of the molecular mechanisms mediating polarization. In particular, positive feedback loops appear to play an important role in amplifying responses at the poles. One such feedback pathway involves phosphatidylinositol 3′-kinase (PI 3-kinase), phosphatidylinositol (3,4,5) trisphosphate (PIP3), and actin polymerization (Weiner et al. 2002; Sasaki et al. 2004). The generation of PIP3 by PI 3-kinase can initiate actin polymerization, which in turn promotes further production of PIP3. Signaling from the microtubule network is also thought to regulate polarity and persistence of pseudopods (Rodriguez et al. 2003; Satulovsky et al. 2008). In fibroblasts and astrocytes, for example, the reorientation of the MTOC (microtubule-organizing center) is an early marker of polarization in wound healing assays (Etienne-Manneville and Hall 2001; Gomes et al. 2005). Furthermore, disruption of microtubules by depolymerizing drugs has been shown to interfere with polarity and movement in several cell types including fibroblasts, keratinocytes and astrocytes (Vasiliev et al. 1970; Etienne-Manneville and Hall 2001; Pegtel et al. 2007).
In this study, we describe a novel mutant that has a strong chemotaxis defect due to an inability to become polarized and correctly orient membrane extensions in chemoattractant gradients. Surprisingly, this mutant, which we designated tsunami, is the D. discoideum homolog of the Fused kinase. This is a highly conserved protein that has been most studied for its role in Hedgehog signaling (Varjosalo and Taipale 2007). The results of this study identify novel functions for a member of this important kinase family.
Results
tsunami is a Fused kinase essential for proper cell–cell aggregation
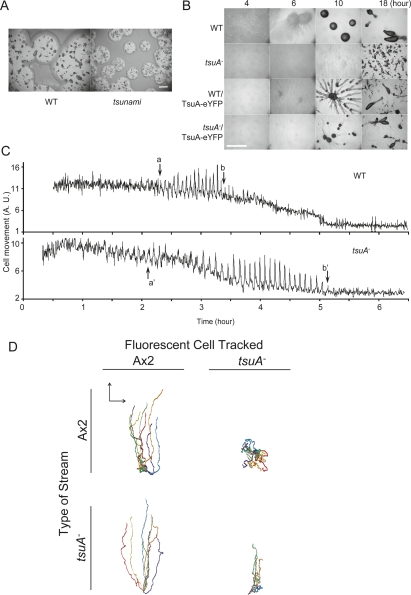
To identify new genes required for chemotaxis in D. discoideum, investigators have carried out genetic screens using restriction endonuclease-mediated integration (REMI) (Kuspa 2006). Typically, wild-type cells are mutagenized, and transformants are plated clonally on agar plates with Klebsiella aerogenes. During 5 d of incubation at 22°C, wild-type cells consume bacteria, form clear plaques, and undergo starvation-induced development to form aggregates called fruiting bodies. We obtained from Dr. Rob Kay (University of Cambridge) a clone that made a small plaque and formed fruiting bodies about one-tenth the size of wild type (Fig. 1A).
Figure 1.
Development and aggregation in wild-type and tsuA− cells. (A) Development on K. aerogenes lawns was assessed by clonally plating wild-type and tsunami cells with bacteria on SM plates. Plaques were photographed after 5 d at 22°C. Bar, 2 mm. (B) To test development on nonnutrient agar, wild-type and tsuA− cells transformed with either vector or TsuA-YFP were plated on nonnutrient agar (DB agar) at 1.25 × 106 cells per square centimeter. Typical fields of view were photographed at the indicated time points. Bar, 2 mm. (C) To monitor the propagation of cAMP waves, the movement of cells on DB agar was recorded by time-lapse microscopy. The images were processed as described in the text. (D) Migration of wild-type and tsuA− cells in chimeric aggregation streams. Unlabeled wild-type (top panels) or tsuA− cells (bottom panels) were mixed with 10% GFP-expressing tsuA− cells and 10% mRFP-expressing AX2 cells. Simultaneous DIC transmitted light images plus red and green fluorescence confocal images were collected at 1-min intervals (Supplemental Movies 3, 4). The movement of eight to 10 cells of each type was tracked and is shown as a Rose plot, which normalizes the starting point of each cell track. Arrows represent 100 μm.
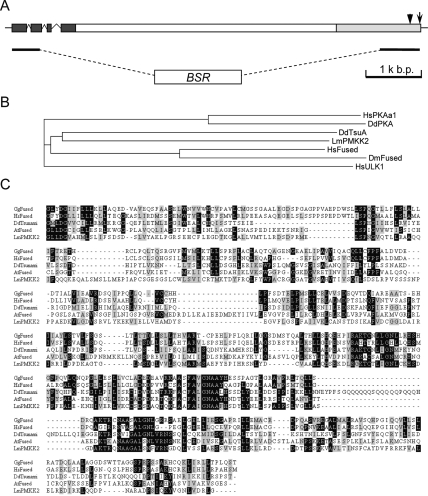
By sequencing genomic fragments flanking the inserted REMI vector retrieved from the tsunami mutant, the insertion was found to have occurred at the 3′ end of a large ORF (Fig. 2A). Two DNA fragments from the tsuA gene, comprising base pairs 24–513 and 6414–7107, respectively, were used to generate a construct for targeting the gene for deletion by homologous recombination. Properly targeted clones were identified by PCR and confirmed by Southern blotting (data not shown). On K. aerogenes lawns, these cell lines recapitulated the small plaque and small fruiting body phenotype shown in Figure 1A while nontargeted clones from the same transformation displayed the wild-type phenotype. One of the cell lines lacking tsuA was used for further investigation and is referred to as tsuA−.
Figure 2.
tsuA gene structure and homology with Fused protein kinase family. (A) Schematic diagram of tsuA gene. The kinase domain is shown by the region shaded in dark gray, and the TH domain is shown by the light-gray shading. The arrow indicates the insertion site of the original REMI mutant, and the arrowhead shows a targeted insertion into the gene. The bars and BSR box illustrate the homologous targeting scheme for deletion of most of the ORF. (B) The phylogenetic tree of the kinase domains from D. discoideum PKA and TsuA; L. major protein kinase (GI: 68125087, arbitrarily annotated Potential MKK2); H. sapiens Unc-51-like kinase 1 (ULK1), PKAa1, and Fused; and D. melanogaster Fused. The tree was generated with ClustalW by the neighbor joining method. (C) Multiple alignment of TH domains of Gallus gallus, H. sapiens, and A. thaliana Fused kinases, as well as those of L. major protein kinase (GI: 68125087) and TsuA. The alignment was generated with ClustalW. Black shows identities; gray shows similarities.
When tsuA− cells were plated on nonnutrient agar, their aggregation was significantly delayed and the final structures formed were greatly reduced in size (Fig. 1B). Under these conditions, wild-type cells aggregate through a series of coordinated chemotactic movements toward diffusing cyclic AMP (cAMP) secreted at 6-min intervals by amoebae at aggregation centers. To monitor the coordinated chemotactic movements in tsuA− cells, synchronously developed cells were plated on nonnutrient agar as a monolayer and recorded by time lapse microscopy (Supplemental Movies 1, 2). Previous studies have shown that, in response to cAMP waves, wild-type cells round up or “cringe” and then migrate toward the center in a movement “step” that covers ∼20 μm in 2 min (Futrelle et al. 1982; Soll et al. 2002; Chen et al. 2003). By the 6-h stage, the migrations develop into stream-like movements, and eventually the waves subside due to coalescence of the monolayer. To enhance the detection of cell movements, we processed the videos by subtracting each frame from the next so that cell movements were converted into changes in intensity. By monitoring changes in signal intensity within a fixed region of the field, we could distinguish the early uncoordinated random movements of individual cells, the periodic waves, as well as the coalescence of the monolayer into aggregates. As shown in Figure 1C, under the conditions used in this experiment the wild-type cells produced well-defined waves from hours 2 to 3.5. Cells lacking tsuA, on the other hand, displayed a significantly prolonged duration of periodic wave propagation. Although the overall length of wave propagation varied with conditions (such as humidity and temperature), wave propagation invariably began at the same time as the wild type but persisted for at least twice as long. We chose the name tsunami to reflect the extended period of wave production.
Careful examination of the tsunami mutant cells revealed why they fail to aggregate. In response to each cAMP wave, the tsunami cells “cringed,” as did wild-type cells. However, the subsequent movement step was greatly attenuated, and the cells advanced no more than a few micrometers toward the center. Ultimately, the waves ceased and tiny aggregates formed without wave propagation, presumably due to the timely expression of extracellular adhesion proteins. Furthermore, the aggregation defects were cell autonomous and specific for chemotaxis and not wave propagation (Fig. 1D; Supplemental Movies 3, 4). The migratory defects of tsuA− cells were not corrected by mixing the mutant cells with an excess of wild-type cells. In wild-type streams under agar, the average speed of wild-type cells was 6.3 μm/min, while that of the tsuA− cells was 4.2 μm/min. More striking was a defect in persistence of movement, which was 0.62 for wild-type cells and 0.2 for the mutant cells (persistence is defined as the distance from start to end position of each cell divided by the total track length). Conversely, wild-type cells migrated normally within the cAMP waves generated by tsuA− cells (Fig. 1D). These observations suggest that cells lacking tsuA function generate cAMP waves, and they can detect these waves; however, their chemotactic response is defective.
The sequence of the tsuA gene product revealed an N-terminal kinase domain homologous to several kinases including Homo sapiens, Drosophila melanogaster, Danio rerio, and Arabidopsis thaliana Fused kinases (GI: 9664225, GI: 1363911, GI: 68360530, and GI: 72004127) as well as a Leishmania major protein kinase (GI: 68125087) and a Trypanosoma cruzi protein kinase (GI: 71424046), with 49% identitical and 74% similar to human Fused in the first 256 amino acid residues. The dendrogram in Figure 2B shows the relationship among these kinase domains. With the exception of D. melanogaster and D. rerio kinases, the Blast-P algorithm detected extensive homology between the same cadre of proteins and TsuA in a ∼400-amino acid C-terminal region that is 23% identical and 43% similar to human Fused (Fig. 2C). We designated this region the Tsunami Homology (TH) domain. Using several iterations of the more sensitive PSI-Blast algorithm, however, some sequence similarity can also be detected in the D. rerio and D. melanogaster proteins (see Supplemental Fig. S1). The TH domain of TsuA also contains putative Armadillo repeats (amino acids 2013–2194) and a region that is similar to the motif in importin-α responsible for binding to nuclear localization signals (amino acids 1831–2088). Between the kinase and TH domains, the D. discoideum protein contains ∼1500 amino acid residues that consist mainly of simple repetitive sequences. In retrospect, the original REMI insertion was in the TH domain at the extreme 3′ end of the gene. The targeting construct we designed replaced most of the ORF, including portions of the kinase and TH domains, with a blasticidin resistance cassette (BSR) (Fig. 2A).
Expression of the full-length tsuA gene fused to YFP rescued the aggregation defects of the deletion strain and the REMI mutant (Fig. 1B, bottom panels). Although the rescued strains formed slightly smaller aggregates, the phenotype matched that of wild-type cells expressing TsuA-YFP. The length of the movement steps resembled those of wild-type cells, indicating that the chemotaxis defects were also reversed (Fig. 1B; data not shown). The differentiated tsuA−/TsuA-YFP cells also elongated and polarized similarly to wild-type cells. These findings suggest that the TsuA-YFP is fully functional and that the phenotypes observed in both the tsuA¯ cells and the original mutant strains are a result of tsuA loss of function.
Cells lacking tsuA have a severe chemotaxis and polarization defect
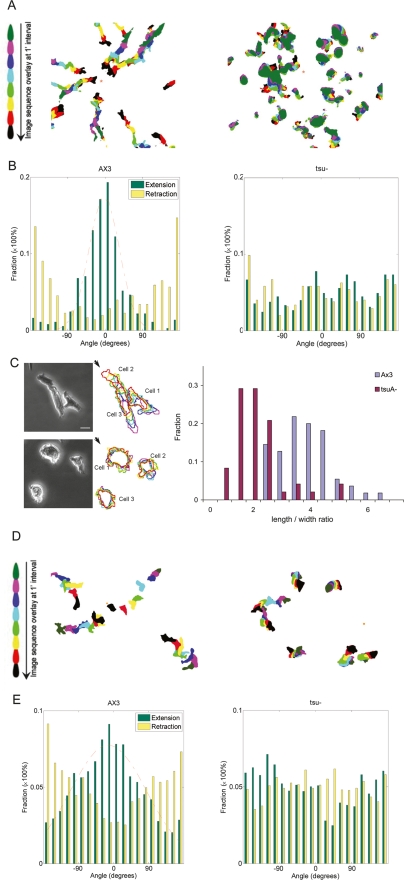
To characterize the chemotactic response, isolated cells were starved for 5–6 h and then presented with a micropipette releasing cAMP, allowing chemotaxis to be assessed by time-lapse microscopy. The wild-type cells displayed coordinated motion toward the micropipette tip, covering several cell lengths over an 8-min period, while the tsuA− cells moved slowly and without consistent direction (Fig. 3A; Supplemental Movies 5, 6). Additionally, striking differences in the direction of pseudopodial extensions and polarization were observed. To evaluate the pseudopod dynamics rigorously, we analyzed the cellular movements using a skeleton representation algorithm (see the Materials and Methods; Kabacoff et al. 2007). By comparing the cell outlines and skeletons between adjacent frames, we were able to detect both pseudopod extensions and retractions accurately. When these were plotted in relation to the angle of the micropipette, it was clear that, whereas wild-type cells preferentially generated membrane extensions toward the cAMP source (centered at 0°) and made retractions at the rear of the cell (centered at 180°), the extensions and retractions in tsuA− cells appeared randomly about the perimeter (Fig. 3B). The time lapse movies also revealed a strong defect in acquisition of the polarized morphology that coincides with high chemotactic efficiency of wild-type cells during aggregation. After 5 h of starvation in nonnutrient buffer, the majority of wild-type cells became “polarized” such that they adopted an asymmetric morphology with a defined leading edge and a slender trailing tail (Fig. 3A,C). We quantified this by measuring the length-to-width ratios of wild-type and mutant cells (Fig. 3C). The average length-to-width ratio in developed Ax3 cells was 3.6 ± 0.13 (mean ± SEM, n = 55), while the tsuA− cells were more rounded and had a mean length to width ratio of 1.9 ± 0.13 (n = 48), similar to that of vegetative wild-type cells, even after 8 h of starvation.
Figure 3.
Chemotactic responses and induced cell polarity. (A) The chemotactic movements of developed wild-type (left) and tsuA− cells (right) expressing PHCRAC-GFP migrating to a micropipette releasing 10 μM cAMP (orange asterisk) were recorded by time lapse microscopy (Supplemental Movies 5, 6). Images from frames at 1-min intervals were processed to outline the cells, color coded for each time point, and the images were overlaid. Bar is 50 μm. (B) Distribution of tip angles for extending (green) and retracting (yellow) pseudopods relative to the direction of the cAMP gradient; 0° points directly to the micropipette. The data represent 13 wild-type cells and 14 tsuA− cells. (C) Histogram of cell length/width ratio distribution. cAMP gradients were applied to developed wild-type Ax3 and tsuA− cells. The distance from the leading edge, as defined by the front of protrusions, to the trailing tail was taken as cell length (l), and the width of the leading edge, as cell width (w). The l/w ratio from Ax3 (n = 55) and tsuA− cells (n = 48) were binned to ranges of 0.5 intervals and the frequency of every ratio range is plotted as a histogram. Representative phase-contrast images are shown on the left, and color-coded cell outlines were made at 30-sec intervals (magenta to red) from time lapse recordings. (D) Chemotactic movements of vegetative wild-type (left) and tsuA− (right) cells expressing PHCRAC-GFP migrating to a micropipette filled with 1 mM folic acid were recorded and processed as described previously (Supplemental Movies 7, 8). (E) Tip angle distributions relative to the folic acid gradient for wild-type (n = 19) and tsuA− (n = 19) cells.
In light of these observations, we wondered whether the inability of tsuA− cells to orient pseudopod extensions in the direction of the gradient was merely a consequence of their failure to become highly polarized. To address this question, we assayed the chemotaxis of vegetative wild-type and tsuA− cells to folic acid (Fig. 3D; Supplemental Movies 7, 8). Previous studies have shown that, in contrast to starved cells, vegetative cells do not become polarized during chemotaxis. By measuring cell “roundness” (see the Materials and Methods) in these experiments, we demonstrated that both wild-type and tsuA− cells were equally round during migration (R = 0.57 vs. 0.61, where R is 4π × Area/Perimeter2), confirming that in this context the contribution of polarization to directional movement is eliminated. In response to a micropipette filled with 1 mM folic acid, wild-type cells effectively moved toward the chemoattractant and preferentially made pseudopod extensions in the direction of the gradient, although to a slightly broader region of the cell than when performing chemotaxis to cAMP. The tsuA− cells, however, did not effectively move toward the folic acid source and generated pseudopod extensions randomly over the entire cell periphery. This indicates that the failure of tsuA− cells to restrict membrane extensions toward the gradient is not simply a result of failing to become polarized. tsuA− cells are also defective in chemotaxis to folic acid in an under agar assay, but in this system, migration is less severely impaired (D. Knecht, pers. comm.).
Cells lacking tsuA differentiate and respond to chemoattractants
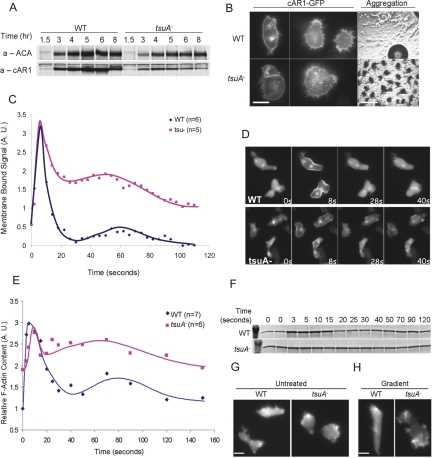
About 50–100 genes are newly expressed as cells gain competence to sense and produce cAMP gradients (Booth et al. 2005). Simultaneously, sensitivity to the vegetative stage chemoattractant, folic acid, is down-regulated (Tillinghast and Newell 1987; Wu and Devreotes 1991). Expression of one of the cAMP receptors, cAR1, is strongly up-regulated after 3 h of development, peaks at 6 h, and declines thereafter. Similarly, the expression of the aggregation stage adenylyl cyclase (ACA) is also increased dramatically during development. Both of these genes are critical for the oscillatory production of cAMP and periodic wave propagation. Wild-type and tsuA− cells had very similar temporal profiles of expression of cAR1 and ACA, although tsuA− cells expressed slightly less cAR1 (Fig. 4A). To investigate whether the reduced levels of receptor could explain the chemotaxis defects seen during development, we assessed the aggregation of tsuA− cells expressing exogenous cAR1-YFP. The exogenous receptors were properly expressed and targeted to the cell membrane but did not alter the small aggregate phenotype of the tsuA− cells, indicating that the tsuA− phenotype is not a result of altered cAR1 expression (Fig. 4B).
Figure 4.
Developmentally regulated gene expression profiles, PHCRAC-GFP translocation, and actin polymerization. (A) Immunoblotting against whole-cell lysates with anti-cAR1 and anti-ACA antibodies. Wild-type and tsuA− cells were lysed in SDS sample buffer at the indicated times after initiation of development. (B) Fluorescence images of cAR1-GFP expressed in wild-type and tsuA− cells (left, midpoint of cells; middle, optical section close to cover slip; bar, 10 μm) and phase-contrast images of wild-type/cAR1-GFP and tsuA−/cAR1-GFP cells developed on DB agar (right; bar, 1 mm). (C) PHCRAC-GFP translocation to cell membrane during uniform cAMP (1 μM) stimulation, as recorded by wide-field epifluorescence microscopy. Images were taken every 4 sec and processed to detect membrane translocation. The intensity of the membrane bound signal is plotted against time in C, and frames from the indicated time points are shown in D. For E and F, developed wild-type and tsuA− cells were treated with 1 μM cAMP. Cells were lysed in 1% Triton X-100 buffer, and the pellet fraction was subsequently analyzed by SDS-PAGE and Coomassie blue staining. Quantification of the amount of actin in the Triton-insoluble pellet is shown in E, and a representative gel is shown in F. G shows the localization of LimEΔcoil-mRFP in randomly migrating vegetative Ax3 and tsuA−, and H shows the localization of the same construct in developed cells exposed to a cAMP gradient. Bars, 12 μm.
The transient accumulation of PIP3 on the cell membrane after a uniform increase in chemoattractant and the local accumulation of PIP3 on the leading edge of cells in cAMP gradients are well-characterized signaling events downstream from receptor activation (Parent et al. 1998). Using the pleckstrin homology (PH) domain of CRAC fused to GFP (PHCRAC-GFP) as a probe, we observed a similar increase in PIP3 in both wild-type and tsuA− cells stimulated by cAMP. As shown in Figure 4, C and D, in both cell types accumulation of PHCRAC-GFP on the cell membrane was detected in the first frame of the 4-sec interval image sequence. The fluorescence signal decreased in the cytosol and the enrichment on membrane peaked at ∼6 sec after the cAMP stimulation. After 20 sec, PHCRAC-GFP returned to the cytosol, and then a second translocation to protruding regions on the membrane ensued from 60 to 80 sec. However, the second phase of accumulation of PIP3 is much more prominent in the tsuA− cells. This is consistent with the observation that these mutant cells have hyperactive protrusions around the cell periphery during this time. A similar correlation was observed in cells lacking the PI 3-phosphatase PTEN (Iijima and Devreotes 2002). Thus, the temporal and spatial regulation of PIP3 indicates that tsuA− cells are able to link receptors to phosphoinositide signaling and suggests that the defect lies downstream or in parallel to this pathway. The enhanced second phase of accumulation and broader localization may reflect the lack of polarity noted previously (see the Discussion).
Chemotactic eukaryotic cells typically display a receptor-mediated actin polymerization response. D. discoideum cells respond to a uniform increase in cAMP with biphasic changes in actin polymerization (Hall et al. 1988; Chen et al. 2003). We measured the levels of F-actin in cells by determining the amount of actin present in 1% Triton X-100-insoluble pellets, typically regarded as the cytoskeleton (McRobbie and Newell 1983; Dharmawardhane et al. 1989). In the “resting” state, ∼30%–40% of the actin is already 1% Triton X-100 insoluble, presumably F-actin in randomly extending pseudopodia and the cortex (Fig. 4E,F). Within 10 sec of applying a cAMP stimulus to wild-type cells, we observed a threefold increase in the amount of F-actin in wild-type cells, which returned to the prestimulus level by 20–30 sec. A second phase of polymerization, peaking at 60–80 sec, which is thought to occur at pseudopodial tips, then ensued. The tsuA− cells, however, displayed a significantly higher relative F-actin content in the “resting” state; nearly 60% of the actin was found in the 1% Triton X-100-insoluble fraction. This may reflect the observed hyperactive extensions around the cell perimeter. Following stimulation, there was an increase in the fractional amount of F-actin with the same time course as wild type; but due to the higher basal levels, the increase was only to 1.3-fold and a distinct second phase was less apparent. F-actin dynamics in live cells were also investigated by expressing the LimE-Δ coil domain fused to GFP as a probe for F-actin (Bretschneider et al. 2004). Consistent with the above findings, unstimulated tsuA−cells indeed showed an increase in the number of protrusions with local enrichment of the LimE-Δ coil (Fig. 4G). Even in chemoattractant gradients, the tsuA− cells made more lateral or rear GFP-labeled actin-filled protrusions (Fig. 4H).
TsuA protein is localized to the microtubule network
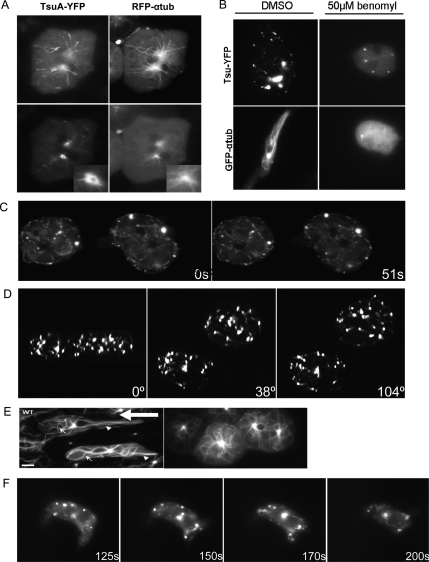
To determine the subcellular localization of the TsuA protein, we studied a full-length TsuA-YFP fusion protein that we had previously shown could rescue tsuA¯ cells. This fusion protein exhibited an intriguing localization pattern. When expressed in either wild-type or tsuA− cells, the TsuA-YFP was found in bright puncta, most often at the periphery of the cell, and in smaller spots along linear structures that emanated from a central focus that was also strongly labeled. This pattern was most clearly revealed in cells that were flattened by overlaying with a thin agar slice and suggested the possibility that TsuA-YFP localizes to microtubules and the MTOC. To test this hypothesis, we cotransformed wild-type cells with mRFP-α-tubulin and TsuA-YFP (Fig. 5A). This revealed extensive colocalization of the two signals along the microtubules and the pericentrosome, indicating that TsuA-YFP indeed localizes to the microtubule network. Further supporting this conclusion, when cells expressing TsuA-YFP were treated with the microtubule depolymerizing drug benomyl, the tubular staining pattern was lost and the level of cytosolic staining increased (Fig. 5B, top panels). Brief benomyl treatment nearly eliminated the microtubule network, as evidenced by imaging of cells expressing GFP-α-tubulin, and severely disrupted polarity, causing even highly polarized streaming cells to round up (Fig. 5B, bottom panels). These data also suggested, however, that TsuA is not an integral component of the centrosome, as when microtubules were disrupted, the pericentrosomal staining was also lost. Interestingly, many of the brighter peripheral puncta remained, suggesting that these regions of TsuA-YFP staining can persist independently from microtubules.
Figure 5.
Subcellular localization of TsuA-YFP. (A) Colocalization of TsuA-YFP and mRFP-α-tubulin. Ax3 cells coexpressing TsuA-YFP were plated on coverglasses and layered with an ∼2-mm slice of 1% agarose to flatten the cells for imaging. Images from two focal planes are shown, and the insets (shown in the bottom panels) magnify the MTOC. Bar, 2 μm. (B) Effect of disrupting the microtubule network with benomyl. Ax3 cells expressing TsuA-YFP (top panel) or GFP–α-tubulin (bottom panel) were treated with 50 μM benomyl or DMSO as a control for 15 min and then flattened by overlaying with DB agarose. (C) TsuA-YFP in vivo dynamics. Ax3 cells expressing TsuA-YFP were flattened by agar overlay and the intracellular movements of TsuA-YFP were recorded by time-lapse fluorescence microscopy (Supplemental Movie 9). Shown in this figure are images from the indicated time points. (D) Three-dimensional reconstruction of TsuA-YFP. Optical sections of unflattened Ax3 expressing TsuA-YFP were taken, and the deconvolved images were made into three-dimensional reconstructions by Metamorph software. Shown are the views from three different angles; a video of the rotation series is available in Supplemental Movie 10. (E) Organization of the microtubule network in developed cells. Ax3 and tsuA− cells expressing GFP-α-tubulin were developed for 6–7 h and plated on coverglass until the wild-type cells formed thin streams. The cells were then flattened by agar overlay and imaged by fluorescent microscopy. Bar, 6 μm. Arrows point to microtubules extending forward; arrowheads point to microtubules stretching to the rear; open large arrow points in the direction of Ax3 cell migration. (F) TsuA-YFP localization in polar ized cells. tsuA− cells rescued with TsuA-YFP were starved for 6 h and plated on coverglass, and the localization of TsuA-YFP was recorded by time-lapse microscopy (Supplemental Movie 11). Shown are images from the indicated time points.
To analyze the dynamics of this localization pattern we monitored TsuA-YFP in timelapse movies. Typically the TsuA-YFP signal did not, except in rare instances, translocate along the microtubules. Instead, it appeared simply to decorate the microtubules and moved in association with them (Fig. 5C; Supplemental Fig. S2; Supplemental Movie 9). We also made three-dimensional reconstructions of TsuA-YFP localization in unflattened cells from deconvolved optical sections (Fig. 5D; Supplemental Movie 10). These images confirmed the impression gained from the flattened cells and showed that the bright puncta were generally distributed to the periphery of the cell, and also showed labeling of microtubules emanating from the MTOC.
The microtubule network in D. discoideum is a dynamic three-dimensional structure. In the anterior portion of wild-type cells, microtubules radiated toward the front and sides, while in the rear they appeared stretched and under tension (Fig. 5E, left). Owing to the polarization defect of the tsuA− cells, there was a striking difference in the microtubule networks of wild-type and tsuA− cells. The network in tsuA− cells radiated symmetrically from the center of the MTOC and seemed loosely attached to the cortex, similar to the distribution found in growing cells (Fig. 5E, right). In tsuA− cells rescued with TsuA-YFP, the TsuA localization conformed to the general distribution of microtubules seen in wild-type cells and did not show any preferential localization to the front or back (Fig. 5F; Supplemental Movie 11).
Both kinase activity and microtubule localization are required for tsuA function
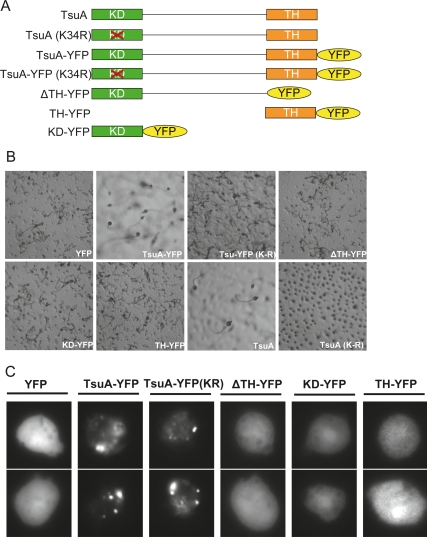
To investigate the contribution of kinase activity and the TH domain to TsuA function, we generated the following series of constructs (Fig. 6A). First, we made kinase dead versions of TsuA by mutating a single residue in the ATP-binding pocket, Lys 34, to arginine (TsuA K34R and TsuA-YFP K34R). Second, we made a C-terminal truncation deleting the TH domain (ΔTH-YFP). Third, we also made constructs that fused either the kinase domain or the TH domain directly to YFP (KD-YFP and TH-YFP). These constructs were then compared with tagged and untagged versions of the wild-type protein (TsuA and TsuA-YFP) for their ability to rescue development on nonnutrient agar and to localize to microtubules. In the developmental assay, only expression of full-length, wild-type proteins complemented the aggregation defects of the tsuA− cells (Fig. 6B). Cells expressing the other constructs formed small aggregates that were similar in size to the control tsuA− cells that expressed YFP alone. Thus, physiological function of TsuA requires both kinase activity and the TH domain. The requirement for the TH domain is consistent with the loss-of-function phenotype of the original tsunami REMI insertion, which was predicted to remove the last six amino acids from the C terminus.
Figure 6.
Deletion analysis of the TsuA protein. (A) Schematic diagram of the constructs made in this study. (B) The indicated constructs were expressed in tsuA− cells and plated on DB agar to assay development. Images were taken after 24 h at 22°C. (C) The localization of the indicated constructs expressed in tsuA− cells was then acquired by fluorescent microscopy. Shown are two representative cells for each construct.
The localization of these constructs was then assessed by fluorescence microscopy. It was clear that the full-length wild-type TsuA-YFP fusion protein as well as the kinase dead version were able to localize to the microtubule network and puncta (Fig. 6C). The ability of the full-length kinase dead molecule to bind to microtubules shows that kinase activity is not required for correct localization and also suggests that this point mutation does not cause gross structural changes. The ΔTH-YFP construct was dispersed throughout the nucleus and cytosol, indicating that the TH domain is essential for microtubule binding. The TH-YFP construct, however, was also cytosolic, suggesting that the fragment used was not sufficient for localization. Together these data indicate that both kinase activity and microtubule localization are required for function.
Discussion
Through a forward genetic screen for morphological mutants in D. discoideum, we uncovered a novel role for a member of the Fused kinase family, TsuA. Cells lacking the tsuA gene have a specific defect in polarization and in orientation of pseudopodia in chemoattractant gradients. The TsuA protein localizes constitutively to the microtubule network via a novel domain in the C terminus, designated the TH domain, which is conserved among Fused kinases. Both kinase activity and proper localization are required for function, suggesting that TsuA acts in association with the microtubule cytosleleton. Consistently, depolymerization of microtubules releases TsuA to the cytosol and causes a loss in cell polarity. These findings and others in divergent organisms suggest that Fused kinases have direct cell biological roles, beyond Hedgehog signaling, and that association with the microtubule network is critical for these functions.
The Fused kinase homolog tsuA regulates chemotaxis
Cells lacking TsuA displayed chemotaxis defects under multiple experimental conditions. The tsuA− cells generate normal cAMP waves but in response to the waves, they only “cringe” but do not subsequently migrate toward the aggregation center. This cell-autonomous defect was not corrected by mixing with wild-type cells. Analysis of the mutant cells exposed to chemoattractant gradients in vitro revealed the basis for these defects. Starved tsuA− cells were unable to become polarized, that is, establish the persistent elongated morphology that characterizes the migration of wild-type cells to cAMP or correctly extend pseudopods in the direction of the gradient. In the context of vegetative stage chemotaxis to folic acid, neither wild-type nor mutant cells became polarized. Yet the tsuA− cells were still compromised in their ability to perform chemotaxis due to their failure to either sense the direction of the gradient or restrict pseudopods to the region of the cell facing the gradient. Together, these defects in chemotaxis can explain key aspects of the tsuA− phenotype. The inability to perform chemotaxis to cAMP during development can account for the persistent wave propagation and reduced fruiting body size. Similarly, the defect in vegetative chemotaxis to bacterial metabolites may explain why, even though the mutant cells grow at the same rate as wild-type cells in axenic culture, they make small plaques when grown on bacterial lawns (data not shown).
The defect in tsuA− is subtle and so is unlikely to be due to a global failure of developmentally regulated gene expression. First, for cells to initiate oscillatory signaling and propagate cAMP waves, cells must express the correct levels of the necessary genes at the appropriate time (Goldbeter and Segel 1980). Significantly, the wild-type and tsuA− cells begin cAMP oscillations at the same time, and the waves are able to properly direct the chemotaxis of wild-type cells (Fig. 1C,D). Second, tsuA− cells are defective in vegetative chemotaxis, a situation that does not require the regulation of genes involved in development. Third, in tsuA− cells cAMP is able to activate the downstream signaling pathways of PIP3 production and actin polymerization.
As PIP3 production and actin polymerization are key downstream signaling events in chemotaxis, we investigated the regulation of these pathways in tsuA− cells. The cells were able to activate these pathways when exposed to cAMP but in comparison with wild type, displayed a prolonged second phase of PIP3 production and a higher basal level of F-actin. The prolonged period of PIP3 accumulation is similar to the phenotype that has previously been reported in pten− cells (Iijima and Devreotes 2002). These cells also have a chemotaxis defect due to the generation of numerous lateral pseudopods. Since there is a well-established feedback loop between F-actin and PIP3 production, one possibility is that the prolonged accumulation of PIP3 is an indirect consequence of the increased basal levels of F-actin found in tsuA− cells. The increased basal level of F-actin is consistent with the polarization and chemotaxis defects. In contrast to the focused pseduopods generated by polarized wild-type cells, tsuA mutants often had multiple pseudopods on the same cells. Furthermore, if cells sequestered most of their actin into filaments in these extraneous pseudopodia, the reduced scope for further polymerization could contribute to the polarization defect by impairing positive feedback loops initiated by actin polymerization (Sasaki et al. 2004).
Tsunami localizes to the microtubule network
There is evidence that the microtubule network can exert both positive and negative effects on the actin cytoskeleton (Rodriguez et al. 2003). First, there are reports of interactions between microtubules and regulators of actin polymerization. In fission yeast, formin regulators, associated with microtubule plus-ends, are required for the proper maintenance of cell shape and polarized growth (Feierbach et al. 2004; Martin et al. 2005; Basu and Chang 2007). In mammalian cells, microtubule dynamics regulate Rac and Rho activities (Wittmann and Waterman-Storer 2001; Krendel et al. 2002; Wittmann et al. 2003). Second, disruption of the microtubule network inhibits migration in a number of cell types. In keratinocytes, nocodazole treatment or perturbations of Par3 and Tiam1 function reduce microtubule stability, impair polarization, and cause cells to frequently change direction (Pegtel et al. 2007). In astrocytes, low doses of nocodazole that interfere with normal microtubule dynamics severely impair membrane extensions and migration (Etienne-Manneville and Hall 2001). In D. discoideum, disruption of microtubules with benomyl severely impairs polarity (Fig. 5B). Similarly, recent studies have shown that D. discoideum cells treated with nocodazole have defects in polarization, pseudopod persistence, and chemotaxis to both cAMP and folic acid (Sroka et al. 2001; Satulovsky et al. 2008). Furthermore, overexpression of dominant-negative Lis1, certain dynein fragments, and DdCP224 lead to detachment of the microtubule network from the cortex and an altered cell shape in amoebae (Hestermann and Graf 2004; Brito et al. 2005; Rehberg et al. 2005). Third, orientation of the MTOC is associated with persistent migration and polarization. In fibroblasts and astrocytes, the orientation of the MTOC toward the anterior of the cell is an early step in establishing cell polarity during wound healing (Etienne-Manneville and Hall 2001; Gomes et al. 2005). In D. discoideum, the ability of a pseudopod to persist and generate cell locomotion depends on MTOC reorientation (Ueda et al. 1997). In contrast to all these positive effects, in leukocytes, inhibitors of microtubule polymerization induce polarization, suggesting that in this context, the microtubule network may inhibit polarization (Niggli 2003; Xu et al. 2005, 2007).
How might TsuA play a role in regulating the cytoskeleton? The constitutive association of TsuA with the microtubule network is essential for function, suggesting that it acts at this location. As noted above, loss of either the microtubule network or TsuA yields similar phenotypes. While the microtubule networks were asymmetric in polarized cells, there was not an additional preferential distribution of TsuA-YFP to either the front or back. It remains to be determined whether TsuA activity is spatially restricted in a manner that could asymmetrically regulate actin polymerization. An important avenue for future research will be to identify how the kinase is regulated and what its substrates are. In light of our studies, we are currently seeking substrates associated with the microtubule network. The critical association of TsuA with the microtubule network may provide important future insights that may be generally applicable to other systems.
Novel role for Fused kinases
Our finding that TsuA regulates polarity and the suppression of lateral pseudopods in D. discoideum suggests a role distinct from Hedgehog signaling and adds to the growing evidence that Fused kinases play divergent roles in different organisms. In D. melanogaster, Fused is a positive mediator of Hedgehog (Hh) signaling (Preat et al. 1990; Varjosalo et al. 2006; Varjosalo and Taipale 2007). In the absence of Hh, a complex consisting of Costal2 (Cos2), Fused, and Suppressor of Fused has been reported to associate with microtubules (Robbins et al. 1997). Interestingly, in in vitro assays, the C terminus but not kinase activity is required for interaction with microtubules. The interaction with microtubules is thought to prevent activation of the pathway by sequestering the complex in the absence of Hh ligand. This sequestering role for microtubule association is in contrast to our data suggesting that the enzyme functions in association with microtubules.
Studies in other systems also suggest that Fused acts unconventionally. In mice, normal embryonic patterning indicates that the Hh pathway is not affected by loss of Fused (Wolff et al. 2003; Chen et al. 2005; Merchant et al. 2005). Instead, the knockout mice fail to thrive after birth and die after ∼2 wk due to hydrocephaly. A recent study in Arabidopsis showed that a mutation in the fused gene causes a cytokinesis defect (Oh et al. 2005). Although very different, these diverse functions could be linked by an association with microtubules. One of the hypotheses proposed by Merchant et al. (2005) that may implicate Fused in regulation of microtubule function is that the hydrocephaly in mice may have a defect in the ciliated tissues that regulate the flow of cerebrospinal fluid. The cytokinesis defect in Arabidopsis may also result from a failure to properly regulate microtubules, since in this organism, Fused localizes to the phragmoplast, a plant-specific structure associated with microtubule-based transport toward the new cell wall. Despite the many suggestions for an association with microtubules, however, ours is the first study to show the localization of a Fused kinase to microtubules in living cells and demonstrate that localization is required for function. Since Fused has been conserved in many organisms that lack the Hh signal transduction pathway, we propose that interactions with microtubules may be a more primitive function of this protein that has been adapted in different organisms.
Materials and methods
Cell culture, development, and chemotaxis assay
The parental line for the REMI screen was DH1, an uracil auxotrophic derivative of axenic laboratory strain Ax3. The parental line for the targeted knockout, tsuA−, was Ax3. Plasmids were introduced into cells by electroporation, and stable transformants were selected with 10 mg/L Blasticidin S sulfate or 20 mg/L G418 sulfate. All strains were grown in HL5 medium at 22°C.
For the nonnutrient agar development assay, exponentially growing cells were rinsed with DB buffer (10 mM phosphate buffer, 2 mM MgSO4, 0.2 mM CaCl2) once and then plated on 1.5% nonnutrient DB agar and 1.5% agar at 1.25 × 106 cells per square centimeter. Time-lapse images were captured using a Nikon Optiphot microscope with 4× magnification. An Olympus SZ-PT dissection scope and a Nikon Coolpix 4700 camera with an adaptor were used for still micrographs.
To monitor the chemotactic motility of wild-type and tsuA− cells in chimeric aggregation streams, development was initiated with a monolayer of 80% unlabeled cells and mixed with 10% GFP-expressing tsuA− cells and 10% mRFP-expressing wild-type cells on glass-bottom Petri dishes (Wilco Wells). After adhering for 20 min, a 1.5% agarose gel made up in a second dish was then added and allowed to settle onto the cells. The excess buffer was then aspirated, and the cells were incubated for about 6 h. Cells were imaged at 60-sec intervals using a Leica SP2 confocal microscope and a dry 20× (0.4 NA) objective. Sequential scanning was used to image the tsuA−-GFP cells with the 488-nm laser and the Ax2-mRFP cells with the 543-nm laser. Images were analyzed by ImageJ using the mTrackJ plug-in (Erik Meijering, Erasmus MC, University Medical Center, Rotterdam) to generate position, speed, and velocity data for each of eight to 10 cells. Persistence was calculated as the ratio of the net distance to the total distance moved.
Chemotactic movements of cells to a micropipette containing cAMP were performed as described in Parent et al. (1998). D. discoideum cells were harvested, rinsed with DB (DB; 10 mM phosphate buffer, 2 mM MgSO4, 0.2 mM CaCl2), resuspended in DB at 2 × 107 cells per milliliter, and put on a shaker at 100 rpm. After 1 h, the cell suspension was pulsed with doses of 50 nM cAMP (Sigma) every 6 min. Samples were taken at the indicated time points, and cells were lysed in sample buffer (3% SDS, 31 mM Tris-HCl at pH 6.8, 5% glycerol, 32 mM DTT, 0.05% bromophenol blue) in order to analyze cAR1 and ACA expression by Western blotting. Cells were spotted onto chambered coverglasses (Lab-tek, NalgeNunc). Once cells were attached, the chambers were filled with 3 mL of DB and presented with a micropipette (Femtotip) filled with 10 μM cAMP was positioned near the cells, and images of moving cells were recorded using a Zeiss inverted microscope (Axiovert 100, Carl Zeiss) fitted with a CCD camera (CoolSNAP, Roper Photometrics) and IPlab software. Adobe Photoshop and ImageJ software packages were used to process data. For the folic acid assay, cells were plated with K. aerogenes on SM agar and incubated for 40 h at 22°C. The amoebae were harvested from the partially cleared bacterial lawn by scraping into DB buffer and washed four times by centrifugation. Cells were then spotted onto the chambered coverglass and presented with a micropipette containing 1 M folic acid (Sigma). Images were collected as above.
Microscopy
To determine the three-dimensional organization of TsuA-YFP in living cells, cells were spotted onto cover slip chambers in DB. Z-section micrographs throughout the cell volume were taken using an Olympus microscope and 60× (NA 1.45) objective with a 1.63× optivar, and deconvolution was performed using Metamorph software (Molecular Devices). Time lapse images and still micrographs were also acquired using an Olympus IX81 microscope with 40× (NA1.3; oil), 60× (NA 1.4; oil), and 100× (NA 1.4; oil) UIS2 objectives. Images were captured with a C9100-02 CCD camera (Hamamatsu Photonics) and IPlab software and processed using IPlab and Adobe Photoshop.
For PHCRAC-GFP membrane translocation studies, wild-type and tsuA− cells were starved for ∼6 h and plated on cover slip chambers. The cells were then stimulated with 1 μM cAMP, and GFP fluorescence was recorded by time-lapse microscopy. To enhance the detection of membrane translocation, images were processed by subtracting from each frame a “blurred” image of the same frame. This subtracts the non-membrane-bound signal, leaving the membrane-associated signal that was then plotted against time to show the dynamics of membrane translocation.
Quantification of chemotaxis and motility
To quantify chemotaxis, we determined the velocity, chemotactic index, directional persistence, and roundness of cells in time-lapse videos, as described previously (Kabacoff et al. 2007). Analysis was performed using a MATLAB Image Processing Toolbox (Mathworks). Individual cells were first segmented and tracked during the course of each video. Branches were classified as extensions or retractions depending on whether they pointed toward regions in which the cell shape was expanding or shrinking. The directions of pseudopod extensions and retractions were defined by the angle formed between a line drawn from the cell center to the needle and the line formed by the cell center and the extending or retracting tip.
Cloning of tsuA and expression constructs
The full-length tsuA gene was assembled by cloning PCR fragments generated using Ax3 genomic DNA as a template and the Phusion DNA polymerase (New England Biolabs). First, Fragment 1 (1–1319 bp) was cloned into pBluescript SKII and then linked to Fragment 2 (1232–2088 bp) by cloning into the internal SwaI site present in the overlap between the two fragments. Fragment 3 (1984–5854 bp) was added using the shared EcoRI site and Fragment 4 (5641–7104 bp) was added using the shared HindIII site. To make the YFP fusion construct, a YFP ORF was generated by PCR that introduced at the 5′ end the sequence matching that of 7090–7104 bp of tsuA, including the internal BglII at 7090 bp, and a XhoI site at the 3′ end. The YFP tag was then fused to tsuA by cloning into the BglII and XhoI sites. The full-length constructs were then transferred to the D. discoideum expression vector pB18X using BamHI and XhoI. The K34R mutant constructs were made by mutagenizing Fragment 1 by the Stratagene QuikChange method and cloning as above. The ΔTH-YFP construct was made inserting the YFP tag at the endogenous HindIII site at 5777 bp. The KD-YFP and TH-YFP constructs were made by fusing the PCR products corresponding to 1–1126 bp and 5660–7120 bp, respectively, to YFP in pB18X.
Acknowledgments
We thank members of the Devreotes, Robinson, and Iglesias laboratories for useful comments. This research was supported by the National Institutes of Health grants GM28007 (to P.N.D.), GM34933 (to P.N.D.), GM40599 (to D.A.K.), and NIGMS 71920 (to P.A.I.).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1694508.
References
- Bagorda A., Mihaylov V.A., Parent C.A. Chemotaxis: Moving forward and holding on to the past. Thromb. Haemost. 2006;95:12–21. [PubMed] [Google Scholar]
- Basu R., Chang F. Shaping the actin cytoskeleton using microtubule tips. Curr. Opin. Cell Biol. 2007;19:88–94. doi: 10.1016/j.ceb.2006.12.012. [DOI] [PubMed] [Google Scholar]
- Booth E.O., Van Driessche N., Zhuchenko O., Kuspa A., Shaulsky G. Microarray phenotyping in Dictyostelium reveals a regulon of chemotaxis genes. Bioinformatics. 2005;21:4371–4377. doi: 10.1093/bioinformatics/bti726. [DOI] [PubMed] [Google Scholar]
- Bretschneider T., Diez S., Anderson K., Heuser J., Clarke M., Muller-Taubenberger A., Kohler J., Gerisch G. Dynamic actin patterns and Arp2/3 assembly at the substrate-attached surface of motile cells. Curr. Biol. 2004;14:1–10. doi: 10.1016/j.cub.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Brito D.A., Strauss J., Magidson V., Tikhonenko I., Khodjakov A., Koonce M.P. Pushing forces drive the comet-like motility of microtubule arrays in Dictyostelium. Mol. Biol. Cell. 2005;16:3334–3340. doi: 10.1091/mbc.E05-01-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Janetopoulos C., Huang Y.E., Iijima M., Borleis J., Devreotes P.N. Two phases of actin polymerization display different dependencies on PI(3,4,5)P3 accumulation and have unique roles during chemotaxis. Mol. Biol. Cell. 2003;14:5028–5037. doi: 10.1091/mbc.E03-05-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.H., Gao N., Kawakami T., Chuang P.T. Mice deficient in the fused homolog do not exhibit phenotypes indicative of perturbed hedgehog signaling during embryonic development. Mol. Cell. Biol. 2005;25:7042–7053. doi: 10.1128/MCB.25.16.7042-7053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devreotes P., Janetopoulos C. Eukaryotic chemotaxis: Distinctions between directional sensing and polarization. J. Biol. Chem. 2003;278:20445–20448. doi: 10.1074/jbc.R300010200. [DOI] [PubMed] [Google Scholar]
- Dharmawardhane S., Warren V., Hall A.L., Condeelis J. Changes in the association of actin-binding proteins with the actin cytoskeleton during chemotactic stimulation of Dictyostelium discoideum. Cell Motil. Cytoskeleton. 1989;13:57–63. doi: 10.1002/cm.970130107. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., Hall A. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCζ. Cell. 2001;106:489–498. doi: 10.1016/s0092-8674(01)00471-8. [DOI] [PubMed] [Google Scholar]
- Feierbach B., Verde F., Chang F. Regulation of a formin complex by the microtubule plus end protein tea1p. J. Cell Biol. 2004;165:697–707. doi: 10.1083/jcb.200403090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franca-Koh J., Kamimura Y., Devreotes P. Navigating signaling networks: Chemotaxis in Dictyostelium discoideum. Curr. Opin. Genet. Dev. 2006;16:333–338. doi: 10.1016/j.gde.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Futrelle R.P., Traut J., McKee W.G. Cell behavior in Dictyostelium discoideum: Preaggregation response to localized cyclic AMP pulses. J. Cell Biol. 1982;92:807–821. doi: 10.1083/jcb.92.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbeter A., Segel L.A. Control of developmental transitions in the cyclic AMP signalling system of Dictyostelium discoideum. Differentiation. 1980;17:127–135. doi: 10.1111/j.1432-0436.1980.tb01090.x. [DOI] [PubMed] [Google Scholar]
- Gomes E.R., Jani S., Gundersen G.G. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 2005;121:451–463. doi: 10.1016/j.cell.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Hall A.L., Schlein A., Condeelis J. Relationship of pseudopod extension to chemotactic hormone-induced actin polymerization in amoeboid cells. J. Cell. Biochem. 1988;37:285–299. doi: 10.1002/jcb.240370304. [DOI] [PubMed] [Google Scholar]
- Hestermann A., Graf R. The XMAP215-family protein DdCP224 is required for cortical interactions of microtubules. BMC Cell Biol. 2004;5:24. doi: 10.1186/1471-2121-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias P.A., Devreotes P.N. Navigating through models of chemotaxis. Curr. Opin. Cell Biol. 2008;20:35–40. doi: 10.1016/j.ceb.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Iijima M., Devreotes P. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell. 2002;109:599–610. doi: 10.1016/s0092-8674(02)00745-6. [DOI] [PubMed] [Google Scholar]
- Kabacoff C., Xiong Y., Musib R., Reichl E.M., Kim J., Iglesias P.A., Robinson D.N. Dynacortin facilitates polarization of chemotaxing cells. BMC Biol. 2007;5:53. doi: 10.1186/1741-7007-5-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krendel M., Zenke F.T., Bokoch G.M. Nucleotide exchange factor GEF-H1 mediates cross-talk between microtubules and the actin cytoskeleton. Nat. Cell Biol. 2002;4:294–301. doi: 10.1038/ncb773. [DOI] [PubMed] [Google Scholar]
- Kuspa A. Restriction enzyme-mediated integration (REMI) mutagenesis. Methods Mol. Biol. 2006;346:201–209. doi: 10.1385/1-59745-144-4:201. [DOI] [PubMed] [Google Scholar]
- Martin S.G., McDonald W.H., Yates J.R., Chang F. Tea4p links microtubule plus ends with the formin for3p in the establishment of cell polarity. Dev. Cell. 2005;8:479–491. doi: 10.1016/j.devcel.2005.02.008. [DOI] [PubMed] [Google Scholar]
- McRobbie S.J., Newell P.C. Changes in actin associated with the cytoskeleton following chemotactic stimulation of Dictyostelium discoideum. Biochem. Biophys. Res. Commun. 1983;115:351–359. doi: 10.1016/0006-291x(83)91011-2. [DOI] [PubMed] [Google Scholar]
- Merchant M., Evangelista M., Luoh S.M., Frantz G.D., Chalasani S., Carano R.A., van Hoy M., Ramirez J., Ogasawara A.K., McFarland L.M., et al. Loss of the serine/threonine kinase fused results in postnatal growth defects and lethality due to progressive hydrocephalus. Mol. Cell. Biol. 2005;25:7054–7068. doi: 10.1128/MCB.25.16.7054-7068.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niggli V. Microtubule-disruption-induced and chemotactic-peptide-induced migration of human neutrophils: Implications for differential sets of signalling pathways. J. Cell Sci. 2003;116:813–822. doi: 10.1242/jcs.00306. [DOI] [PubMed] [Google Scholar]
- Oh S.A., Johnson A., Smertenko A., Rahman D., Park S.K., Hussey P.J., Twell D. A divergent cellular role for the FUSED kinase family in the plant-specific cytokinetic phragmoplast. Curr. Biol. 2005;15:2107–2111. doi: 10.1016/j.cub.2005.10.044. [DOI] [PubMed] [Google Scholar]
- Parent C.A., Blacklock B.J., Froehlich W.M., Murphy D.B., Devreotes P.N. G protein signaling events are activated at the leading edge of chemotactic cells. Cell. 1998;95:81–91. doi: 10.1016/s0092-8674(00)81784-5. [DOI] [PubMed] [Google Scholar]
- Pegtel D.M., Ellenbroek S.I., Mertens A.E., van der Kammen R.A., de Rooij J., Collard J.G. The Par–Tiam1 complex controls persistent migration by stabilizing microtubule-dependent front–rear polarity. Curr. Biol. 2007;17:1623–1634. doi: 10.1016/j.cub.2007.08.035. [DOI] [PubMed] [Google Scholar]
- Preat T., Therond P., Lamour-Isnard C., Limbourg-Bouchon B., Tricoire H., Erk I., Mariol M.C., Busson D. A putative serine/threonine protein kinase encoded by the segment-polarity fused gene of Drosophila. Nature. 1990;347:87–89. doi: 10.1038/347087a0. [DOI] [PubMed] [Google Scholar]
- Rehberg M., Kleylein-Sohn J., Faix J., Ho T.H., Schulz I., Graf R. Dictyostelium LIS1 is a centrosomal protein required for microtubule/cell cortex interactions, nucleus/centrosome linkage, and actin dynamics. Mol. Biol. Cell. 2005;16:2759–2771. doi: 10.1091/mbc.E05-01-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A.J., Schwartz M.A., Burridge K., Firtel R.A., Ginsberg M.H., Borisy G., Parsons J.T., Horwitz A.R. Cell migration: Integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Robbins D.J., Nybakken K.E., Kobayashi R., Sisson J.C., Bishop J.M., Therond P.P. Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein costal2. Cell. 1997;90:225–234. doi: 10.1016/s0092-8674(00)80331-1. [DOI] [PubMed] [Google Scholar]
- Rodriguez O.C., Schaefer A.W., Mandato C.A., Forscher P., Bement W.M., Waterman-Storer C.M. Conserved microtubule–actin interactions in cell movement and morphogenesis. Nat. Cell Biol. 2003;5:599–609. doi: 10.1038/ncb0703-599. [DOI] [PubMed] [Google Scholar]
- Sasaki A.T., Chun C., Takeda K., Firtel R.A. Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J. Cell Biol. 2004;167:505–518. doi: 10.1083/jcb.200406177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satulovsky J., Lui R., Wang Y.L. Exploring the control circuit of cell migration by mathematical modeling. Biophys. J. 2008;94:3671–3683. doi: 10.1529/biophysj.107.117002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll D.R., Wessels D., Heid P.J., Zhang H. A contextual framework for characterizing motility and chemotaxis mutants in Dictyostelium discoideum. J. Muscle Res. Cell Motil. 2002;23:659–672. doi: 10.1023/a:1024459124427. [DOI] [PubMed] [Google Scholar]
- Sroka J., Madeja Z., Galanty A., Michalik M., Przestalski S., Rakoczy L., Korohoda W. Trimethyltin inhibits the chemotaxis of Dictyostelium discoideum amoebae. Eur. J. Protistol. 2001;37:313–326. [Google Scholar]
- Tillinghast H.S., Newell P.C. Chemotaxis towards pteridines during development of Dictyostelium. J. Cell Sci. 1987;87:45–53. doi: 10.1242/jcs.87.1.45. [DOI] [PubMed] [Google Scholar]
- Ueda M., Graf R., MacWilliams H.K., Schliwa M., Euteneuer U. Centrosome positioning and directionality of cell movements. Proc. Natl. Acad. Sci. 1997;94:9674–9678. doi: 10.1073/pnas.94.18.9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varjosalo M., Taipale J. Hedgehog signaling. J. Cell Sci. 2007;120:3–6. doi: 10.1242/jcs.03309. [DOI] [PubMed] [Google Scholar]
- Varjosalo M., Li S.P., Taipale J. Divergence of hedgehog signal transduction mechanism between Drosophila and mammals. Dev. Cell. 2006;10:177–186. doi: 10.1016/j.devcel.2005.12.014. [DOI] [PubMed] [Google Scholar]
- Vasiliev J.M., Gelfand I.M., Domnina L.V., Ivanova O.Y., Komm S.G., Olshevskaja L.V. Effect of colcemid on the locomotory behaviour of fibroblasts. J. Embryol. Exp. Morphol. 1970;24:625–640. [PubMed] [Google Scholar]
- Weiner O.D., Neilsen P.O., Prestwich G.D., Kirschner M.W., Cantley L.C., Bourne H.R. A PtdInsP(3)- and Rho GTPase-mediated positive feedback loop regulates neutrophil polarity. Nat. Cell Biol. 2002;4:509–513. doi: 10.1038/ncb811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard S.S., Devreotes P.N. Signaling pathways mediating chemotaxis in the social amoeba, Dictyostelium discoideum. Eur. J. Cell Biol. 2006;85:897–904. doi: 10.1016/j.ejcb.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Wittmann T., Waterman-Storer C.M. Cell motility: Can Rho GTPases and microtubules point the way? J. Cell Sci. 2001;114:3795–3803. doi: 10.1242/jcs.114.21.3795. [DOI] [PubMed] [Google Scholar]
- Wittmann T., Bokoch G.M., Waterman-Storer C.M. Regulation of leading edge microtubule and actin dynamics downstream of Rac1. J. Cell Biol. 2003;161:845–851. doi: 10.1083/jcb.200303082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff C., Roy S., Ingham P.W. Multiple muscle cell identities induced by distinct levels and timing of hedgehog activity in the zebrafish embryo. Curr. Biol. 2003;13:1169–1181. doi: 10.1016/s0960-9822(03)00461-5. [DOI] [PubMed] [Google Scholar]
- Wu L.J., Devreotes P.N. Dictyostelium transiently expresses eight distinct G-protein α-subunits during its developmental program. Biochem. Biophys. Res. Commun. 1991;179:1141–1147. doi: 10.1016/0006-291x(91)91690-e. [DOI] [PubMed] [Google Scholar]
- Xu J., Wang F., Van Keymeulen A., Rentel M., Bourne H.R. Neutrophil microtubules suppress polarity and enhance directional migration. Proc. Natl. Acad. Sci. 2005;102:6884–6889. doi: 10.1073/pnas.0502106102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Van Keymeulen A., Wakida N.M., Carlton P., Berns M.W., Bourne H.R. Polarity reveals intrinsic cell chirality. Proc. Natl. Acad. Sci. 2007;104:9296–9300. doi: 10.1073/pnas.0703153104. [DOI] [PMC free article] [PubMed] [Google Scholar]