Abstract
The AKT–mTOR pathway harbors several known and putative oncogenes and tumor suppressors. In a phenotypic screen for lymphomagenesis, we tested candidate genes acting upstream of and downstream from mTOR in vivo. We find that Rheb, a proximal activator of mTORC1, can produce rapid development of aggressive and drug-resistant lymphomas. Rheb causes mTORC1-dependent effects on apoptosis, senescence, and treatment responses that resemble those of Akt. Moreover, Rheb activity toward mTORC1 requires farnesylation and is readily blocked by a pharmacological inhibitor of farnesyltransferase (FTI). In Pten-deficient tumor cells, inhibition of Rheb by FTI is responsible for the drug’s anti-tumor effects, such that a farnesylation-independent mutant of Rheb renders these tumors resistant to FTI therapy. Notably, RHEB is highly expressed in some human lymphomas, resulting in mTORC1 activation and increased sensitivity to rapamycin and FTI. Downstream from mTOR, we examined translation initiation factors that have been implicated in transformation in vitro. Of these, only eIF4E was able to enhance lymphomagenesis in vivo. In summary, the Rheb GTPase is an oncogenic activity upstream of mTORC1 and eIF4E and a direct therapeutic target of farnesyltransferase inhibitors in cancer.
Keywords: Mouse model, Akt, translation, oncogenesis
The PI3K/AKT pathway is a frequent target for mutational activation in human cancer. Several genetic lesions upstream of mTOR have been identified that can activate the AKT pathway, and these include mutations in PI3k, AKT/PKB, or loss of the tumor suppressors PTEN, TSC1, or TSC2 (for review, see Vivanco and Sawyers 2002). These upstream lesions affect a variety of cellular processes including protein translation via activation of mTOR. Similarly, translation factors acting downstream from mTOR are highly expressed in many cancers and sometimes correlate with poor prognosis (Mamane et al. 2006). Moreover, expression of individual translation factors can transform rodent fibroblasts, and, in particular, the eIF4E initiation factor can drive tumor formation in vivo (Lazaris-Karatzas et al. 1990; Ruggero et al. 2004; Wendel et al. 2004). Thus, genetic lesions acting both upstream of and downstream from mTOR can promote tumorigenesis and are important in human cancer.
mTOR integrates protein translation with cellular nutrient status and growth signals. The mTOR kinase can participate in two biochemically and functionally distinct complexes, mTORC1 and mTORC2. The mTORC1 complex is sensitive to inhibition with rapamycin and signals downstream to activate translation initiation. The second complex, mTORC2, is resistant to rapamycin and signals upstream to activate Akt via Ser 473 phosphorylation (for review, see Guertin and Sabatini 2007). The GTPase Rheb (Ras homolog enriched in brain) is a proximal activator of mTORC1 and translation initiation. In addition, Rheb has the opposite effect on mTORC2 and produces inhibition of the upstream Akt pathway, most likely this involves a feedback mechanism (Tee et al. 2002; Inoki et al. 2003).
The mTORC1 complex activates key components of the translation initiation machinery. For example, mTORC1 acts to liberate the eIF4E and eIF4A initiation factors from their inhibitory binding proteins, thus allowing them to enter the initiation complex (Hay and Sonenberg 2004; Dorrello et al. 2006). While eIF4E is considered the rate-limiting factor, efficient translation requires that eIF4E act in concert with other factors. These include the RNA helicase eIF4A and its activator eIF4B, which act to unwind the 5′ untranslated region (UTR) of certain mRNAs. The eIF4G protein serves as a scaffold for assembly of the initiation complex (for review, see Sonenberg and Hinnebusch 2007), and eIF2α is a key negative regulator that can block translation in response to phosphorylation by PKR (Protein kinase RNA-activated) and other kinases (Clemens 2004). The eIF4E translation factor can affect general translation, but most importantly, eIF4E facilitates the translation of subsets of mRNAs characterized by complex and structured 5′UTRs, and these include transformation-relevant genes and regulators of apoptosis like Bcl-xL, Mcl1 or Survivin (Polunovsky et al. 2000; Mamane et al. 2004, 2007; Larsson et al. 2007; Silva and Wendel 2007).
The significance of genetic lesions acting upstream in the Akt pathway in cancer is established (Vivanco and Sawyers 2002). In contrast, the malignant potential of direct activation of mTORC1 in the absence of additional Akt signals is controversial. For example, increased expression or mutational activation of mTOR has not been reported in cancer, and moreover, activation of mTORC1 can induce feedback mechanisms that block pro-oncogenic signals emanating from Akt (Manning et al. 2005).
Downstream from Rheb and mTORC1, the eIF4E translation factor can act as an oncogene in vitro and in vivo; moreover, its expression has been associated with poor prognosis in human cancer (Lazaris-Karatzas et al. 1990; Ruggero et al. 2004; Wendel et al. 2004). A similar in vitro transforming activity has been proposed for other translation factors as well; however, clear in vivo evidence of their oncogenic activity is lacking. Examples include the eIF4G factor and also a mutant form of eIF2α/S51A, which eludes regulation by the PKR kinase. Enforced expression of these proteins can cause malignant transformation of rodent cells. Also, the RNA helicase eIF4A is controlled by the tumor suppressor gene Pdcd4 (programmed cell death-4) and has been implicated in cancer (Donze et al. 1995; Fukuchi-Shimogori et al. 1997; Avdulov et al. 2004; Zakowicz et al. 2005; Hilliard et al. 2006).
We previously examined the oncogenic activities of Akt and eIF4E using the Eμ-Myc mouse lymphoma model and adoptive transfer of retrovirally modified hematopoietic progenitor cells (HPCs) (Wendel et al. 2004, 2007). We have now chosen a candidate screening approach to assess the in vivo oncogenic function of certain factors acting either upstream of or downstream from mTOR. Our results identify the tumor-promoting ability of Rheb and direct mTORC1 activation and pinpoint the initiation factor eIF4E as the key oncogenic activity downstream from mTORC1. Moreover, Rheb activity depends on farnesylation, and in some tumors, inhibition of Rheb mediates the anti-tumor effect of farnesyltransferase inhibitors (FTIs).
Results
We selected genes acting upstream of and downstream from mTOR that have been implicated in transformation in vitro to assess their in vivo oncogenic potential. For example, we tested direct mTORC1 activation by different Rheb mutants and examined a series of translation factors. Specifically, we looked at myristoylated Akt (Akt) (Wendel et al. 2004), wild-type Rheb, Rheb/Q64L, a previously characterized mutant with twofold increased activity toward mTORC1, and Rheb/5A, an inactive Rheb mutant (Inoki et al. 2003). Downstream from mTOR, we tested the translation initiation factors eIF4E, eIF4GI, eIF4AI, eIF4B, and eIF2α/S51A, a mutant of eIF2α that disables the PKR phosphorylation site (Donze et al. 1995). The cDNAs were cloned in an identical manner into the MSCV-IRES-GFP retroviral vector. Efficient protein expression from the constructs was confirmed in 3T3 fibroblasts (Supplemental Fig. 2A,B) and the resulting tumors (Fig. 1D).
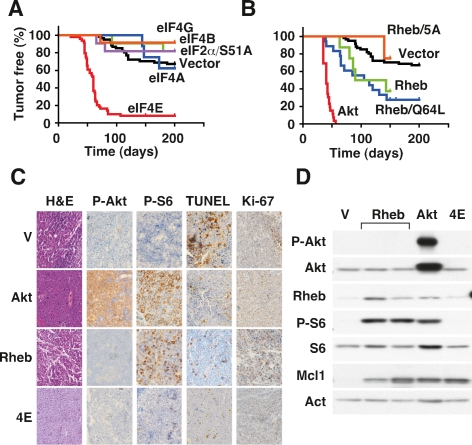
Figure 1.
In vivo candidate screen for lymphomagenesis identifies the tumorigenic activity of Rheb GTPase. (A,B) Kaplan-Meier plots showing the latency to tumor development in mice reconstituted with retrovirally transduced Eμ-Myc HPCs, where day 0 is the day of HPC transplantation. A shows eIF4E (red; n = 50), eIF4B (green; n = 11), eIF4AI (blue; n = 8), eIF2α/S51A (purple; n = 11), eIF4GI (orange; n = 12) and vector control (black; n = 62); includes mice analyzed in parallel and historic controls.B shows myrAkt (Akt; red; n = 24), Rheb (green; n = 9); Rheb/Q64L (blue; n = 18), Rheb/5A (orange; n = 6), and vector control (as in A). (C) Representative micrographs of the indicated Eμ-Myc lymphomas (Control: V; Rheb/Q64L: Rheb; myrAkt: Akt; eIF4E: 4E) stained with hematoxylin and eosin (H&E), antibodies against phosphorylated Akt (Ser473) (P-Akt), phosphorylated ribosomal S6 protein (Ser240/244) (P-S6), or TUNEL and Ki-67. (D) Immunoblot of lysates prepared from Eμ-Myc lymphomas (V) and Eμ-Myc lymphomas expressing Rheb/Q64L (Rheb), myrAkt (Akt), and eIF4E (4E) probed with the indicated antibodies.
In order to test the ability of these candidate genes to promote lymphomagenesis, we used adoptive transfer of virally transduced Eμ-Myc transgenic HPCs into lethally irradiated, wild-type, syngeneic animals, monitored the animals by palpation and blood analysis, and tested arising tumors for expression of the GFP reporter (schematic in Supplemental Fig. 1; Wendel et al. 2004).
As expected downstream from mTOR, eIF4E was able to promote tumorigenesis in vivo (Fig. 1A; Wendel et al. 2004). In contrast, the other translation factors—eIF4GI, eIF4AI, eIF4B, and eIF2α/S51A—were unable to produce an acceleration of tumor development compared with vector controls (Vector vs. 4G, 4A, 4B, 2α; P > 0.5) (Fig. 1A). Nearly all tumors arising from eIF4E-transduced HPCs expressed the GFP reporter, while the few tumors that arose in the other animals were uniformly GFP negative, indicating that they had arisen from nontransduced Eμ-Myc HPCs (Supplemental Fig. 3). Apparently, eIF4E is unique among the translation initiation factors we analyzed in its ability to drive tumor development in vivo.
The small GTPase Rheb was able to promote lymphomagenesis in vivo and clearly had oncogenic potential. Mice receiving HPCs expressing wild-type Rheb or Rheb/Q64L, an allele with increased activity toward mTORC1, developed tumors with high penetrance and short latency compared with vector controls or an inactive Rheb mutant (Rheb/5A) (Fig. 1B) (P < 0.0001 for Rheb and Rheb/Q64L vs. Vector or Rheb/5A). The tumorigenic potency of Rheb was less than that of myristoylated Akt, and Rheb/Q64L-induced tumors with a median latency of 109.5 d compared with only 40 d for myristoylated Akt (Rheb/Q64L vs. Akt, P < 0.001). Moreover, Rheb- and Akt-expressing HPCs had a selective advantage and were enriched during tumorigenesis. For example, with only ∼25% of HPCs infected with the retroviral constructs coexpressing the GFP reporter (data not shown), we observed that tumors arising from Akt-expressing HPCs were almost invariably GFP-positive. Similarly, Rheb and Rheb/Q64L expression resulted in 67% and 80% GFP-positive tumors compared with 0%–23% for vector and other constructs (Supplemental Fig. 3). Thus, Rheb can act as an oncogene and cooperate with c-Myc in tumorigenesis in vivo.
Next we wanted to explore the mechanisms and consequences of Rheb action in cancer. First, surface marker expression revealed that Rheb-driven lymphoma had a mature B-cell phenotype, marked by B220, CD19, and IgM expression, which most closely resembled tumors expressing eIF4E and was clearly distinct from Akt-driven lymphoma (Supplemental Table 1). Consistent with Rheb’s ability to activate mTORC1 and not mTORC2, the Rheb-expressing tumors showed extensive phosphorylation of ribosomal S6 protein without detectable Ser 473 phosphorylation of Akt (Fig. 1C). All lymphomas were highly proliferative and irrespective of genotype >80% of viable lymphoma cells stained positive for Ki-67. Moreover, Rheb tumors resembled the tumors expressing Akt or eIF4E in that they displayed a very low frequency of apoptotic cells as determined by TUNEL assay (Fig. 1C). Immunoblot analysis confirmed the signaling effects of Rheb and Akt in vivo and showed high expression of the anti-apoptotic Mcl1 protein, whose translation is controlled in part by mTORC1 and eIF4E (Fig. 1D; Wendel et al. 2007). Endogenous Rheb protein levels were somewhat heterogeneous across tumors and did not appear to correlate with Akt pathway activating lesions in vivo or expression of Akt in vitro (Supplemental Fig. 4A,B). Thus, Rheb activates mTORC1 in vivo and can suppress apoptosis in a manner comparable with Akt or eIF4E.
c-Myc initiated lymphomagenesis is largely limited by apoptosis (Strasser et al. 1990; Schmitt et al. 2002). The rapid onset of Rheb-expressing tumors, their low levels of apoptosis, and high expression of the anti-apoptotic Mcl1 protein suggest that Rheb may promote tumor formation by opposing c-Myc-induced apoptosis. We wanted to directly examine how Rheb affected apoptosis in hematopoietic cells where apoptosis can be triggered by cytokine withdrawal (Plas et al. 2001; Edinger and Thompson 2004). Briefly, we introduced vectors coexpressing either Rheb, Akt, or Mcl1 along with a GFP reporter into populations of FL5-12 cells such that the transduced cells would compete with nontransduced cells. Upon cytokine depletion, we observed massive cell death and concomitant enrichment in the populations transduced with Rheb, Akt, or Mcl1 but not vector controls (Fig. 2A). Next we tested if Rheb could oppose apoptosis in c-Myc-expressing mouse embryo fibroblasts (MEFs), where apoptosis is readily triggered by serum withdrawal (Evan et al. 1992). Analysis of TUNEL (Fig. 2B) and propidium iodide uptake by flow cytometry (Supplemental Fig. 5A,B) revealed that MEFs coinfected with c-Myc and Rheb, Akt, or Mcl1 were relatively protected from apoptosis compared with vector controls. Importantly, Rheb’s ability to block apoptosis required activation of mTORC1 and treatment with 100 nM rapamycin completely reversed the anti-apoptotic effects of Rheb and Akt with little consequence in Mcl1-expressing cells (Fig. 2B; Supplemental Fig. 5A,B). Thus, in vitro, Rheb can oppose apoptosis triggered by factor depletion or c-Myc activation in an mTORC1-dependent and rapamycin-sensitive manner.
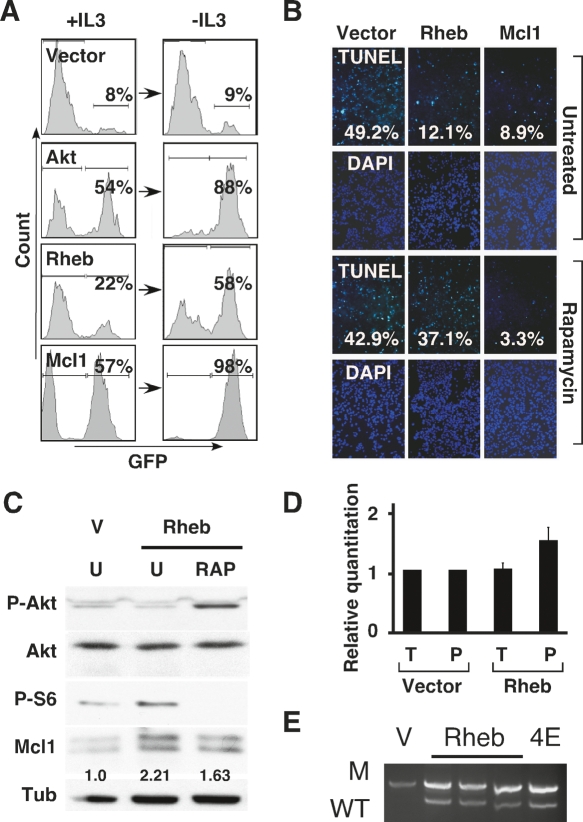
Figure 2.
Rheb opposes apoptosis in an mTORC1-dependent manner. (A) Flow cytometric analysis of mixed FL5-12 cell populations partially transduced with vectors coexpressing GFP (vector) and either myrAkt (Akt), Rheb/Q64L (Rheb), or Mcl1. Indicated are the percentages of GFP expressing cells before (+IL3) and after (−IL3) IL3-withdrawal. (B) TUNEL and DAPI stains of primary MEFs transduced with c-Myc and either vector, Rheb/Q64L (Rheb), or Mcl1 24 h after apoptosis was triggered by serum withdrawal in the presence or absence of 100 nM rapamycin; quantification of TUNEL positive cells is indicated. (C) Immunoblot of lysates prepared from MEFs transduced with empty vector or Rheb/Q64L (Rheb), either untreated (U) or rapamycin-treated (RAP), and probed with antibodies against phosphorylated Akt (Ser473), phosphorylated ribosomal S6 (Ser240/244), Mcl1, or tubulin. Indicated is the relative quantification of Mcl1 protein levels. (D) Quantitative real-time RT–PCR of Mcl1 normalized to β-actin from cDNA samples prepared from MEFs transduced with vector or Rheb/Q64L (Rheb). Shown is a “relative quantification” whereby the Mcl1 expression in triplicates of the total and polysome samples from vector transduced cells is set to 1 and compared with the total and polysome fraction from cells expressing Rheb/Q64L. (E) Allele-specific PCR to detect the wild-type p53 (WT) and mutant (M) allele in tumors derived from Eμ-Myc/p53+/− HSCs transduced with vector (V), Rheb/Q64L (Rheb) or eIF4E (4E).
The anti-apoptotic Bcl2 family protein, Mcl1, is controlled through several mechanisms including translation. Accordingly, Mcl1 is up-regulated in cells expressing eIF4E and, moreover, Mcl1 can massively accelerate lymhomagenesis in the Eμ-Myc lymphoma model (Wendel et al. 2007). Consistent with Rheb acting upstream of mTORC1 and eIF4E, we observed similar increases in Mcl1 protein expression in both Rheb- and eIF4E-expressing lymphomas (Fig. 1D). Acute expression of Rheb in MEFs produced twofold increased Mcl1 protein levels that were somewhat sensitive to rapamycin (Fig. 2C). As expected, rapamycin treatment completely blocked mTORC1 activation and phosphorylation of ribosomal S6 protein. Rapamycin treatment produced an increase in Akt phosphorylation that is consistent with previously reported feedback mechanisms and may involve S6 kinase and Irs1 (Jaeschke et al. 2002; Sun et al. 2005). Increased Akt activity can impede Mcl1 degradation and may explain the partial effect of rapamycin treatment on Mcl1 protein levels (Fig. 2C; Maurer et al. 2006). The ability of Rheb to increase Mcl1 protein was partly due to translation, since cells expressing Rheb contained on average 1.6-fold more Mcl1 mRNA in the polysome fraction compared with controls, and which remained unchanged in total mRNA (Fig. 2D). While small, this increase was consistent and resembled in magnitude the effect of eIF4E expression on Mcl1 translation (Wendel et al. 2007). Collectively, these data indicate that increased production of Mcl1 may contribute to Rheb’s anti-apoptotic action in vitro and in tumors.
To assess if Rheb could oppose c-Myc induced apoptosis in vivo, we tested whether Rheb could prevent p53 loss in lymphomas derived from p53+/− cells. Briefly, tumors arising from Eμ-Myc/p53+/− HPCs invariably undergo loss of heterozygosity (LOH) and inactivate the remaining p53 allele. Expression of potent anti-apoptotic activities, like Akt, Bcl-2, or eIF4E, obviates the need for p53 loss, and tumors remain heterozygous (Schmitt et al. 2002; Wendel et al. 2004). Rheb expression in Eμ-Myc/p53+/− HPCs produced tumors that retained the wild-type p53 allele and, like eIF4E-expressing cells, did not undergo LOH in the p53 locus (Fig. 2E). Thus, Rheb can attenuate the apoptotic effects of c-Myc activation in vivo in a manner comparable with Akt or eIF4E.
Cellular senescence is an important failsafe mechanism against transformation and is directly induced by certain oncogenes including Ras, Akt, and eIF4E (Serrano et al. 1997; Ruggero et al. 2004; Chen et al. 2005). We found Rheb can activate senescence in primary MEFs just like Akt or Ras. Rheb expression causes morpho logical changes (data not shown), induction of senescence-associated β-galactosidase (SA β-Gal) (Fig. 3A), early growth arrest (Vector vs. Rheb, P < 0.0001) (Fig. 3C), and p16 protein induction (Fig. 3D). As expected, these effects depended on p53 and did not occur in the context of c-Myc expression (Fig. 3A,C; Ruggero et al. 2004). Notably, pharmacologic inhibition of mTORC1 with rapamycin could prevent many phenotypic changes associated with senescence induced by Akt and Rheb and had a partial effect on Ras-induced senescence (Fig. 3B,D). Thus, Rheb can block c-Myc-induced apoptosis, and conversely, c-Myc may interfere with senescence activation by Rheb—likely, these interactions underlie the observed oncogenic cooperation of c-Myc and Rheb.
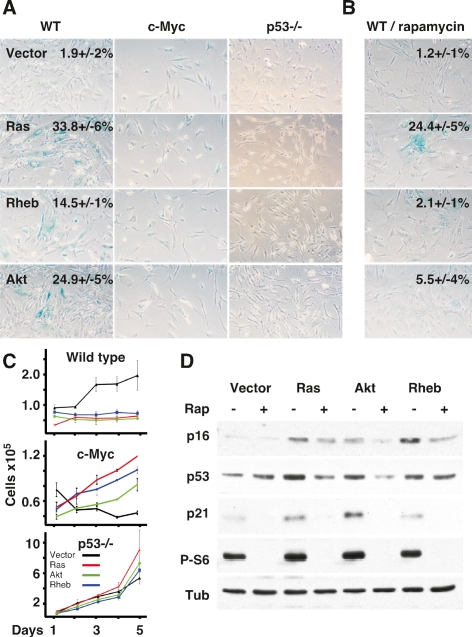
Figure 3.
Rheb induces cellular senescence in a p53, mTORC1-dependent, and c-Myc-sensitive manner. (A) Representative micrographs of wild-type (WT), p53−/−, and c-Myc-expressing MEFs transduced with vector, Ras/V12 (Ras), Rheb/Q64L (Rheb), or myrAkt (Akt) and stained for senescence-associated β-galactosidase (SA β-Gal) activity. The mean and standard deviation of the percentages of SA-β Gal-positive cells are indicated; no number indicates mean <1%. (B) Wild-type MEFs (WT) transduced with the same expression constructs and treated with 100 nM rapamycin. Mean and standard deviation (n = 3) of SA β-Gal-positive cells for untreated and rapamycin-treated wild-type MEFs is indicated. (C) Growth curves of wild-type, p53−/−-, and c-Myc-expressing MEFs expressing vector (black line), Rheb/Q64L (Rheb; blue line), Ras/V12 (Ras; red line), or myrAkt (Akt; green line). (D) Immunoblot of lysates prepared from wild-type MEFs transduced with the indicated alleles and either left untreated (−) or treated with 100 nM rapamycin (+) and probed for the indicated proteins.
Mutational activation of the AKT pathway can have profound effects on treatment responses in cancer (Dilling et al. 1994; Wendel et al. 2004). To examine how Rheb would affect the long-term effects of therapy in vivo, we transplanted tumors expressing Akt, Rheb, or chemo-sensitive controls (Eμ-Myc/Arf−/−) into recipient animals. Upon tumor formation, we initiated treatment with doxorubicin, a chemotherapeutic topoisomerase-2 poison and principle agent in lymphoma therapy, the mTORC1 inhibitor rapamycin, and a combination of both drugs (Wendel et al. 2004). We monitored mice for complete remissions, defined as the absence of detectable disease by palpation and blood smear, and determined the time to relapse (Supplemental Fig. 6) and survival to preterminal condition (Fig. 4). As expected, Akt reduced the effectiveness of treatment with doxorubicin, which induced complete remissions and prolonged survival in animals harboring control lymphoma (Fig. 4A). Rheb expression had similar effects on tumor-free survival as Akt (Control vs. Akt or Rheb, P = 0.007) (Supplemental Fig. 6A) and resulted in a significant survival difference compared with controls (Control vs. Akt, P = 0.02; Control vs. Rheb, P = 0.004) (Fig. 4A). Consistent with previous observations in Akt-expressing tumors (Wendel et al. 2004), single agent treatment with rapamycin was unable to produce significant effects in Rheb or Akt tumors and did not improve survival (Control vs. Akt or Rheb, P > 0.1 for tumor-free and overall survival) (Fig. 4B; Supplemental Fig. 6B). In contrast, combination treatment more than doubled the mean survival times in Akt (47 d) and Rheb (44 d) tumor-bearing mice compared with chemo-sensitive controls (21 d) (Control vs. Akt or Rheb, P < 0.0001) (Fig. 4C) and induced correspondingly longer remissions (Supplemental Fig. 6C). Thus, Rheb can reduce the effectiveness of chemotherapy, and this consequence of increased Rheb expression is readily reversed by inhibition of mTORC1.
Figure 4.

Rapamycin reverses Rheb-mediated resistance to chemotherapy. Kaplan-Meier analyses of survival to preterminal condition in mice that were injected with control (Eμ-Myc/Arf−/−; black lines), Rheb/Q64L (Rheb; green lines), and myrAkt lymphoma cells (Akt; red lines). Treatment was initiated upon detection of well-palpable tumors on day 0. (A) Doxorubicin treatment (10 mg/kg) on day 1 of mice bearing control (n = 19), Akt (n = 16), and Rheb (n = 8) lymphoma. (B) Rapamycin treatment (4 mg/kg) on days 1, 3, and 5 of control (n = 11), Akt (n = 14), and Rheb lymphoma (n = 8). (C) Combination therapy with doxorubicin (10 mg/kg on day 1) and rapamycin (4 mg/kg on days 1, 3, and 5) of mice harboring control (n = 22), Akt (n = 15), and Rheb (n = 10) tumors. Treatment with rapamycin and doxorubicin significantly prolonged survival in mice bearing Akt- and Rheb-expressing tumors.
Rheb is a homolog of Ras and requires post-translational prenylation for proper localization and activity (Castro et al. 2003; Buerger et al. 2006). However, unlike Ras, Rheb is not a substrate for geranylgeranyltransferases and can only undergo farnesylation (Gau et al. 2005). We speculated that this specific requirement for farnesylation should allow therapeutic inhibition of Rheb with FTIs. To assess if FTI could block mTORC1 activation by Rheb, we transduced lymphoma cells with an activated allele (Rheb/Q64L) and a mutant that does not require farnesylation and instead undergoes geranylgeranylation (Rheb/Q64L/M184L; referred to throughout the figures as Rheb/M184L) (Gau et al. 2005). FTI (FTI-277 at 20 μM) was able to block mTORC1 activation by Rheb/Q64L as indicated by decreased phosphorylation of ribosomal S6 protein in lymphoma cells (Fig. 5A). In contrast, mTORC1 activation by the geranylgeranylated mutant (Rheb/Q64L/M184L) was insensitive to FTI (Fig. 5A). The effect of FTI on Rheb signaling was independent of genotype and occurred equally in Arf−/− lymphocytes, that do not activate the Akt pathway, and also in Pten-deficient cells (Eμ-Myc/Pten+/−), which engage the Akt pathway (Supplemental Fig. 7). Similarly, FTI-277 treatment was able to induce cell death in both the chemosensitive EμMyc/Arf−/− and the relatively chemo-resistant Eμ-Myc/Pten+/− cells (Fig. 5B; Wendel et al. 2006). Thus, FTI treatment can block Rheb activity and induce anti-tumor effects in lymphoma cells.
Figure 5.
Rheb inhibition by FTIs is responsible for FTI’s anti-tumor effect in Pten-deficient lymphoma. (A) Immunoblot of lysates prepared from Arf−/− lymphocytes expressing Rheb/Q64L or Rheb/Q64L/M184L (Rheb/M184L) and either untreated (U) or treated with 100nM rapamycin (R) for 6 h, or 20 μM FTI-277 for 24 h (F) and probed with antibodies against phosphorylated (Ser240/244) and total ribosomal S6 and tubulin. (B) Flow cytometric analysis of Eμ-Myc/Pten+/− and Eμ-Myc/Arf−/− tumor cells at different times after treatment with FTI-277 (20 μM). Indicated is the fraction of viable cells of all nucleated cells. (C) Flow cytometric analysis of mixed PTEN+/− or Arf−/− lymphoma cell populations partially transduced with vectors coexpressing GFP and either Rheb/Q64L, the farnesylation-independent mutant Rheb/Q64L/M184L (Rheb/M184L) or Mcl1. Indicated are the percentages of GFP-expressing cells before and after treatment with FTI-277 (30 μM/48 h).
A variety of proteins undergo farnesylation, and it has been unclear which of these is the relevant target of FTI therapy. We wondered if Rheb inhibition might be responsible for the anti-tumor effects of farnesyltransferase inhibition. Using retroviral vectors that coexpress the GFP reporter, we transduced subpopulations of lymphoma cells with the active and FTI-sensitive Rheb mutant (Rheb/Q64L) and the farnesylation-independent Rheb allele (Rheb/Q64L/M184L), along with empty vector and Mcl1, and monitored the percentage of cells expressing the GFP reporter before and after FTI treatment. In this “competition assay,” an increase in GFP indicates that cells expressing a certain allele have a selective advantage compared with the uninfected population. Briefly, in Eμ-Myc/Pten+/− tumor cells, the geranylgeranylated Rheb/Q64L/M184L allele was able to bypass the anti-proliferative effect of the FTI, such that cells expressing this allele were massively enriched under FTI treatment (Fig. 5C). A similar resistance to FTI was seen with the anti-apoptotic Mcl1, while enforced expression of the FTI-sensitive Rheb/Q64L allele was unable to protect against FTI and behaved like the empty vector (Fig. 5C). The ability of Rheb/Q64L/M184L to confer FTI resistance was genotype-dependent, and we did not observe a similar enrichment of Rheb/Q64L/M184L-expressing cells in the Eμ-Myc/Arf−/− background (Fig. 5C). This likely indicates that different mechanisms contribute to FTI drug action in genetically distinct tumors. In summary, Rheb inhibition by FTIs is responsible for their activity against tumors harboring activating lesions in the Akt pathway.
Our results establish that Rheb can have profound effects in a murine model of lymphoma. To understand its relevance to human lymphoma, we examined the expression of RHEB across a panel of Burkitts’ and diffuse large B-cell lymphoma (DLBCL). We found a substantial heterogeneity and a third, 8 of 24 cases, showed modest (between threefold and 10-fold) increases in RHEB expression compared with reactive tonsils, and individual cases had 20-fold and even 43-fold elevation of RHEB mRNA levels (Fig. 6A). As seen in the analysis of murine lymphoma (Fig. 1C,D), high RHEB expression in human tumors produced activation of mTORC1 and phosphorylation of the ribosomal S6 and inhibitory eIF4E-binding proteins (4E-BP1) (Fig. 6B). The reason for overexpression in these tumors is not clear. We observed only a twofold increase in gene copy number in the highest RHEB-expressing tumors (Supplemental Fig. 8). The RHEB promoter region and first exon do not contain significant CpG islands (data not shown). And while the promoter contains potential c-Myc-binding sites (Li et al. 2003), we saw no correlation between c-MYC and RHEB expression levels in our lymphoma samples (Supplemental Fig. 9A,B). The expression of a second RHEB gene (RHEB L1) showed little variation between normal and tumor samples (Supplemental Fig. 10). And while a recent large-scale effort has identified tumor-associated mutations in RHEB (Wood et al. 2007), we did not detect mutations in the RHEB coding sequence (data not shown). Taken together, RHEB is highly expressed in some human lymphomas, and while the mechanism is not clear, RHEB expression corresponds to increased mTORC1 activation in these cases.
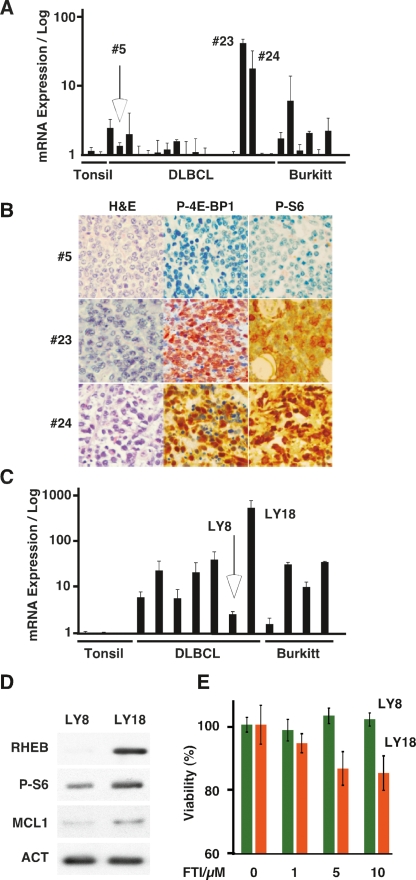
Figure 6.
RHEB expression and drug sensitivity in human lymphoma. (A) Quantitative real-time RT–PCR analysis of RHEB expression from cDNAs prepared from reactive tonsils (tonsil), a collection of DLBCL, and some cases of Burkitt’s (Burkitt) lymphoma. (B) Representative micrographs of lymphomas #5, #23, and #24 representing low and high RHEB mRNA-expressing lymphoma in A. Samples are stained with hematoxylin and eosin (H&E), and antibodies against the indicated antigens. (C) Quantitative real-time RT–PCR analysis of RHEB expression in human lymphoma cells lines representing DLBCL and Burkitt’s lymphoma compared with reactive tonsils. (D) Immunoblot of lysates prepared from low (LY8) and high (LY18) RHEB-expressing DLBCL lines probed with the indicated antibodies. (E) Mean and standard deviation of viability of LY8 and LY18 human lymphoma lines treated with FTI-277 at the indicated concentrations for 48 h.
To assess if this heterogeneous RHEB expression could also impact drug action in human lymphoma, we analyzed RHEB expression in several well-characterized human cell lines derived from Burkitts’ and DLBCL lymphoma (Mehra et al. 2002). As in the primary tumors, we observed significant heterogeneity and overall higher RHEB expression levels (Fig. 6C). For example, the LY18 line showed >100-fold increase in RHEB mRNA compared with the LY8 line. Similar to the murine lymphoma, these changes were accompanied by increased expression of the RHEB and MCL1 proteins and phosphorylation of ribosomal S6 (Fig. 6D). The LY8 and LY18 cell lines are not syngeneic, both have inactivating mutations in the p53 tumor suppressor, and while both carry translocations affecting the c-MYC locus, they differ in c-MYC expression (Supplemental Fig. 9A; Mehra et al. 2002). Upon in vitro treatment of LY8 and LY18 with doxorubicin or rapamycin alone, we observed no significant differences (LY8 vs. LY18, P > 0.5) (Supplemental Fig. 11A,B). In contrast, the high RHEB-expressing LY18 line and the low RHEB LY8 cell lines significantly differed in their susceptibility to the chemo-sensitizing effect of rapamycin (P < 0.01) (Supplemental Fig. 11C). Similarly, the high Rheb-expressing LY18 cells were somewhat more sensitive to FTI-277 compared with the LY8 cell line (P = 0.002 by students t-test) (Fig. 6E; Supplemental Fig. 12). These results indicate that heterogeneous RHEB expression in human lymphoma may pinpoint tumors with an increased sensitivity to pharmacologic inhibition of RHEB farnesylation and mTORC1 activity.
Discussion
Our study illustrates the potential of genetically versatile mouse models and adoptive transfer strategies to functionally evaluate candidate oncogenes and explore rational therapeutic strategies in vivo. For example, we surveyed the in vivo oncogenic potential of genes acting upstream of and downstream from mTOR and in translation initiation. We identify Rheb, a proximal activator of mTORC1, as a novel oncogenic activity capable of enhancing lymphomagenesis in vivo. Notably, it has been controversial if mTORC1 activation can affect cancer phenotypes in the absence of additional Akt signals. For example, neither activating mutations nor increased expression of mTOR have been reported in cancer, and the functional consequence of a recently reported, tumor-associated RHEB mutation is unclear (Wood et al. 2007). Similarly, genetic inactivation of TSC1 and TSC2, which act upstream of RHEB and mTORC1, but also affect mTORC2, is associated only with benign tumor syndromes (Manning et al. 2005; Huang et al. 2008). However, other findings suggest the potential importance of mTORC1 in cancer. For example, inhibition of mTORC1 with rapamycin can produce anti-tumor effects in some tumors (Dilling et al. 1994; Wendel et al. 2006), and conversely, mTORC1 activation can blunt the anti-proliferative effects of genetic Akt inactivation in vitro (Skeen et al. 2006). We find that mTORC1 activation by Rheb is able to drive tumorigenesis and affect apoptosis, cellular senescence, and treatment responses. Moreover, RHEB is highly expressed in some human lymphoma and possibly other cancers (Lassman et al. 2005). Our findings indicate that Rheb contributes to the malignant phenotype and may affect clinical behavior and treatment responses.
Downstream from mTOR we looked at certain components of the translation initiation complex that have been implicated previously in cellular transformation in vitro. Specifically, we examined the initiation factors eIF4AI, eIF4B, and eIF4GI, which act in concert with the oncogenic eIF4E factor and also a mutant form of eIF2α (eIF2α/S51A) that is resistant to PKR-mediated translation inhibition. At least in the Eμ-Myc context, we find that only eIF4E is able to promote tumor development in vivo. While it remains possible that the other translation factors can contribute to cancer in other settings, our results underscore the unique role of eIF4E as a translation factor capable of driving tumor development in vivo and are consistent with a rate-limiting role of eIF4E in the translation of transformation-relevant proteins (Polunovsky et al. 2000; Hay and Sonenberg 2004; Wendel et al. 2004; Mamane et al. 2007).
Oncogenesis driven by c-Myc is largely opposed by apoptosis (Strasser et al. 1990; Schmitt et al. 2002; Wendel et al. 2004), and we find that Rheb, like eIF4E and Akt, is able to counter the pro-apoptotic activity of c-Myc. Conversely, c-Myc can interfere with the induction of cellular senescence by Rheb and Akt as well as eIF4E (Ruggero et al. 2004). These interactions likely explain the oncogenic cooperation between c-Myc and distinct activating lesions in Akt-mTOR signaling and translation initiation. Besides these broad similarities, there are notable differences between oncogenic factors acting at different points in the Akt pathway. For example, myristoylated Akt has a relatively stronger oncogenic effect compared with Rheb and produces tumors that express a distinct pattern of surface markers possibly indicating a different cellular origin. Likely this reflects additional tumor-promoting activities and evasion of negative feedback controls by this constitutively active form of Akt. For example, mTORC1 activation has been shown to produce feedback inhibition of Akt, which can limit the transforming potential of TSC1/2 inactivation (Ma et al. 2005; Manning et al. 2005). We can speculate that similarly, disruption of feedback controls as seen upon loss of Pten may further potentiate Rheb’s oncogenicity.
Acting downstream from mTORC1, eIF4E produces tumors that are phenotypically very similar to Rheb-driven tumors. However, eIF4E tumors are insensitive to rapamycin. Moreover, eIF4E expression is sufficient to bypass the anti-tumor effects of rapamycin, indicating that eIF4E is a critical downstream mediator of mTORC1 activity (Wendel et al. 2004). And while Rheb certainly has additional targets (Clark et al. 1997; Im et al. 2002), we find that Rheb’s transformation-relevant effects on apoptosis, cellular senescence, and also chemotherapy responses are strictly dependent on mTORC1 activation and are sensitive to rapamycin. Together, these genetic findings suggest an epistatic relation between the pro-oncogenic activities of Rheb, mTORC1, and eIF4E.
Activation of Rheb and mTORC1 or increased abundance of the initiation factor eIF4E can affect the translation of a wide range of proteins; however, there are also specific effects on the translation of proteins involved in cellular transformation and apoptosis (Sonenberg and Hinnebusch 2007). For example, the anti-apoptotic Mcl1 protein is a transformation-relevant protein whose translation is increased upon expression of eIF4E or Rheb. In addition, Mcl1 is highly expressed in murine and human lymphomas driven by Rheb or eIF4E and is itself capable of enhancing c-Myc-induced lymphomagenesis (Wendel et al. 2007). Clearly, Mcl1 is regulated through nontranslational mechanisms as well (Le Gouill et al. 2004), and in addition to Mcl1, other regulators of apoptosis including Bcl-xL or Survivin can be affected by changes in translational regulation (Polunovsky et al. 2000; Rajasekhar et al. 2003; Larsson et al. 2007; Mamane et al. 2007). We can conclude that Mcl1 may exemplify how changes in translational efficiencies of specific proteins can contribute to the anti-apoptotic and tumor-promoting effects of eIF4E or Rheb.
Our findings on oncogenic activities upstream of and downstream from mTOR have implications for cancer therapy. For example, the unique oncogenic potency of the eIF4E translation factor provides a further rationale for targeting eIF4E in cancer therapy. In particular, eIF4E’s dual regulation by the Akt and Mapk pathways may be amenable to targeted inhibitors of these signaling pathways (Silva and Wendel 2007; Wendel et al. 2007). Rheb’s transformation-relevant activities depend on mTORC1 activation. Accordingly, mTORC1 inhibition by rapamycin may present a rational therapeutic strategy in Rheb-expressing cancers. Supporting this conclusion, we observe that rapamycin improves treatment responses in Rheb-driven tumors in vivo and also sensitizes human lymphoma cells that express high levels of RHEB to DNA-damaging agents in vitro. However, targeted inhibition of signaling pathways can have broad effects on cellular and organismal physiology, and some effects may counter their therapeutic purpose. For example, rapamycin may enhance upstream oncogenic signals through feedback mechanisms (Sun et al. 2005), and, at least in cells that have not disabled this failsafe mechanism, rapamycin can affect aspects of the senescence phenotype and potentially enhance proliferation of cells. In addition, rapamycin is a potent immunosuppressive. Thus, markers are needed to identify patients for whom the anti-tumor effects outweigh unwanted treatment consequences. RHEB expression in tumors may be a predictor of sensitivity to rapamycin or direct Rheb inhibition.
FTI can directly block Rheb activity and induce cytotoxic effects in human and murine lymphoma cells. Notably, FTIs were first devised to inhibit Ras; however, alternative prenylation of K- and N-Ras hinders these drugs’ ability to affect Ras oncogenes (for review, see Braun and Fenaux 2008). Nonetheless, FTIs are less toxic than chemotherapy and have notable activity in acute leukemias, where they produce responses in up to 30% and complete remissions in some 10%–15% of patients. These clinical effects do not appear to be linked to Ras mutations or inhibition of Ras effectors (Kurzrock et al. 2004; Burnett and Kell 2007; Lancet et al. 2007). It is therefore likely that other substrates of farnesyltransferase contribute to the anti-cancer effects of FTI therapy, and several farnesylated proteins have been implicated including Rheb, H-Ras, Lamin A, Lamin B, RhoB, and the centromere proteins CENP-E and CENP-F (Du and Prendergast 1999; Castro et al. 2003; Basso et al. 2005; Gau et al. 2005; Karbowniczek et al. 2008). We find that FTIs can inhibit Rheb in lymphoma cells, and that inhibition of Rheb is responsible for the anti-tumor effect of FTIs in Pten-deficient tumor cells. Accordingly, a farnesylation-independent mutant of Rheb can confer resistance to FTI in tumors that activate Akt signaling. In contrast, in tumor cells that do not engage the Akt pathway, the constitutive Rheb allele is unable to bypass the cytotoxic effect of the inhibitor. It is well established that FTIs can affect the activity of several signaling molecules (Braun and Fenaux 2008), and that the relative importance of these pathways differs between genetically distinct tumors (Wendel et al. 2004). In this way, the genetic context can determine FTI activity, and also which mechanism is responsible for FTI anti-tumor action.
Together, our study defines Rheb as an oncogenic activity that is amenable to rational therapeutic strategies using rapamycin or direct inactivation with inhibitors of farnesyltransferase. RHEB is highly expressed in some human cancers, including lymphoma, and may pinpoint patients most likely to benefit from such targeted strategies.
Materials and methods
Generation of mice
Lymphomas were generated and analyzed as described (Schmitt et al. 2002). The retroviruses used to produce tumors and used in cell assays were based on MSCV-IRES-GFP and expressed cDNAs encoding eIF4E, eIF4B, eIF4AI, eIF2α/S51A, eIF4GI, and myristoylated Akt (Wendel et al. 2004), and Rheb, Rheb/Q64L, and Rheb/5A (Inoki et al. 2003). Mice were monitored by blood count and palpation, and lymphomas were harvested and either fixed for histological evaluation, rendered single cell suspensions and frozen in 10% DMSO, or transplanted directly into normal mice for treatment studies.
Treatment studies
Treatment studies were performed as described previously (Wendel et al. 2004). Briefly, 1 × 106 DMSO frozen or primary lymphoma cells were injected into the tail vein of 6- to 8-wk-old female C57BL/6 mice. Upon the formation of well-palpable tumors, the animals were treated with rapamycin (LC Labs; 4 mg/kg, i.p.), doxorubicin (Sigma; 10 mg/kg, i.p.), or a combination of both. In treatment studies, control lymphomas were Eμ-Myc/Arf−/− tumors, which are homogeneous with respect to p53 status. Following treatment, the mice were monitored by twice weekly palpation and blood smears stained with Giemsa (Fisher Diagnostics). Tumor-free and overall survival data were analyzed in the Kaplan-Meier format using the log-rank (Mantel-Cox) test for statistical significance.
Histopathology
Preparation of samples, H&E stain, and detection of antibody stains were as described previously (Wendel et al. 2004). Antibodies were against phosphorylated ribosomal S6 protein (Ser240/244; Cell Signaling, #2215 and #4857), phosphorylated Akt (Ser473; Cell Signaling, #9271), phosphorylated 4E-BP1 (Cell Signaling, #2855), and Ki67 (NovoCastra, NCL-Ki67-P). TUNEL was performed using a published protocol (McCurrach and Lowe 2001).
Immunophenotyping
Tumor cell suspensions of representative tumors of each genotype were stained as described (Wendel et al. 2004). The antibodies were B220/CD45R (PharMingen, #553089), CD19 (PharMingen, catalog no. 557399), IgM/IgK (PharMingen, #555584), Thy1/CD90 (Cedarlane, #CL8610PE), CD4 (PharMingen, #557308), CD8 (PharMingen, #553032), Sca-1 (PharMingen, #553108), or c-kit (PharMingen, #553869) conjugated with phycoerythrin and analyzed with a Guava EasyCyte Plus system.
Cell culture and viability
Primary MEFs were cultured as described (McCurrach and Lowe 2001). FL5-12 cells were maintained in RPMI-1640 medium supplemented with 10% FBS, 10% WEHI-3B conditioned medium, and interleukin-3 (400 pM) (Plas et al. 2001). For apoptosis assays, MEFs were transduced with pBabe-Myc, selected with puromycin (2.5 μg/mL), and subjected to serum withdrawal for 12–24 h in the presence or absence of rapamycin during the same period. For cytokine depletion, FL5-12 cells were washed four times in PBS and resuspended in growth media without interleukin-3 or WEHI-3B conditioned media (Plas et al. 2001). FTI-277 (Calbiochem) was dissolved in DMSO and used at no more than 0.1% of final volume; for immunoblots, cells were treated for 24 h and 48 h in viability and competition experiments. Viability was determined by trypan blue stain and TUNEL assay as described (McCurrach and Lowe 2001). Flow cytometric analysis of cell death was by Viacount, which contains a cell-permeable dye (LDS-751) and the non-membrane-permeable propidium iodide (Guava #4500-0160).
Cell senescence assay
Early passage MEFs were infected and selected for growth curves; cells were plated at 104 cells, and cell numbers were determined at various time points. Cells were untreated or treated twice with rapamycin 100 nM and collected 7 d post-selection for immunoblotting and senescence-associated β-galactosidase (SA β-Gal) as described (Serrano et al. 1997).
Western blot analysis
Immunoblots were performed as described (Wendel et al. 2004) using antibodies against Rheb (1:1000; Cell Signaling, #4935), total and phosphorylated (Ser473) Akt (1:1000; Cell Signaling, #9272, #9271), total and phosphorylated (Ser240/244) ribosomal S6 (1:1000; Cell Signaling, #2317, #2215), p16 (1:1000; Santa Cruz Biotechnologies, M-156), p53 (1:500; Novocastra, NCL-p53-505), p21 (1:1000; Santa Cruz Biotechnologies, c-19), mouse Mcl1 (1:1000; Abcam, ab32087-100), human MCL1 (1:1000; Santa Cruz Biotechnologies, S-19), Tubulin (1:5000; Sigma, B-5-1-2), Actin (1:5000; Sigma, AC-15), and anti-HA (1:5000; Roche, #12013819).
Polysome fractionation
As described in detail in Xi et al. (2004), cells were treated with cycloheximide before harvesting and lysing. The nuclei were pelleted, and the ribosomal components were separated from the soluble fraction by ultracentrifugation. RNA was isolated using Trizol (Invitrogen), and cDNA synthesis was performed using the SuperScriptIII kit (Invitrogen). RT–PCR primers were Mcl1F, AGCCTGACTTCCCAGCTCAC; Mcl1R, GCACTCA GACCACATGCTTC; ActinF, AAGCTAACCGGGAGAAGA TG; and ActinR, GTAGTCAGTGAGGTCGCGAC.
Expression and gene copy number analyses
Total RNA was extracted using Allprep DNA/RNA/Protein kit (Promega, #80004). cDNA synthesis was by SuperScript III Reverse Transcriptase (Invitrogen, #18080-400), and PCR was performed in triplicate using the DNA Engine 7900HT FAST Real time PCR system (Applied Biosystems) and TaqMan Universal PCR Master Mix (Roche, 4304437). The human RHEB (assay ID: Hs00950803_m1), MYC (assay ID: Hs00153408_m1), GUSB (#4333767T), and PGK1 (#4333765T) primers were from TaqMan Gene Expressions Assays. Target gene expression was quantified by normalizing against endogenous controls GUSB or PGK using the ΔΔ Ct method. The gene copy number of RHEB was determined by quantitative PCR using RHEB forward/reverse primers (Forward, 5′-CTGAGGCCGCCAAG-3′; reverse, 5′-CTCCAAACTTCGCAGGCC-3′) and a TaqMan dual-labeled MGB probe corresponding to intronic sequence AAGTCCCGG AAGATCGCGATCCTG. The relative quantification of RHEB was referenced to RNase P and compared with male genomic DNA.
Sequence analysis
RHEB cDNA was amplified from DLBCL first strand synthesized cDNA by using the following RHEB forward/reverse primers: Forward, 5′-CAGGAGCCACCGCCGCCGCGGTT-3′; Reverse, 5′-GAATATATTCCCAGTGTCCTCAGGC-3′). For the sequencing reaction, the following nested primers were used: Forward, 5′-TGTGGTTGGGCCGGGGCTGAGGAGGC-3′; Reverse, 5′-TCAGGCTTTGCAGCAGAATCA-3′. Analysis was performed on an ABI 3730 Capillary Sequencer.
Acknowledgments
We thank Mayumi Isaka of the MSKCC animal core and Agnes Viale of the genomics core. We thank Joan Massague for critical review and editorial advice. H.G.W’s laboratory is supported by a Special Fellowship from the Leukemia and Lymphoma Society; funds from the Special Projects Committee of Memorial Sloan-Kettering, the Costas Lemos foundation, the New York Community Trust, and the Geoffrey Beene Cancer Center; grants CA13106 and CA87497 from the National Cancer Institute (to S.W.L); and a CHIR grant (MOP-79385) (to J.P.).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1690808.
References
- Avdulov S., Li S., Michalek V., Burrichter D., Peterson M., Perlman D.M., Manivel J.C., Sonenberg N., Yee D., Bitterman P.B., et al. Activation of translation complex eIF4F is essential for the genesis and maintenance of the malignant phenotype in human mammary epithelial cells. Cancer Cell. 2004;5:553–563. doi: 10.1016/j.ccr.2004.05.024. [DOI] [PubMed] [Google Scholar]
- Basso A.D., Mirza A., Liu G., Long B.J., Bishop W.R., Kirschmeier P. The farnesyl transferase inhibitor (FTI) SCH66336 (lonafarnib) inhibits Rheb farnesylation and mTOR signaling. Role in FTI enhancement of taxane and tamoxifen anti-tumor activity. J. Biol. Chem. 2005;280:31101–31108. doi: 10.1074/jbc.M503763200. [DOI] [PubMed] [Google Scholar]
- Braun T., Fenaux P. Farnesyltransferase inhibitors and their potential role in therapy for myelodysplastic syndromes and acute myeloid leukaemia. Br. J. Haematol. 2008;141:576–586. doi: 10.1111/j.1365-2141.2008.07099.x. [DOI] [PubMed] [Google Scholar]
- Buerger C., DeVries B., Stambolic V. Localization of Rheb to the endomembrane is critical for its signaling function. Biochem. Biophys. Res. Commun. 2006;344:869–880. doi: 10.1016/j.bbrc.2006.03.220. [DOI] [PubMed] [Google Scholar]
- Burnett A.K., Kell J. Tipifarnib in acute myeloid leukemia. Drugs Today (Barc) 2007;43:795–800. doi: 10.1358/dot.2007.43.11.1088617. [DOI] [PubMed] [Google Scholar]
- Castro A.F., Rebhun J.F., Clark G.J., Quilliam L.A. Rheb binds tuberous sclerosis complex 2 (TSC2) and promotes S6 kinase activation in a rapamycin- and farnesylation-dependent manner. J. Biol. Chem. 2003;278:32493–32496. doi: 10.1074/jbc.C300226200. [DOI] [PubMed] [Google Scholar]
- Chen Z., Trotman L.C., Shaffer D., Lin H.K., Dotan Z.A., Niki M., Koutcher J.A., Scher H.I., Ludwig T., Gerald W., et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark G.J., Kinch M.S., Rogers-Graham K., Sebti S.M., Hamilton A.D., Der C.J. The Ras-related protein Rheb is farnesylated and antagonizes Ras signaling and transformation. J. Biol. Chem. 1997;272:10608–10615. doi: 10.1074/jbc.272.16.10608. [DOI] [PubMed] [Google Scholar]
- Clemens M.J. Targets and mechanisms for the regulation of translation in malignant transformation. Oncogene. 2004;23:3180–3188. doi: 10.1038/sj.onc.1207544. [DOI] [PubMed] [Google Scholar]
- Dilling M.B., Dias P., Shapiro D.N., Germain G.S., Johnson R.K., Houghton P.J. Rapamycin selectively inhibits the growth of childhood rhabdomyosarcoma cells through inhibition of signaling via the type I insulin-like growth factor receptor. Cancer Res. 1994;54:903–907. [PubMed] [Google Scholar]
- Donze O., Jagus R., Koromilas A.E., Hershey J.W., Sonenberg N. Abrogation of translation initiation factor eIF-2 phosphorylation causes malignant transformation of NIH 3T3 cells. EMBO J. 1995;14:3828–3834. doi: 10.1002/j.1460-2075.1995.tb00052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrello N.V., Peschiaroli A., Guardavaccaro D., Colburn N.H., Sherman N.E., Pagano M. S6K1- and βTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467–471. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- Du W., Prendergast G.C. Geranylgeranylated RhoB mediates suppression of human tumor cell growth by farnesyltransferase inhibitors. Cancer Res. 1999;59:5492–5496. [PubMed] [Google Scholar]
- Edinger A.L., Thompson C.B. An activated mTOR mutant supports growth factor-independent, nutrient-dependent cell survival. Oncogene. 2004;23:5654–5663. doi: 10.1038/sj.onc.1207738. [DOI] [PubMed] [Google Scholar]
- Evan G.I., Wyllie A.H., Gilbert C.S., Littlewood T.D., Land H., Brooks M., Waters C.M., Penn L.Z., Hancock D.C. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- Fukuchi-Shimogori T., Ishii I., Kashiwagi K., Mashiba H., Ekimoto H., Igarashi K. Malignant transformation by overproduction of translation initiation factor eIF4G. Cancer Res. 1997;57:5041–5044. [PubMed] [Google Scholar]
- Gau C.L., Kato-Stankiewicz J., Jiang C., Miyamoto S., Guo L., Tamanoi F. Farnesyltransferase inhibitors reverse altered growth and distribution of actin filaments in Tsc-deficient cells via inhibition of both rapamycin-sensitive and -insensitive pathways. Mol. Cancer Ther. 2005;4:918–926. doi: 10.1158/1535-7163.MCT-04-0347. [DOI] [PubMed] [Google Scholar]
- Guertin D.A., Sabatini D.M. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Hay N., Sonenberg N. Upstream and downstream of mTOR. Genes & Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- Hilliard A., Hilliard B., Zheng S.J., Sun H., Miwa T., Song W., Goke R., Chen Y.H. Translational regulation of autoimmune inflammation and lymphoma genesis by programmed cell death 4. J. Immunol. 2006;177:8095–8102. doi: 10.4049/jimmunol.177.11.8095. [DOI] [PubMed] [Google Scholar]
- Huang J., Dibble C., Matsuzaki M., Manning B. The TSC1–TSC2 complex is required for proper activation of mTOR complex 2. Mol. Cell. Biol. 2008;28:4104–4115. doi: 10.1128/MCB.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im E., von Lintig F.C., Chen J., Zhuang S., Qui W., Chowdhury S., Worley P.F., Boss G.R., Pilz R.B. Rheb is in a high activation state and inhibits B-Raf kinase in mammalian cells. Oncogene. 2002;21:6356–6365. doi: 10.1038/sj.onc.1205792. [DOI] [PubMed] [Google Scholar]
- Inoki K., Li Y., Xu T., Guan K.L. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes & Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeschke A., Hartkamp J., Saitoh M., Roworth W., Nobukuni T., Hodges A., Sampson J., Thomas G., Lamb R. Tuberous sclerosis complex tumor suppressor-mediated S6 kinase inhibition by phosphatidylinositide-3-OH kinase is mTOR independent. J. Cell Biol. 2002;159:217–224. doi: 10.1083/jcb.jcb.200206108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowniczek M., Spittle C.S., Morrison T., Wu H., Henske E.P. mTOR is activated in the majority of malignant melanomas. J. Invest. Dermatol. 2008;128:980–987. doi: 10.1038/sj.jid.5701074. [DOI] [PubMed] [Google Scholar]
- Kurzrock R., Albitar M., Cortes J.E., Estey E.H., Faderl S.H., Garcia-Manero G., Thomas D.A., Giles F.J., Ryback M.E., Thibault A., et al. Phase II study of R115777, a farnesyl transferase inhibitor, in myelodysplastic syndrome. J. Clin. Oncol. 2004;22:1287–1292. doi: 10.1200/JCO.2004.08.082. [DOI] [PubMed] [Google Scholar]
- Lancet J.E., Gojo I., Gotlib J., Feldman E.J., Greer J., Liesveld J.L., Bruzek L.M., Morris L., Park Y., Adjei A.A., et al. A phase 2 study of the farnesyltransferase inhibitor tipifarnib in poor-risk and elderly patients with previously untreated acute myelogenous leukemia. Blood. 2007;109:1387–1394. doi: 10.1182/blood-2006-04-014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson O., Li S., Issaenko O.A., Avdulov S., Peterson M., Smith K., Bitterman P.B., Polunovsky V.A. Eukaryotic translation initiation factor 4E induced progression of primary human mammary epithelial cells along the cancer pathway is associated with targeted translational deregulation of oncogenic drivers and inhibitors. Cancer Res. 2007;67:6814–6824. doi: 10.1158/0008-5472.CAN-07-0752. [DOI] [PubMed] [Google Scholar]
- Lassman A.B., Rossi M.R., Raizer J.J., Abrey L.E., Lieberman F.S., Grefe C.N., Lamborn K., Pao W., Shih A.H., Kuhn J.G., et al. Molecular study of malignant gliomas treated with epidermal growth factor receptor inhibitors: Tissue analysis from North American Brain Tumor Consortium Trials 01-03 and 00-01. Clin. Cancer Res. 2005;11:7841–7850. doi: 10.1158/1078-0432.CCR-05-0421. [DOI] [PubMed] [Google Scholar]
- Lazaris-Karatzas A., Montine K.S., Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- Le Gouill S., Podar K., Harousseau J.L., Anderson K.C. Mcl-1 regulation and its role in multiple myeloma. Cell Cycle. 2004;3:1259–1262. doi: 10.4161/cc.3.10.1196. [DOI] [PubMed] [Google Scholar]
- Li Z., Van Calcar S., Qu C., Cavenee W.K., Zhang M.Q., Ren B. A global transcriptional regulatory role for c-Myc in Burkitt’s lymphoma cells. Proc. Natl. Acad. Sci. 2003;100:8164–8169. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Teruya-Feldstein J., Behrendt N., Chen Z., Noda T., Hino O., Cordon-Cardo C., Pandolfi P.P. Genetic analysis of Pten and Tsc2 functional interactions in the mouse reveals asymmetrical haploinsufficiency in tumor suppression. Genes & Dev. 2005;19:1779–1786. doi: 10.1101/gad.1314405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamane Y., Petroulakis E., Rong L., Yoshida K., Ler L.W., Sonenberg N. eIF4E–from translation to transformation. Oncogene. 2004;23:3172–3179. doi: 10.1038/sj.onc.1207549. [DOI] [PubMed] [Google Scholar]
- Mamane Y., Petroulakis E., LeBacquer O., Sonenberg N. mTOR, translation initiation and cancer. Oncogene. 2006;25:6416–6422. doi: 10.1038/sj.onc.1209888. [DOI] [PubMed] [Google Scholar]
- Mamane Y., Petroulakis E., Martineau Y., Sato T.A., Larsson O., Rajasekhar V.K., Sonenberg N. Epigenetic activation of a subset of mRNAs by eIF4E explains its effects on cell proliferation. PLoS ONE. 2007;2:e242. doi: 10.1371/journal.pone.0000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning B.D., Logsdon M.N., Lipovsky A.I., Abbott D., Kwiatkowski D.J., Cantley L.C. Feedback inhibition of Akt signaling limits the growth of tumors lacking Tsc2. Genes & Dev. 2005;19:1773–1778. doi: 10.1101/gad.1314605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer U., Charvet C., Wagman A.S., Dejardin E., Green D.R. Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Mol. Cell. 2006;21:749–760. doi: 10.1016/j.molcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- McCurrach M.E., Lowe S.W. Methods for studying pro- and antiapoptotic genes in nonimmortal cells. Methods Cell Biol. 2001;66:197–227. doi: 10.1016/s0091-679x(01)66010-2. [DOI] [PubMed] [Google Scholar]
- Mehra S., Messner H., Minden M., Chaganti R.S. Molecular cytogenetic characterization of non-Hodgkin lymphoma cell lines. Genes Chromosomes Cancer. 2002;33:225–234. doi: 10.1002/gcc.10025. [DOI] [PubMed] [Google Scholar]
- Plas D.R., Talapatra S., Edinger A.L., Rathmell J.C., Thompson C.B. Akt and Bcl-xL promote growth factor-independent survival through distinct effects on mitochondrial physiology. J. Biol. Chem. 2001;276:12041–12048. doi: 10.1074/jbc.M010551200. [DOI] [PubMed] [Google Scholar]
- Polunovsky V.A., Gingras A.C., Sonenberg N., Peterson M., Tan A., Rubins J.B., Manivel J.C., Bitterman P.B. Translational control of the antiapoptotic function of Ras. J. Biol. Chem. 2000;275:24776–24780. doi: 10.1074/jbc.M001938200. [DOI] [PubMed] [Google Scholar]
- Rajasekhar V.K., Viale A., Socci N.D., Wiedmann M., Hu X., Holland E.C. Oncogenic Ras and Akt signaling contribute to glioblastoma formation by differential recruitment of existing mRNAs to polysomes. Mol. Cell. 2003;12:889–901. doi: 10.1016/s1097-2765(03)00395-2. [DOI] [PubMed] [Google Scholar]
- Ruggero D., Montanaro L., Ma L., Xu W., Londei P., Cordon-Cardo C., Pandolfi P.P. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat. Med. 2004;10:484–486. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- Schmitt C.A., Fridman J.S., Yang M., Baranov E., Hoffman R.M., Lowe S.W. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 2002;1:289–298. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
- Serrano M., Lin A.W., McCurrach M.E., Beach D., Lowe S.W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- Silva R.L., Wendel H.G. MNK, EIF4E and targeting translation for therapy. Cell Cycle. 2007;7:553–555. doi: 10.4161/cc.7.5.5486. [DOI] [PubMed] [Google Scholar]
- Skeen J.E., Bhaskar P.T., Chen C.C., Chen W.S., Peng X.D., Nogueira V., Hahn-Windgassen A., Kiyokawa H., Hay N. Akt deficiency impairs normal cell proliferation and suppresses oncogenesis in a p53-independent and mTORC1-dependent manner. Cancer Cell. 2006;10:269–280. doi: 10.1016/j.ccr.2006.08.022. [DOI] [PubMed] [Google Scholar]
- Sonenberg N., Hinnebusch A.G. New modes of translational control in development, behavior, and disease. Mol. Cell. 2007;28:721–729. doi: 10.1016/j.molcel.2007.11.018. [DOI] [PubMed] [Google Scholar]
- Strasser A., Harris A.W., Bath M.L., Cory S. Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature. 1990;348:331–333. doi: 10.1038/348331a0. [DOI] [PubMed] [Google Scholar]
- Sun S.Y., Rosenberg L.M., Wang X., Zhou Z., Yue P., Fu H., Khuri F.R. Activation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer Res. 2005;65:7052–7058. doi: 10.1158/0008-5472.CAN-05-0917. [DOI] [PubMed] [Google Scholar]
- Tee A.R., Fingar D.C., Manning B.D., Kwiatkowski D.J., Cantley L.C., Blenis J. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc. Natl. Acad. Sci. 2002;99:13571–13576. doi: 10.1073/pnas.202476899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivanco I., Sawyers C.L. The phosphatidylinositol 3-kinase AKT pathway in human cancer. Nat. Rev. Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- Wendel H.G., De Stanchina E., Fridman J.S., Malina A., Ray S., Kogan S., Cordon-Cardo C., Pelletier J., Lowe S.W. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- Wendel H.G., Malina A., Zhao Z., Zender L., Kogan S.C., Cordon-Cardo C., Pelletier J., Lowe S.W. Determinants of sensitivity and resistance to rapamycin-chemotherapy drug combinations in vivo. Cancer Res. 2006;66:7639–7646. doi: 10.1158/0008-5472.CAN-06-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel H.G., Silva R.L., Malina A., Mills J.R., Zhu H., Ueda T., Watanabe-Fukunaga R., Fukunaga R., Teruya-Feldstein J., Pelletier J., et al. Dissecting eIF4E action in tumorigenesis. Genes & Dev. 2007;21:3232–3237. doi: 10.1101/gad.1604407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood L.D., Parsons D.W., Jones S., Lin J., Sjoblom T., Leary R.J., Shen D., Boca S.M., Barber T., Ptak J., et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- Xi Q., Cuesta R., Schneider R.J. Tethering of eIF4G to adenoviral mRNAs by viral 100k protein drives ribosome shunting. Genes & Dev. 2004;18:1997–2009. doi: 10.1101/gad.1212504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakowicz H., Yang H.S., Stark C., Wlodawer A., Laronde-Leblanc N., Colburn N.H. Mutational analysis of the DEAD-box RNA helicase eIF4AII characterizes its interaction with transformation suppressor Pdcd4 and eIF4GI. RNA. 2005;11:261–274. doi: 10.1261/rna.7191905. [DOI] [PMC free article] [PubMed] [Google Scholar]