Abstract
The high mass measurement accuracy and precision available with recently developed mass spectrometers is increasingly used in proteomics analyses to confidently identify tryptic peptides from complex mixtures of proteins, as well as post-translational modifications and peptides from non-annotated proteins. To take full advantage of high mass measurement accuracy instruments it is necessary to limit systematic mass measurement errors. It is well known that errors in the measurement of m/z can be affected by experimental parameters that include e.g., outdated calibration coefficients, ion intensity, and temperature changes during the measurement. Traditionally, these variations have been corrected through the use of internal calibrants (well-characterized standards introduced with the sample being analyzed). In this paper we describe an alternative approach where the calibration is provided through the use of a priori knowledge of the sample being analyzed. Such an approach has previously been demonstrated based on the dependence of systematic error on m/z alone. To incorporate additional explanatory variables, we employed multidimensional, nonparametric regression models, which were evaluated using several commercially available instruments. The applied approach is shown to remove any noticeable biases from the overall mass measurement errors, and decreases the overall standard deviation of the mass measurement error distribution by 1.2- to 2-fold, depending on instrument type. Subsequent reduction of the random errors based on multiple measurements over consecutive spectra further improves accuracy and results in an overall decrease of the standard deviation by 1.8- to 3.7-fold. This new procedure will decrease the false discovery rates for peptide identifications using high accuracy mass measurements.
INTRODUCTION
Liquid chromatography-mass spectrometry (LC-MS) is increasingly used to broadly identify and quantify proteins over a large range of abundances in biological samples. Analysis of MS-based proteomic data generally uses search engines that compare the mass to charge ratios (m/z) of observed ions to those predicted from a finite set of candidates to infer the peptides (e.g., from proteolytic digestion of intact proteins) or proteins that may be present in a sample. Therefore, the confidence in the results depends significantly on the mass measurement error (MME) achieved. Lower MME allows superior discrimination e.g., between true and false peptide or protein identifications, as demonstrated by several groups over the last few years 1-4
While most mass spectrometers can exhibit quite impressive MME (sub- to low-ppm) for single compounds and when properly calibrated (e.g. using an “internal standard”), such accuracy is problematic with complex biological samples due to limitations on resolution of different species as well as a range of experimental details. The latter issues and their effects vary for different instruments or platforms. For example, temperature changes during LC separations can cause drift of the power supply voltage and consequently lead to systematic MME using time-of-flight (TOF) instruments 5. Space charge effects resulting from the trapped ion population is the major cause of systematic mass measurement errors for Fourier transform ion cyclotron resonance (FTICR) instruments. Outdated calibration coefficients can also contribute to systematic mass measurement errors.
Several experimental methodologies have been developed to reduce the systematic errors of mass measurement. For example, some FTICR instruments are equipped with an automated gain control (AGC) system that aims to control the number of ions trapped in the ICR cell. The most effective approach is internal calibration, where known substances are measured in the same spectrum with the analyte(s) of interest. It can be implemented by either introducing calibrants to the sample prior ionization 6, by introducing calibrants into MS through an additional electrospray source 7, 8, as well as by using ubiquitious chemical contaminants as internal calibrants 2, 3. This approach generally works well, but is most effective when the m/z of both the analyte and calibrant are similar. Thus, for proteomic applications it is desirable to have multiple internal calibrants to extensively cover m/z range. However, such internal calibrants can overlap with species of interest during MS scans and preclude their detection, as well as limit the analyte ion population that can be addressed (e.g. due to space charge limitations), and thus limit the dynamic range achievable for measurements.
One approach for addressing the latter issues was to introduce a calibrant mixture immediately before and after the LC separation and use multivariate regression approach to find the calibration coefficients 9. However, this approach is limited in that it assumes a uniform drift of the calibration coefficients. It also fails to account for the influence of the total ion signal as well as the individual ion intensities on MME during the analysis. The latter is actually an important factor for a certain type of time-of-flight instruments, as we are going to show later in this report. This work also proposed the use of highly expressed proteins commonly present in a given type of sample as effective internal calibrants in addition to external calibrants, which was subsequently demonstrated using abundant peptides in Deinococcus radiodurans samples 10.
There are few approaches have been developed, which avoid the use of internal calibrants and rely solely on the information from the sample itself. One of the earliest approaches is based on observation multiply charged states of the same peptides during MS scans in FTICR instruments. That allowed for correction for space charge effects to produce an improved calibration function, a process referred to as deconvolution of Coulombic affected linearity, or DeCAL 11.
It is also worth mentioning similar efforts for recalibration of fragment ions in MS/MS spectra acquired on high-resolution instrumentation 12, 13. It has been shown, that for MS/MS scan recalibration one can use all the theoretical fragments of a putatively identified peptide.
Recently, based upon the use of new hybrid instrumentation, it has been proposed to use confident peptide identifications from MS/MS spectra in a post hoc strategy for recalibration of parent ions 14. In one implementation of this approach 15 MS spectra from LC-MS/MS measurements are calibrated individually using peptide identifications from the neighboring MS/MS scans, that is several scans acquired right before or after the considered MS scan. However, this implementation did not take into account the effect of individual ion intensities on mass measurement errors. In particular, the peptides used for calibration are sampled from the most highly-abundant peptides, and the obtained calibration coefficients are applied to the entire set of the observed species. Moreover, there is no guarantee that peptides identified by MS/MS fragmentation will cover as extensive m/z range as the peptides present in the MS scan. Nevertheless, we believe that with adjustment of the method for selection of parent ions for fragmentation, this approach holds a potential for use with appropriate hybrid instruments.
Another approach used a priori knowledge about the sample content instead of deriving it from the MS/MS data 16. Masses of ions in a sample are matched using a wide mass tolerance against all the masses in a database of peptides expected to be detected in the sample. However, since the matching used only the monoisotopic masses, it is expected that the matches contain a significant portion of false identifications for complex samples. Thus, the matches were modeled as a mixture of correct and incorrect matches. The entire set of matches was recalibrated using a Bayesian statistical technique assuming certain distributions of true and false peptide matches, and the systematic bias of mass measurement error on m/z value was removed. The presence of a significant proportion of incorrect assignments in the initial set prohibited the authors from using regression models or scatter smoothing techniques. A robust scatter smoothing technique was used in another study 17 as a part of three-stage approach combining external calibration, single “lock-mass” internal correction with a robust scatter smoothing approach to reduce any systematic dependencies of the mass measurement errors along the m/z parameter. Similarly, work from our lab attempted to account for the mass measurement error dependency on a number of parameters like elution time, m/z, TIC and individual ion intensity by partitioning the parameter space into multiple equally sized regions, with each region recalibrated independently 18. The recalibration was achieved by fitting a linear transformation functions from observed m/z values to theoretical m/z values. The merit of each transformation function was judged based on the height of the peak of the mass error histogram relative to the random errors of the false matches. However, that implementation provided no statistical basis to defend the use of certain parameters or their combinations as dimensions for breaking the data into the regions. In addition, there were no global constraints imposed and each region was recalibrated independently of each other. Those two factors can potentially lead to overfitting of the data, and overestimation of the performance of the recalibration procedure. Ideally, we wish to describe the systematic error trends with continuous differentiable functions, as opposed to the discontinuous dependencies used previously.
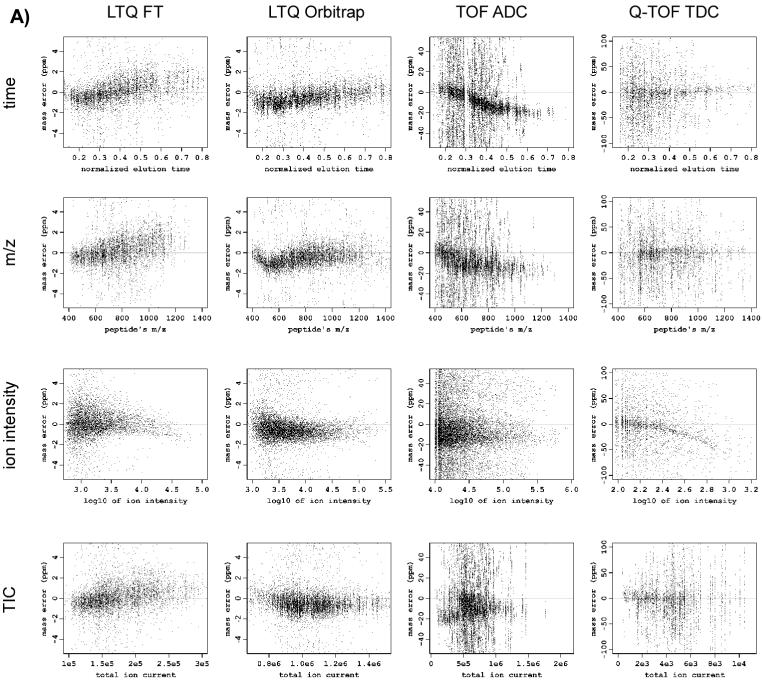
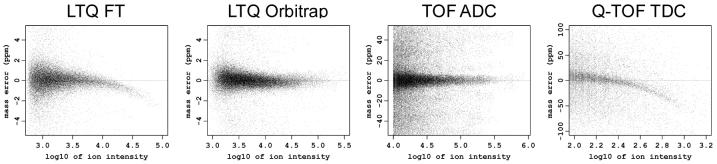
To address the aforementioned issues we developed a regression approach that uses a set of candidate matches between masses of peptide peaks observed in a measurement and a database of possible candidate peptide identifications, in order to detect and remove functional dependencies that may be observed between mass measurement error and predefined parameters. Such dependencies are often present in the data and are easily revealed by exploratory data analysis, such as plotting the mass measurement errors as a function of elution time, m/z, ion intensity, or other explanatory variables (Figure 1A). However, regression analysis is complicated by several factors. The set of candidate matches used in the regression analysis is composed of correct and incorrect matches, which have different error distributions. Because of the significant fraction of false matches that can be initially obtained, regression analysis is often unsuccessful with simple least-squares fitting. Thus, our approach involves an analysis using robust regression techniques with a subset of matches that is enriched with true matches, and then applies the regression function to the entire dataset. The trends observed may reveal nonparametric dependencies on multiple explanatory variables, such as m/z, LC elution time, etc. As a result, we use multidimensional projection pursuit regression 19, 20 with smoothing splines 21 as a ridge function. To select the optimal regression models and the set of relevant explanatory variables for each instrument type we applied a 10-fold cross-validation approach.
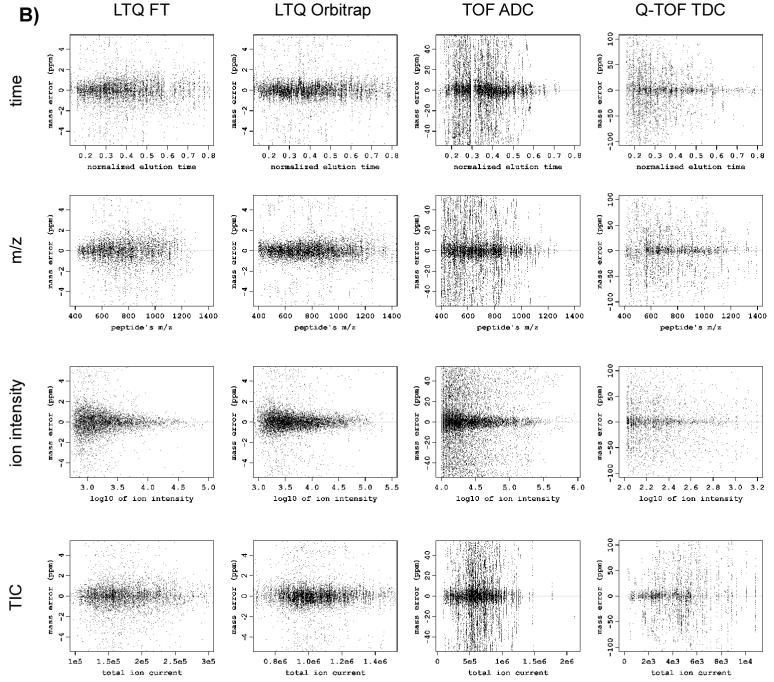
Figure 1.
Scatter plots showing mass measurement error versus different parameters for different instruments before (A) and after (B) applying the systematic error correction procedure. Note different scales of mass measurement error for different instruments.
In addition to the elimination of systematic errors, we explored the possibility of reducing random mass measurement errors using the multiplicity of peptide ion observations. Generally, in an LC-MS analysis, most ions are observed over several consecutive spectra with similar, but not identical, masses because of variation in mass measurement accuracy. Repeated measurements of the same peptide in multiple spectra can be grouped together, and the representative mass of each ion group can be calculated as the average or median value of individual ion masses. The latter procedure was suggested in a recent publication 3 and has always been an inherent part of the AMT tag approach developed at our laboratory 22, 23. Here we provide quantitative measures of reducing the random component of the MME by using the mean of the groups as opposed to using individual peptide ion masses from individual scans.
The current approach for reduction of systematic and random mass measurement errors was experimentally validated by spiking known tryptic peptides into a sample of tryptic peptides from mouse brain. Cross validation was performed by training the regression models for the mouse brain tryptic peptides and evaluated using spiked control peptides. Consecutive application of the systematic and random error reduction procedures is shown to remove any noticeable bias and provide a significant and consistent decrease in the standard deviation of the mass measurement errors for both mouse brain and spiked-in peptides. After reducing both systematic and random error components, we find the standard deviation of the mass error distribution is decreased by 1.8- to 3.7-fold, depending on the instrument type. The decrease in the standard deviation provides a proportional reduction the mass error tolerances used to provide confident peptide identifications without any loss of correct identifications, thus reducing false peptide identifications and providing improved false discovery rates.
Here we present the analysis of systematic MME dependencies for four major types of commercially available high accuracy mass spectrometers. First, LTQ FT (Thermo Fisher Scientific Inc., Waltham, MA) is a hybrid instruments having ion-trap coupled with FTICR mass analyzer. Second, LTQ Orbitrap (Thermo Fisher Scientific Inc., Waltham, MA) is also a hybrid mass spectrometer having ion-trap coupled with novel mass analyzer, called Orbitrap. Both of the instruments equipped with AGC for controlling the total ion charge in the ion-trap and mass analyzer. Third, Agilent MSD TOF (Agilent Technologies, Santa Clara, CA) utilizes time-of-flight (TOF) approach for mass analyzer and analog-to-digital converter (ADC) for detection of ions. Forth, Micromass Q-TOF Ultima (Waters, Milford, MA) also a TOF instrument uses time-to-digital converter (TDC) for ion detector. Further for the most part of the text, to highlight the main instrumental differences in regards of ion mass measurement and detection, we are going to refer Agilent MSD TOF and Micromass Q-TOF Ultima as TOF ADC and Q-TOF TDC, respectively.
METHODS
Sample Preparation and LC-MS analysis
Mouse brain tryptic peptide sample preparation and LC-MS analyses are described elsewhere 24, 25. Here we also spiked mouse brain samples with peptides obtained by tryptic digest of 11 standard proteins: bovine serum albumin (Swiss Prot Accession number PO2769, Sigma-Aldrich cat. No. A7638), bovine carbonic anhydrase (Q865Y7, C3934), bovine beta-lactoglobulin (P02754, L3908), bovine serotransferrin (Q29443, T1408), rabbit glyceraldehyde-3-phosphate dehydrogenase (P46406, G2267), E.coli beta-galactosidase (P00722, G5635), bovine alpha-lactalbumin (P00711, L6010), equine skeletal muscle myoglobin (P02188, M0630), chicken ovalbumin (P01012, A2512), bovine cytochrome c (P00006, C2037), and rabbit phosphorylase b (P00489, P6635). An aliquot of 100 μg of mouse brain tryptic peptides was spiked with 0.5-2 μg of tryptic peptides from each of 11 known proteins.
The 10 μg aliquots were analyzed on several different mass spectrometers: LTQ FT (Thermo Fisher Scientific Inc., Waltham, MA), LTQ Orbitrap (Thermo Fisher Scientific Inc., Waltham, MA), Agilent MSD TOF (Agilent Technologies, Santa Clara, CA) and Micromass Q-TOF Ultima (Waters, Milford, MA). The LC-MS gradient length was 100 min for the LTQ FT, LTQ Orbitrap, and Agilent MSD TOF, and was 30 min for the Micromass Q-TOF. The LTQ FT and LTQ Orbitrap were operated with 5×105 and 1×106 ion population AGC setting, respectively. The resolution setting was 1×105 at 400-2000 m/z range for both instruments. Both TOF instruments had 1 sec signal integration time and were operated in the V-mode.
LC-MS data analysis
The spectra deisotoping followed by peak matching was done using in-house developed software ICR-2LS and VIPER 26, 27, respectively. The following regression analysis was done using scripts written in the R language for statistical computing 28. A demo R script is available in the supplement. For real-world data analysis, a variation of the present approach is embedded into the VIPER software.
The LC-MS features were defined as clusters of ion observations observed in consecutive MS spectra and having similar m/z values within certain tolerances. The m/z value of each LC-MS feature was computed as a median of the m/z values of the constituting ions. The elution time of the feature was defined as the time of the mass spectrum with the maximum ion abundance. Peptide identification was done by matching detected LC-MS features against a peptide database, which contains theoretical monoisotopic masses and consistently observed LC elution times. In this study, we used a database consisting of ∼44,000 peptides (corresponding to ∼8,000 proteins) confidently identified in the mouse brain 25, 29 and ∼1,200 confidently identified peptides from a tryptic digest of 11 spiked-in proteins. For the sake of simplicity, peptides common to both mouse brain and spiked-in standards were ignored.
Regression Analysis
The MME of an ion can be described as the combination of systematic and random measurement errors:
| (1) |
The systematic error in ion mass measurement is the result of variation in several known and unknown physical parameters whose combined effect is difficult to predict. Some parameters like m/z, ion intensity, total ion current, and temperature within the mass analyzer are known to affect MME. MME can depend on mass to charge ratio if the instrument calibration is outdated. It is also known that MME depends on TIC and ion intensity values because of electrostatic interactions between the ions that cause shifts of rotation frequencies in the ICR cells. Other physical parameters such as elution time are seen to affect the MME, but these relationships are only correlational, rather than causational, and represent gross changes in unknown physical parameters that change during the course of an experiment. While some of the fundamental parameters such as m/z and ion intensity are captured during the course of the experiment, others like temperature are not. Correspondingly, other gross correlational parameters such as elution time are captured and implicitly represent some information about unknown parameters.
In this report, we attempted to model the systematic errors, εsystematic, as a function of the four variables m/z, elution time, ion intensity and TIC. Since the ion intensity and TIC values reported by the Xcalibur software (Thermo Finnigan, CA) are normalized by the AGC accumulation times to 1 sec, we multiplied them by the accumulation times of the corresponding spectra to get the actual values in the ICR or Orbitrap cells. While it is extremely challenging to universally characterize systematic errors for all experiments using a function of these four parameters, it is possible to fit models separately for each experiment by observing the trend between MME and the parameters (Figure 1A). A statistical technique for modeling nonlinear trends in the space of multiple variables is the projection pursuit regression 20. The model expresses εsystematic in terms of a sum of univariate functions of linear combinations of the explanatory variables, translating the model (1) to
| (2) |
The proposed approach reduces systematic MME by estimating model (2) and subtracting off the fitted values from the mass measurements of individual ions, leaving only the random error component.
The scatter plots of MME vs. the relevant variables in Figure 1A reveal a combination of dense bands that represent true matches and scattered points that represent false matches. Dependency between MME and a variable can be seen from the band of true matches. The width of the dense bands reflects the amplitude of the random mass measurement error. The deviation of the trend of a dense band from the horizontal zero line serves as an indication of the systematic error. It can be seen that in some cases the contribution of the systematic error is comparable or even larger than the contribution of the random error into the deviation of individual mass measurement error residuals from the zero line. For example, the amplitude of the systematic MME trend for the TOF ADC dataset in response to the elution time parameter, or for the Q-TOF TDC dataset in response to the ion intensity parameter, exceeds the width of the band itself.
To reduce the interference of false identifications on the regression analysis, we filtered the data based on elution time agreement criterion and retained only peptide identifications having less than 1% difference with database elution time values. After performing regression analysis on this subset, the trained function was recursively applied to the entire dataset. Although the peptide identifications were enriched with true identifications using strict filtering criterion based on peptide elution times, there are still enough false identifications to potentially have a detrimental effect on standard least-squares regression approaches. Thus we employed a robust two step regression approach. In the first pass, we applied Tukey’s running median (3) 30, 31 to reduce the effect of outlying false identifications. The Tukey’s running median smoothed the error values by taking medians of the measurement errors εi in a sliding window of odd width 2k+1. We then additionally smoothed the results of the running median into continuous differentiable function with a cubic smoothing spline (4) 21 (Figure 2). Fitting of the smoothing spline g is based on minimization of the objective function (PRSS), which is the residual sum of squares plus the penalty for roughness weighted by the smoothing parameter λ.
| (3) |
| (4) |
Projection pursuit regression is an iterative process. Here for each of the iterations we considered points only within a certain interval around the zero line. For the first iteration those intervals were set at ±5 ppm, ±5 ppm, ±50 ppm and ±100 ppm for LTQ FT, LTQ Orbitrap, TOF ADC and Q-TOF TDC instruments, respectively. For subsequent iterations we either kept the intervals fixed or dynamically adjusted them to 4 standard deviations of the MME distribution of the true identifications, derived with an expectation-maximization (EM) algorithm. However, in practice there was no noticeable difference for the final results. The procedure was applied iteratively until convergence was achieved or a predefined maximum number of iterations were performed.
Figure 2.
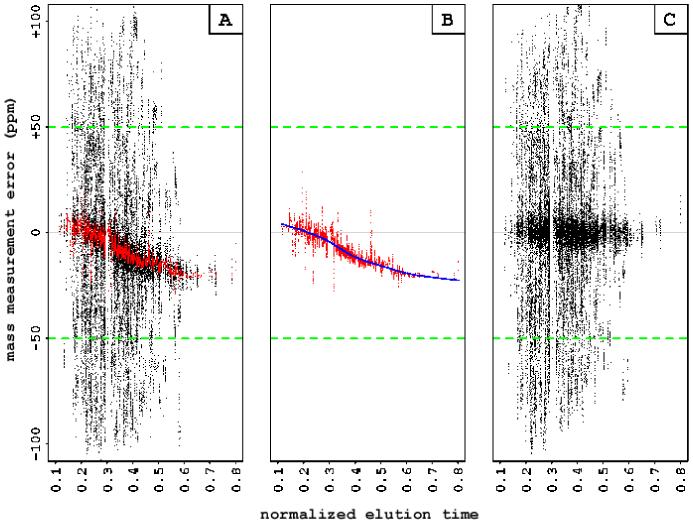
An illustration of the iterative regression-based systematic error elimination approach. The data shows dependency of the mass measurement errors on elution time for the TOF ADC instrument. We considered only data points that fell within a certain tolerance window (+/- 50 ppm, in the case of TOF ADC, indicated by the dashed green line). (A) At the first step, the data are smoothed using Tukey’s running median (red points). (B) At the second step, the running medians are fitted with a smoothing spline (blue line). (C) The predicted systematic errors are subtracted from the observed mass measurement errors for the entire dataset.
To fit model (2), we must estimate the β parameters and the function g. To estimate the β parameters, we used a variant of a hill climbing optimization algorithm with fixed direction set along the coordinate axis, and successive reduction of the learning step length along the dimension used for each move (Figure 3). For each step the ridge function g was fit and the goodness of fit evaluated as the average error within the initial fixed interval. As a ridge function g we used Tukey’s running median followed by smoothing spline as described above (Figure 2). We used average error or mean absolute error (MAE) instead of commonly used mean squared error (MSE) as a more robust goodness of fit measure in the presence of outlying false identifications.
Figure 3.
Outline of the algorithm for minimizing the systematic component of mass measurement error residuals based on elution time, m/z, ion intensities and TIC. The objective of the algorithm is to minimize the average error within a predefined error tolerance window using the projection pursuit regression approach. (A) Main iteration loop for minimization of the MAE value. (B) Variation of the hill climbing optimization algorithm for finding the optimal linear combination of the explanatory variables. This function is declared as FindOptimalProjection in the main iteration loop.
Model Selection
Besides potentially complex dependencies, there is some degree of correlation between the four selected explanatory variables under consideration. For example, the m/z of peptides is correlated with their elution times, with higher m/z peptides generally eluting later in an LC separation. Thus, although some variables may reveal trends in the mass measurement errors, they may be largely explained by correlation with other variables. To select the parameter set that sufficiently explains the systematic error, we searched for the model with the fewest variables and with the lowest prediction error.
To estimate the prediction errors of all 15 models with all possible parameter combinations as explanatory variables, we applied a 10-fold cross-validation approach. Unfortunately the mass measurement error residuals do not follow the normal distribution, as it is contain for example residuals of false matches. Thus, instead of relying on some sort of information criteria, we had to use computationally intensive cross-validation procedure, which does not require the assumption of normality of the error residual distribution. In the situation when the error residual do not follow any distribution model, the assessment of statistical significance becomes a non-trivial task. Nevertheless, we can certainly rank the performance of the models using the results of 10-fold cross validation and choose the best one. In such an approach the dataset is randomly split into 10 parts and then 9 parts used to fit (train) the regression model; however the performance (prediction error) is estimated based on the error residuals for the rest part not used for model training. Such cross validation approach penalizes overfitting. Particularly, increasing the complexity of the model by adding extra parameter, which indeed have no affect on the mass accuracy only increases the prediction error since the training and testing of the regression model performed on separate pieces of the dataset. The procedure repeated 10 times, so each of the 10 parts is used for model testing and the overall prediction error is taken as an average. The model having the minimal prediction error was considered the optimal solution (Figure 4).
Figure 4.
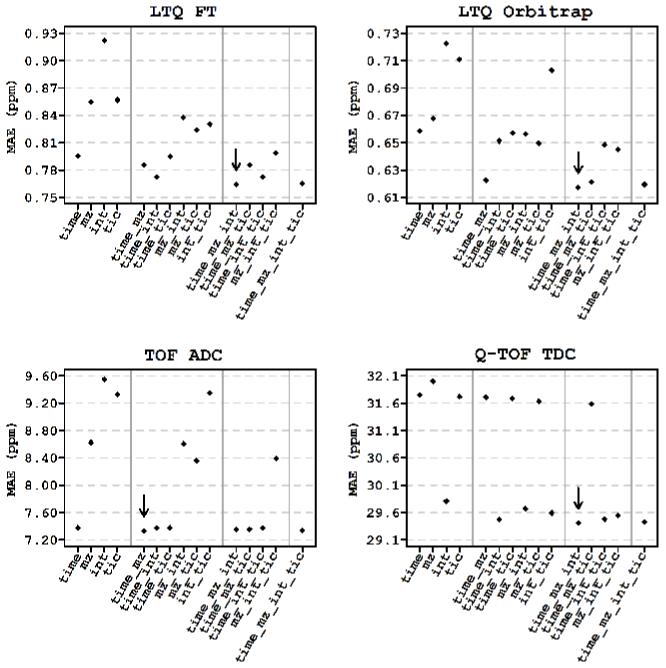
Selection of the optimal regression model by a 10-fold cross-validation approach. The lables “time”, “mz”, “int” and “tic” correspond to the explanatory variables elution time, m/z, ion intensity, and TIC, respectively. Grey vertical lines separate regression models of different complexity. Diamonds represent the average error value for the testing samples. The parameter combination of the best regression model is indicated by the arrow.
RESULTS
Figure 4 shows the performance of regression models having different combinations of explanatory variables for different LC-MS platforms. The results indicate that the systematic error depends on elution time, m/z and ion intensities in all platforms except TOF ADC. The systematic MME in the TOF ADC could be sufficiently explained by just two variables: elution time and m/z. Table 1 shows the results of applying the optimal models, along with the contributions of the individual variables to the systematic errors observed for each platform. After correction with the optimal models (Figure 1B), no apparent dependency was observed between MME and any of the four variables. The improvement in the mass measurement accuracy and precision was estimated by comparing the mean and the standard deviation of mass measurement error residuals of the correct matches before and after the application of the model. To estimate those parameters, we assumed a mixture model for the errors: normal distribution with relatively small standard deviation for the correct matches and another normal distribution with large standard deviation (∼50 ppm 32) for incorrect matches. We used the expectation-maximization algorithm to find maximum likelihood estimates of the means and standard deviations of the distributions (Table 1).
Table 1.
Results of applying the regression model to eliminate the systematic MME component
| LTQ FT | LTQ Orbitrap | TOF ADC | Q-TOF TDC | |
|---|---|---|---|---|
| mean before/aftera) | 0.056/-0.009 | -0.580/0.001 | -9.86/0.06 | -1.13/0.26 |
| SD before/aftera) | 0.93/0.71 | 0.69/0.54 | 8.2/4.0 | 8.5/5.5 |
| elution timeb) | 81.4% | 66.0% | 98.7% | 8.3% |
| m/zb) | 4.7% | 30.6% | 1.3% | 6.3% |
| ion intensityb) | 13.9% | 3.4% | --- | 85.4% |
| TICb) | --- | --- | --- | --- |
Reduction of the bias (ppm) and the standard deviation (ppm) after removing the systematic MME component using the optimal regression model.
Estimated contribution of individual parameters into reduction of the average error associated with systematic MME. The contributions were estimated by applying models with increasing number of parameters and scaling the decrease of the average error from 0 to 100%. Where 0% is the average error of zero-centered MME, and 100% is the average error after applying the optimized model. The order at which the parameters were added is derived from the results of model performance estimation (Figure 4) and starts from the most important to the least important. The order is elution time, ion intensity, m/z for LTQ FT, elution time, m/z, ion intensity for LTQ Orbitrap, elution time, m/z for TOF ADC and ion intensity, elution time, m/z for Q-TOF TDC.
Evaluation of Regression Models by Spiking a Set of Known Peptides
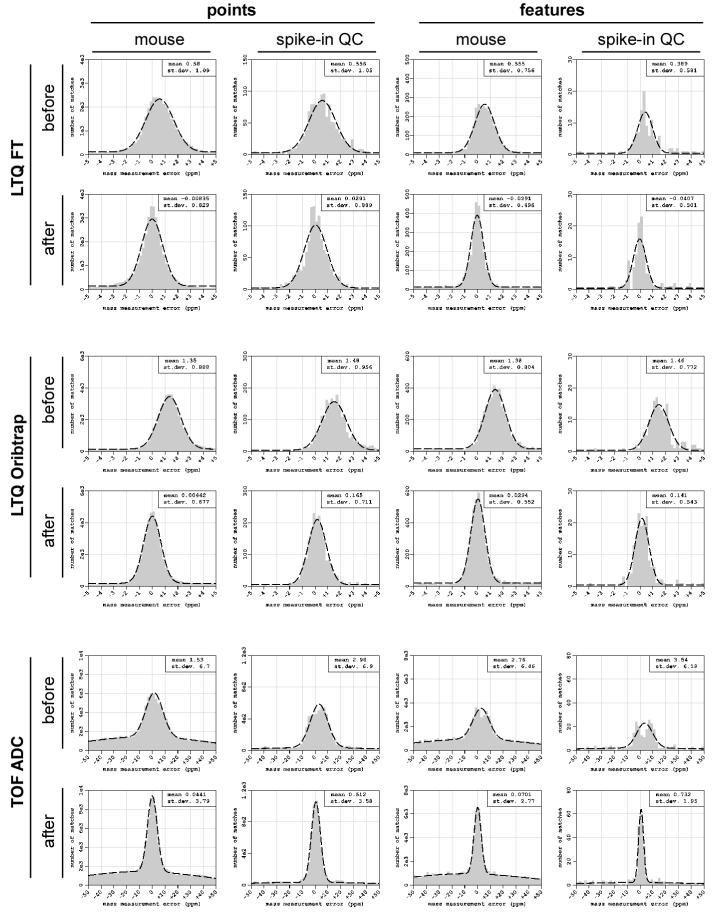
We used a mouse brain tryptic peptide sample spiked with tryptic peptides from a set of known purified proteins to validate the approach. The regression model was trained using only the mouse brain peptides, and the fitted model was applied to all the peptides, including the spiked set of peptides. It was expected that if the procedure performed correctly, both mouse brain peptides and the spiked peptides would show similar distributions of MME after correction. Indeed, Figure 5 shows that the reduction of the standard deviation of MME of both the brain tryptic peptides and peptides from the added protein are comparable. Besides elimination of the overall bias, the reduction of the standard deviations based on spiked-in sample is about 1.3-fold for LTQ FT, 1.2-fold for LTQ Orbitrap instruments and about 1.9-fold for the TOF ADC MS.
Figure 5.
Validation of the systematic error elimination approach using a set of known, spiked-in peptides. Mass error distribution histograms were plotted before and after the procedure. The mean and the standard deviation of the correct matches were estimated using the EM algorithm. The regression model was trained only on tryptic peptides from mouse brain, but applied to all peptides including spiked-in QC peptides. The reduction in MME of the spiked peptides was comparable to that observed for the mouse peptides.
Reduction of the Random Error by Averaging the Mass Error Residuals
In LC-MS measurements, the elution profile of an individual peptide usually consists of several (1-100) consecutive spectra and depends on the peak capacity of the LC column, the gradient length, and the time required for the instrument to perform one scan, i.e. the duty cycle. As mentioned earlier for the AMT tag approach, an ion seen in multiple consecutive spectra with similar m/z values will have its observations grouped together. The difference between the theoretical monoisotopic masses of the peptides and the representative mass of the groups of ions is one of the main criteria for discrimination between the true and false peptide identifications. The m/z value of each group is computed as a median of the m/z values of the individual measurements. Here we explored the extent of reduction of the mass measurement error by observing the reduction in MME between the median of the m/z values of the groups and the individual m/z values. For the analyzed datasets, we observed an additional 1.8-, 1.3- and 1.8-fold reduction of the standard deviation of the peptide/group mass matching errors for the LTQ FT, LTQ Orbitrap and TOF ADC instruments, respectively (Figure 5).
DISCUSSION
The present report emphasizes the importance of selecting parameters that actually affect the systematic mass measurement errors for a given MS instrument. Retaining only significant parameters allows the reduction of data dimensionality, and thus avoiding overfitting and reducing computational time. Our approach does not rely on any specific knowledge about the factors affecting MME for each instrument type. The significance of the factors is discovered based solely on a statistical analysis of the mass error residuals of peaks matching a set of “expected” peptides. Most of the factors found to affect the MME for the studied datasets have previously been reported or commonly expected to have an affect on MME. For example, it is known that TOF instruments may have a pronounced drift in the mass measurement accuracy with time. It should be noted that this drift is likely to be attributed to the temperature-dependent changes of the power supply voltage and the expansion of the flight tube during the analysis, but not the LC elution time per se. One widely applied solution to this problem involves the introduction of calibrants e.g. from a separate electrospray ionization source into the instrument. In this report we did not consider the actual temperature of the power supply and the flight tube during the spectrum acquisition as a parameter for each mass spectrum, but including additional temperature variables in the regression analysis might be helpful in decoupling temperature from other unknown parameters and could potentially provide a more precise understanding of systematic mass measurement errors. Similar considerations apply for the LTQ Orbitrap instrument for which temperature change has been reported to be a primary factor causing systematic mass measurement errors 3.
Although we have successfully applied the projection pursuit regression approach with robust splines as ridge functions for post hoc systematic error elimination, we would like to discuss some potential limitations and corresponding improvements of the approach. For example, as we mentioned before LC elution time may be a statistically good explanatory variable for systematic error. However, elution time itself is not a physical factor directly affecting the mass accuracy, but simply reflects the changes of some other parameter with time, for example thermal expansion of the flight tube on the TOF instruments, change of the power supply voltage on Orbitrap and TOF instruments or, as we mentioned above, ion composition in the ICR cell. Potentially a problem may arise if any of these parameters (e.g. power supply voltage) undergo sudden change in time such that the systematic mass measurement error caused by the sudden change appears comparable or large than standard deviation of the random errors. If the change is step-like and not smooth enough it may not be easily captured with the spline or some other continues function in the time domain. However such change is likely to be well fit with a smooth continues function on the domain of the actual physical parameter directly affecting the mass measurement errors (e.g. power supply voltage). Thus it would helpful if developers of the commercial mass spectrometers make possible to log such an information such as temperature of the flight tube and power supply voltage for the acquire scans and make it user accessible. In addition it would also be helpful if such information could be stored in widely accepted mass spectrometry data storage and exchange formats like mzXML 33 and mzData 34. Moreover, neither of the formats currently contains information on AGC accumulation times for hybrid instrumentation like LTQ FT and LTQ Orbitrap, thus it becomes impossible to account for the actual ion intensities and TIC parameters in recalibration, since we used that accumulation time to compute the mentioned parameter values in the ICR and Orbitrap cells because Xcalibur software returns those values normalized to 1 sec. Inclusion of more information into both of those formats should facilitate the development and dissemination of the software employing the described and perhaps other recalibration strategies accounting for multiple parameters affecting systematic mass measurement errors.
The present approach provides some new insights into aspects of platform performance. For example, the observed dependency of the LTQ FT systematic error on the LC elution time can not be explained just by correlation of elution time with m/z values. Indeed the performance of the regression model noticeably improves if elution time parameter accounted in addition to m/z and ion intensity (Figure 4). Such a dependency is somewhat unexpected and does not have a straightforward explanation. The drift of the magnetic field is an almost negligible factor for mass measurement accuracy and is typically < 0.05 ppm/hour for commercial instruments, and thus should not be more than 0.1 ppm for a 2 hour LC-MS measurement. It is likely that the observed dependency reflects a dependency on other factors. For example, although the LTQ FT instrument is equipped with AGC, which acts to minimize variation the total ion current level, it is apparent that control is imperfect and that the actual stored ion populations vary with time (Figure 6A), although certainly much greater variation would be observed without the use of AGC. We tentatively attribute the observation to the changes of the average peptide mass (and m/z) during the course of a separation (Figure 6B). Heavier species, with higher charge states, tend to elute later from the reversed phase LC column. The average ion charge of the peptides coming out of the LC column changes from ∼2 to ∼3, if to compare beginning and the end of the gradient. Thus, the total signal, m/z, and average ion charge values all have some correlation with LC elution time, and potentially making it a better explanatory variable.
Figure 6.
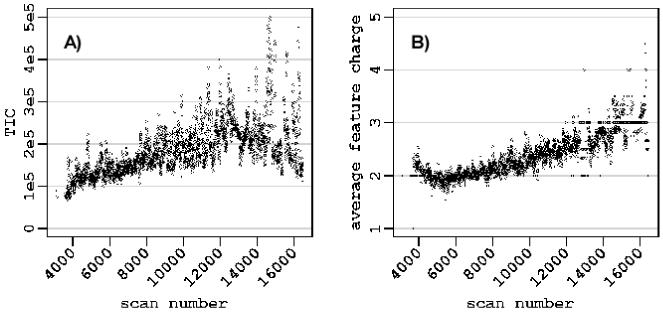
Profiles of TIC (A) and averaged LC-MS feature charge (B) for the LC-MS analysis using the LTQ FT.
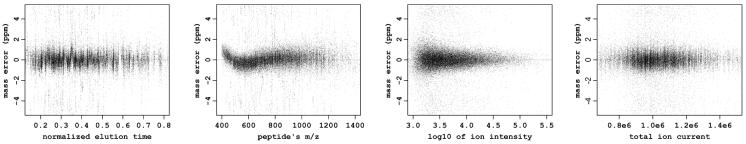
Another interesting finding is that in certain cases, in particular for LTQ Orbitrap instrument, there are clearly non-linear trends in dependency of systematic mass measurement errors on m/z (Figure 1A). To exclude the influence of other parameters we applied a regression model taking into account only the elution time, ion intensity and TIC and plot the resulting error residuals (Figure 7). We would like to emphasize that the dependency of the systematic error on the m/z has still remained non-linear and rather complicated. Thus, such complicated behavior can not be explained by correlation of m/z with other parameters like elution time. This raises an important issue, that in this case calibration curve can not be perfectly fitted with commonly used linear function. In the presented procedure it is well approximated with non-parametric spline function (Figure 1B). However, for other three types of instruments we did not notice any significant deviations from linear dependency of the mass measurement errors on m/z.
Figure 7.
Scatter plots showing the dependencies of mass measurement errors on elution time, m/z, ion intensity and TIC for LTQ Orbitrap instrument after apply systematic error elimination procedure taking into account all parameters other than m/z. The complicated non-linear dependency of mass measurement error on m/z still remained while none of other parameters used for regression model reveals any residual trend.
The m/z and LC elution time variables are also correlated with each other. For example, for the 100 min LC-MS analysis using the TOF instrument, the dependency trend of the error residuals on the m/z value primarily can be explained by the drift of mass measurement accuracy with LC elution time. This observation suggests some caution in using the entire LC-MS measurement dataset for calibrations schemes which does not take into account the elution time 4, 16, as the dependency on m/z may be primarily due to other factors (e.g. elution time) that correlate with m/z. The contribution of m/z to the systematic mass measurement error is highly variable between instruments, and is likely to depend on how long ago the instrument was calibrated, how stable the calibration is, and the length of the LC-MS analysis.
Ion intensity was the most significant factor affecting mass measurement accuracy for the Q-TOF TDC. A change of one order of magnitude of the ion intensity caused a shift of the measured mass errors by about 50 ppm. Such a dependency has a been previously observed, and a correction procedure has been proposed 35. This difference in behavior of the MME between the TOF ADC and Q-TOF TDC instruments in response to the ion intensity value stems from the differences in the ion detectors. The ADC takes the maximum of a signal’s peak as an ion arrival time; whereas the TDC detects an ion arrival time as soon as its signal crosses certain preset threshold level. This behavior causes mass measurements by TDC-based detectors to be very sensitive to ion intensities, since multiple ions create a signal that cross the threshold faster, effectively making the ion masses appear to be of slightly lower m/z. To isolate and visualize the effects of ion intensity parameter on the mass measurement errors for all four instruments we applied the regression models with elution time, m/z and TIC, to make sure all dependencies on other parameters are removed, and plotted the error residuals versus ion intensity (Figure 8). The effect of ion intensity on the mass measurement errors is quite pronounced for LTQ FT and rather subtle for LTQ Obritrap. Indeed, the effect of individual ion intensity on mass accuracy is well known for FTICR instruments, and recently a recalibration procedure has been proposed accounting for individual peak intensities 36. Intensity dependent effect for mass accuracy has also been observed for LTQ Orbitrap instrumentation 3.
Figure 8.
Scatter plots showing the dependency of mass measurements error residuals on ion intensity after applying systematic error elimination procedure taking into account only elution time and m/z parameters. The dependency of systematic mass measurement error on ion intensity is highly pronounced for Q-TOF TDC and LTQ FT instruments as a bias of high intense ions towards lighter masses. For LTQ Orbitrap instrument the dependency is significantly less and present as a bias of low intense ions towards heavier masses. Mass measurement error for the TOF ADC does not exhibit any dependency on ion intensity.
Another important observation is the variation of the random error component (width of the trend) across some parameters. This effect is most apparent from the scatter plot of mass measurement error residuals vs. ion intensity for the LTQ FT after the systematic error elimination. We estimated the standard deviations for the subsets of correct matches for low and high intensity ions that have log10 ion intensity values of 3.0±0.1 and 4.0±0.1, respectively. The standard deviation values estimated with the EM algorithm for low and high intensity ions have a 2.5-fold difference, with values of 0.935 ppm and 0.365 ppm, respectively. The observation can be explained by a shorter transient signal in the ICR cells for low intensity ions. Precision of the mass measurement is also known to linearly decrease with m/z of an ion for ICR instruments and as a square root of m/z for the Orbitrap. These factors indicate heteroscedasticity and a mixture distribution for the random mass measurement errors, with standard deviations being a function of ion intensity and m/z. The issue is more complicated for the groups of ions representing the same peptide, as the amplitude of the reduction of the random error depends on the number of observations in a group. However, in the EM algorithm reported here, we used only a single normal distribution for correct matches and another normal distribution for incorrect matches. Having one normal distribution for correct matches greatly reduces the computational burden and simplifies comparisons of dataset measurements. Overall, the two mixture model proved effective (Figure 5), noticeable deviation was observed only for the LTQ FT, which exhibited the strongest heteroscedasticity along the intensity and m/z parameters.
In this work we demonstrated a method for correction of systematic MME based on statistics of peptides identified with AMT tag approach. It is important that the systematic mass measurement error component is minimized or eliminated from LC-MS datasets, since they have a potential to cause artificial bias in the assignment of the confidence of the peptide identifications. Since accurately measured peptide mass is one of the parameters used for peptide identification in AMT tag approach, the benefit of using this method is apparent as it helps to reduce the maximum allowable mass deviation and better discriminate between correct and incorrect identifications.
However, the reported algorithm can also benefit another widely used bottom-up proteomic approach based on MS/MS fragmentation patterns if accurately measured parent ion mass is one of the parameters used for discrimination of correct and incorrect peptide identifications. For instance, for the instruments equipped both with fragmentation cells and high accuracy mass analyzers, the subset of peptides enriched with true identifications can be selected based on the goodness of matching of the observed fragmentation patterns with expected ones. XCorr and ΔCn values from SEQUEST 37 or E-values from X!Tandem 38 are couple of typical scores used for discrimination between true and false peptide identifications based on their fragmentation patterns. The subset of peptides enriched with true identification can be used for elimination of the systematic MME based on dependencies on m/z, elution time, ion intensity and other parameters as described in this report. To mitigate the effect of random errors, the peptide’s mass can be estimate as a mean of peptide’s ion masses from few consecutive scans 3, 22. Finally, after elimination of systematic and reducing the random errors, parent ion MME can be used as additional criteria to reduce the number of false identifications to achieve lower FDR and higher confidence. Based on the results from the current work, we expect that the presented procedure should results in a 2-4 fold decrease of the number of false positive identifications as to compare to simply using the raw, unrefined MME values.
Projection pursuit regression is computationally intensive procedure, especially given the size of the proteomic datasets (on average ∼105 detected isotopic envelops in this work). A reasonable compromise could be to use a simple additive model (5), which avoids the search for optimal projections and performs regressions along the parameter dimensions. Thus, it is drastically less computationally intensive, but may not potentially capture complicated inter-parameter dependencies as a possible trade off.
| (5) |
The simple additive regression model has been implemented in the software developed at our laboratory 26, 27, 39 and currently allows for correction of the systematic mass measurement errors as a function of elution time and m/z of the LC-MS features (Figure 9).
Figure 9.
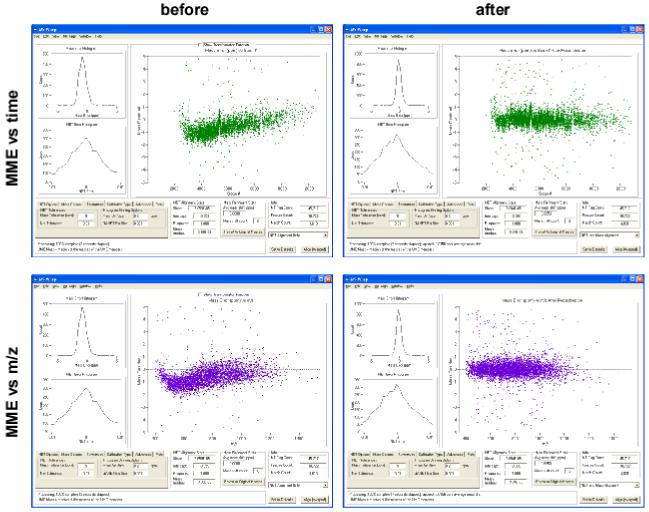
One of the user interfaces of the VIPER software. Scatter plots show the mass measurement error residuals of the LC-MS features before and after correction. The example shows data obtained using an LTQ Orbitrap mass spectrometer.
The presented algorithm for reduction of the MME based on statistics from errors for putatively identified components will be applied in future efforts at our laboratory and also expect it to be adopted for a wide range of related LC-MS approaches (e.g. top-down proteomics, metabolomics) to increase the confidence of identifications for the data acquired on the high-resolution instrumentation.
AKNOWLEDGEMENTS
The research was supported by the NIH National Center for Research Resources (RR18522 to RDS). Proteomic analyses were performed in the Environmental Molecular Sciences Laboratory, a U.S. Department of Energy (DOE) national scientific user facility located at the Pacific Northwest National Laboratory (PNNL) in Richland, Washington. PNNL is a multi-program national laboratory operated by Battelle Memorial Institute for the DOE under Contract DE-AC05-76RL01830.
REFERENCES
- (1).Qian WJ, Camp DG, 2nd, Smith RD. Expert Rev Proteomics. 2004;1:87–95. doi: 10.1586/14789450.1.1.87. [DOI] [PubMed] [Google Scholar]
- (2).Haas W, Faherty BK, Gerber SA, Elias JE, Beausoleil SA, Bakalarski CE, Li X, Villen J, Gygi SP. Mol Cell Proteomics. 2006;5:1326–1337. doi: 10.1074/mcp.M500339-MCP200. [DOI] [PubMed] [Google Scholar]
- (3).Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M. Mol Cell Proteomics. 2005;4:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- (4).Jaffe JD, Mani DR, Leptos KC, Church GM, Gillette MA, Carr SA. Mol Cell Proteomics. 2006;5:1927–1941. doi: 10.1074/mcp.M600222-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Loboda AV, Krutchinsky AN, Bromirski M, Ens W, Standing KG. Rapid Commun Mass Spectrom. 2000;14:1047–1057. doi: 10.1002/1097-0231(20000630)14:12<1047::AID-RCM990>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- (6).Palmer ME, Clench MR, Tetler LW, Little DR. Rapid Communications in Mass Spectrometry. 1999;13:256–263. [Google Scholar]
- (7).Belov ME, Zhang R, Strittmatter EF, Prior DC, Tang K, Smith RD. Anal Chem. 2003;75:4195–4205. doi: 10.1021/ac0206770. [DOI] [PubMed] [Google Scholar]
- (8).Herniman JM, Bristow TW, O’Connor G, Jarvis J, Langley GJ. Rapid Commun Mass Spectrom. 2004;18:3035–3040. doi: 10.1002/rcm.1729. [DOI] [PubMed] [Google Scholar]
- (9).Strittmatter EF, Rodriguez N, Smith RD. Anal Chem. 2003;75:460–468. doi: 10.1021/ac026057g. [DOI] [PubMed] [Google Scholar]
- (10).Strittmatter EF, Ferguson PL, Tang K, Smith RD. J Am Soc Mass Spectrom. 2003;14:980–991. doi: 10.1016/S1044-0305(03)00146-6. [DOI] [PubMed] [Google Scholar]
- (11).Bruce JE, Anderson GA, Brands MD, Pasa-Tolic L, Smith RD. J Am Soc Mass Spectrom. 2000;11:416–421. doi: 10.1016/S1044-0305(00)00096-9. [DOI] [PubMed] [Google Scholar]
- (12).Matthiesen R, Trelle MB, Hojrup P, Bunkenborg J, Jensen ON. J Proteome Res. 2005;4:2338–2347. doi: 10.1021/pr050264q. [DOI] [PubMed] [Google Scholar]
- (13).Matthiesen R. Proteomics. 2007;7:2815–2832. doi: 10.1002/pmic.200700116. [DOI] [PubMed] [Google Scholar]
- (14).Zubarev R, Mann M. Mol Cell Proteomics. 2006 doi: 10.1074/mcp.M600380-MCP200. [DOI] [PubMed] [Google Scholar]
- (15).Palmblad M, Bindschedler LV, Gibson TM, Cramer R. Rapid Commun Mass Spectrom. 2006;20:3076–3080. doi: 10.1002/rcm.2707. [DOI] [PubMed] [Google Scholar]
- (16).Yanofsky CM, Bell AW, Lesimple S, Morales F, Lam TT, Blakney GT, Marshall AG, Carrillo B, Lekpor K, Boismenu D, Kearney RE. Anal Chem. 2005;77:7246–7254. doi: 10.1021/ac050640q. [DOI] [PubMed] [Google Scholar]
- (17).Becker CH, Kumar P, Jones T, Lin H. Anal Chem. 2007;79:1702–1707. doi: 10.1021/ac061359u. [DOI] [PubMed] [Google Scholar]
- (18).Tolmachev AV, Monroe ME, Jaitly N, Petyuk VA, Adkins JN, Smith RD. Anal Chem. 2006;78:8374–8385. doi: 10.1021/ac0606251. [DOI] [PubMed] [Google Scholar]
- (19).Härdle W. Applied nonparametric regression. Cambridge University Press; Cambridge [England] ; New York: 1990. [Google Scholar]
- (20).Friedman JH, Stuetzle W. Journal of the American Statistical Association. 1981;76:817–823. [Google Scholar]
- (21).Hastie T, Tibshirani R, Friedman JH. The elements of statistical learning : data mining, inference, and prediction : with 200 full-color illustrations. Springer; New York: 2001. [Google Scholar]
- (22).Zimmer JS, Monroe ME, Qian WJ, Smith RD. Mass Spectrom Rev. 2006;25:450–482. doi: 10.1002/mas.20071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Lipton MS, Pasa-Tolic L, Anderson GA, Anderson DJ, Auberry DL, Battista JR, Daly MJ, Fredrickson J, Hixson KK, Kostandarithes H, Masselon C, Markillie LM, Moore RJ, Romine MF, Shen Y, Stritmatter E, Tolic N, Udseth HR, Venkateswaran A, Wong KK, Zhao R, Smith RD. Proc Natl Acad Sci U S A. 2002;99:11049–11054. doi: 10.1073/pnas.172170199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Wang H, Qian WJ, Mottaz HM, Clauss TR, Anderson DJ, Moore RJ, Camp DG, 2nd, Khan AH, Sforza DM, Pallavicini M, Smith DJ, Smith RD. J Proteome Res. 2005;4:2397–2403. doi: 10.1021/pr050160f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Petyuk VA, Qian WJ, Chin MH, Wang H, Livesay EA, Monroe ME, Adkins JN, Jaitly N, Anderson DJ, Camp DG, 2nd, Smith DJ, Smith RD. Genome Res. 2007;17:328–336. doi: 10.1101/gr.5799207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26). http://ncrr.pnl.gov/software.
- (27).Monroe ME, Tolic N, Jaitly N, Shaw JL, Adkins JN, Smith RD. Bioinformatics. 2007 doi: 10.1093/bioinformatics/btm281. [DOI] [PubMed] [Google Scholar]
- (28). http://www.r-project.org.
- (29).Wang H, Qian WJ, Chin MH, Petyuk VA, Barry RC, Liu T, Gritsenko MA, Mottaz HM, Moore RJ, Camp Ii DG, Khan AH, Smith DJ, Smith RD. J Proteome Res. 2006;5:361–369. doi: 10.1021/pr0503681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Tukey JW. Exploratory data analysis. Addison-Wesley Pub. Co.: Reading, Mass.; 1977. [Google Scholar]
- (31).Hardle W, Steiger W. Applied Statistics-Journal of the Royal Statistical Society Series C. 1995;44:258–264. [Google Scholar]
- (32).Wolski WE, Farrow M, Emde AK, Lehrach H, Lalowski M, Reinert K. Proteome Sci. 2006;4:18. doi: 10.1186/1477-5956-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Pedrioli PG, Eng JK, Hubley R, Vogelzang M, Deutsch EW, Raught B, Pratt B, Nilsson E, Angeletti RH, Apweiler R, Cheung K, Costello CE, Hermjakob H, Huang S, Julian RK, Kapp E, McComb ME, Oliver SG, Omenn G, Paton NW, Simpson R, Smith R, Taylor CF, Zhu W, Aebersold R. Nat Biotechnol. 2004;22:1459–1466. doi: 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- (34).Orchard S, Taylor C, Hermjakob H, Zhu W, Julian R, Apweiler R. Expert Rev Proteomics. 2004;1:179–183. doi: 10.1586/14789450.1.2.179. [DOI] [PubMed] [Google Scholar]
- (35).Kofeler HC, Gross ML. J Am Soc Mass Spectrom. 2005;16:406–408. doi: 10.1016/j.jasms.2004.11.022. [DOI] [PubMed] [Google Scholar]
- (36).Masselon C, Tolmachev AV, Anderson GA, Harkewicz R, Smith RD. J Am Soc Mass Spectrom. 2002;13:99–106. doi: 10.1016/S1044-0305(01)00333-6. [DOI] [PubMed] [Google Scholar]
- (37).Yates JR, 3rd, Eng JK, McCormack AL, Schieltz D. Anal Chem. 1995;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- (38).Fenyo D, Beavis RC. Anal Chem. 2003;75:768–774. doi: 10.1021/ac0258709. [DOI] [PubMed] [Google Scholar]
- (39).Jaitly N, Monroe ME, Petyuk VA, Clauss TR, Adkins JN, Smith RD. Anal Chem. 2006;78:7397–7409. doi: 10.1021/ac052197p. [DOI] [PubMed] [Google Scholar]