Abstract
The terminal differentiation of B cells into antibody-secreting plasma cells is tightly regulated by a complex network of transcription factors. Here we evaluated the role of the Ets factor Spi-B during terminal differentiation of human B cells. All mature tonsil and peripheral blood B-cell subsets expressed Spi-B, with the exception of plasma cells. Overexpression of Spi-B in CD19+ B cells inhibited, similar to the known inhibitor BCL-6, the expression of plasma cell–associated surface markers and transcription factors as well as immunoglobulin production, ie, in vitro plasma cell differentiation. The arrest in B-cell differentiation enforced by Spi-B was independent of the transactivation domain, but dependent on the Ets-domain. By chromatin immunoprecipitation and assays using an inducible Spi-B construct BLIMP1 and XBP-1 were identified as direct target genes of Spi-B mediated repression. We propose a novel role for Spi-B in maintenance of germinal center and memory B cells by direct repression of major plasma cell factors and thereby plasma cell differentiation.
Introduction
Germinal centers (GCs) are specialized areas in the follicles of lymphoid organs, where B cells on antigen challenge undergo multiple rounds of proliferation, accompanied by somatic hypermutation and immunoglobulin (Ig) class-switch recombination,1 generating memory B cells, or, alternatively, plasma cells (PCs). Memory B cells retain a high-affinity B-cell receptor (BCR) at their cell surface, do not secrete antibody, and have the intrinsic ability to respond rapidly and proliferate strongly on secondary encounter with antigen.2 The formation of nondividing antibody-producing PCs is controlled by a complex network of transcription factors.3 BLIMP1, encoded by the PRDM1 gene, is essential for PC formation and Ig secretion4 by initiating a gene regulation cascade, which leads to cessation of the cell cycle, repression of genes that are required for the identity of mature and GC B cells, and induction of the Ig secretory program.5 Furthermore XBP-1, which is controlling the secretory machinery of PCs,6,7 and IRF-4 play an essential role in PC differentiation.8,9 Induction of PC differentiation requires an active suppression of the B-cell phenotype, ie, of factors that are expressed in GC B cells, most importantly BCL-6 and PAX-5.3,10 These factors have been shown to inhibit differentiation of activated B cells, allowing sufficient time for affinity maturation and class-switch recombination to occur in response to antigen and T-cell signals. The proteins act predominantly by repression of the factors required for PC differentiation,11–16 resulting in a double-negative feedback mechanism that ensures maintenance of different developmental states in a mutually exclusive manner.3
In addition to BCL-6 and PAX-5, the Ets factor Spi-B is directly repressed by BLIMP1 in murine B cells,5 suggesting that the regulation of Spi-B is important in PC differentiation. Spi-B– deficient mice,17 which have normal B-cell numbers, show a defect in GC formation and maintenance, precluding the assessment of the role of Spi-B during later stages of B-cell differentiation. Other cells that express Spi-B include early T lineage cells and plasmacytoid dendritic cells (pDCs).18–20 Spi-B is crucial for development of human pDCs19,21 but not for human B-cell development,21 consistent with data from Spi-B–deficient mice.17 Furthermore, it was recently shown that the Spi-B locus is translocated in the activated B cell–like (ABC) diffuse large B-cell lymphoma (DLBCL) cell line OCI-Ly3,22 leading to increased expression of the transcription factor. To determine whether the overexpression of Spi-B is linked to the pathophysiology of this lymphoma subtype, it is required to understand the function of Spi-B in human B-cell differentiation.
Our data suggest a role for Spi-B in controlling differentiation of human B cells by repressing the induction of the plasma cell gene expression program. Spi-B bound the regulatory elements of PRDM1 and XBP1, encoding BLIMP1 and XBP-1, respectively, and directly repressed expression of these 2 factors.
Methods
B-cell isolation
Tonsillectomies were performed at the Department of Otolaryngology at the Academic Medical Center, Amsterdam. The use of this tissue was approved by the medical ethical committees of the institution and was contingent on informed consent. Studies were conducted in accordance with the Declaration of Helsinki for the use of human tissue. Buffy coats were obtained from Sanquin bloodbank (Amsterdam, The Netherlands). CD19+ B cells were isolated by positive selection using MACS CD19-coupled microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany), resulting in purities of more than 98%. Tonsil and peripheral blood subpopulations were separated by FACSAria sorting.
Flow cytometry
Monoclonal antibodies against human CD3, CD19, CD20, CD27, CD38, CD40, CD123, IgD, IgG, and NGFR directly conjugated with FITC, PE, APC, PE-Cy7, or APC-Cy7 were purchased from BD Biosciences PharMingen (San Diego, CA), CD138-PE from Dako North America (Carpinteria, CA), BDCA2-APC from Miltenyi Biotec, and CD40-PE from Beckman Coulter (Fullerton, CA). Samples were analyzed by flow cytometry on a LSRII (BD Biosciences PharMingen) and analyzed using FlowJo software (TreeStar, Ashland, OR).
B-cell cultures
For PC differentiation assay, B cells were cocultured on irradiated CD40L expressing, stably transfected L cells, together with interleukin-2 (IL-2; 40 U/mL; R&D Systems, Abdingdon, United Kingdom) and murine (m) IL-21 (50 ng/mL, R&D Systems) in Iscove modified Dulbecco medium with 8% fetal calf serum. After 72 hours, L cells were withdrawn and B cells were cultured with cytokines only.
pDC cultures
CD34+CD1a− human postnatal thymus progenitors were cultured on a layer of OP9 stromal cells in the presence of 5 ng/mL of IL-7 (PeproTech, Rocky Hill, NJ) and Flt3L (PeproTech)23 and analyzed after 7 days.
Constructs and retroviral transductions
Retroviral constructs Spi-B and BCL-6 LZRS-IRES-GFP were described previously.19,24 Spi-B/fl∼ER and Spi-B/ΔEts∼ER fusion protein vectors were constructed by fusing the C-terminus of full-length or ΔEts Spi-B, respectively, to a truncated murine estrogen receptor.24 Spi-B/ΔTAD was obtained from Spi-B PCR amplification of cDNA generated from CD34+CD38+ fetal liver cells. By sequencing, the location and length of the truncation (215 bp) of Spi-B/ΔTAD were determined. For virus production, the constructs were transfected into the Phoenix-GalV packaging cells.24 Control cells were transduced with empty LZRS-IRES-GFP constructs. Before transduction, isolated CD19+ B cells were cocultured with L cells and mIL-21 (50 ng/mL) for a minimal duration of 36 hours. Cells were transferred to plates coated with fibronectin (30 μg/mL, Takara, Kyoto, Japan) and incubated with virus for 6 to 8 hours. To induce translocation of ER fusion proteins, cells were treated with 0.5 μM 4-hydroxytamoxifen (4HT; Sigma-Aldrich, St Louis, MO). De novo protein translation was inhibited by preincubation with 2.8 μg/mL cycloheximide (CHX; Sigma-Aldrich).
Immunoblot analysis
Western blotting was performed as described.19 Membranes were incubated with antibodies against human Spi-B20,25 (kindly provided by Lee Ann Sinha, State University of New York, Buffalo, NY, and F. Moreau-Gachelin, Institute Curie, Paris, France), BLIMP126 (kindly provided by R. Tooze, Leeds Institute of Molecular Medicine, Leeds, United Kingdom), PAX-527 (kindly provided by Stephen Nutt, Walter and Eliza Hall Institute of Medical Research, Parkville, Australia), BCL-6 (C-19), IRF-4 (M-17), estrogen receptor (ER) (MC-20), or actin (I-19, all Santa Cruz Biotechnology, Santa Cruz, CA).
Quantitative real-time PCR
Total mRNA was isolated from cells using RNeasy mini kit (Qiagen, Valencia, CA) and reverse-transcribed into cDNA with first strand buffer, superscript II reverse transcriptase (Invitrogen, Carlsbad, CA), dNTP (Roche Diagnostics, Mannheim, Germany), and Oligo(dT) (Promega, Madison, WI). For quantitative polymerase chain reaction (PCR), we used an iCycler and SYBR green Supermix (Bio-Rad, Hercules, CA). Each sample was analyzed in duplicates, and expression levels were normalized to β-actin expression. Primer sequences for BCL-6, BLIMP1, and β-actin24 and Spi-B23 are published; other primers are listed in Tables S1 and S2 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Chromatin immunoprecipitation
A total of 8 × 106 SpiB∼ER−GFP+RAJI cells were incubated with or without 4HT for 4 hours. Chromatin immunoprecipitation (ChIP) was performed according to an adapted version of the Upstate ChIP kit protocol (Upstate Biotechnology, Charlottesville, VA). Immunoprecipitation was performed with either 3 μg polyclonal anti-ER antibody (Santa Cruz Biotechnology) or 3 μg normal rabbit IgG (Invitrogen). Precipitated chromatin was purified with QIAmp DNA mini kit (Qiagen) analyzed by icycler PCR. Primers are listed as supplemental data. Each ChIP was performed in triplicates and each PCR reaction in duplicates.
Enzyme-linked immunosorbent assay
Plates were coated with capture Abs antihuman IgG or IgM (Dako) at 10 μg/mL washed in enzyme-linked immunosorbent assay (ELISA) wash buffer; 10% fetal calf serum in phosphate-buffered saline was used as blocking agent and diluent for cell supernatants and for enzyme-conjugated detection antibodies. TMB substrate/stop solution (BioSource International, Camarillo, CA) was used for development of IgG and IgM ELISAs.
Results
Spi-B is expressed in all human B-cell subsets but not in PCs
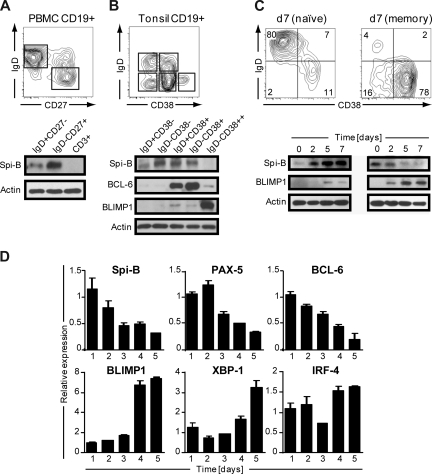
Human B cells express Spi-B,19 but its role during terminal B-cell differentiation has not been investigated. Analysis of the role of Spi-B in terminal differentiation of Spi-B–deficient B cells was precluded because of impaired BCR signaling, leading to defects in maturation and maintenance of GCs.17 Here we analyzed Spi-B protein expression in various mature B-cell subsets from blood and tonsil representing different stages in B-cell development. Peripheral blood (PB) CD19+CD3− B cells, including naive (IgD+CD27−) and switched memory (IgD−CD27+) B cells, were analyzed by immunoblotting. Spi-B protein was expressed both in naive and memory B cells, but with higher expression in memory B cells (Figure 1A). In line with our earlier observation, Spi-B was not detected in CD3+ T cells from the same blood donor.19 Human CD19+CD3− tonsil B cells were discriminated as described previously28 into naive B cells (IgD+CD38−), class-switched memory cells (IgD−CD38−), early nonswitched GC B cells (IgD+CD38+), GC B cells (IgD−CD38+), and PCs (PC, IgD−CD38++). Spi-B protein was expressed in all tonsil subsets except in PCs (Figure 1B). Higher expression levels were observed in GC B cells and resting memory cells than in naive cells. The same lysates were also analyzed for expression of BCL-6 and BLIMP1 protein. As expected, BCL-6 protein was highly expressed in early and late GC B-cell subsets and only weakly in PCs (Figure 1B),29 whereas BLIMP1 was highly expressed in IgD−CD38++ PCs only (Figure 1B).30
Figure 1.
Spi-B is expressed in human B-cell subsets, but not in plasma cells. Equal numbers of CD19+ B cells of (A) peripheral blood or (B) tonsil were sorted into subpopulations as indicated by gating and analyzed by immunoblotting. Actin levels were determined as loading control. One representative experiment of 3 is shown. (C,D) CD19+ B cells purified from peripheral blood were cocultured under plasma cell promoting conditions. (C) Naive (left panel) or memory (right panel) B cells were cultured for 7 days and analyzed for CD20 and CD38 expression by flow cytometry. Numbers in the quadrants indicate percentages of cells. For immunoblotting, cells were collected at the indicated time points and analyzed for Spi-B and BLIMP1 expression. Actin levels were determined to ensure equal loading of the samples. (D) Memory B cells were cultured for the time periods indicated, and expression levels were assessed by quantitative real-time PCR. Mean plus or minus SD values of PCR duplicates are shown. One representative experiment of 2 is shown.
Next we analyzed Spi-B expression levels during in vitro PC generation. We adapted previously described culture conditions that drive proliferation and differentiation of human B cells into antibody-secreting PCs,31,32 accompanied by down-regulation of cell surface BCR, CD19, and CD20 expression and up-regulation of the activation marker CD38 as well as the PC marker Syndecan-1 (CD138) on B cells.33 Naive and memory PB B cells were cultured for 3 days on CD40-ligand–transfected (CD40L)–L cells in the presence of IL-2 and IL-21, followed by 4 days of culture only in the presence IL-2 and IL-21. In line with previous findings,34,35 we observed that the generation of CD38+CD− PCs from memory B cells was much faster than from naive B cells (Figure 1C). Less than 15% of naive B cells differentiated into PCs within 7 days; and consistent with this, we observed only low levels of BLIMP1 protein. Within 2 days, naive B cells strongly increased Spi-B protein levels after activation in vitro with CD40L and cytokines (Figure 1C). When starting from memory B cells, BLIMP1 was readily and strongly induced, consistent with the observation that the majority of B cells acquired a CD38+CD20− PC phenotype within 7 days of culture. Notably, Spi-B protein levels rapidly decreased during PC differentiation.
During PC differentiation, B cells completely and irreversibly switch their gene expression profile. To get further insight in Spi-B regulation during PC differentiation, cultured memory PB cells were analyzed at consecutive days for Spi-B mRNA levels by quantitative PCR. In addition, we analyzed expression of factors known to be regulated during B-cell differentiation, including PAX-5 and BCL-6, which are down-regulated, and BLIMP1, IRF-4, and XBP-1, which are induced and mediate PC formation.3 Consistent with results obtained by immunoblotting, in vitro PC differentiation of PB memory B cells was associated with a decrease in Spi-B transcript levels (Figure 1D). Interestingly, the kinetics in reduction of Spi-B expression levels paralleled that of the repressors PAX-5 and BCL-6. As expected, BLIMP1, XBP-1, and IRF-4 expression levels increased during in vitro PC differentiation. Taken together, these results show that Spi-B is expressed in all human PB and tonsil B-cell subsets but not in PCs. Spi-B mRNA and protein levels decreased during in vitro differentiation of PB memory B cells into PCs. The differential expression of this transcription factor in memory and plasma cells suggests an active regulation and a role for Spi-B during human B-cell differentiation.
Ectopic overexpression of Spi-B arrests differentiation of PB B cells into immunoglobulin-secreting PCs
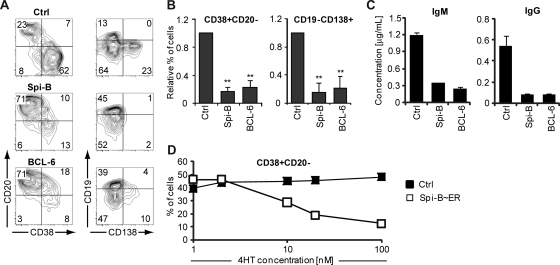
To elucidate the reason for down-regulation of Spi-B in PCs, Spi-B was constitutively expressed in human B cells, using the LZRS retroviral vector harboring the Spi-B coding sequence upstream of the IRES-GFP cassette (Figure S1),19 and cultured under plasma cell-inducing conditions. In parallel, we transduced B cells with BCL-6, which arrests human PC differentiation in vitro as previously reported.24 Seven days after transduction, a vast proportion of control transduced cells had differentiated into PCs (Figure 2A), a proportion of which expressed CD138. Strikingly, ectopic expression of Spi-B prevented PC differentiation, as we observed significantly less formation of CD38+CD− and CD19−CD138+ cells compared with control cultures (Figure 2A,B). As expected, also BCL-6−GFP+ cells were blocked in PC differentiation. AnnexinV/7-amino-actinomycin D labeling of transduced cells showed that Spi-B did not affect cell survival (data not shown). Transduction of sorted naive as well as memory B cells with Spi-B revealed that Spi-B abrogates PC differentiation of both B-cell subsets (data not shown). In addition, we observed that Spi-B was not only able to arrest in vitro plasma cell differentiation induced by IL-21, but also by IL-1031 (data not shown).
Figure 2.
Impaired human B-cell differentiation into Ig-secreting plasma cells by ectopic expression of Spi-B. CD19+ B cells isolated from peripheral blood were transduced with Spi-B, BCL-6, Spi-B∼ER, or control vectors and cultured in conditions promoting plasma cell differentiation (as in Figure 1). (A,B) After 7 days of culture, GFP+ cells were analyzed for CD19, CD20, CD38, and CD138 surface expression by flow cytometry. (A) Contour plots of one representative experiment of 10 are shown. Numbers in the quadrants indicate percentages of cells. (B) Percentages of CD38+CD20− and CD19−CD138+ cells in Spi-B– and BLC-6–transduced cultures were normalized to control cultures. Mean plus or minus SD values of 10 independent experiments are shown (Student t test, ** P < .01). (C) Five days after transduction, GFP+ cells were sorted, and equal numbers of cells (50 000) were cultured with IL-2 and IL-21 for an additional 48 hours. The supernatants were collected, and IgM and IgG protein levels were analyzed by ELISA. Mean plus or minus SD values of ELISA triplicates are shown. One representative experiment of 3 is displayed. (D) After 7 days of culture, the percentages of CD38+CD20− cells within the GFP+ population in Spi-B∼ER and control transduced cultures supplemented with the indicated concentrations of 4HT were assessed. One representative experiment of 2 is shown.
Terminal differentiation of B cells is intimately associated with increased Ig secretion. Based on the strong inhibitory effect of Spi-B on phenotypically defined PC differentiation, we speculated that ectopic expression of Spi-B would inhibit antibody secretion under PC permissive conditions. Conformed to this hypothesis, PB B cells transduced with Spi-B secreted 4- to 6-fold less IgM and IgG than controls as determined by ELISA in the supernatant of transduced cell cultures (Figure 2C). As expected, also BCL-6–transduced B cells were impaired in Ig production, similar to Spi-B–transduced B cells.
To exclude that our observations are merely the result of high levels of Spi-B overexpression, we used a Spi-B∼ER fusion construct, which allows nuclear transport of the ER-fusion protein in a 4HT concentration-dependent manner.36 Immunoblot analysis confirmed the presence of the fusion protein, which was detectable by antibodies directed against Spi-B and ER (Figure S2; data not shown). Whereas PB B cells transduced with a control vector were not affected by 4HT, in Spi-B∼ER–transduced cell cultures the percentage of CD38+CD− PCs generated correlated with the 4HT concentration, indicating a Spi-B dose-dependent regulation of PC formation. Thus, forced expression of Spi-B during in vitro PC generation inhibited the differentiation of PB B cells into PCs in a dose-dependent manner. Spi-B blocked PC formation as efficient as BCL-6, a known PC repressor. Consequently, antibody production of the pre-GC isotype IgM as well as the switched isotype IgG were strongly reduced after forced expression of Spi-B in PB B cells.
Spi-B represses the induction of the PC gene expression program
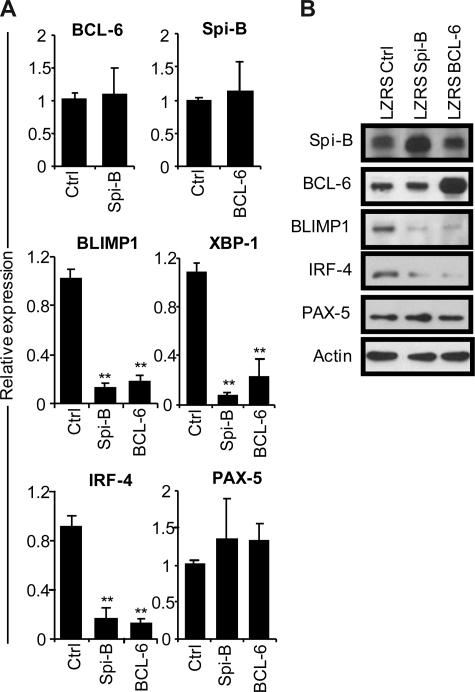
To gain insight into the mechanism of the Spi-B enforced PC differentiation block, we assessed the gene expression levels of BCL-6, BLIMP1, XBP-1, IRF-4, and PAX-5 in B cells after ectopic Spi-B expression (Figure 3A). In parallel, we analyzed the expression levels of these genes in BCL-6–transduced B cells. Five days after culture in PC-inducing conditions, which induced the expression of PC genes (Figure 1D), GFP+ cells were sorted and analyzed by real-time PCR. Notably, forced expression of Spi-B did not affect BCL-6 levels in PB B cells and, vice versa, overexpression of BCL-6 had no effect on Spi-B expression levels (Figure 3A), which suggests that these genes do not act in a sequential pathways. However, 6- to 10-fold lower levels of BLIMP1, IRF-4, and XBP-1 mRNA were detected both in Spi-B– and BCL-6–transduced cells compared with controls (Figure 3A). As assessed by conventional RT-PCR, the reduction of XBP-1 expression resulted in decreased levels of the spliced form of XBP-1 mRNA that gives rise to the transcriptionally active XBP-1(s) protein (Figure S3). Expression levels of PAX-5 did not significantly change in cells overexpressing Spi-B or BCL-6 compared with controls. Protein levels in transduced cells reflected mRNA levels of the analyzed transcription factors as forced expression of Spi-B inhibited expression of BLIMP1 and IRF-4 protein to a similar extent as forced expression of BCL-6, whereas Spi-B did not affect BCL-6 or PAX-5 protein levels (Figure 3B). Decreasing Spi-B expression levels by shRNA21 did not increase PC formation or affect expression levels of the PC transcription factors, leaving open the possibility that multiple transcriptional repressors have to be down-regulated in parallel (data not shown). Thus, forced expression of Spi-B prevents the induction of the PC gene program under culture conditions that permit PC differentiation.
Figure 3.
Repression of the plasma cell gene expression program by Spi-B. CD19+ B cells were retrovirally transduced with constructs expressing Spi-B, BCL-6, or control-GFP. Five days after transduction and culturing in conditions promoting plasma cell differentiation (as in Figure 1), GFP+ cells were sorted. (A) Gene expression levels of Spi-B, BCL-6, BLIMP-1, XBP-1, IRF-4, and PAX-5 were analyzed by quantitative RT-PCR. Expression levels in Spi-B– and BCL-6–transduced cells were normalized to expression levels in control transduced cells. Mean plus or minus SD values of 4 independent experiments are shown (Student t test, **P < .01). (B) Cell lysates from sorted GFP+ cells were subjected to immunoblot analysis using antibodies directed against Spi-B, BCL-6, BLIMP-1, IRF-4, or PAX-5. Actin levels were determined as loading control. Representative results of 2 independent experiments are shown.
The Spi-B transactivation domain is not required to arrest PC differentiation
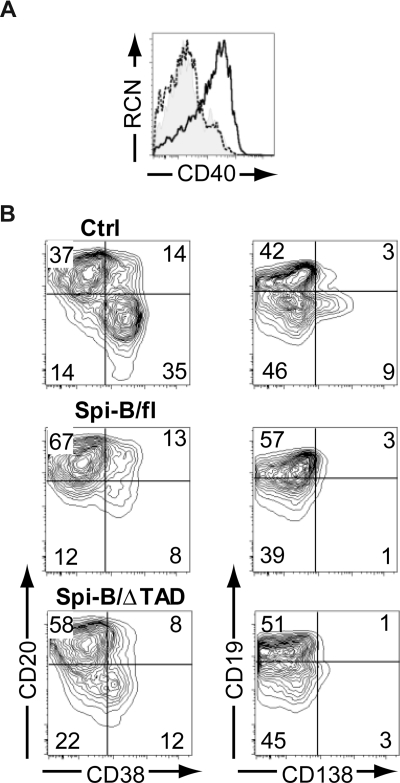
It has been well accepted that Spi-B is able to act as a transcriptional activator.37–42 We were, therefore, intrigued by the findings described in Figure 3 suggesting a repressor role for Spi-B. Structurally, Spi-B possesses an amino-terminal transactivation domain (TAD), which is required for activating transcription.39 Next we aimed to assess the contribution of this domain to the regulatory activity of Spi-B by transducing cells with a truncated Spi-B protein lacking TAD (Spi-B/ΔTAD) (Figure S1). As a positive control, we determined the expression level of CD40, a direct target of Spi-B transactivation activity.37 Overexpression of Spi-B full-length (fl) in hematopoietic precursors cultured on OP9 cells with Flt3L, which drives development of pDCs,19 resulted in higher surface expression levels of CD40 on pDCs (Figure 4A), whereas Spi-B/ΔTAD did not alter CD40 levels compared with controls. This indicates that the TAD domain of Spi-B is crucial for the induction of CD40 gene transcription. Interestingly, both Spi-B/fl– as well as Spi-B/ΔTAD–transduced B cells were arrested in differentiation into CD38+CD− and CD19−CD138+ PCs (Figure 4B) when analyzed at day 5 of a PC differentiation assay. These data support the notion that transactivation activity by TAD is largely dispensable for the repression of PC differentiation by Spi-B. Collectively, these data suggest that Spi-B uses distinct mechanisms to promote expression of CD40 on one hand and to regulate expression of PC genes on the other, and that different Spi-B domains are required to either exert transactivation or repression, respectively.
Figure 4.
The transactivation domain of Spi-B is not required for regulation of plasma cell differentiation. (A) CD34+CD1a− progenitors isolated from human postnatal thymus were transduced with control, Spi-B/full-length (fl), or Spi-B/ΔTAD constructs and cultured on OP9 with Flt3L and IL-7. The histogram shows surface expression level of CD40 on CD123+BDCA2+pDCs transduced with control (shaded histogram), Spi-B/fl (solid line), and Spi-B/ΔTAD (dashed line) constructs. (B) Peripheral blood CD19+ B cells were transduced with control, Spi-B/fl, or Spi-B/ΔTAD constructs and cultured in conditions promoting plasma cell differentiation (as in Figure 1). After 7 days of culture, GFP+ cells were analyzed for CD19, CD20, CD38, and CD138 surface expression. Contour plots of one representative experiment of 2 are shown. Numbers in the quadrants indicate percentages of cells.
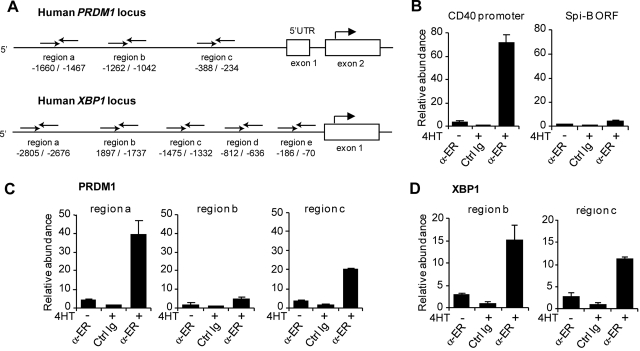
Spi-B binds to the promoter of human PRDM1 and XBP-1
Spi-B–transduced B cells expressed lower levels of the plasma cell genes BLIMP1, XBP-1, and IRF-4 (Figure 3), but whether these were direct target genes was not resolved. Therefore, we investigated whether Spi-B directly binds to the promoter regions of these genes by using ChIP assays. Immunoprecipitation was performed using an antibody directed against ER (α-ER) or unspecific rabbit Ig on lysates derived from Spi-B∼ER–transduced Raji cells that were either left untreated or incubated with 4HT for 4 hours. Chromatin abundance was measured by quantitative PCR using primer pairs in the promoter regions of BLIMP1, XBP-1, and IRF-4. Primers were designed to randomly amplify several regions within the potential regulatory elements of PRDM1, XBP1, and IRF4 (Figure 5A; data not shown). Based on previous reports,15,43,44 we assessed Spi-B binding to the human PRDM1 locus upstream of the transcription start as well as in intron 4, which corresponds to intron 5 of the murine PRDM1 locus. Furthermore, we analyzed Spi-B binding to a region up to 3 kb upstream of the transcription start and the proximal −200 to −700 region of the human XBP1 and IRF4 loci, respectively, which have been reported to control promoter activity.12,45,46 As a positive and negative control, we analyzed binding of Spi-B to the CD40 promoter37 and the ORF of Spi-B, respectively (Figure 5B). Notably, we observed binding of Spi-B to the PRDM1 promoter at 2 of the 3 regions investigated (Figure 5C). For these regions, termed region a (−1660 to −1467 relative to the BLIMP1 transcription start) and region c (−388 to −234), chromatin levels after ChIP with α-ER were 10- and 6-fold increased, respectively, on treatment with 4HT, and 40- and 20-fold higher, respectively, compared with nonspecific Ig precipitation. No binding of Spi-B to any of the regions tested for intron 4 of the PRDM1 locus was detected (Figure 5A; data not shown). Furthermore, binding of Spi-B was observed at 2 of 5 regions analyzed within the XBP1 promoter (Figure 5D; data not shown). Using primers amplifying region b (−1897 to −1737) and region c (−1475 to −1332), chromatin abundance was 15- and 10-fold increased in IP samples with α-ER versus unspecific Ig, respectively. Binding to the IRF-4 promoter was analyzed with 3 primer sets: region a (−103 to −330), region b (−834 to −1064), and region c (−1212 to −1350). However, no binding of Spi-B was detected (data not shown).
Figure 5.
Binding of Spi-B to the promoter regions of the human PRDM1 and the XBP-1 locus. (A) Schematic representation of the 5′ region of the human PRDM1 and XBP1 locus. Binding sites of primers used for analysis of chromatin immunoprecipitation (ChIP) are indicated relative to the transcription start site. (B-D) ChIP analysis for Spi-B binding. SpiB∼ER-GFP+ RAJI cells were cultured in the presence or absence of 4HT for 4 hours and subjected to ChIP using α-ER antibody or, as control, normal rabbit IgG (Ctrl Ig). (B) As positive control for Spi-B binding, precipitated chromatin was analyzed by real-time PCR for abundance of CD40 promoter DNA. The Spi-B open reading frame (ORF) served as irrelevant gene control for Spi-B binding. (C) Precipitated chromatin was analyzed for abundance of 3 different regions (regions a, b, c) of the PRDM1 promoter. (D) Precipitated chromatin was analyzed with primers binding to regions b and locus c upstream of the transcription start in the XBP1 gene. Values are normalized to chromatin levels in control Ig samples. Mean plus or minus SD values of precipitation triplicates are shown. One representative experiment of 2 is shown.
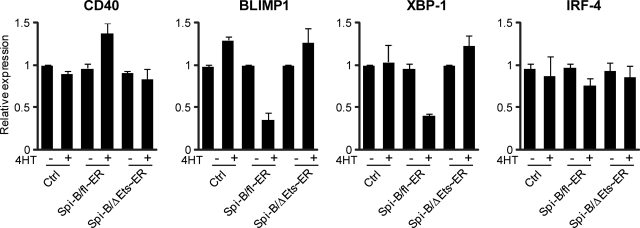
Spi-B directly represses expression of BLIMP1 and XBP-1
We used a 4HT-inducible Spi-B/fl∼ER fusion construct to further investigate whether binding of Spi-B to the promoters of BLIMP1 and XBP-1 also directly controls expression of these genes. As an additional negative control, we made use of a truncated Spi-B protein lacking the DNA binding domain (ΔEts),19,25 which we also fused to ER (Spi-B/ΔEts∼ER, Figure S2) and which did not affect PC differentiation on induction by 4HT addition (data not shown). Spi-B/fl∼ER, Spi-B/ΔEts∼ER, and control transduced B cells were cultured under conditions permitting PC development and sorted on day 4 after transduction for GFP expression. Cells were cultured for an additional 48 hours, after which the cells were first preincubated with CHX, impeding de novo protein translation, before treatment with 4HT for 4 hours. As expected, we detected higher levels of CD40 expression in Spi-B/fl∼ER–transduced cells cultured with 4HT compared with untreated cells as measured by quantitative PCR, whereas 4HT treatment alone did not affect CD40 levels in cells transduced with a control or Spi-B/ΔEts∼ER vector (Figure 6). Within 4 hours after 4HT addition, Spi-B/fl∼ER reduced the expression levels of both BLIMP1 and XBP-1 mRNA, which were 2.5- to 3-fold lower compared with control cells, whereas induction of the mutated Spi-B/ΔEts∼ER by 4HT did not affect the expression of these PC factors (Figure 6). This suggests direct regulation of BLIMP1 and XBP-1 by Spi-B that is dependent on the Ets domain of Spi-B. In contrast, IRF-4 levels were similar in all conditions, indicating that IRF-4 expression is indirectly controlled by Spi-B (Figure 3). Importantly, treatment of cells with 4HT for 4 hours did not affect the number or percentages of living cells in control, Spi-B/fl∼ER, or Spi-B/ΔEts∼ER–transduced cultures, including GFP+ cells, CD38+CD− cells or CD138+ cells (data not shown), excluding the possibility that 4HT had a toxic effect on Spi-B/fl∼ER–transduced cells.
Figure 6.
Direct repression of BLIMP1 and XBP-1 by Spi-B. Peripheral blood CD19+ B cells transduced with Spi-B/fl∼ER-GFP, Spi-B/ΔEts∼ER, or control GFP vector were cultured in conditions allowing for plasma cell differentiation (as in Figure 1) and sorted for GFP expression. After sorting, the cells were preincubated with cycloheximide (CHX) before addition of 4-hydroxytamoxifen (4HT) for 4 hours. Gene expression levels were analyzed by quantitative RT-PCR. Values are normalized to expression in samples without 4HT treatment. Mean plus or minus SD values of PCR duplicates are shown. One representative experiment of 2 is shown.
Direct binding and repression of the BLIMP1 promoter by Spi-B was confirmed by a luciferase reporter assay (Figure S4). We used a pGL3 vector containing a 2-kb fragment of the PRDM1 locus upstream of the transcription start,43 which was cotransfected with pcDNA3.1 control or Spi-B expression vectors into NIH3T3 cells. As expected, treatment of the cells with phorbol-12-myristate-13-acetate/ionomycin for 6 hours induced luciferase expression.43 Concomitant expression of Spi-B reduced luciferase expression by 30%, indicating that Spi-B directly repressed BLIMP1 promoter activity. A restrictive DNA consensus sequence for Spi-B binding is not known; Spi-B binds to the short GGAA/T motif47,48 as well as the noncanonical AGAA motif,41 with a rule for flanking nucleotides.41 Within the 2-kb BLIMP1 promoter region used in our experiments, we found numerous (58) GGAA and AGAA motifs, which precluded assessment of the exact Spi-B binding site(s).
Taken together, our results show that Spi-B directly binds to the promoters of 2 important plasma cells factors BLIMP1 and XBP-1 and favor the concept that Spi-B directly represses the transcription of these PC-inducing genes.
Discussion
This report reveals a novel role of Spi-B in late stages of human B-cell differentiation. Primary naive and memory B cells from tonsil and peripheral blood expressed Spi-B at the mRNA and protein levels. In contrast, CD38++CD20− PCs isolated from tonsils or generated in vitro from PB B cells lacked expression of Spi-B. We demonstrated that down-regulation of Spi-B is required for efficient differentiation of human PB B cells into PCs, as enforced expression of Spi-B completely blocked PC differentiation and Ig secretion.
Several factors have been reported to regulate PC differentiation, including PAX-5, BCL-6, Bach2, and Mitf (reviewed by Shapiro-Shelef and Calame3). PAX-5 represses a number of genes involved in PC differentiation, including BLIMP1 and possibly XBP-1.11,13,49 Similarly, BLIMP1 gene expression is further repressed by BCL-6 and Bach2.14,16 It is doubtful that Spi-B inhibits PC differentiation through induction of PAX-5, BCL-6, or Bach2 expression because transcript and protein levels of these genes did not significantly increase on forced expression of Spi-B (Figure 3; data not shown). In contrast, expression levels of both BLIMP1 and XBP-1 were reduced shortly after nuclear translocation of Spi-B in the presence of the protein synthesis blocker CHX. This, together with the observation that Spi-B bound to the regulatory elements of PRDM1 and XBP1 as revealed by ChIP analysis, argues in favor of a role for Spi-B in directly repressing transcription of BLIMP-1 and XBP-1 providing an explanation for the observed block in PC formation and Ig secretion in Spi-B–transduced B cells.
To date, Spi-B has only been implicated in transactivation of gene expression. Target genes include, in addition to CD40,37 the adaptor protein Grap2,38 the heptahelical receptor P2Y10,39 the Rel/NF-kappa B family member c-rel,40 and the tyrosine kinases c-fes/c-fps41 and Btk.42 To our knowledge, this is the first report that attributes repressor activity to Spi-B. Other Ets family members, including PU.1, which is a close homolog of Spi-B,50 have been reported to display repressor activity previously.51,52 PU.1 negatively regulates expression of its targets by recruiting several proteins, including a histone deacetylase.51 Repressor activity of PU.1 depends on the Ets domain53 and was pinpointed to 2 lysine-rich acetylation motifs, which when mutated strongly affected the repressor, but not the activator function of PU.1.54 Interestingly, a similar acetylation motif as in PU.1 can be identified in the Ets domain of Spi-B (data not shown). In line with a role for the Ets domain of Spi-B in conferring repressor activity, we show here that this domain is required to arrest PC formation, whereas the TAD domain can be omitted. In contrast, however, the TAD domain of Spi-B is required to positively regulate CD40 expression. Taken together, it is tempting to speculate that a similar mechanism as used by PU.1, mediating an activator as well as a repressor function, controls the dual role of Spi-B.
All our studies investigating the effects of Spi-B and its DNA binding were done using overexpression of Spi-B. We have conclusively shown that Spi-B can repress BLIMP1 and XBP-1 expression and consequently PC differentiation when ectopically expressed. These expression levels are probably at higher than endogenous levels in B cells. We did observe, however, a block in plasma cell formation already when Spi-B∼ER expression was induced at very low concentrations of 4HT, suggesting that even low nuclear levels of Spi-B are sufficient to repress plasma cell formation. Still we cannot exclude the possibility that ectopically expressed Spi-B is binding to low affinity sites, thereby blocking the ability of adjacent activators to bind or induce transcription. Whether these putative low affinity sites are recognized by endogenous levels of Spi-B remains elusive.
It is evident from our results that Spi-B needs to be down-regulated for proper PC formation, but it is unclear what regulates Spi-B expression. We exclude the possibility that BCL-6 controls Spi-B levels because overexpression of BCL-6 did not increase mRNA or protein expression levels of Spi-B. It was reported that in mice Spi-B is a direct target of BLIMP1, suggesting that Spi-B expression is repressed by this factor.5 Our data, however, do not support the notion that BLIMP1 is the factor that initiates down-regulation of Spi-B during PC differentiation because we observed that Spi-B levels had already decreased before induction of BLIMP1 (Figure 1D). This is in line with recent findings in the mouse indicating that BLIMP1 is not required for the initiation of PC differentiation and that the initial decrease of PAX-5 levels occurs before BLIMP1 expression.27 These new findings challenge the current model in which the silencing of the B-cell transcription program and the induction of the plasma cell program is initiated by BLIMP1.55 It is yet unclear which factor(s) is(are) responsible for the initial decrease of PAX-5 and Spi-B.
We observed that shRNA-mediated down-regulation of Spi-B did not promote spontaneous Ig secretion or PC formation. This is in line with a previous report in Spi-B–deficient mice, which do not have increased numbers of plasma cells or elevated serum Ig titers.17 It suggests that Spi-B may not be required to inhibit the spontaneous differentiation of naive B cells, but rather to inhibit premature differentiation of proliferating B cells in the GC. If true, then GC B cells deficient in Spi-B expression might prematurely differentiate, which would be consistent with the kinetics of GC reactions seen in the Spi-B knockout mouse.17 Such early termination of the GC reaction may be the cause of decreased antigen-specific antibodies produced in Spi-deficient mice, especially if loss of Spi-B actually promotes plasma cell formation from GC B cells. These data are especially intriguing given the potential role of another Ets family member, Ets-1, in regulating spontaneous Ig secretion from naive B cells.56 Thus, it is possible that Ets-1 may play a dominant role in controlling the premature differentiation of naive B cells, whereas Spi-B plays a dominant role in regulating the premature differentiation of GC B cells. This hypothesis is supported by the observation that expression of Ets-1 is low in GC B cells,57 whereas expression of Spi-B is high (present study).57 Alternatively, our observations raise the possibility that Spi-B is required for maintenance of the memory B cell fate, consistent with the high expression levels of Spi-B protein in human tonsil and PB memory B cells compared with naive B cells.
Aberrant expression of Spi-B has been implicated in tumorigenesis. It has been shown recently that Spi-B is expressed at higher levels in ABC DLBCL than in germinal center B cell–like DLBCL.58 The gene profile of ABC DLBCL suggests that this lymphoma subgroup is derived from B cells that are blocked in the process of differentiating from GC B cells to PCs.59 In one of these cell lines, the Spi-B locus was translocated and inserted in proximity to the Ig 3′α enhancer, resulting in relatively high Spi-B transcription levels.22 Therefore, it was suggested that Spi-B might play a role in the pathophysiology of the ABC DLBCL subtype. In line with this notion, we observed here that forced Spi-B expression in primary human B cells prevented plasmacytic differentiation. Thus, aberrantly expressed, Spi-B may contribute to the formation of B-cell lymphomas by blocking terminal differentiation and in this context represent a valuable marker for diagnosis of this tumor type.
In conclusion, in this paper, we describe 2 novel findings: (1) Spi-B may function as a transcriptional repressor via a mechanism independent of the protein's transactivation domain; and (2) Spi-B prevents transcription of 2 of the main PC factors BLIMP1 and XBP-1, which ultimately results in preserving human B cells in an undifferentiated state and thus preventing PC differentiation.
Supplementary Material
Acknowledgments
The authors thank Berend Hooibrink (Academic Medical Center, University of Amsterdam, Amsterdam, The Netherlands) for expert cell sorting, Derk Amsen for helpful scientific input, Lee Ann Garrett-Sinha (State University of New York at Buffalo, Buffalo, NY), F. Moreau-Gachelin (Institute Curie, Paris, France) for α-Spi-B antibodies, Reuben Tooze (Leeds Institute of Molecular Medicine, Leeds, United Kingdom) for α-BLIMP1 antibody, and Stephen Nutt (Walter and Eliza Hall Institute of Medical Research, Parkville, Australia) for α-PAX-5 antibody.
H. Schmidlin is supported by the National Institutes of Health (grant R01-AI52002). S.A.D. is supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (grant F32-AI0638-46).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: H. Schmidlin and S.A.D. designed research, performed experiments, analyzed data, and wrote the paper; M.N. performed experiments; F.A.S. designed research; R.S. performed experiments and analyzed data; C.H.U. and H. Spits analyzed data; and B.B. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bianca Blom, Department of Cell Biology and Histology, Academic Medical Center, University of Amsterdam, Meibergdreef 15, 1105AZ Amsterdam, The Netherlands; e-mail: b.blom@amc.uva.nl.
References
- 1.McHeyzer-Williams MG. B cells as effectors. Curr Opin Immunol. 2003;15:354–361. doi: 10.1016/s0952-7915(03)00046-3. [DOI] [PubMed] [Google Scholar]
- 2.Tangye SG, Avery DT, Deenick EK, Hodgkin PD. Intrinsic differences in the proliferation of naive and memory human B cells as a mechanism for enhanced secondary immune responses. J Immunol. 2003;170:686–694. doi: 10.4049/jimmunol.170.2.686. [DOI] [PubMed] [Google Scholar]
- 3.Shapiro-Shelef M, Calame K. Regulation of plasma-cell development. Nat Rev Immunol. 2005;5:230–242. doi: 10.1038/nri1572. [DOI] [PubMed] [Google Scholar]
- 4.Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, et al. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19:607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 5.Shaffer AL, Lin KI, Kuo TC, et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 6.Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 7.Reimold AM, Iwakoshi NN, Manis J, et al. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 8.Klein U, Casola S, Cattoretti G, et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- 9.Sciammas R, Shaffer AL, Schatz JH, et al. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 2006;25:225–236. doi: 10.1016/j.immuni.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8:22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- 11.Delogu A, Schebesta A, Sun Q, et al. Gene repression by Pax5 in B cells is essential for blood cell homeostasis and is reversed in plasma cells. Immunity. 2006;24:269–281. doi: 10.1016/j.immuni.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 12.Lin L, Gerth AJ, Peng SL. Active inhibition of plasma cell development in resting B cells by microphthalmia-associated transcription factor. J Exp Med. 2004;200:115–122. doi: 10.1084/jem.20040612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nera KP, Kohonen P, Narvi E, et al. Loss of Pax5 promotes plasma cell differentiation. Immunity. 2006;24:283–293. doi: 10.1016/j.immuni.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Ochiai K, Katoh Y, Ikura T, et al. Plasmacytic transcription factor Blimp-1 is repressed by Bach2 in B cells. J Biol Chem. 2006;281:38226–38234. doi: 10.1074/jbc.M607592200. [DOI] [PubMed] [Google Scholar]
- 15.Tunyaplin C, Shaffer AL, Angelin-Duclos CD, et al. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J Immunol. 2004;173:1158–1165. doi: 10.4049/jimmunol.173.2.1158. [DOI] [PubMed] [Google Scholar]
- 16.Shaffer AL, Yu X, He Y, et al. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13:199–212. doi: 10.1016/s1074-7613(00)00020-0. [DOI] [PubMed] [Google Scholar]
- 17.Su GH, Chen HM, Muthusamy N, et al. Defective B-cell receptor-mediated responses in mice lacking the Ets protein, Spi-B. EMBO J. 1997;16:7118–7129. doi: 10.1093/emboj/16.23.7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen HM, Zhang P, Voso MT, et al. Neutrophils and monocytes express high levels of PU. 1 (Spi-1) but not Spi-B. Blood. 1995;85:2918–2928. [PubMed] [Google Scholar]
- 19.Schotte R, Rissoan MC, Bendriss-Vermare N, et al. The transcription factor Spi-B is expressed in plasmacytoid DC precursors and inhibits T-, B-, and NK-cell development. Blood. 2003;101:1015–1023. doi: 10.1182/blood-2002-02-0438. [DOI] [PubMed] [Google Scholar]
- 20.Su GH, Ip HS, Cobb BS, et al. The Ets protein Spi-B is expressed exclusively in B cells and T cells during development. J Exp Med. 1996;184:203–214. doi: 10.1084/jem.184.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schotte R, Nagasawa M, Weijer K, Spits H, Blom B. The ETS transcription factor Spi-B is required for human plasmacytoid dendritic cell development. J Exp Med. 2004;200:1503–1509. doi: 10.1084/jem.20041231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lenz G, Nagel I, Siebert R, et al. Aberrant immunoglobulin class-switch recombination and switch translocations in activated B cell-like diffuse large B cell lymphoma. J Exp Med. 2007;204:633–643. doi: 10.1084/jem.20062041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dontje W, Schotte R, Cupedo T, et al. Delta-like1-induced Notch1 signaling regulates the human plasmacytoid dendritic cell versus T-cell lineage decision through control of GATA-3 and Spi-B. Blood. 2006;107:2446–2452. doi: 10.1182/blood-2005-05-2090. [DOI] [PubMed] [Google Scholar]
- 24.Scheeren FA, Naspetti M, Diehl S, et al. STAT5 regulates the self-renewal capacity and differentiation of human memory B cells and controls Bcl-6 expression. Nat Immunol. 2005;6:303–313. doi: 10.1038/ni1172. [DOI] [PubMed] [Google Scholar]
- 25.Ray-Gallet D, Tavitian A, Moreau-Gachelin F. An alternatively spliced isoform of the Spi-B transcription factor. Biochem Biophys Res Commun. 1996;223:257–263. doi: 10.1006/bbrc.1996.0881. [DOI] [PubMed] [Google Scholar]
- 26.Tooze RM, Stephenson S, Doody GM. Repression of IFN-gamma induction of class II transactivator: a role for PRDM1/Blimp-1 in regulation of cytokine signaling. J Immunol. 2006;177:4584–4593. doi: 10.4049/jimmunol.177.7.4584. [DOI] [PubMed] [Google Scholar]
- 27.Kallies A, Hasbold J, Fairfax K, et al. Initiation of plasma-cell differentiation is independent of the transcription factor Blimp-1. Immunity. 2007;26:555–566. doi: 10.1016/j.immuni.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 28.Pascual V, Liu YJ, Magalski A, et al. Analysis of somatic mutation in five B-cell subsets of human tonsil. J Exp Med. 1994;180:329–339. doi: 10.1084/jem.180.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cattoretti G, Chang CC, Cechova K, et al. BCL-6 protein is expressed in germinal-center B cells. Blood. 1995;86:45–53. [PubMed] [Google Scholar]
- 30.Cattoretti G, Angelin-Duclos C, Shaknovich R, et al. PRDM1/Blimp-1 is expressed in human B-lymphocytes committed to the plasma cell lineage. J Pathol. 2005;206:76–86. doi: 10.1002/path.1752. [DOI] [PubMed] [Google Scholar]
- 31.Arpin C, Dechanet J, Van Kooten C, et al. Generation of memory B cells and plasma cells in vitro. Science. 1995;268:720–722. doi: 10.1126/science.7537388. [DOI] [PubMed] [Google Scholar]
- 32.Ettinger R, Sims GP, Fairhurst AM, et al. IL-21 induces differentiation of human naive and memory B cells into antibody-secreting plasma cells. J Immunol. 2005;175:7867–7879. doi: 10.4049/jimmunol.175.12.7867. [DOI] [PubMed] [Google Scholar]
- 33.Liu YJ, Arpin C. Germinal center development. Immunol Rev. 1997;156:111–126. doi: 10.1111/j.1600-065x.1997.tb00963.x. [DOI] [PubMed] [Google Scholar]
- 34.Arpin C, Banchereau J, Liu YJ. Memory B cells are biased towards terminal differentiation: a strategy that may prevent repertoire freezing. J Exp Med. 1997;186:931–940. doi: 10.1084/jem.186.6.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tangye SG, Avery DT, Hodgkin PD. A division-linked mechanism for the rapid generation of Ig-secreting cells from human memory B cells. J Immunol. 2003;170:261–269. doi: 10.4049/jimmunol.170.1.261. [DOI] [PubMed] [Google Scholar]
- 36.Kurata H, Lee HJ, O'Garra A, Arai N. Ectopic expression of activated Stat6 induces the expression of Th2-specific cytokines and transcription factors in developing Th1 cells. Immunity. 1999;11:677–688. doi: 10.1016/s1074-7613(00)80142-9. [DOI] [PubMed] [Google Scholar]
- 37.Nguyen VT, Benveniste EN. Involvement of STAT-1 and ets family members in interferon-gamma induction of CD40 transcription in microglia/macrophages. J Biol Chem. 2000;275:23674–23684. doi: 10.1074/jbc.M002482200. [DOI] [PubMed] [Google Scholar]
- 38.Garrett-Sinha LA, Hou P, Wang D, et al. Spi-1 and Spi-B control the expression of the Grap2 gene in B cells. Gene. 2005;353:134–146. doi: 10.1016/j.gene.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 39.Rao S, Garrett-Sinha LA, Yoon J, Simon MC. The Ets factors PU. 1 and Spi-B regulate the transcription in vivo of P2Y10, a lymphoid restricted heptahelical receptor. J Biol Chem. 1999;274:34245–34252. doi: 10.1074/jbc.274.48.34245. [DOI] [PubMed] [Google Scholar]
- 40.Hu CJ, Rao S, Ramirez-Bergeron DL, et al. PU. 1/Spi-B regulation of c-rel is essential for mature B cell survival. Immunity. 2001;15:545–555. doi: 10.1016/s1074-7613(01)00219-9. [DOI] [PubMed] [Google Scholar]
- 41.Ray-Gallet D, Mao C, Tavitian A, Moreau-Gachelin F. DNA binding specificities of Spi-1/ PU. 1 and Spi-B transcription factors and identification of a Spi-1/Spi-B binding site in the c-fes/c-fps promoter. Oncogene. 1995;11:303–313. [PubMed] [Google Scholar]
- 42.Muller S, Sideras P, Smith CI, Xanthopoulos KG. Cell specific expression of human Bruton's agammaglobulinemia tyrosine kinase gene (Btk) is regulated by Sp1- and Spi-1/ PU. 1-family members. Oncogene. 1996;13:1955–1964. [PubMed] [Google Scholar]
- 43.Vasanwala FH, Kusam S, Toney LM, Dent AL. Repression of AP-1 function: a mechanism for the regulation of Blimp-1 expression and B lymphocyte differentiation by the B cell lymphoma-6 protooncogene. J Immunol. 2002;169:1922–1929. doi: 10.4049/jimmunol.169.4.1922. [DOI] [PubMed] [Google Scholar]
- 44.Parekh S, Polo JM, Shaknovich R, et al. BCL6 programs lymphoma cells for survival and differentiation through distinct biochemical mechanisms. Blood. 2007;110:2067–2074. doi: 10.1182/blood-2007-01-069575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ponath PD, Fass D, Liou HC, Glimcher LH, Strominger JL. The regulatory gene, hXBP-1, and its target, HLA-DRA, utilize both common and distinct regulatory elements and protein complexes. J Biol Chem. 1993;268:17074–17082. [PubMed] [Google Scholar]
- 46.Sharma S, Grandvaux N, Mamane Y, et al. Regulation of IFN regulatory factor 4 expression in human T cell leukemia virus-I-transformed T cells. J Immunol. 2002;169:3120–3130. doi: 10.4049/jimmunol.169.6.3120. [DOI] [PubMed] [Google Scholar]
- 47.Nye JA, Petersen JM, Gunther CV, Jonsen MD, Graves BJ. Interaction of murine ets-1 with GGA-binding sites establishes the ETS domain as a new DNA-binding motif. Genes Dev. 1992;6:975–990. doi: 10.1101/gad.6.6.975. [DOI] [PubMed] [Google Scholar]
- 48.Wang CY, Petryniak B, Ho IC, Thompson CB, Leiden JM. Evolutionarily conserved Ets family members display distinct DNA binding specificities. J Exp Med. 1992;175:1391–1399. doi: 10.1084/jem.175.5.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reimold AM, Ponath PD, Li YS, et al. Transcription factor B cell lineage-specific activator protein regulates the gene for human X-box binding protein 1. J Exp Med. 1996;183:393–401. doi: 10.1084/jem.183.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ray D, Bosselut R, Ghysdael J, et al. Characterization of Spi-B a transcription factor related to the putative oncoprotein Spi-1/PU. 1. Mol Cell Biol. 1992;12:4297–4304. doi: 10.1128/mcb.12.10.4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kihara-Negishi F, Yamamoto H, Suzuki M, et al. In vivo complex formation of PU.1 with HDAC1 associated with PU. 1-mediated transcriptional repression. Oncogene. 2001;20:6039–6047. doi: 10.1038/sj.onc.1204756. [DOI] [PubMed] [Google Scholar]
- 52.Hwang CK, Kim CS, Choi HS, McKercher SR, Loh HH. Transcriptional regulation of mouse mu opioid receptor gene by PU. 1. J Biol Chem. 2004;279:19764–19774. doi: 10.1074/jbc.M400755200. [DOI] [PubMed] [Google Scholar]
- 53.Suzuki M, Yamada T, Kihara-Negishi F, Sakurai T, Oikawa T. Direct association between PU.1 and MeCP2 that recruits mSin3A-HDAC complex for PU. 1-mediated transcriptional repression. Oncogene. 2003;22:8688–8698. doi: 10.1038/sj.onc.1207182. [DOI] [PubMed] [Google Scholar]
- 54.Kihara-Negishi F, Suzuki M, Yamada T, Sakurai T, Oikawa T. Impaired repressor activity and biological functions of PU. 1 in MEL cells induced by mutations in the acetylation motifs within the ETS domain. Biochem Biophys Res Commun. 2005;335:477–484. doi: 10.1016/j.bbrc.2005.07.098. [DOI] [PubMed] [Google Scholar]
- 55.Shapiro-Shelef M, Lin KI, Savitsky D, Liao J, Calame K. Blimp-1 is required for maintenance of long-lived plasma cells in the bone marrow. J Exp Med. 2005;202:1471–1476. doi: 10.1084/jem.20051611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.John SA, Clements JL, Russell LM, Garrett-Sinha LA. Ets-1 regulates plasma cell differentiation by interfering with the activity of the transcription factor blimp-1. J Biol Chem. 2008;283:951–962. doi: 10.1074/jbc.M705262200. [DOI] [PubMed] [Google Scholar]
- 57.Bhattacharya D, Cheah MT, Franco CB, et al. Transcriptional profiling of antigen-dependent murine B-cell differentiation and memory formation. J Immunol. 2007;179:6808–6819. doi: 10.4049/jimmunol.179.10.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosenwald A, Wright G, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 59.Wright G, Tan B, Rosenwald A, et al. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci U S A. 2003;100:9991–9996. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.