Abstract
Ectopic C/EBPα expression in p210BCR/ABL-expressing hematopoietic cells induces granulocytic differentiation, inhibits proliferation, and suppresses leukemogenesis. To assess the underlying mechanisms, C/EBPα targets were identified by microarray analyses. Upon C/EBPα activation, expression of c-Myb and GATA-2 was repressed in 32D-BCR/ABL, K562, and chronic myelogenous leukemia (CML) blast crisis (BC) primary cells but only c-Myb levels decreased slightly in CD34+ normal progenitors. The role of these 2 genes for the effects of C/EBPα was assessed by perturbing their expression in K562 cells. Ectopic c-Myb expression blocked the proliferation inhibition– and differentiation-inducing effects of C/EBPα, whereas c-Myb siRNA treatment enhanced C/EBPα-mediated proliferation inhibition and induced changes in gene expression indicative of monocytic differentiation. Ectopic GATA-2 expression suppressed the proliferation inhibitory effect of C/EBPα but blocked in part the effect on differentiation; GATA-2 siRNA treatment had no effects on C/EBPα induction of differentiation but inhibited proliferation of K562 cells, alone or upon C/EBPα activation. In summary, the effects of C/EBPα in p210BCR/ABL-expressing cells depend, in part, on transcriptional repression of c-Myb and GATA-2. Since perturbation of c-Myb and GATA-2 expression has nonidentical consequences for proliferation and differentiation of K562 cells, the effects of C/EBPα appear to involve dif-ferent transcription-regulated targets.
Introduction
The transcription factor C/EBPα, the founding member of the CCAAT/enhancer binding protein family, plays essential roles in regulating proliferation and differentiation of myeloid progenitor cells1,2; structural-functional relationship studies have shown that DNA binding and transcription activation function are required for C/EBPα-mediated differentiation of myeloid progenitors.3 In other models, interaction of C/EBPα with members of the cell cycle regulator E2F family has been shown to be required for differentiation as C/EBPα mutants lacking the E2F interaction domain failed to induce differentiation.4,5 In many types of myeloid leukemia, C/EBPα is mutated or its expression is altered, suggesting that loss of C/EBPα expression/activity is an important requirement for leukemogenesis.6 The mechanisms of C/EBPα inactivation in myeloid leukemia include mutations in the N- and C-terminus that reduce the functional levels of the protein and have the potential to generate molecules with dominant-negative activity,7 transcriptional and posttranscriptional down-regulation,8,9 and posttranslational modification (ie, MAP kinase–dependent phosphorylation) causing reduced protein activity.10 In myeloid cells transformed by the p210BCR/ABL oncoprotein, expression of C/EBPα is repressed at the translational level by a mechanism involving MAP kinase–dependent phosphorylation and stabilization of the RNA binding protein hnRNPE2, which binds the 5′ UTR of C/EBPα mRNA and inhibits mRNA translation.9,11
Regardless of the mechanism responsible for C/EBPα loss of function, ectopic expression of C/EBPα in myeloid leukemia lines and in primary blast cells induces differentiation and inhibits proliferation,8,9,12,13 further emphasizing the importance of genetic and/or functional inactivation of C/EBPα for leukemogenesis and the therapeutic potential of strategies restoring C/EBPα expression in leukemic cells.
In a previous study, we found that activation of C/EBPα in myeloid precursor 32Dcl3 cells transformed by the p210BCR/ABL oncoprotein induced differentiation, inhibited proliferation, and suppressed leukemogenesis.13 Instead, a DNA binding/transcription activation-deficient mutant (K298E C/EBPα) failed to induce differentiation of 32D-BCR/ABL cells, but was able to inhibit proliferation and to suppress in vivo leukemogenesis, although less efficiently than wild-type C/EBPα.13 Mechanistically, it is unclear how activation of C/EBPα exerts its antileukemia effects in p210BCR/ABL-expressing cells, although a number of potential C/EBPα targets were identified by assessing differentially expressed genes in wild-type and C/EBPα knockout fetal liver cells and upon activation of C/EBPα in U937 cells14,15; conceivably, changes in the expression of some of these genes (including c-Myc) may be responsible for the leukemia-suppressive effects of C/EBPα.14,15
To identify bona fide C/EBPα transcriptional targets whose changes in expression may suppress p210BCR/ABL-dependent leukemogenesis, we performed microarray hybridization analyses to search for genes regulated in a DNA binding–dependent manner at early time points after C/EBPα activation in 32D-BCR/ABL cells. In addition to several differentiation-regulated genes whose expression was enhanced by C/EBPα, levels of some were rapidly down-regulated by transcriptionally competent C/EBPα but not by the DNA binding–deficient K298E C/EBPα mutant. Two of these genes are the transcription factors c-Myb and GATA-2. We report here that expression of c-Myb and GATA-2 is transcriptionally repressed by C/EBPα in a DNA binding–dependent manner in p210BCR/ABL-expressing cell lines; levels of c-Myb and GATA-2 were also markedly down-regulated by C/EBPα in chronic myelogenous leukemia (CML) blast crisis (BC) but not in normal CD34+ cells. Modulation of c-Myb and GATA-2 levels in K562 cells by ectopic expression or RNA interference had nonidentical consequences on C/EBPα-induced differentiation and proliferation inhibition, suggesting that different transcription-regulated targets are involved in the biologic effects of C/EBPα in p210BCR/ABL-expressing cells.
Methods
Plasmids
p42C/EBPα-ERTAM, K298E C/EBPα-ERTAM, and Δ177-191 C/EBPα-ERTAM all cloned in the XhoI/EcoRI-digested MigRI vector were previously described.13 pMSCV Δ(358-452) c-Myb was obtained by releasing the Δ(358-452) c-Myb insert from MigRI Δ(358-452) c-Myb16 and cloning it at the XhoI/EcoRI sites of pMSCV. pMSCV/IRES/YFP GATA-2 was generated by polymerase chain reaction (PCR) amplification of the human GATA-2 cDNA with 5′ and 3′ primers including the EcoRI restriction site at the 5′ and the HA-tag followed by the stop codon and the XhoI restriction site at the 3′ and ligation of the EcoRI-XhoI PCR product into the EcoRI/XhoI-digested pMSCV/IRES/YFP vector.
Cell cultures and retroviral infections
32-BCR/ABL and derivative cell lines were cultured in Iscove modified Dulbecco medium (IMDM) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM l-glutamine, and penicillin/streptomycin (100 μg/mL each). CML-blast crisis K562 cells were cultured in IMDM supplemented with 10% heat-inactivated FBS. BCR/ABL-expressing primary cells were obtained from the peripheral blood of an imatinib-resistant (M351T mutation) CML-BC patient. Peripheral blood (granulocyte colony-stimulating factor [G-CSF] mobilized) normal CD34+ cells were purchased from StemCell Technologies (Vancouver, BC). Cells were cultured in StemSpan SFEM medium (StemCell Technologies) supplemented with StemSpan CC100 (20 ng/mL IL-3, 20 ng/mL IL-6, 100 ng/mL KL, 100 ng/mL Flt-3 ligand; StemCell Technologies) and 50 ng/mL thrombopoietin (StemCell Technologies). For retroviral infections, Phoenix cells (kind gift of G. P. Nolan, Stanford University School of Medicine, Stanford, CA) were transiently transfected with the indicated plasmids. The infectious supernatant was collected 48 hours later and was used to infect (a 48-hour procedure) p210BCR/ABL-expressing cells. Twenty-four hours later, infected cells were sorted (EPICS Profile Analyzer; Coulter, Hialeah, FL) for green florescent protein (GFP) or yellow fluorescent protein (YFP) expression or selected in the presence of puromycin and kept in culture as described. Images were visualized using an Olympus CK2 microscope with a 40×/0.65 numeric aperture objective, and were photographed using an Olympus SC35 type 12 camera (Olympus, Melville, NY). JPEG images were viewed using Adobe Photoshop (Adobe Systems, San Jose, CA) and contrast adjustments were made.
Cell proliferation and differentiation assays
For proliferation and differentiation assays of p210BCR/ABL cells transduced with wild-type or mutant C/EBPα-ER with or without Δ(358-432) c-Myb or the full-length GATA-2 cDNA, cells were washed with phosphate-buffered saline (PBS) and treated with 4-hydroxytamoxifen (4-HT, 100 nM; Sigma, St Louis, MO) in the presence of 10% FBS. For in vitro differentiation assays of MigRI-C/EBPα-ER retrovirally transduced normal CD34+ and primary CML-BC cells, 4-HT treatment (1 μM) was carried out in medium supplemented with IL-3 (20 ng/mL), KL (20 ng/mL), IL-6 (4 ng/mL), and G-CSF (10 ng/mL). Viable cells were counted by trypan blue exclusion. Differentiation was monitored by May-Grünwald/Giemsa staining and by detection of differentiation-related markers (Gr-1, CD11b, CD14, CD15) with specific phycoerythrin (PE)–conjugated mouse monoclonal antibodies (PharMingen, San Diego, CA). In some experiments, CD11b was also detected with a specific allophycocyanin (APC)–conjugated mouse monoclonal antibody (PharMingen).
Western blotting and reverse-transcription–PCR analyses
For Western blotting, cells were lysed (2 × 105 cells/20 μL in Laemmli buffer) and proteins of interest were detected with anti–c-Myb monoclonal antibody (clone 1-1; Upstate Biotechnology, Lake Placid, NY), anti–GATA-2 rabbit polyclonal antibody (H-116; Santa Cruz Biotechnology, Santa Cruz, CA), anti-PU.1 rabbit polyclonal antibody (Spi-1, sc-352; Santa Cruz Biotechnology), anti-HA monoclonal antibody (HA.11; Covance, Berkeley, CA), or anti-GRB2 monoclonal antibody (610112; BD Transduction Laboratories, Lexington, KY).
For reverse-transcription (RT)–PCR analysis of mRNA expression, total RNA of untreated or 4-HT–treated cells was extracted using RNAeasy Mini Handbook (Qiagen, Valencia, CA) according to the manufacturer's protocol. RT-PCR was performed with 200 ng RNA using the ONE STEP RT-PCR KIT (Qiagen) protocol according to the manufacturer's instructions. Oligodeoxynucleotides specific for the granulocyte colony-stimulating factor (G-CSF) receptor cDNA were upstream primer 5′-ACCTGGGCACAGCTGGAGTGG-3′ and downstream primer 5′-GCTGCTGTGAGCTGGGTCTGG-3′ and for macrophage-colony stimulating factor (M-CSF) receptor cDNA were upstream primer 5′-CAAGTTCATTCAGAGCCAGG-3′ and downstream primer 5′-GGAAATCTACTTGATCGAGG-3′. As internal control, all cDNA samples were adjusted to yield relatively equal amplification of GAPDH. For RT-PCR analysis of pre-mRNA expression, nuclear RNA was extracted from untreated or 4-HT–treated 32D-BCR/ABL-C/EBPα-ER cells using the PARIS kit (Ambion, Austin, TX) and nuclear RNA was DNase-treated using TurboDNase treatment and Removal Reagents (Ambion), reverse transcribed, and PCR-amplified with a pair of intron 1 primers for c-Myb or GATA-2. Oligodeoxynucleotides specific for c-Myb were upstream primer 5′-GATGAATGGAAAACTCTGCG-3′ and downstream primer 5′-CCCTAAAATCTTTGCAGCTGC-3′, whereas those for GATA-2 were 5′-GAATCTGAGCAGCTGCGAAATCG-3′ and 5′-CACACAGGATCACAACTCAG-3′. As internal control, all cDNA samples were adjusted to yield relatively equal amplification of GAPDH (upstream primer 5′-AAGGTCAGCGCTCGGACCTG-3′ and downstream primer 5′-CATTCATTTCCTTCCCG-3′). Control PCR reactions were run with-out template, and duplicates of each reaction were performed without reverse transcriptase.
siRNA transfection
For transfection of siRNAs we used the Amaxa cell optimization kit V (Amaxa, Gaithersburg, MD) according to Amaxa guidelines. Briefly, cells were resuspended in the nucleofector V solution and 100 μL of cell suspension at a density of 106/mL was mixed with 5 μg siRNA. The solution was added to Amaxa electrode cuvettes and electroporated in an Amaxa Nucleofector II, using program T-16 for K562 and U-08 for EM-3 cells. Immediately afterward, cells were diluted in 2 mL IMEM (supplemented with 10% FBS, 100 U/mL penicillin, 0.1 mg/mL streptomycin, 2 mM l-glutamine) at room temperature (5 × 105 cells/mL). The siRNA pools used were ON-TARGET plus SMART pool directed against c-Myb (ThermoFisherScientific, Dharmacon, Waltham, MA) and the control siRNA (ThermoFisherScientific, Dharmacon). The siRNA used for GATA-2 was Silencer Pre-designer siRNA (Ambion).
Untreated cells and cells in nucleofector solution with control siRNA, both subjected to the electroporation protocol, were used as negative controls.
Cell-cycle analysis
Cells (0.2-0.5 × 106/sample) were washed in 1 mL PBS and fixed by slowly adding ice-cold 100% ethanol to reach a final concentration of 70% and incubated on ice for 20 minutes. Cells were washed in PBS and incubated in 0.5 mL PBS containing 200 μg/mL DNase-free RNase A for 30 minutes at room temperature. Propidium iodide was then added to a final concentration of 50 μg/mL and incubated for at least 15 minutes at room temperature in the dark, and samples were then analyzed by flow cytometry (EPICS Profile Analyzer; Coulter).
Results
Activation of C/EBPα suppresses c-Myb and GATA-2 expression in 32D-BCR/ABL cells
In a previous study, we found that activation of the C/EBPα-ER chimeric protein by tamoxifen (4-HT) induced differentiation, inhibited proliferation, and suppressed leukemogenesis of 32D-BCR/ABL cells.13 Induction of differentiation was dependent on the DNA binding and transcription activation function of C/EBPα, while inhibition of proliferation and suppression of leukemogenesis was also achieved by a C/EBPα DNA binding–deficient mutant. However, leukemogenesis was more potently suppressed by activation of wild-type C/EBPα or of Δ(177-191) C/EBPα, a transcription activation competent mutant lacking the CDK2/CDK4 interaction domain.13
To identify potential mechanisms involved in the leukemia-suppressive effects of C/EBPα, microarray hybridization studies were performed using RNA from 4-HT–treated 32D-BCR/ABL cells transduced with the MigRI empty vector, MigRI WT C/EBPα-ER, MigRI K298E C/EBPα-ER (a DNA binding–deficient mutant), or MigRI Δ(177-191) C/EBPα-ER (a mutant deficient in CDK2/CDK4 interaction). Microarray data have been deposited at Gene Expression Omnibus (GEO) under accession number GSE11766.17
Total RNA was extracted at 6, 9, and 12 hours after 4-HT treatment in 3 separate experiments and hybridized onto the Affymetrix MOE430_2 array (Santa Clara, CA), as described.18,19
We identified several genes whose expression was up-regulated or down-regulated in a DNA binding–dependent manner. The transcription factors c-Myb and GATA-2 were among those specifically down-regulated upon activation of WT or Δ (177-191) C/EBPα: c-Myb and GATA-2 mRNA levels were down-regulated approximately 3- to 4-fold by WT or Δ(177-191) C/EBPα, compared with MigRI-transduced or K298E C/EBPα-ER–expressing cells (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
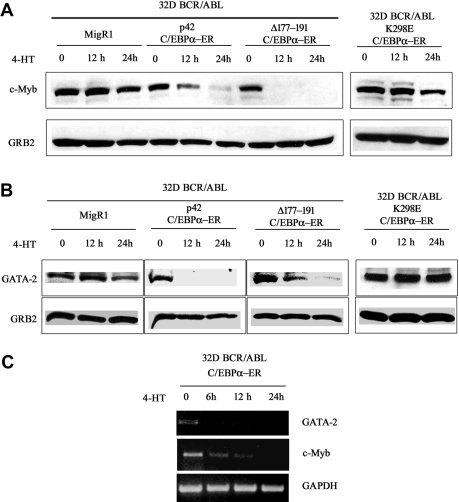
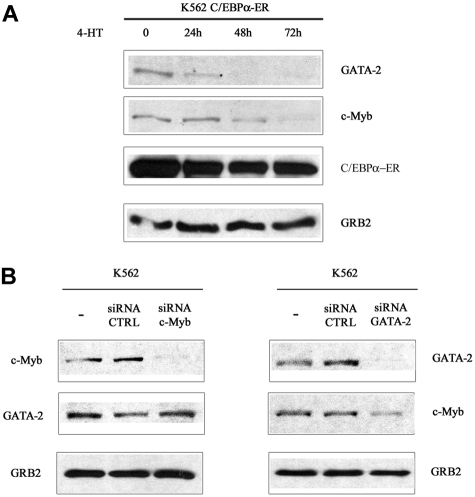
To assess whether the decrease in mRNA levels correlated with reduced protein levels, c-Myb and GATA-2 expression was assessed by Western blotting in lysate of 32D-BCR/ABL cells expressing WT or mutant C/EBPα and treated with 4-HT for 12 and 24 hours. c-Myb expression was markedly down-regulated upon activation of WT C/EBPα or Δ(177-191) C/EBPα; 4-HT treatment of cells transduced with the MigRI empty vector or expressing the K298E C/EBPα mutant induced only a slight decrease of c-Myb levels (Figure 1A). Likewise, GATA-2 expression was rapidly down-regulated upon activation of WT or Δ (177-191) C/EBPα-ER; GATA-2 levels did not change in 4-HT–treated MigRI-transduced cells or in cells expressing the K298E C/EBPα mutant (Figure 1B). To assess whether activation of C/EBPα suppressed c-Myb and GATA-2 expression at the transcriptional level, we performed semiquantitative RT-PCR amplification of c-Myb or GATA-2 pre-mRNA transcripts in 32D-BCR/ABL-C/EBPα-ER cells before and after 4-HT treatment using c-Myb– or GATA-2–specific intron 1 primers. As shown in Figure 1C, expression of c-Myb and GATA-2 pre-mRNA was rapidly down-regulated upon C/EBPα activation, indicating that the effect of C/EBPα on c-Myb and GATA-2 expression is due to transcriptional repression. Rapid down-regulation of c-Myb and GATA-2 expression was also observed upon 4-HT treatment of Philadelphia-positive K562 cells expressing WT C/EBPα-ER (Figure 2A). C/EBPα could repress transcription of both genes independently, or only of c-Myb or GATA-2, which, in turn, could inhibit transcription of the other. To distinguish between these 2 possibilities, K562 cells were transfected with c-Myb or GATA-2 siRNAs and the levels of c-Myb and GATA-2 were tested at different times after transfection. Expression of c-Myb or GATA-2 was completely downmodulated by treatment with gene-specific siRNAs 24 hours after transfection, but GATA-2 levels were not affected in c-Myb siRNA–transfected cells (Figure 2B); by contrast, c-Myb levels were reduced in GATA-2 siRNA–transfected cells (Figure 2B). Together, these studies indicate that the transcription of GATA-2 is suppressed by a C/EBPα-dependent mechanism, although the effect on c-Myb could be, in part, indirect via GATA-2.
Figure 1.
c-Myb and GATA-2 expression in 4-HT–treated 32D-BCR/ABL-C/EBPα-ER cells. Western blots (representative of 3 experiments) show c-Myb (A) and GATA-2 (B) expression in 4-HT–treated 32D-BCR/ABL cells expressing wild-type or mutant C/EBPα-ER. (C) RT-PCR amplification of c-Myb or GATA-2 pre-mRNA in 4-HT–treated 32D-BCR/ABL-C/EBPα-ER cells using pairs of intron 1 c-Myb– or GATA-2–specific primers. RT-PCR amplification of pre-GAPDH RNA was used as control. Representative of 4 different experiments.
Figure 2.
c-Myb and GATA-2 expression in 4-HT/siRNA-treated parental or C/EBPα-ER–expressing K562 cells. (A) Western blots show c-Myb and GATA-2 expression in 4-HT–treated wild-type C/EBPα-ER–expressing K562 cells. (B) Western blots show c-Myb or GATA-2 levels in K562 cells transfected with c-Myb or GATA-2 siRNAs. Data are representative of 3 experiments.
Activation of C/EBPα suppresses c-Myb and GATA-2 expression in CML-BC primary cells but not in CD34+ normal progenitors
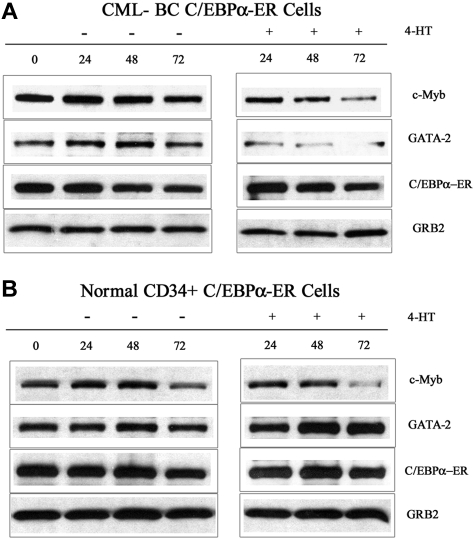
To exclude the possibility that C/EBPα-induced suppression of c-Myb and GATA-2 expression are limited to BCR/ABL-expressing cell lines, we tested the effects of C/EBPα in CML-BC and in normal CD34+ primary cells. Peripheral blood mononuclear cells (80% CD34+) from a CML-BC patient (M351T mutation) and G-CSF–mobilized normal CD34+ cells were retrovirally transduced with MigRI-C/EBPα-ER, GFP-sorted, and treated with 4-HT to activate C/EBPα. After 24, 48, and 72 hours, lysates were prepared and used for Western blotting to assess c-Myb and GATA-2 levels. Expression of c-Myb was markedly down-regulated in CML-BC cells (2- and 6-fold at 48 and 72 hours after 4-HT treatment, respectively), whereas the decrease was modest in normal CD34+ cells (1.3- and 1.4-fold at 48 and 72 hours, respectively) (Figure 3); by contrast, GATA-2 levels were decreased in CML-BC cells (> 6-fold at 48 and 72 hours after 4-HT treatment) but did not change in normal CD34+ cells (Figure 3). Expression of endogenous p42 C/EBPα slightly increased in normal CD34+ cells at 72 hours after 4-HT treatment, but was barely detectable and did not change in CML-BC cells (not shown). Expression of p30 C/EBPα was detectable at low levels in normal and CML-BC cells and did not change upon 4-HT treatment (not shown).
Figure 3.
c-Myb and GATA-2 expression in 4-HT–treated C/EBPα-ER–transduced normal and CML-BC CD34+ cells. Western blots show c-Myb and GATA-2 levels and in normal CD34+ cells (A) and in CML-BC CD34-enriched (80%) cells (B). Expression of C/EBPα-ER and GRB2 was also assessed as control. Data are representative of 2 experiments. CD34+ normal cells were from 2 donors.
Effects of ectopic c-Myb or GATA-2 expression on C/EBPα-induced differentiation and cell-cycle arrest of p210BCR/ABL-expressing cells
To assess whether the effects of C/EBPα on differentiation and proliferation were, in part, c-Myb dependent, we transduced 32D-BCR/ABL WT C/EBPα-ER and 32D-BCR/ABL K298E C/EBPα-ER cells with Δ(358-452) c-Myb, a mutant expressing a c-Myb protein with longer half-life.16 Upon 4-HT treatment, WT C/EBPα-ER but not K298E C/EBPα-ER induced differentiation of 32D-BCR/ABL cells as indicated by the increased expression of the granulocytic differentiation marker Gr-1; however, in cells transduced with Δ(358-452) c-Myb, treatment with 4-HT failed to induce Gr-1 expression (Figure S1A). Of interest, expression of endogenous c-Myb was rapidly down-regulated in both cell lines, whereas levels of Δ(358-452) c-Myb decreased only after 72 hours of 4-HT treatment (Figure S1B).
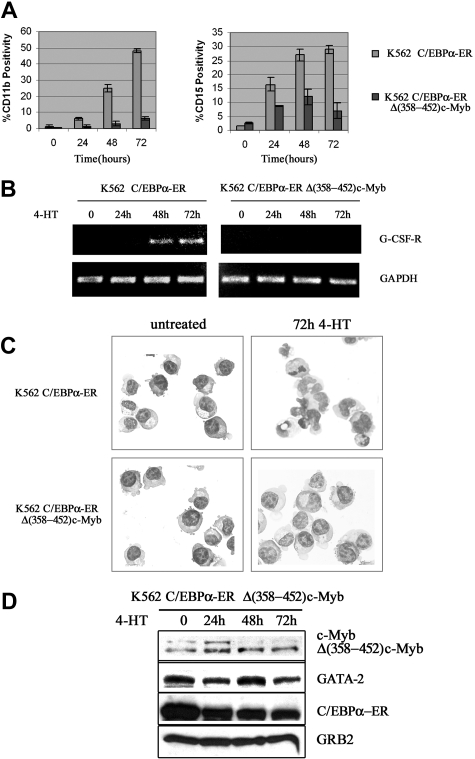
We also tested whether c-Myb suppressed C/EBPα-induced differentiation of K562 cells. Upon 4-HT treatment, expression of CD11b and CD15, 2 markers of myeloid differentiation, was rapidly induced in K562-C/EBPα-ER cells (Figure 4A); by contrast, in cells expressing Δ(358-452) c-Myb, CD11b expression was markedly suppressed (75%-80% inhibition) and levels of CD15 were induced much less (Figure 4A).
Figure 4.
Effect of ectopic c-Myb expression on C/EBPα-induced differentiation of K562 cells. CD11b and CD15 levels (A); G-CSFR mRNA levels (B); morphology (C); and protein levels (D) in 4-HT–treated K562 cells. Data are representative of 3 experiments.
Likewise, expression of the G-CSFR mRNA was induced by 4-HT treatment of K562-C/EBPα-ER cells, but there was no induction in cells expressing Δ(358-452) c-Myb (Figure 4B). Morphologically, 4-HT–treated K562-C/EBPα-ER cells showed clear features of granulocytic differentiation, including the presence of segmented nuclei, whereas 4-HT–treated K562-C/EBPα-ER/Δ(358-452) c-Myb cells were essentially indistinguishable from untreated K562 cells (Figure 4C). C/EBPα activation induced a decrease in the expression of endogenous c-Myb in C/EBPα-ER/Δ(358-452) c-Myb cells; by contrast, C/EBPα had no effect on the levels of Δ(358-452) c-Myb (Figure 4D).
Levels of GATA-2 showed a modest decrease 24 hours after 4-HT treatment but expression was essentially unchanged at 48 and 72 hours (Figure 4D), probably because the cells continued to proliferate (see “Discussion”).
Induction of differentiation was also blocked in 4-HT–treated K562-C/EBPα-ER cells ectopically expressing full-length c-Myb as indicated by CD11b and CD15 levels and by morphology (Figure S2).
The effects of ectopic expression of GATA-2 on C/EBPα-induced differentiation of K562 cells were less dramatic: C/EBPα-dependent induction of CD11b and CD15 (Figure 5A) and expression of G-CSFR (Figure 5B) were partially blocked by GATA-2, in agreement with incomplete reversal of C/EBPα-induced morphologic differentiation (Figure 5C).
Figure 5.
Effect of ectopic GATA-2 expression on C/EBPα-induced differentiation of K562 cells. CD11b and CD15 levels (mean plus SD); G-CSFR mRNA levels (B); morphology (C); and protein levels (D) of 4-HT–treated K562-C/EBPα-ER cells; representative of 3 different experiments.
As expected, expression of endogenous c-Myb remained constant upon 4-HT treatment of GATA-2–expressing K562 cells (Figure 5D).
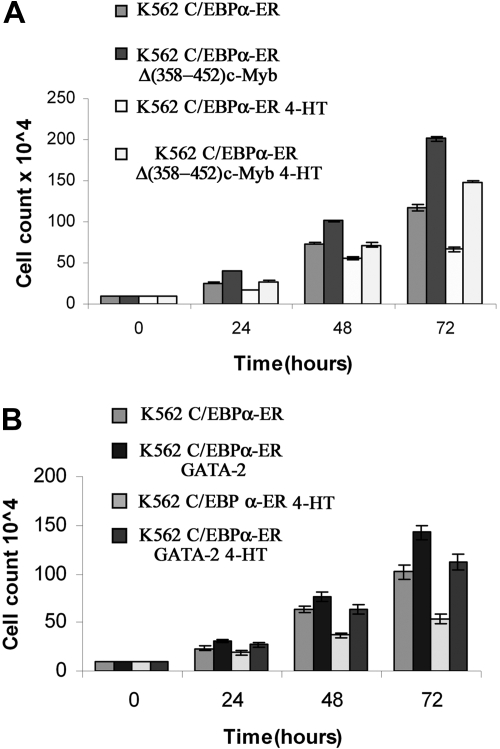
We also tested whether ectopic c-Myb or GATA-2 expression blocked the proliferation inhibitory effects induced by C/EBPα. Compared with untreated cells, K562-C/EBPα-ER cells treated with 4-HT to activate C/EBPα showed a 43% decrease in cell counts at 72 hours (Figure 6A); by contrast, in K562 cells coexpressing C/EBPα-ER and Δ(358-452) c-Myb, inhibition of proliferation induced by C/EBPα was completely reversed (Figure 6A). DNA content analysis of cells treated with 4-HT for 72 hours was consistent with cell counts: C/EBPα activation in parental K562 cells induced a 22% increase in G1 cells and a 30% decrease in S phase cells; by contrast, in c-Myb–expressing K562 cells, activation of C/EBPα did not induce changes in cell-cycle distribution (Figure 7). Likewise, activation of C/EBPα did not inhibit proliferation of K562 cells coexpressing C/EBPα and GATA-2 (Figure 6B), and there were modest effects (7% increase in G1, 10% decrease in S phase) on the cell-cycle distribution of these cells (Figure 7).
Figure 6.
Effect of ectopic c-Myb or GATA-2 expression on C/EBPα-dependent proliferation inhibition of K562 cells. (A) Cell counts of untreated and 4-HT–treated K562-C/EBPα-ER and Δ(358-452) c-Myb/K562-C/EBPα-ER cells. (B) Cell counts of untreated and 4-HT–treated C/EBPα-ER and GATA-2/K562-C/EBPα-ER cells. Mean plus SD (3 different experiments performed in triplicate).
Figure 7.
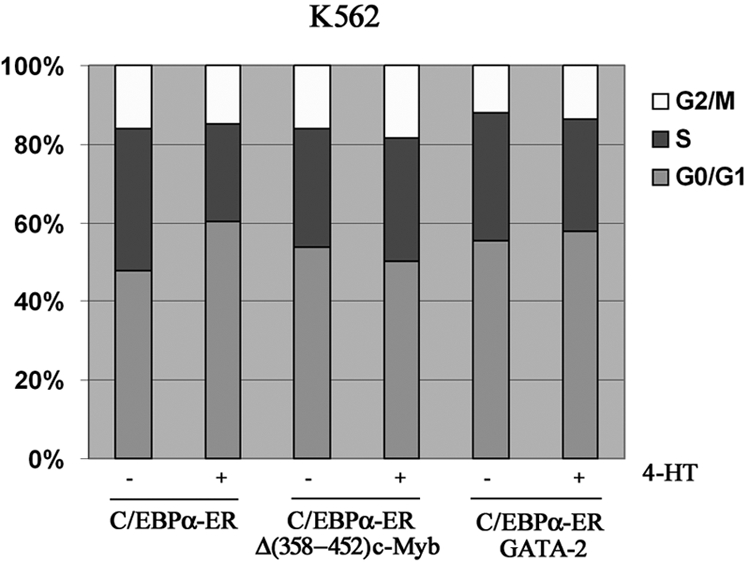
Cell-cycle distribution of K562-C/EBPα-ER, K562-C/EBPα-ER-Myb, and GATA-2/K562-C/EBPα-ER cells. Histogram shows cell-cycle distribution (DNA content of propidium iodide–stained nuclei) of untreated and 4-HT–treated cells. Representative of 2 experiments.
Down-regulation of c-Myb expression by siRNAs modulates the effects of C/EBPα in p210BCR/ABL-expressing cells
Since expression of c-Myb is down-regulated upon C/EBPα activation and ectopic c-Myb expression blocks the biologic effects of C/EBPα in p210BCR/ABL-expressing cells, we reasoned that silencing c-Myb expression by RNA interference might enhance the effects of C/EBPα in p210BCR/ABL cells. Thus, K562-C/EBPα-ER cells were transfected with control or c-Myb siRNAs, and the effects on proliferation and differentiation were tested after treatment with 4-HT to activate C/EBPα.
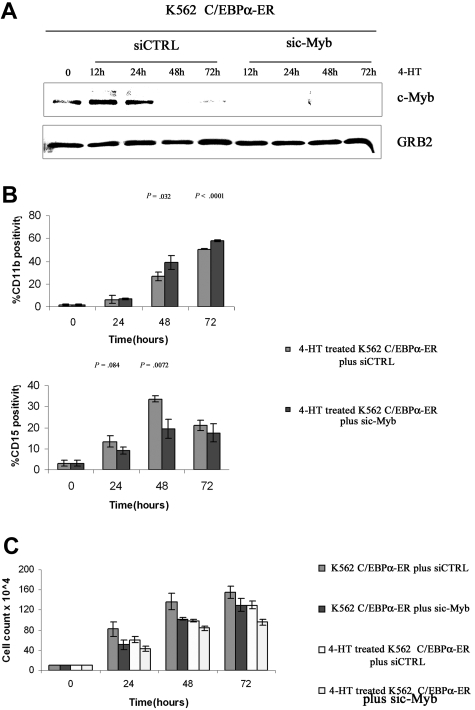
Simultaneous treatment with 4-HT and c-Myb siRNA led to rapid and sustained disappearance of c-Myb expression in K562-C/EBPα-ER cells; by contrast, expression of c-Myb decreased more slowly in cells treated with 4-HT and a control siRNA (Figure 8A). By flow cytometry, cells expressing the myelomonocytic marker CD11b were more numerous 48 and 72 hours after 4-HT treatment (from 28% to 45% and from 50% to 60%, respectively) in c-Myb siRNA– than in control siRNA–treated cultures (Figure 8B); by contrast, treatment with c-Myb siRNA blocked, in part, the increase in the number of cells expressing the CD15 granulocytic antigen induced by 4-HT in K562-C/EBPα-ER cells (10% vs 15%; 20% vs 33%; 18% vs 23%; at 24, 48, and 72 hours after C/EBPα activation, respectively), but the effect was statistically significant only at 48 hours (Figure 8B). The effect of c-Myb down-regulation on C/EBPα-induced differentiation of K562 cells was further analyzed by assessing number of double-positive CD11b/CD15 cells and single-positive CD11b, CD15, and CD14 cells and expression of PU.1, G-CSFR, and M-CSFR. Compared with cells treated with 4-HT and a control siRNA, cotreatment with c-Myb siRNAs and 4-HT induced an increase of single-positive CD11b cells and a decrease of both double-positive CD11b/CD15 cells and single-positive CD15 cells (Figure S3A), further suggesting that 4-HT– and c-Myb siRNA–treated cells have a decreased granulocytic differentiation capability. Consistent with these findings, expression of G-CSFR mRNA was also reduced (Figure S3B). To assess whether the decrease in granulocytic differentiation was compensated by a proportional increase in monocytic differentiation, we assessed levels of monocytic differentiation markers CD14 and M-CSFR and expression of PU.1, a transcription factor essential for monocytic differentiation.20 As shown in Figure S3C, expression of PU.1 was induced upon C/EBPα activation and was more abundant after cotreatment with c-Myb siRNAs; in agreement with these findings, M-CSFR expression was induced earlier in c-Myb siRNA–treated cells (Figure S3D), but expression of CD14, a late marker of monocytic differentiation, was not detectable (not shown). Both 4-HT–induced activation of C/EBPα and c-Myb siRNA treatment of K562-C/EBPα-ER cells induced a decrease of proliferation. The effect of c-Myb siRNA was essentially identical to that of 4-HT–induced C/EBPα activation. In combination, treatment with 4-HT and c-Myb siRNAs was slightly more potent than that with c-Myb siRNA alone (Figure 8C).
Figure 8.
Effect of c-Myb silencing on C/EBPα-induced differentiation and proliferation inhibition in K562 cells. c-Myb (A) and CD11b and CD15 (B) levels in K562-C/EBPα-ER cells treated with 4-HT and a control or c-Myb siRNAs. (C) Cell counts of siRNA-, 4-HT–, or siRNA and 4-HT–treated K562-C/EBPα-ER cells. Mean plus SD (3 different experiments, performed in triplicate).
The effects of c-Myb siRNA and C/EBPα activation were also assessed in p210BCR/ABL-expressing EM-3 CML-blast crisis cells. In this cell line, treatment with c-Myb siRNAs alone was more potent than 4-HT–induced C/EBPα activation in suppressing cell proliferation, and the combined treatment with 4-HT and c-Myb siRNA was slightly more inhibitory than c-Myb siRNA alone (not shown).
Down-regulation of GATA-2 expression by siRNAs enhances C/EBPα-dependent proliferation inhibition but not differentiation of K562 cells
We also assessed the effects of GATA-2 down-regulation on C/EBPα-induced differentiation and proliferation inhibition of K562 cells. Treatment of K562 cells with GATA-2 siRNAs completely suppressed GATA-2 expression (Figure S4A) and blocked, in part, the increase of CD11b levels induced by C/EBPα activation but had no effect on those of CD15 (Figure S4B). As may have been expected by the decrease in the number of CD11b-positive cells upon GATA-2 siRNA treatment, PU.1 levels were slightly reduced 72 hours after C/EBPα activation (Figure S4A), in marked contrast with the effects induced by c-Myb siRNAs (Figure S3C). GATA-2 siRNA treatment suppressed proliferation of K562 cells and further enhanced proliferation inhibition induced by C/EBPα (Figure S4C).
Discussion
This study was undertaken to gain insight on the mechanisms whereby expression of functional C/EBPα induces differentiation and suppresses proliferation of p210BCR/ABL-expressing cells.
To identify transcriptionally regulated targets potentially required for the effects of C/EBPα, we performed microarray hybridization studies using RNA from 32D-BCR/ABL cells treated with 4-HT for conditional activation of transcriptionally competent or DNA binding–deficient C/EBPα.
The majority of the C/EBPα-regulated genes showed increased expression, but levels of a gene subset that included c-Myb and GATA-2, 2 transcription factors with important regulatory roles in proliferation, survival, and differentiation of normal and leukemic hematopoietic cells,21–25 were markedly reduced.
The ability of C/EBPα to repress gene expression at the transcriptional level is not unprecedented: in U937 cells, transcription of c-Myc is negatively regulated by C/EBPα indirectly, probably by its interaction with E2F proteins that prevent E2F-dependent activation of the c-Myc promoter via an E2F binding site.15 Our microarray expression studies did not reveal changes in the expression of c-Myc, raising the possibility that this dif-ference reflects cell type– or context-specific (transformation by p210BCR/ABL) effects of C/EBPα. Of interest, expression of c-Myb was also repressed by C/EBPα in U937 cells.15
Although little is known regarding GATA-2 expression in CML cells, there is evidence that BCR/ABL-transformed cell lines and primary CML-BC cells express high levels of c-Myb16; although this may be due to several mechanisms,16 it is conceivable that loss of C/EBPα-dependent negative regulation of c-Myb transcription plays some role.
Upon C/EBPα activation, levels of GATA-2 and c-Myb pre-mRNA were both down-regulated but those of GATA-2 decreased more rapidly, suggesting that the transcriptional down-regulation of c-Myb was, in part, dependent on suppression of GATA-2 expression. Indeed, silencing of c-Myb expression in K562 cells had no effect on GATA-2 levels, whereas silencing of GATA-2 led to reduced c-Myb expression.
Since the 5′ flanking region of the human c-Myb gene contains putative GATA-2 binding sites,26 it will be important to assess whether GATA-2 binds to the promoter of the c-Myb gene to activate its transcription. Down-regulation of c-Myb and GATA-2 expression was also observed after C/EBPα activation in retrovirally transduced CD34-enriched (80% positivity) primary cells from a patient with myeloid CML-BC. However, in CD34+ normal progenitors, activation of C/EBPα induced only a modest decrease of c-Myb levels, whereas GATA-2 expression was increased even at 48 and 72 hours. The reasons for such differences in the response to C/EBPα activation in normal and p210BCR/ABL-expressing progenitors are unclear. We noted, however, that activation of C/EBPα induced expression of CD11b in both normal and CML-BC CD34+ cells (Figure S5); by contrast, it slowed the proliferation of CML-BC primary cells, whereas it enhanced markedly (3-fold increase at 72 hours) that of normal CD34+ cells.
Although it is unknown whether GATA-2 protein levels change during the cell cycle of primitive hematopoietic progenitors, GATA-2+/− progenitors cycle less than the GATA-2+/+ counterpart,24 a finding consistent with the increase in GATA-2 expression upon the C/EBPα-induced early expansion of normal CD34+ cells. Even if C/EBPα activation did not block proliferation of normal CD34+ cells because it failed to down-regulate GATA-2 expression, it is unclear why the effects were different in normal and CML-BC CD34+ progenitors and whether they may be due to differences in the proliferative potential of these progenitors.
Expression of endogenous C/EBPα was higher in normal than in CML-BC CD34+ cells, a finding that raises the possibility that higher threshold levels of functional C/EBPα might be required for proliferation inhibition of CML-BC cells.
The requirements of c-Myb and GATA-2 for the biologic effects of C/EBPα were tested in K562 cells by ectopic expression or siRNA-mediated down-regulation.
C/EBPα-induced differentiation of K562 cells was completely blocked by c-Myb expression, whereas the effects of GATA-2 were much less striking.
In contrast to the effects on differentiation, c-Myb or GATA-2 was essentially indistinguishable in the ability to prevent the inhibition of proliferation induced by C/EBPα.
Based on these findings, c-Myb and GATA-2 may have different roles in modulating the biologic effects of C/EBPα: changes of c-Myb levels may be more important for the effects of C/EBPα in differentiation, whereas changes of GATA-2 levels may be more relevant for the proliferation inhibitory effect of C/EBPα.
These issues were further addressed by assessing the effects of C/EBPα in K562 cells in which expression of c-Myb or GATA-2 was specifically downmodulated by siRNAs. Down-regulation of either c-Myb or GATA-2 or activation of C/EBPα was sufficient to inhibit the proliferation of K562 cells; however, treatment with GATA-2 siRNAs was more effective than that with c-Myb siRNAs, and the combination with 4-HT (to activate C/EBPα) further inhibited proliferation.
Since GATA-2 expression was not completely down-regulated in K562 cells treated with c-Myb siRNAs and 4-HT, and c-Myb expression was not detectable in K562 cells treated with GATA-2 siRNAs and 4-HT (not shown), GATA-2 down-regulation may lead to inhibition of K562 cell proliferation not only via GATA-2–dependent effects but also indirectly via c-Myb down-regulation.
On examining the effects of c-Myb downmodulation on C/EBPα-induced differentiation of K562 cells, we found that silencing of c-Myb expression induced a modest increase in the number of CD11b+ cells and a comparable decrease of CD15+ cells. Enhanced expression of CD11b was also observed upon C/EBPα activation in c-Myb siRNA–treated CML-BC primary cells (Figure S5).
Since CD11b and CD15 are expressed at distinct stages of granulocytic differentiation and CD11b is also expressed during monocytic differentiation,27 the effect of c-Myb downmodulation on the expression of granulocyte- versus monocyte-specific markers was investigated in more detail.
Interestingly, cotreatment of K562-C/EBPα-ER cells with 4-HT and c-Myb siRNAs caused a decrease of double-positive CD11b/CD15 cells and single-positive CD15 cells and induced an earlier increase of M-CSFR expression but failed to activate CD14 expression, suggesting that the complete down-regulation of c-Myb expression led to an increase in the number of cells progressing along the monocytic lineage, although these cells may not have reached a differentiation stage permissive for the expression of a late marker such as CD14. This interpretation is supported by the higher levels of PU.1 expression in 4-HT– and c-Myb siRNA–treated K562 cells compared with cells treated with 4-HT alone. The mechanism underlying such an increase is unclear since c-Myb siRNA treatment alone had no effects on PU.1 levels (not shown).
Whereas expression of PU.1 is activated by C/EBPα and complete loss of PU.1 expression prevents C/EBPα-induced granulocytic differentiation,28–30 an increase in the expression of PU.1 over C/EBPα31,32 may favor monocytic differentiation and would be consistent with the higher propensity of c-Myb siRNA–treated K562 cells to initiate monocytic differentiation, as indicated by the earlier induction of M-CSFR expression and the increase in the number of CD11b+ cells. Of interest, PU.1 activates the expression of both CD11b and M-CSFR.33,34
In summary, we have shown that c-Myb and GATA-2 are 2 novel transcription targets of C/EBPα with important roles in mediating the proliferation inhibition– and differentiation-inducing effects of C/EBPα in p210BCR/ABL cells. However, the effect of C/EBPα appears to be context dependent because it was not observed in normal CD34+ cells. Since c-Myb and GATA-2 transcription repression depends on the DNA binding and transcription regulatory activity of C/EBPα, our findings indicate that C/EBPα-mediated inhibition of cell proliferation does not depend entirely on protein-protein interactions with cell-cycle regulators.4,5 In vivo studies in mouse models of p210BCR/ABL leukemogenesis are now being performed to determine whether downmodulation of c-Myb or GATA-2 expression enhances the antileukemia effects of C/EBPα.
Supplementary Material
Acknowledgments
We thank Deborah Swerloff for editorial assistance.
This study was supported by National Institutes of Health (Bethesda, MD) grants RO1 95111 and PO1 78890, and by DOD grant W81XWH-06-1-0558 (B.C). M.R.L. was supported by a postdoctoral fellowship of the American-Italian Cancer Foundation (AICF, New York, NY).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.R.S. performed most of the work and wrote parts of the paper; G.F.-A. established the C/EBPα-expressing lines; M.R.L. helped with flow cytometric analyses; M.P. generated some plasmids; Y.Z. and R.V.M. performed and analyzed microarray studies; N.J.D. provided the primary CML-BC sample and discussed experiments and results; and B.C. designed the experiments and wrote the final version of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bruno Calabretta, Department of Cancer Biology, Kimmel Cancer Center, Thomas Jefferson Medical College, Philadelphia, PA 19107; e-mail: B_Calabretta@mail.jci.tju.edu.
References
- 1.Zhang DE, Zhang P, Wong ND, Hetherington CJ, Darlington GJ, Tenen DG. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT/enhancer binding protein alpha-deficient mice. Proc Natl Acad Sci U S A. 1997;94:569–574. doi: 10.1073/pnas.94.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Radomska HS, Huettner CS, Zhang P, Cheng T, Scadden DT, Tenen DG. CCAAT/enhancer binding protein alpha is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol Cell Biol. 1998;18:4301–4314. doi: 10.1128/mcb.18.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keeshan K, Santilli G, Corradini F, Perrotti D, Calabretta B. Transcription activation function of C/EBPα is required for induction of granulocytic differentiation. Blood. 2003;102:1267–1275. doi: 10.1182/blood-2003-02-0477. [DOI] [PubMed] [Google Scholar]
- 4.Porse BT, Pedersen T, Xu X, et al. E2F repression by C/EBPα is required for adipogenesis and granulopoiesis in vivo. Cell. 2001;107:247–258. doi: 10.1016/s0092-8674(01)00516-5. [DOI] [PubMed] [Google Scholar]
- 5.D'Alo F, Johansen LM, Nelson EA, et al. The amino terminal and E2F interaction domains are critical for C/EBPα-mediated induction of granulopoietic development of hematopoietic cells. Blood. 2003;102:3163–3171. doi: 10.1182/blood-2003-02-0479. [DOI] [PubMed] [Google Scholar]
- 6.Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer. 2003;3:89–101. doi: 10.1038/nrc989. [DOI] [PubMed] [Google Scholar]
- 7.Leroy H, Roumier C, Huyghe P, Biggio V, Fenaux P, Preudhomme C. CEBPA point mutations in hematological malignancies. Leukemia. 2005;19:329–334. doi: 10.1038/sj.leu.2403614. [DOI] [PubMed] [Google Scholar]
- 8.Pabst T, Mueller BU, Harakara N, et al. ETO downregulates the granuloycytic differentiation factor C/EBPα in t(8;21) myelod leukemia. Nat Med. 2001;7:444–451. doi: 10.1038/86515. [DOI] [PubMed] [Google Scholar]
- 9.Perrotti D, Cesi V, Trotta R, et al. BCR-ABL suppresses C/EBPα expression through inhibitory action of hnRNP E2. Nat Genet. 2002;30:48–58. doi: 10.1038/ng791. [DOI] [PubMed] [Google Scholar]
- 10.Radomska HS, Baneres DS, Zheng R, et al. Block of C/EBPα alpha function by phosphorylation in acute myeloid leukemia with FLT3 activating mutations. J Exp Med. 2006;203:371–381. doi: 10.1084/jem.20052242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang JS, Santhanam R, Trotta R, et al. High levels of the BCR/ABL oncoprotein are required for the MAPK-hnRNPE2-dependent expression of C/EBPα-driven myeloid differentiation. Blood. 2007;110:994–1003. doi: 10.1182/blood-2007-03-078303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tavor S, Park DJ, Gery S, Vuong PT, Gombart AF, Koeffler HP. Restoration of C/EBPα expression in a BCR-ABL+ cell line induces terminal granuloycytic differentiation. J Biol Chem. 2003;278:52651–52659. doi: 10.1074/jbc.M307077200. [DOI] [PubMed] [Google Scholar]
- 13.Ferrari-Amorotti G, Keeshan K, Zattoni M, et al. Leukemogenesis induced by wild-type and STI571-resistant BCR/ABL is potently suppressed by C/EBPα. Blood. 2006;108:1353–1362. doi: 10.1182/blood-2006-01-011833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwama A, Zhang P, Darlington GJ, McKerchen SR, Maki R, Tenen DG. Use of RDA analysis of knockout mice to identify myeloid genes regulated in vivo by PU. 1 and C/EBPα. Nucleic Acids Res. 1998;26:3034–3043. doi: 10.1093/nar/26.12.3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johansen LM, Iwama A, Lodie TA, et al. c-Myc is a critical target for C/EBPα in granulopoiesis. Mol Cell Biol. 2001;21:3789–3806. doi: 10.1128/MCB.21.11.3789-3806.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corradini F, Cesi V, Bartella V, Bussolari R, Candini O, Calabretta B. Enhanced proliferative potential of hematopoietic cells expressing degradation-resistant c-Myb mutants. J Biol Chem. 2005;280:30254–30262. doi: 10.1074/jbc.M504703200. [DOI] [PubMed] [Google Scholar]
- 17.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 19.Armstrong SA, Stanton JE, Silverman LB, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 20.Friedman AD. Transcriptional regulation of granulocyte and monocyte development. Oncogene. 2002;13:3377–3390. doi: 10.1038/sj.onc.1205324. [DOI] [PubMed] [Google Scholar]
- 21.Gewirtz AM, Calabretta B. A c-myb antisense oligodeoxynucleotide inhibits normal human hematopoiesis in vitro. Science. 1988;242:1303–1306. doi: 10.1126/science.2461588. [DOI] [PubMed] [Google Scholar]
- 22.Caracciolo D, Venturelli D, Valtieri M, Peschle C, Gewirtz AM, Calabretta B. Stage-related proliferative activity determines c-myb functional requirements during normal hematopoiesis. J Clin Invest. 1990;85:55–61. doi: 10.1172/JCI114433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emambokus N, Vegiopoulos A, Harmon B, Jerikman E, Anderson G, Frampton J. Progression through key stages of hematopoiesis is dependent on distinct threshold levels of c-Myb. EMBO J. 2003;22:4478–4488. doi: 10.1093/emboj/cdg434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai FY, Orkin SH. The transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not erythroid and myeloid terminal differentiation. Blood. 1997;84:3636–3643. [PubMed] [Google Scholar]
- 25.Rodrigues NP, Janzen V, Forkert R, et al. Haploinsufficiency of GATA-2 perturbs adult hematopoietic stem-cell homeostasis. Blood. 2005;106:477–484. doi: 10.1182/blood-2004-08-2989. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, Scott E, Sawyers CL, Friedman AD. C/EBPα bypasses G-CSF signals to rapidly induce PU. 1 gene expression, stimulate granulocytic differentiation, and limit proliferation in 32Dcl3 myeloblasts. Blood. 1999;94:560–571. [PubMed] [Google Scholar]
- 27.Nicolaides NC, Gualdi R, Casadevall C, Manzella L, Calabretta B. Positive autoregulation of c-myb expression via Myb binding sites in the 5′ flanking region of the human c-myb gene. Mol Cell Biol. 1991;11:6166–6176. doi: 10.1128/mcb.11.12.6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charrad R-S, Gadhoum Z, Qi J, et al. Effects of anti-CD44 monoclonal antibodies on differentiation and apoptosis of human myeloid leukemia cell lines. Blood. 2002;99:290–299. doi: 10.1182/blood.v99.1.290. [DOI] [PubMed] [Google Scholar]
- 29.Wang D, D'Costa J, Civin CI, Friedman AD. C/EBPα directs monocytic commitment of primary myeloid progenitors. Blood. 2006;108:1223–1224. doi: 10.1182/blood-2005-12-008763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeamans C, Wang D, Paz-Priel I, Torbett BE, Tenen DG, Friedman AD. C/EBPα binds and activates the PU. 1 distal enhancer to induce monocyte lineage commitment. Blood. 2007;110:3136–3142. doi: 10.1182/blood-2007-03-080291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dahl R, Walsh JC, Lancki D, et al. Regulation of macrophage and neutrophil cell fates by the PU. 1: C/EBPα ratio and granulocyte colony-stimulating factor. Nat Immunol. 2003;6:1024–1036. doi: 10.1038/ni973. [DOI] [PubMed] [Google Scholar]
- 32.Laslo P, Spooner CJ, Warmflash A, et al. Multi-lineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126:755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- 33.Zhang DE, Hetherington CJ, Chen HM, Tenen DG. The macrophage transcription factor PU. 1 directs tissue-specific expression of the macrophage colony-stimulating factor receptor. Mol Cell Biol. 1994;14:373–381. doi: 10.1128/mcb.14.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pahl HL, Scheibe RJ, Zhang DE, et al. The proto-oncogene PU. 1 regulates expression of the myeloid-specific CD11b promoter. J Biol Chem. 1993;268:5014–5020. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.