Abstract
Microtubule-associated proteins (MAPs) bind to and stabilize microtubules (MTs) both in vitro and in vivo and are thought to regulate MT dynamics during the cell cycle. It is known that p220, a major MAP of Xenopus, is phosphorylated by p34cdc2 kinase as well as MAP kinase in mitotic cells, and that the phosphorylated p220 loses its MT-binding and -stabilizing abilities in vitro. We cloned a full-length cDNA encoding p220, which identified p220 as a Xenopus homologue of MAP4 (XMAP4). To examine the physiological relevance of XMAP4 phosphorylation in vivo, Xenopus A6 cells were transfected with cDNAs encoding wild-type or various XMAP4 mutants fused with a green fluorescent protein. Mutations of serine and threonine residues at p34cdc2 kinase-specific phosphorylation sites to alanine interfered with mitosis-associated reduction in MT affinity of XMAP4, and their overexpression affected chromosome movement during anaphase A. These findings indicated that phosphorylation of XMAP4 (probably by p34cdc2 kinase) is responsible for the decrease in its MT-binding and -stabilizing abilities during mitosis, which are important for chromosome movement during anaphase A.
INTRODUCTION
Microtubules (MTs)1 are dynamic polymers composed of 13 protofilaments of α- and β-tubulin heterodimers (Kirschner and Mitchison, 1986). In interphase cells, the heterodimers constitute relatively stable and long MTs. At the onset of mitosis, under the control of p34cdc2 kinase (Hunt, 1989; Murray and Kirschner, 1989; Maller, 1990; Nurse, 1990; King et al., 1994), the frequencies of transition in MTs from growth to shrinkage (catastrophe) increase (Belmont et al., 1990), resulting in the organization of the dynamic and unstable MTs into the mitotic spindle (Salmon et al., 1984; Saxton et al., 1984; Verde et al., 1990; McNally, 1996; Hyman and Karsenti, 1996).
The dynamic character of MTs is thought to be important in several processes during mitosis. First, the abrupt decrease of MT polymer, i.e., the elimination of interphase MTs, before the mitotic spindle formation is attributed to the increase of MT dynamics concomitantly associated with nuclear envelope breakdown (Zhai et al., 1996). Second, the poleward flux of kinetochore MTs during metaphase, which is characterized by accelerated depolymerization of kinetochore MTs from their minus ends at the spindle pole, requires the increase of MT dynamics (Mitchison, 1989; Rodionov et al., 1994; Zhai et al., 1995). Third, chromosome segregation during anaphase A is also based on MT dynamics. Although many processes in mitotic spindle assembly and chromosome segregation require ATP-dependent motor activities (Vernos and Karsenti, 1996; Hirokawa et al., 1998), anaphase A has been suggested to be an ATP-independent process (Cande, 1982; Koshland, 1994). Recent studies have shown that depolymerization of MTs at their plus ends, which are linked to the kinetochore by a kinesin-like protein, CENP-E, drives chromosomes toward the spindle pole in an ATP-independent manner (Desai and Mitchison, 1995; Lombillo et al., 1995a,b).
The factors responsible for regulation of MT dynamics are subclassified into three groups. The first and second of these are MT-severing factors such as p56, katanin, and elongation factor 1α (Shiina et al., 1992a, 1994; McNally and Vale, 1993) and catastrophe factors such as Op18 (Belmont and Mitchison, 1996; Larsson et al., 1997) and XKCM1 (for Xenopus kinesin central motor 1)/MCAK (for mitotic centromere-associated kinesin) (Walczak et al., 1996). The third group is the MT-associated proteins (MAPs). Various MAPs have been identified, and these have been shown to promote MT assembly and to be localized on MTs both in vitro and in vivo (Olmsted, 1991; Mandelkow and Mandelkow, 1995). p220 (= XMAP230), a ubiquitous Xenopus MAP, was reported to be phosphorylated specifically in mitosis, which was associated with a decrease in the MT-binding and -stabilizing activities (Shiina et al., 1992b; Andersen et al., 1994). Kinases responsible for p220 phosphorylation have been identified as p34cdc2 kinase and MAP kinase (Shiina et al., 1992b), both of which are known to be activated in mitosis during oocyte maturation (Kosako et al., 1994; Gotoh et al., 1995). In mammalian tissues and cells, MAP4 is known to be a thermostable, ubiquitously expressed MAP with a molecular mass of ∼200 kDa (Olmsted, 1991; Mandelkow and Mandelkow, 1995). MAP4 is localized along MT networks in vivo, and recently MAP4–green fluorescent protein (GFP) fusion proteins have been used to examine the behavior and functions of MAP4 in vivo (Olson et al., 1995). MAP4 is differentially phosphorylated at the onset of mitosis (Vandre et al., 1991), and phosphorylation by p34cdc2 kinase decreases the ability of MAP4 to stabilize MTs in vitro (Ookata et al., 1995). Phosphorylation of MAP4 by other kinases such as MARK (for MAP/microtubule affinity-regulating kinase) has also been reported to decrease its MT-stabilizing ability (Illenberger et al., 1996; Drewes et al., 1997).
In the present study, we cloned a cDNA encoding Xenopus p220, and its deduced amino acid sequence revealed that it is a Xenopus homologue of MAP4 (XMAP4). Using this isolated XMAP4 cDNA, we constructed GFP–XMAP4 fusion proteins and examined the physiological relevance of the phosphorylation of XMAP4 during mitosis.
MATERIALS AND METHODS
Cells and Antibodies
Xenopus A6 cells were grown in A6 medium (L-15 [Life Technologies, Gaithersburg, MD]/H2O/FCS at a ratio of 50:40:10) at 23°C. A monoclonal anti-p220 (anti-XMAP4) antibody was previously produced against p220 purified from Xenopus eggs (Shiina et al., 1992b).
Cell Staining
For DAPI staining of living cells, A6 cells were incubated in A6 medium containing 2.5 μg/ml DAPI for 1 h, followed by washing with the medium three times. The cells did not appear to be damaged by UV illumination (1 sec three times). Actin was stained by rhodamine–phalloidin (Molecular Probes, Eugene, OR).
cDNA Cloning and Sequencing
A λgt11 expression cDNA library of Xenopus oocyte (Clontech Laboratories, Palo Alto, CA) was screened with the monoclonal anti-p220 antibody according to the method described previously (Huynh et al., 1985). From 8 × 105 plaques, 13 positive clones were isolated. Inserts of these clones were subcloned into pBluescript SK(−) (Stratagene, La Jolla, CA), sequenced with a Taq Dye-Deoxy Terminator cycle sequencing kit (Applied Biosystems, Foster City, CA), and all clones were found to be overlapping. A 0.64-kb NotI–EcoRV fragment of one of the clones, F12 (corresponding to nucleotides 1499–3920), was labeled using a digoxygenin labeling kit (Boehringer Mannheim, Indianapolis, IN) and used to screen the same cDNA library using a digoxygenin detection kit (Boehringer Mannheim). Fifteen cDNA clones were isolated, one of which, clone S36 (corresponding to nucleotides 1–2350), and F12 were sequenced on both strands.
Mutagenesis and GFP–XMAP4 Constructs
The full-length cDNA for p220 was cloned into pBluescript SK(−) and subjected to mutagenesis. Site-directed mutations were generated using primers that introduced the desired alanine point mutations using a Transformer site-directed mutagenesis kit (Clontech). All mutants were confirmed by sequencing.
The ORF of wild-type or mutant XMAP4 was amplified by PCR with the 5′ primer 5′-GACTAGTATGGCGGACCTTGGACAA-3′ and the 3′ primer 5′-GACTAGTGATGCTTGTCTCTGGTAT-3′, which produced SpeI sites at both ends of the XMAP4 ORF. The resultant SpeI fragment was cloned into pQBI25 (Quantum Biotechnologies, Montreal, Quebec, Canada) to produce an expression vector for a GFP fusion protein. All constructs were reconfirmed by sequencing.
Transfection and Selection of Stable Cell Lines
A6 cells were grown to ∼30% confluence in 60-mm dishes and transfected with pQBI25-XMAP4 plasmid DNA according to the Lipofectin protocol (Life Technologies)
A6 cells with stably integrated plasmids were selected by treatment with 1 mg/ml Geneticin, a neomycin analogue (Life Technologies). Clones stably expressing GFP–XMAP4 were picked up under an inverted fluorescence microscope (IX70; Olympus, Tokyo, Japan) followed by limiting dilution in 96-well plates. Stable transfectants were used for all experiments.
Observation of Living Cells
Living cells grown on glass coverslips were mounted in 20 μl of growth medium on chambers comprising a square bounded by four strips of transparent tape on a glass slide with silicone grease (Beckman Instruments, Fullerton, CA) just inside the square. Photographs were taken using a Zeiss Axiophot 2 microscope (Carl Zeiss, Thornwood, NY) with a 63× Plan-Apochromat objective lens. For extended time lapse observation, living cells grown in six-well plates were observed under an inverted fluorescence microscope (Olympus IX70) with a 40× LCPlanFl objective lens.
Extracts from Mitotic and Interphase Cells
Mitotic A6 cells were selectively collected by pipetting after culture in A6 medium containing 0.4 μg/ml nocodazole for 6 h. Collected cells were washed with MBS (88 mM NaCl, 1 mM KCl, 2.4 mM NaHCO3, and 15 mM Tris-HCl, pH 7.6) and extracted with NP-40 lysis buffer (150 mM NaCl, 1% Nonidet P-40, 50 mM Tris-HCl, pH 8.0, 10 μg/ml leupeptin, 10 μg/ml pepstatin, 100 μg/ml aprotinin, 1 mM PMSF, and 1 mM DTT) on ice for 30 min. After centrifugation for 10 min at 10,000 × g at 4°C, the supernatant was recovered as “mitotic extract.” For preparation of “interphase extracts,” the cells remained on the culture plates after pipetting were extracted with NP-40 lysis buffer.
Immunoblotting
After SDS-PAGE (5%), proteins were transferred onto Immobilon membranes (Millipore, Bedford, MA), which were blocked with 5% skimmed milk and then incubated with the anti-p220 (anti-XMAP4) antibody. For antibody detection, a blotting detection kit (Amersham, Arlington Heights, IL) was used.
In Vitro Phosphorylation and Dephosphorylation
Heat-stable XMAP4 fractions were prepared by boiling the mitotic or interphase extracts for 3 min after addition of 0.6 M NaCl and 0.5% β-mercaptoethanol. After centrifugation at 12,000 × g for 30 min at 4°C, the supernatants were used for phosphorylation and dephosphorylation reactions. Heat-stable XMAP4 was phosphorylated by purified p34cdc2 kinase (Shiina et al., 1992b) for 180 min at 25°C in 2 mM EGTA, 10 mM MgCl2, 30 mM β-glycerophosphate, 20 mM Tris-HCl, pH 7.5, and 0.2 mM ATP. Dephosphorylation of heat-stable XMAP4 was performed by incubation with bacterial alkaline phosphatase (Toyobo, Tokyo, Japan) for 60 min at 25°C in 1 mM MgCl2 and 50 mM Tris-HCl, pH 8.0. The reactions were terminated by addition of SDS-PAGE sample buffer.
MT Sedimentation Assay
A 75-μl aliquot of Taxol-stabilized MTs prepared as described previously (Shiina et al., 1992b) was added to 25 μl of cell extract, incubated for 10 min at 25°C, and then centrifuged through a 30% sucrose cushion (200 μl) at 20,000 × g for 30 min at 25°C. The precipitate was suspended in 100 μl of 20PME buffer (Shiina et al., 1992b) containing 20 μM Taxol and centrifuged again. The supernatant of the first centrifugation and the precipitate of the second centrifugation were analyzed by immunoblotting.
RESULTS
Molecular Cloning of XMAP4
Using a monoclonal antibody against p220 (Shiina et al., 1992b), we screened a λgt11 expression cDNA library of Xenopus oocytes. The longest cDNA obtained was 3920 nucleotides in length and encoded a 1224-amino acid protein with a predicted molecular mass of 130 kDa (Figure 1). The deduced amino acid sequence contained one of the internal peptide sequences directly determined from purified XMAP230 (Andersen and Karsenti, 1997), indicating that XMAP230 is identical to p220 (Figure 1A).
Figure 1.
Cloning and sequencing of p220 cDNA. (A) Alignment of the predicted amino acid sequence of p220 with human MAP4 (West et al., 1991). Identical residues are shaded in black, and similar residues are shaded in gray. The dashed line above the sequence indicates an internal peptide sequence from XMAP230. Potential phosphorylation sites for p34cdc2 kinase are indicated by black circles; MAP kinase is indicated by lines; and MARK is indicated by white circles. (B) Structural comparison of Xenopus and human MAP4. The domains are N, highly conserved amino terminus; a, acidic region; RPT, repetitive domain; b, acidic region; P, highly conserved proline-rich domain; SP, serine- and proline-rich domain; PGGG, highly conserved domain containing the MT-binding domain; and C, acidic tail (see West et al., 1991). Potential phosphorylation sites for p34cdc2 kinase are indicated by black circles; MAP kinase is indicated by squares; and MARK is indicated by white circles. Overall XMAP4 and human MAP4 are 30% identical and 65% similar.
A database search revealed that p220 was a Xenopus homologue of MAP4 (XMAP4), and XMAP4 shared the following common structural features with mammalian MAP4 (Figure 1B). First, XMAP4 contained a PGGG domain (aa 995-1141), which is a well-characterized MT-binding domain highly conserved among mammalian MAPs such as MAP4, MAP2, and tau (Aizawa et al., 1990; Chapin and Bulinski, 1991; West et al., 1991). Second, XMAP4 bore two domains (aa 1–94 and 733–800) corresponding to domains N and P in mammalian MAP4, respectively, which are highly conserved at the amino acid sequence level among human, mouse, and bovine MAP4 (West et al., 1991). Third, the NH2-terminal half of XMAP4 was characterized by a repetitive domain containing 20 consecutive repeats of 10 amino acids each: PEAEVL(T/S)(S/A)PI (aa 431–640). This repetitive domain has been found neither in mammalian MAP2 nor in tau but in mammalian MAP4, although the repeat unit of mammalian MAP4 consists of 14 amino acids (Aizawa et al., 1990; West et al., 1991). XMAP4 contained six potential phosphorylation sites (S437, T752, S771, T795, T823, and T877) for p34cdc2 kinase, (S/T)PX(K/R), two sites (T630 and T756) for MAP kinase, PX(S/T)P and PXX(S/T)P, and two sites (S741 and S827) for both kinases (Figure 1). Among these, S741 and S827 were conserved between Xenopus and human, but the other potential phosphorylation motifs were not conserved. Four potential phosphorylation sites (S1001, S1075, S1111, and S1132) for MARK (KXGS) (Drewes et al., 1997) were also found in the PGGG domain, three of which were conserved between Xenopus and human (Figure 1).
Behavior of XMAP4 in Cultured A6 Cells
To examine the behavior of XMAP4, we introduced a GFP–XMAP4 fusion protein into cultured Xenopus A6 cells. As shown in Figure 2, GFP–XMAP4 was distributed along MTs throughout the cell cycle. In interphase cells, GFP–XMAP4 was localized on the MT network (Figure 2A), as previously shown for GFP–human MAP4 (Olson et al., 1995). This localization was the same as that revealed by immunofluorescence microscopy with an anti-p220 monoclonal antibody (Shiina et al., 1992b). In mitotic cells, GFP–XMAP4 was abundantly found on the mitotic spindle and faintly on the astral MTs (Figure 2B). When mitotic cells were fixed with formaldehyde, the majority of GFP–XMAP4 appeared to be diffusely distributed in the cytoplasm, leaving only a small amount on spindle MTs (our unpublished results), which was consistent with the immunofluorescence images of formaldehyde-fixed A6 cells with the anti-p220 antibody (Shiina et al., 1992b).
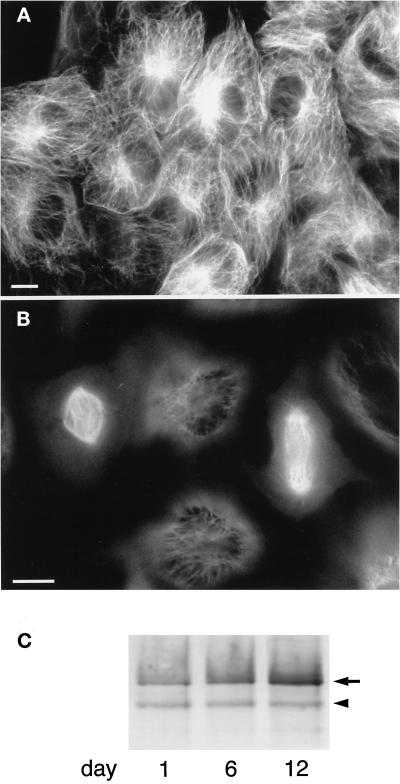
Figure 2.
Expression of GFP–XMAP4 in A6 cells. (A and B) Fluorescence images of living A6 cells expressing GFP–XMAP4. A, interphase cells; B, mitotic cells in metaphase (left) and late anaphase (right). Bars, 10 μm. (C) Immunoblotting of A6 cell extracts with anti-XMAP4 antibody. A6 cells expressing GFP–XMAP4 were extracted at 1, 6, and 12 d after they were sparsely plated (∼1% of confluence) in 100-mm dishes (cells became confluent in 12 d) and subjected to immunoblotting with anti-XMAP4 antibody. The arrowhead and arrow denote endogenous XMAP4 and GFP–XMAP4, respectively. Aliquots of ∼10 μg of protein were loaded in each lane. Because cells were not synchronized, the mobility shift (see Figure 3) was not clearly detectable.
To confirm the expression of GFP–XMAP4 in the transfected cells, the cell extracts were immunoblotted with the anti-p220 (anti-XMAP4) antibody (Figure 2C). Interestingly, the expression level of GFP–XMAP4 was almost the same as that of endogenous XMAP4 in exponentially proliferating cells, whereas the GFP–XMAP4 level increased up to approximately fivefold that of endogenous XMAP4 in confluent cells (Figure 2C). In mitotic extracts, the electrophoretic mobility was decreased in GFP–XMAP4 as well as endogenous XMAP4 bands (Figure 3), suggesting their mitosis-specific phosphorylation.
Figure 3.
Electrophoretic mobility shift of XMAP4 in mitosis. Interphase (I) and mitotic (M) cell extracts of nontransfected A6 cells (−) and A6 cells expressing wild-type GFP–XMAP4 (WT) or GFP–CM10A (CM10A) at subconfluence were immunoblotted with anti-XMAP4 antibody. Arrowheads and arrows denote endogenous XMAP4 and wild-type GFP–XMAP4 and GFP–CM10A, respectively. Electrophoretic mobility shift in mitosis was observed within two arrows or two arrowheads. This antibody recognized upwardly shifted XMAP4 bands more intensely, although the reasons for this are not known.
XMAP4 Mutants and Their Overexpression in A6 Cells
To examine the physiological relevance of XMAP4 phosphorylation in vivo, we constructed a mutated GFP–XMAP4 (GFP–CM10A) in which all of the 10 potential phosphorylation serine/threonine residues for p34cdc2 kinase and/or MAP kinase were mutated to alanine and transfected it to A6 cells. Immunoblotting with the anti-XMAP4 antibody revealed that the expression level of GFP–CM10A was similar to that of GFP–wild-type XMAP4 (GFP–WT), but in contrast GFP–CM10A did not show the mobility shift in mitotic cells (Figure 3). This finding suggested that, as expected, CM10A was nonphosphorylatable by p34cdc2 kinase and/or MAP kinase during mitosis.
The stable transfectants expressing GFP–CM10A proliferated normally when plated sparsely. Under such a culture condition, the expression level of GFP–CM10A was relatively low (almost the same as that of endogenous XMAP4), which may be the reason why mitosis proceeds normally and the stable transfectants can be maintained. However, for unknown reason, when the cell density reached subconfluence, the GFP–CM10A expression level increased up to fivefold of endogenous XMAP4, and the cells began to show abnormal mitosis; the cells entered into mitosis with normal mitotic spindle formation, but furrowing was initiated without spindle elongation, followed by the formation of multiple cleavage furrows. For example, the cell shown in Figure 4A bore two cleavage furrows (a and b) at the beginning of the observation period. At 3 min, furrowing proceeded at b, and the spindle was pushed leftward, which might have triggered the further furrowing at a at 12 min, resulting in the rightward shift of the spindle. We called this series of movement “back-and-forth peristaltic movement” (BFP movement). At 21, 33, 36, and 39 min, the cell repeated the BFP movement with two furrows, and the movement continued throughout the observation period (129 min) without spindle elongation or normal cytokinesis. Another cell in Figure 4B also started BFP movement with two cleavage furrows (a and b). At ∼36–45 min, the spindle appeared to trigger the third furrow (c) and the fourth furrow (d) at ∼60–69 min, and then the spindle began to elongate. Furrowing at c at 75 and 81 min was unsuccessful, but that at d at 90 min led to abnormal cytokinesis at 102 min. In the cells exhibiting BFP movement, actin filaments were concentrated at furrows similarly to normal cleavage furrows (Figure 4C).
Figure 4.
Examples of BFP movement in living cells expressing GFP–CM10A. (A and B) Two A6 cells expressing GFP–CM10A were observed by phase contrast (left) and GFP fluorescence (right). The time after the beginning of observation is indicated in the top right of each image. In the cells expressing wild-type GFP–XMAP4, cytokinesis occurred ∼30 min after spindle formation. (C) An A6 cell exhibiting BFP movement was observed by differential interference contrast (left) and rhodamine–phalloidin fluorescence (right). Cleavage furrows are indicated by arrowheads.
To examine the phenotypic changes of the GFP–CM10A-expressing cells in more detail, chromosomes were visualized by DAPI (Figure 5). In cells expressing GFP–WT, chromosomes were aligned at the spindle equator in metaphase (Figure 5A) and separated toward spindle poles without spindle elongation in anaphase A (Figure 5B), followed by spindle elongation and cytokinesis. In cells expressing GFP–CM10A, chromosomes appeared to be aligned at the spindle equator (Figure 5C). Surprisingly, in these cells, the spindle elongated without chromosome separation (Figure 5D); i.e., chromosomes failed to move toward the spindle poles during anaphase A. Moreover, cytokinesis appeared to proceed without chromosome segregation, occasionally resulting in forced segregation of chromosomes into two daughter cells (Figure 5E). In some cases, cytokinesis failed to proceed, and chromosomes began to decondense without segregation (Figure 5F).
Figure 5.
Impaired anaphase chromosome separation in A6 cells expressing GFP–CM10A. Living A6 cells expressing GFP–WT (A and B) or GFP–CM10A (C–F) were observed. Left, differential interference contrast images; middle, GFP fluorescence images; right, DNA staining images with DAPI (2.5 μg/ml). (A) normal metaphase; (B) normal anaphase A; (C) BFP movement; (D) spindle elongation without chromosome separation; (E) cytokinesis without chromosome separation; (F) incomplete cytokinesis with decondensed chromosomes. Arrowheads in E and F denote cleavage furrows. Bar, 10 μm.
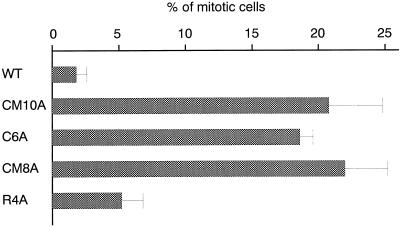
We constructed three more GFP–XMAP4 mutants: C6A (mutated in six potential phosphorylation sites specific for p34cdc2 kinase, S437, T752, S771, T795, T823, and T877), CM8A (mutated in two potential phosphorylation sites for both p34cdc2 kinase and MAP kinase, S741 and S827, in addition to the above six sites), and R4A (mutated in four potential phosphorylation sites for MARK, S1001, S1075, S1111, and S1132) (Table 1). As summarized in Figure 6, transfectants expressing GFP–C6A or GFP–CM8A, as well as GFP–CM10A-expressing cells, exhibited BFP movement without chromosome separation more frequently than control transfectants expressing GFP–WT. This finding indicated that mutations in p34cdc2 kinase-specific phosphorylation sites (C6A) were sufficient to induce the BFP movement without chromosome separation. On the other hand, transfection with GFP–R4A did not increase the frequency of BFP movement and showed normal mitosis.
Table 1.
Mutations of serine and threonine residues in potential phosphorylation sites to alanine
| Consensus
|
p34cdc2 kinase [(S/T)PX(K/R)]
|
MAP kinase [PX(S/T)P, PXX(S/T)P]
|
p34cdc2 kinase and MAP kinase
|
MARK [KXGS]
|
|---|---|---|---|---|
| Residues | S437,T752,S771, T795,T823,T877 | T630,T756 | S741,S827 | S1001,S1075, S1111,S1132 |
| Wild type | ||||
| WT | – | – | – | – |
| Mutant | ||||
| CM10A | A | A | A | – |
| C6A | A | – | – | – |
| CM8A | A | – | A | – |
| R4A | – | – | – | A |
Four XMAP4 mutants were produced by site-directed mutagenesis. A, serine and threonine residues were mutated to alanine; –, not mutated.
Figure 6.
BFP movement in A6 cells expressing GFP–CM10A, GFP–C6A, or GFP–CM8A. Transfectants expressing GFP–WT, GFP–CM10A, GFP–C6A, GFP–CM8A, or GFP–R4A were plated in six-well plates to form ∼50 colonies per well. After 12 d, mitotic cells were identified from phase contrast and GFP fluorescence images. Mitotic cells of three individual cell lines were scored (∼300 cells × 3), and the mean ratio of cells showing BFP movement to mitotic cells is shown for transfectants expressing each GFP–XMAP4 fusion protein. It should be noted that colony growth and proportion of mitotic cells to interphase cells were similar among the transfectants.
Phosphorylation of XMAP4 In Vivo and In Vitro
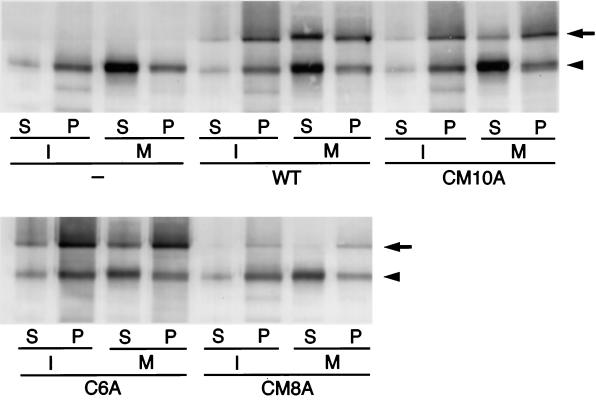
We then checked whether endogenous XMAP4 and introduced GFP–WT, but not GFP–CM10A, GFP–C6A, and GFP–CM8A, were phosphorylated in mitotic cells. As shown in Figure 7A (and also in Figure 3), in mitotic cells, the bands of GFP–WT as well as endogenous XMAP4 were shifted upward. In contrast, those of GFP–CM10A, GFP–C6A, and GFP–CM8A did not show any mobility shift in mitosis. Furthermore, when the XMAP4 fractions were treated with alkaline phosphatase, these mitosis-specific upward shifts of bands were completely suppressed (Figure 7B). These findings indicated that endogenous XMAP4 and introduced GFP–WT were phosphorylated in a mitosis-specific manner, and that GFP–CM10A, GFP–C6A, and GFP–CM8A were nonphosphorylatable during mitosis.
Figure 7.
Phosphorylation of XMAP4 and mutants in mitosis. (A) Mobility shift of XMAP4 and mutants in mitosis. Interphase (I) and mitotic (M) extracts of A6 cells expressing GFP–WT, GFP–CM10A, GFP–C6A, GFP–CM8A, or GFP–R4A were immunoblotted with anti-XMAP4 antibody. Arrowheads and arrows denote endogenous XMAP4 and GFP–XMAP4, respectively (see Figure 3). Note that the bands of GFP–WT and GFP–R4A as well as endogenous XMAP4 were shifted upward up to the top arrow and arrowhead in mitosis, whereas those of GFP–CM10A, GFP–C6A, and GFP–CM8A did not show any mobility shift. (B) Mobility shift of XMAP4 by phosphorylation. Heat-stable XMAP4 fractions were prepared from interphase (I) and mitotic (M) A6 cells expressing GFP–WT. The fractions were treated with alkaline phosphatase in the presence or absence of the phosphatase inhibitor β-glycerophosphate (30 mM) and then immunoblotted with anti-XMAP4 antibody. (C) XMAP4 phosphorylation by p34cdc2 kinase. Heat-stable XMAP4 fractions were prepared from interphase (I) and mitotic (M) A6 cells expressing GFP–WT or GFP–CM10A and then phosphorylated by p34cdc2 kinase (+). Immunoblotting was with anti-XMAP4 antibody. Arrowheads and arrows denote endogenous XMAP4 and GFP–XMAP4, respectively.
Next, we incubated the interphase XMAP4 fractions with p34cdc2 kinase in vitro (Figure 7C). This incubation shifted the bands of endogenous XMAP4 as well as introduced GFP–WT upward, but not those of GFP–CM10A. These findings were consistent with the expectation that the CM10A mutant was nonphosphorylatable by p34cdc2 kinase.
As for GFP–R4A, the band was shifted upward in mitotic cells, indicating that MARK-specific phosphorylation sites are not involved in the mitosis-specific phosphorylation in this system (Figure 7A).
MT Affinity of XMAP4 and Mutants during Mitosis
Interphase and mitotic cell extracts were incubated with Taxol-stabilized MTs, and the MT-binding abilities of endogenous XMAP4 and various GFP–XMAP4 fusion proteins were evaluated by a cosedimentation experiment. As shown in Figure 8, endogenous XMAP4 bound to MTs in large amounts in interphase but showed little binding and remained in the supernatant in mitosis, as previously shown for Xenopus egg p220 (Shiina et al., 1992b; Andersen et al., 1994). Similarly to endogenous XMAP4, ∼70% of GFP–WT was recovered in the supernatant in mitosis, indicating that GFP–WT also reduced its MT-binding ability in mitosis. In contrast, most of the GFP–CM10A, GFP–C6A, and GFP–CM8A were bound to MTs even in mitosis. These findings indicated that XMAP4 mutants in p34cdc2 kinase-specific phosphorylation sites did not reduce their MT-binding ability in mitosis.
Figure 8.
MT affinity of XMAP4 and mutants. Interphase (I) and mitotic (M) cell extracts of nontransfected A6 cells (−) and A6 cells expressing GFP–WT, GFP–CM10A, GFP–C6A, or GFP–CM8A were incubated with Taxol-stabilized MTs and then centrifuged. XMAP4 coprecipitated with MTs (P) or remaining in the supernatant (S) was detected by immunoblotting with anti-XMAP4 antibody. The arrowhead and arrow denote endogenous XMAP4 and GFP–XMAP4, respectively. Approximately 70% of GFP–WT remained in the supernatant in mitosis, whereas ∼90% of GFP–CM10A, GFP–C6A, and GFP–CM8A coprecipitated with MTs.
DISCUSSION
In our previous study, Xenopus MAP p220 was shown to be similar to mammalian MAP4 in terms of its heat stability, apparent molecular mass, and ubiquitous expression (Shiina et al., 1992b). However, the lack of cross-reactivity of antibodies has hampered further assessment of the relationship between these MAPs. In this study, cDNA cloning conclusively showed that p220 is a Xenopus homologue of MAP4 because of their overall similarity in amino acid sequence and structural features. Furthermore, p220 was shown to be identical to XMAP230 (Andersen et al., 1994), because the previously reported peptide sequence of XMAP230 was found in the p220 sequence. Taken together, we designate p220 (XMAP230) XMAP4.
Using Xenopus eggs and cultured Chinese hamster ovary cells, it was previously shown that p220 (XMAP230) as well as mammalian MAP4 were phosphorylated specifically during mitosis (Vandre et al., 1991; Shiina et al., 1992b; Andersen et al., 1994), and their MT-binding as well as their MT-stabilizing abilities were coincidentally reduced along with the phosphorylation (Shiina et al., 1992b; Andersen et al., 1994). In this study, p220 (XMAP4) was also found to be phosphorylated in a mitosis-specific manner and concomitantly lost its MT-binding ability in cultured Xenopus epithelial A6 cells. Furthermore, a mutant XMAP4–GFP fusion protein, CM10A, in which 10 serine/threonine residues of potential p34cdc2 kinase- and/or MAP kinase-specific phosphorylation sites were mutated to alanine, was not phosphorylated during mitosis. Interestingly, in this nonphosphorylatable XMAP4 mutant, the MT-binding ability was not reduced during mitosis. These findings indicated that mitosis-specific phosphorylation of XMAP4 is required for reduction of its MT affinity. Fluorescence studies of mitotic cells, which have shown that the majority of GFP–XMAP4 and endogenous XMAP4 are diffusely distributed throughout the cell after formaldehyde fixation (Shiina et al., 1992b), were consistent with this reduction of XMAP4 activity during mitosis. However, GFP–WT appeared to be associated with spindle MTs in living cells, and the localization of (X)MAP4 on spindle MTs was also previously observed in methanol-fixed Xenopus and mammalian cells (Andersen et al., 1994; Ookata et al., 1995). As the density of spindle MTs is very high in the cytoplasm, (X)MAP4 may be detectable on MTs even if the amount of (X)MAP4 molecule on the MTs is decreased by phosphorylation. Our observation that GFP fluorescence of GFP–CM10A on mitotic spindles was much brighter than that of GFP–WT (our unpublished results) supported this notion.
The high affinity of the mutant XMAP4 to MTs during mitosis may induce hyperstabilization of spindle MTs. For instance, this mutant affected chromosome separation during anaphase and induced characteristic BFP movement during the mitotic phase when overexpressed in A6 cells. In contrast, in cells overexpressing wild-type XMAP4, mitosis proceeded normally. These findings suggested that nonphosphorylatable mutant XMAP4, but not wild-type XMAP4, hyperstabilized spindle MTs and inhibited chromosome movement driven by MT depolymerization. BFP movement may be attributed to delay in the spindle processes in reference to the cleavage furrow formation during anaphase. The molecular mechanism behind BFP movement should be examined in the future.
The dynamic character of MTs is considered to be important in several processes during mitosis such as elimination of interphase MTs before mitotic spindle formation (Zhai et al., 1996), the poleward flux of spindle MTs during metaphase (Mitchison, 1989; Rodionov et al., 1994; Zhai et al., 1995), and chromosome movement toward spindle poles during anaphase A (Desai and Mitchison, 1995; Lombillo et al., 1995a,b). Because nonphosphorylated (X)MAP4 suppresses MT dynamics (Andersen et al., 1994; Ookata et al., 1995), these processes may require the phosphorylation-dependent inactivation of (X)MAP4. Interestingly, among these processes, in cultured A6 cells, the expression of nonphosphorylatable XMAP4 such as CM10A, C6A, or CM8A appeared to primarily affect the chromosome movement during anaphase A. It remains unclear why other processes that are considered to require MT dynamics were not significantly affected by these XMAP4 mutants. In particular, the phosphorylation of MAP4 has been suggested to be responsible for the regulation of MT dynamics at the G2-M phase transition, i.e., the disappearance of interphase-type long, stable MTs. However, the interphase-type stable MT network normally disappeared at the G2-M phase transition in A6 transfectants expressing nonphosphorylatable XMAP4 mutants (our unpublished results). To determine the regulatory mechanism of MT dynamics, further detailed analyses on other MT regulatory factors such as MT catastrophe factors (Belmont and Mitchison, 1996; Walczak et al., 1996), MT-severing factors (Shiina et al., 1992a, 1994; McNally and Vale, 1993), and other MAPs (Vasquez et al., 1994) are required.
The question has naturally arisen of which kinase is responsible for the mitosis-specific phosphorylation of XMAP4. p34cdc2 kinase and MAP kinase were thought to be responsible for this mitosis-specific phosphorylation of p220, partly because these kinases were activated with the same time course as p220 phosphorylation during the cell cycle and partly because the phosphopeptide mapping pattern of p220 phosphorylated in vivo was identical to that of p220 phosphorylated by these kinases in vitro (Shiina et al., 1992b). In this study, we showed that mutation in six serine/threonine residues of potential p34cdc2 kinase-specific phosphorylation sites in C6A was sufficient to affect anaphase chromosome separation and to induce BFP movement. Judging from the upward shift of bands in SDS-PAGE, C6A was not phosphorylated during mitosis in vivo or by p34cdc2 kinase in vitro, and it is therefore likely that p34cdc2 kinase is primarily responsible for the mitosis-specific phosphorylation of XMAP4. Of course, the possibility that MAP kinase is also involved has not been completely excluded. Because MAP kinase induces the interphase-M phase transition of MT dynamics in cell-free extracts of Xenopus eggs (Gotoh et al., 1991), and because it phosphorylates XMAP4 as well as mammalian MAP4 efficiently in vitro, resulting in down-regulation of their MT-binding ability (Hoshi et al., 1992; Shiina et al., 1992b), the possible involvement of MAP kinase in the mitosis-specific phosphorylation of XMAP4 in vivo should be further evaluated. Another candidate for the kinase responsible for XMAP4 phosphorylation is MARK. Previous studies revealed that MAP4 phosphorylation by MARK markedly reduces its MT-stabilizing ability (Illenberger et al., 1996), and that overexpression of MARK in cells results in disruption and disappearance of MTs (Drewes et al., 1997). In this study, however, we showed that mutation in all of potential MARK-specific phosphorylation sites in R4A did not affect its mitosis-specific phosphorylation in cells or anaphase chromosome separation. Furthermore, R4A was phosphorylated by p34cdc2 kinase in vitro with concomitant reduction in its MT-binding ability (our unpublished results). These lines of evidence indicated that MARK is not responsible for the mitosis-specific phosphorylation of XMAP4 at least in the Xenopus system. Taken together, these findings favored the notion that XMAP4 phosphorylation at p34cdc2 kinase-specific sites is principally responsible for its mitosis-specific phosphorylation and reduction in its MT-binding ability, which is important for chromosome separation in anaphase.
ACKNOWLEDGMENTS
We greatly thank Dr. E. Nishida (Kyoto University) for encouragement and many suggestions. We thank Dr. A. Asano for critical reading of the manuscript. We express appreciation to M. Irie for excellent technical assistance.
Abbreviations used:
- BFP
back-and-forth peristaltic
- GFP
green fluorescent protein
- MAP
microtubule-associated protein
- MARK
MAP/microtubule affinity-regulating kinase
- MT
microtubule
- WT
wild-type XMAP4
- XMAP
Xenopus MAP
REFERENCES
- Aizawa H, Emori Y, Murofushi H, Kawasaki H, Sakai H, Suzuki K. Molecular cloning of a ubiquitously distributed microtubule-associated protein with Mr 190,000. J Biol Chem. 1990;265:13849–13855. [PubMed] [Google Scholar]
- Andersen SSL, Buendia B, Domínguez JE, Sawyer A, Karsenti E. Effect on microtubule dynamics of XMAP230, a microtubule-associated protein present in Xenopus laevis eggs and dividing cells. J Cell Biol. 1994;127:1289–1299. doi: 10.1083/jcb.127.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen SSL, Karsenti E. XMAP310: a Xenopus rescue-promoting factor localized to the mitotic spindle. J Cell Biol. 1997;139:975–983. doi: 10.1083/jcb.139.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont LD, Hyman AA, Sawin KE, Mitchison TJ. Real-time visualization of cell cycle-dependent changes in microtubule dynamics in cytoplasmic extracts. Cell. 1990;62:579–589. doi: 10.1016/0092-8674(90)90022-7. [DOI] [PubMed] [Google Scholar]
- Belmont LD, Mitchison TJ. Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell. 1996;84:623–631. doi: 10.1016/s0092-8674(00)81037-5. [DOI] [PubMed] [Google Scholar]
- Cande WZ. Nucleotide requirements for anaphase chromosome movements in permeabilized mitotic cells: anaphase B but not anaphase A requires ATP. Cell. 1982;28:15–22. doi: 10.1016/0092-8674(82)90370-1. [DOI] [PubMed] [Google Scholar]
- Chapin SJ, Bulinski JC. Nonneuronal 210 × 103 Mr microtubule-associated protein (MAP4) contains a domain homologous to the microtubule-binding domains of neuronal MAP2 and tau. J Cell Sci. 1991;98:27–36. doi: 10.1242/jcs.98.1.27. [DOI] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ. A new role for motor proteins as couplers to depolymerizing microtubules. J Cell Biol. 1995;128:1–4. doi: 10.1083/jcb.128.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewes G, Ebneth A, Preuss U, Mandelkow E-M, Mandelkow E. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell. 1997;89:297–308. doi: 10.1016/s0092-8674(00)80208-1. [DOI] [PubMed] [Google Scholar]
- Gotoh Y, Masuyama N, Dell K, Shirakabe K, Nishida E. Initiation of Xenopus oocyte maturation by activation of the mitogen-activated protein kinase cascade. J Biol Chem. 1995;270:25898–25904. doi: 10.1074/jbc.270.43.25898. [DOI] [PubMed] [Google Scholar]
- Gotoh Y, Nishida E, Matsuda S, Shiina N, Kosako H, Shiokawa K, Akiyama T, Ohta K, Sakai H. In vitro effects on microtubule dynamics of purified Xenopus M phase-activated MAP kinase. Nature. 1991;349:251–254. doi: 10.1038/349251a0. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Noda Y, Okada Y. Kinesin and dynein superfamily proteins in organelle transport and cell division. Curr Opin Cell Biol. 1998;10:60–73. doi: 10.1016/s0955-0674(98)80087-2. [DOI] [PubMed] [Google Scholar]
- Hoshi M, Ohta K, Gotoh Y, Mori A, Murofushi H, Sakai H, Nishida E. Mitogen-activated-protein-kinase-catalyzed phosphorylation of microtubule-associated proteins, microtubule-associated protein 2 and microtubule-associated protein 4, induced an alteration in their function. Eur J Biochem. 1992;203:43–52. doi: 10.1111/j.1432-1033.1992.tb19825.x. [DOI] [PubMed] [Google Scholar]
- Hunt T. Maturation promoting factor, cyclin and the control of M-phase. Curr Opin Cell Biol. 1989;1:268–274. doi: 10.1016/0955-0674(89)90099-9. [DOI] [PubMed] [Google Scholar]
- Huynh TV, Toung RA, Davis RW. Construction and screening cDNA libraries in λgt10 and λgt11. In: Glover DM, editor. DNA Cloning: A Practical Approach. Oxford, United Kingdom: IRL Press; 1985. pp. 49–78. [Google Scholar]
- Hyman AA, Karsenti E. Morphogenetic properties of microtubules and mitotic spindle assembly. Cell. 1996;84:401–410. doi: 10.1016/s0092-8674(00)81285-4. [DOI] [PubMed] [Google Scholar]
- Illenberger S, Drewes G, Trinczek B, Biernat J, Meyer HE, Olmsted JB, Mandelkow EM, Mandelkow E. Phosphorylation of microtubule-associated proteins MAP2 and MAP4 by the protein kinase p110mark. J Biol Chem. 1996;271:10834–10843. doi: 10.1074/jbc.271.18.10834. [DOI] [PubMed] [Google Scholar]
- King RW, Jackson PK, Kirschner MW. Mitosis in transition. Cell. 1994;79:563–571. doi: 10.1016/0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- Kirschner M, Mitchison T. Beyond self-assembly: from microtubules to morphogenesis. Cell. 1986;45:329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- Kosako H, Gotoh Y, Nishida E. Requirement for the MAP kinase kinase/MAP kinase cascade in Xenopus oocyte maturation. EMBO J. 1994;13:2131–2138. doi: 10.1002/j.1460-2075.1994.tb06489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D. Mitosis: back to the basics. Cell. 1994;77:951–954. doi: 10.1016/0092-8674(94)90432-4. [DOI] [PubMed] [Google Scholar]
- Larsson N, Marklund U, Gradin HM, Brattsand G, Gullberg M. Control of microtubule dynamics by oncoprotein 18: dissection of the regulatory role of multisite phosphorylation during mitosis. Mol Cell Biol. 1997;17:5530–5539. doi: 10.1128/mcb.17.9.5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombillo VA, Nislow C, Yen TJ, Gelfand VI, McIntosh JR. Antibodies to the kinesin motor domain and CENP-E inhibit microtubule depolymerization-dependent motion of chromosomes in vitro. J Cell Biol. 1995a;128:107–115. doi: 10.1083/jcb.128.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombillo VA, Stewart RJ, McIntosh JR. Minus-end-directed motion of kinesin-coated microspheres driven by microtubule depolymerization. Nature. 1995b;373:161–164. doi: 10.1038/373161a0. [DOI] [PubMed] [Google Scholar]
- Maller JL. Xenopus oocytes and the biochemistry of cell division. Biochemistry. 1990;29:3157–3166. doi: 10.1021/bi00465a001. [DOI] [PubMed] [Google Scholar]
- Mandelkow E, Mandelkow E-M. Microtubules and microtubule-associated proteins. Curr Opin Cell Biol. 1995;7:72–81. doi: 10.1016/0955-0674(95)80047-6. [DOI] [PubMed] [Google Scholar]
- McNally FJ. Modulation of microtubule dynamics during the cell cycle. Curr Opin Cell Biol. 1996;8:23–29. doi: 10.1016/s0955-0674(96)80044-5. [DOI] [PubMed] [Google Scholar]
- McNally FJ, Vale RD. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell. 1993;75:419–429. doi: 10.1016/0092-8674(93)90377-3. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ. Polewards microtubule flux in the mitotic spindle: evidence from photoactivation of fluorescence. J Cell Biol. 1989;109:637–652. doi: 10.1083/jcb.109.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW, Kirschner MW. Dominoes and clocks: the union of two views of the cell cycle. Science. 1989;246:614–621. doi: 10.1126/science.2683077. [DOI] [PubMed] [Google Scholar]
- Nurse P. Universal control mechanism regulating onset of M-phase. Nature. 1990;344:503–507. doi: 10.1038/344503a0. [DOI] [PubMed] [Google Scholar]
- Olmsted JB. Nonmotor microtubule-associated proteins. Curr Opin Cell Biol. 1991;3:52–58. doi: 10.1016/0955-0674(91)90165-u. [DOI] [PubMed] [Google Scholar]
- Olson KR, McIntosh JR, Olmsted JB. Analysis of MAP4 function in living cells using green fluorescent protein (GFP) chimeras. J Cell Biol. 1995;130:639–650. doi: 10.1083/jcb.130.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ookata K, Hisanaga S, Bulinski JC, Murofushi H, Aizawa H, Itoh TJ, Hotani H, Okumura E, Tachibana K, Kishimoto T. Cyclin B interaction with microtubule-associated protein 4 (MAP4) targets p34cdc2 kinase to microtubules and is a potential regulator of M-phase microtubule dynamics. J Cell Biol. 1995;128:849–862. doi: 10.1083/jcb.128.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov VI, Lim S-S, Gelfand VI, Borisy GG. Microtubule dynamics in fish melanophores. J Cell Biol. 1994;126:1455–1464. doi: 10.1083/jcb.126.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon ED, Leslie RJ, Saxton WM, Karow ML, McIntosh JR. Spindle microtubule dynamics in sea urchin embryos: analysis using a fluorescein-labeled tubulin and measurements of fluorescence redistribution after laser photobleaching. J Cell Biol. 1984;99:2165–2174. doi: 10.1083/jcb.99.6.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton WM, Stemple DL, Leslie RJ, Salmon ED, Zavortink M, McIntosh JR. Tubulin dynamics in cultured mammalian cells. J Cell Biol. 1984;99:2175–2186. doi: 10.1083/jcb.99.6.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina N, Gotoh Y, Kubomura N, Iwamatsu A, Nishida E. Microtubule severing by elongation factor 1α. Science. 1994;266:282–285. doi: 10.1126/science.7939665. [DOI] [PubMed] [Google Scholar]
- Shiina N, Gotoh Y, Nishida E. A novel homo-oligomeric protein responsible for an MPF-dependent microtubule-severing activity. EMBO J. 1992a;11:4723–4731. doi: 10.1002/j.1460-2075.1992.tb05577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina N, Moriguchi T, Ohta K, Gotoh Y, Nishida E. Regulation of a major microtubule-associated protein by MPF and MAP kinase. EMBO J. 1992b;11:3977–3984. doi: 10.1002/j.1460-2075.1992.tb05491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandre DD, Centonze VE, Peloquin J, Tombes RM, Borisy GG. Proteins of the mammalian mitotic spindle: phosphorylation/dephosphorylation of MAP-4 during mitosis. J Cell Sci. 1991;98:577–588. doi: 10.1242/jcs.98.4.577. [DOI] [PubMed] [Google Scholar]
- Vasquez RJ, Gard DL, Cassimeris L. XMAP from Xenopus eggs promotes rapid plus end assembly of microtubules and rapid microtubule polymer turnover. J Cell Biol. 1994;127:985–993. doi: 10.1083/jcb.127.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F, Labbé J-C, Dorée M, Karsenti E. Regulation of microtubule dynamics by cdc2 protein kinase in cell-free extracts of Xenopus eggs. Nature. 1990;343:233–238. doi: 10.1038/343233a0. [DOI] [PubMed] [Google Scholar]
- Vernos I, Karsenti E. Motors involved in spindle assembly and chromosome segregation. Curr Opin Cell Biol. 1996;8:4–9. doi: 10.1016/s0955-0674(96)80041-x. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Mitchison TJ, Desai A. XKCM1: a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- West RR, Tenbarge KM, Olmsted JB. A model for microtubule-associated protein 4 structure. J Biol Chem. 1991;266:21886–21896. [PubMed] [Google Scholar]
- Zhai Y, Kronebusch PJ, Borisy GG. Kinetochore microtubule dynamics and the metaphase-anaphase transition. J Cell Biol. 1995;131:721–734. doi: 10.1083/jcb.131.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y, Kronebusch PJ, Simon PM, Borisy GG. Microtubule dynamics at the G2/M transition: abrupt breakdown of cytoplasmic microtubules at nuclear envelope breakdown and implications for spindle morphogenesis. J Cell Biol. 1996;135:201–214. doi: 10.1083/jcb.135.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]