Abstract
Knowledge of the mitochondrial DNA (mtDNA) sequence of divergent murine species is critical from both a phylogenetic perspective and in understanding nuclear-mitochondrial interactions, particularly as the latter influences our xenocybrid models of mitochondrial disease. To this end, the sequence of the mitochondrial genome of the murine species Mus terricolor (formerly Mus dunni) is reported and compared with the published sequence for the common laboratory mouse Mus musculus domesticus strain C57BL/6J. These species are of interest because xenomitochondrial cybrid mice were created that harbor Mus terricolor mtDNA in a M. m. domesticus nuclear background. Although the total of 1763 nucleotide substitutions represents striking heterogeneity, the majority of these are silent, leading to highly conserved protein sequences with only 159 amino acid differences. Moreover, 58% of these amino acid differences represented conservative substitutions. All of the tRNA genes and rRNA genes have homology of 91% or greater. The control region shows the greatest heterogeneity, as expected, with 85% homology overall. Regions of 100% homology were found for Conserved Sequence Block I, the Conserved Sequence Block III and the L-strand Origin of Replication. Complex I genes showed the greatest degree of difference among protein-coding genes with amino acid homology of 91–97% among the seven mitochondrial genes. Complexes III and IV genes show high homology ranging from 98–100%. From these data, Complex I differences appear most critical for the viability of M. m. domesticus: M. terricolor cybrids. Moreover, the sequence information reported here should be useful in identifying critical regions for mitochondrial transfer between species, for furthering the understanding of mitochondrial dynamics and pathology in transmitochondrial organisms, and for the study of Mus genus origins.
Keywords: mitochondria, mtDNA, phylogeny, Mus terricolor, Mus dunni, C57BL/6, mouse
INTRODUCTION
The mitochondrial (mt) genomes of murine species are of increasing interest for both phylogenetic analysis and disease modeling. The large number of mouse species available enables the study of intra-genus and intra-species evolution, but complete mtDNA sequences are relatively lacking. The emerging field of transmitochondrial murine modeling, that is, creating mice with mtDNA from one species, genus or strain with a nuclear DNA background from another species, genus or strain, is being used to model mtDNA-based human diseases. Nuclear-encoded mitochondrial gene knockouts (e.g., Tfam, Ant1, MnSOD, and GPx1) have provided information about the role that these nuclear genes play in respiratory chain deficiency, mitochondrial myopathy, cardiomyopathy, liver dysfunction, and neuronal degeneration (Larsson et al., 1998; Wallace, 1999, 2001; Pinkert and Trounce, 2002). Yet, the creation of animal models with mtDNA mutations has proven more elusive. The high mtDNA copy number, unknown threshold barriers for heteroplasmy, and lack of reliable mitochondrial DNA recombination processes have provided technical barriers, not only to the creation of mitochondrial gene knockout organisms, but to the development of cultured animal cell lines bearing specific mtDNA mutations. Therefore, creation of xenomitochondrial cybrids provides a means to monitor mitochondrial segregation and fate.
Xenomitochondrial mice were created by fusion of mouse donor cytoplasts from Mus terricolor (previously designated Mus dunni) with 129S6/SVEvTac (129S6) strain embryonic stem (ES) cells to create cybrids; these cells were then injected into donor M. m. domesticus embryos (C57BL/6 strain; McKenzie et al., 2004). Chimeric female offspring were mated with M. m. domesticus males to create homoplasmic transmitochondrial offspring.
In addition to in vivo mouse studies, cells from other animal species were used in creating xenomitochondrial cybrids in vitro (Pinkert and Trounce, 2002 and references therein). A common observation is that as species diverge, an increasing deficiency gradient in Complex I- and other oxidative phosphorylation (OXPHOS) proteins occurs that hinders cybrid viability and survival (Kenyon and Moraes 1997; Barrientos et al., 1998; Barrientos and Moraes, 1999; Dey et al., 2000; McKenzie and Trounce, 2000; Yamaoka et al., 2000; McKenzie et al., 2003). Experiments such as these are helping to elucidate interactions between nuclear and mtDNA gene products and are proving valuable as models of human mitochondrial disease. However, progress is limited by the lack of mitochondrial DNA sequence information available for species other than the Mus musculus group and Rattus norvegicus. Therefore, to facilitate future studies, the mitochondrial genome of the Asian mouse species M. terricolor was sequenced; the mtDNA source used to create the first xenomitochondrial mice (McKenzie et al., 2004; Trounce et al., 2004).
M. m. domesticus is the common inbred laboratory mouse of western European origin. It belongs to the Mus musculus species group that is comprised of the five subspecies M. domesticus, M. musculus, M. castaneus, M. molossinus and M. bactrianus (see Trounce et al., 2004). Interestingly, the mtDNA of these classic laboratory models is nearly identical and is believed to originate from a common M. m. domesticus maternal ancestor (Yonekawa et al., 1980; Ferris et al., 1982; Goios et al. 2007). In contrast, wild mice show considerably more variation in mtDNA sequence than these laboratory species (Guénet and Bonhomme, 2003).
The species whose mtDNA sequence is detailed in this study, the Indian pigmy mouse M. terricolor, cannot interbreed with the M. musculus complex, presumably because of heterochromatin repatterning (Sharma et al., 2003). This species appears to have originated on the Indian peninsula and is reported to have separated from the M. musculus ancestor approximately 3 to 4 million years before present (Guénet and Bonhomme, 2003; Suzuki et al., 2004). In this report, we examine allelic diversity in the mitochondrial genome of M. terricolor and compare results with published sequence for the M. musculus group.
MATERIALS AND METHODS
mtDNA Templates
Genomic DNA was isolated from M. terricolor fibroblasts (ATCC CRL-2017) using phenol-chloroform extraction and ethanol precipitation as previously described (McKenzie et al., 2003).
PCR Amplification of mtDNA
A series of primer sets was designed, initially based on comparison of the published mtDNA sequences of M. m. domesticus (accession #NC001569) and Rattus norvegicus (accession #001665). After an initial round of sequencing was performed, primers specific to the newly obtained M. terricolor mtDNA sequence were designed to eliminate the risk of contamination with non-M. terricolor murine DNA. These primers and their annealing positions are shown in Supplemental Data (Table 5). Primers amplified fragments ranging from 400–950 bp in length. Some primers were used only for sequencing and not for PCR amplification. Fragments were amplified in 0.2 mM dNTPs, 15 mM MgCl2, 1X PCR buffer, and 0.5 U Taq Polymerase (Life Technologies, Gaithersburg, MD). Thirty rounds of amplification included 1 min at 94°C, 1.5 min at 49°C and 1.5 min at 72°C, followed by an additional 7-min extension at 72°C. DNA fragments were checked for the proper length by electrophoretic separation on an agarose gel stained with ethidium bromide. Fragments were purified by QiaQuick PCR Clean-up spin columns (Qiagen, Valencia, CA).
DNA Sequencing and Sequence Alignment
Sequencing was performed using primers specific for M. terricolor using overlapping fragments to provide redundancy. In cases where overlapping sequence data were not in agreement, several additional amplification and sequencing rounds were performed. Sequencing was performed at the State University of New York College at Geneseo using a Li-Cor Model 4300 Genetic Analyzer, and at the University of Rochester and Auburn University using Big Dye Termination Kits and an ABI Prism 3100 Genetic Analyzer. Analysis, fragment assembly and sequence translations were performed using the software package Vector NTI (Invitrogen). Additional sequence alignments were performed using BLAST analysis (http://www.ncbi.nlm.nih.gov/BLAST/). Sequence data for M. terricolor were compared with sequences for M. musculus cell lines (GenBank). Although sequence information was available from ten or more live animals in the M. musculus group, the reference sequence used in the tables here was from the M. m. domesticus strain C57BL/6J (Accession #AY172335), a group E inbred strain (http://www.informatics.jax.org; Silver, 1995; Beck et al., 2000).
RESULTS
Complete mtDNA sequences are available for sixteen laboratory strains of Mus musculus (see Goios et al., 2007). The M. m. domesticus mouse strain used for comparison, C57BL/6J, was chosen because it is one of the most highly used models in biomedical research and was the source of DNA used for sequencing the mouse nuclear genome at the Sanger Institute (Bayona-Bafaluy et al., 2003).
The mitochondrial genome sequence of M. terricolor (16310 base pairs) was generated and compared with that of M. m. domesticus strain C57BL/6J (16299 base pairs), a total of 1802 differences were identified. A single base addition or deletion was counted as one difference, while two additions or deletions were counted as two. Each substitution was counted as a single difference.
Mitochondrial Genome Features
The mouse mitochondrial genome has a structure and organization remarkably similar to that of humans and other mammals. Its genes are contiguous or overlapping and it has 13 open reading frames that represent protein-encoding genes (Bibb et al., 1981; Anderson et al., 1982). Twelve of these reading frames come from the so-called heavy (H) strand that contains more G and C bases. The light (L)-strand produces 8 tRNAs and the reading frame for ND6 (Michael et al., 1984). The approximately 800-bp control region (15423–16310), also known as the D-loop, is bordered by tRNAPhe and tRNAPro. It contains promoters for both H-strand and L-strand transcription (PH and PL) and the H-strand origin of replication (Clayton, 1992). The transcription promoters are believed to contain binding sites for Tfam; a nuclear-encoded transcription factor (Dairaghi et al., 1995).
Table 1–Table 4 compare homology in nucleotides and amino acids for individual tRNA, rRNA and protein-coding genes as well as non-coding elements. As expected, despite the high number of nucleotide changes, most of the codon differences in protein-coding genes were silent. Yet, 157 of the differences did result in an amino acid substitution and 82 (52%) were conservative substitutions.
Table 1.
Nucleotide Homology in tRNA Genes – M. terricolor vs. M. m. domesticus (C57BL/6).
| Gene |
*Position in AY172335 (M. m. domesticus) |
±Position in M. terricolor |
Changes in M. terricolor |
Identities |
|---|---|---|---|---|
| tRNA-Phe | 1–68 68 nt |
1–66 66 nt |
1 substitution 2 deletions |
65/68 96% |
| tRNA-Val | 1025–1093 69 nt |
1021–1089 69 nt |
3 substitutions | 66/69 96% |
| tRNA-Leu | 2676–2750 75 nt |
2672–2746 75 nt |
2 substitutions | 73/75 97% |
| tRNA-Ile | 3706–3774 69 nt |
3702–3770 69 nt |
5 substitutions | 64/69 93% |
| tRNA-Gln |
#C3772–3842 71 nt |
C3768–3838 71 nt |
1 substitution | 70/71 99% |
| tRNA-Met | 3845–3913 69 nt |
3843–3911 69 nt |
0 changes | 69/69 100% |
| tRNA-Trp | 4950–5016 67 nt |
4948–5014 67 nt |
3 substitutions | 64/67 96% |
| tRNA-Ala |
#C5018–5086 69 nt |
C5016–5084 69 nt |
4 substitutions | 65/69 94% |
| tRNA-Asn |
#C5089–5159 71 nt |
C5088–5158 71 nt |
5 substitutions | 65/71 92% |
| tRNA-Cys |
#C5192–5257 66 nt |
C5190–5256 67 nt |
1 insertion | 66/67 99% |
| tRNA-Tyr |
#C5260–5326 67 nt |
C5258–5324 67 nt |
1 substitution | 66/67 99% |
| tRNA-Ser |
#C6870–6938 69 nt |
C6868–6936 69 nt |
2 substitutions | 67/69 97% |
| tRNA-Asp | 6942–7011 70 nt |
6940–7008 69 nt |
1 deletion 3 substitutions |
66/70 94% |
| tRNA-Lys | 7700–7764 65 nt |
7697–7760 64 nt |
1 deletion | 64/65 98% |
| tRNA-Gly | 9391–9458 68 nt |
9387–9454 68 nt |
4 substitutions | 64/68 94% |
| tRNA-Arg | 9808–9875 68 nt |
9804–9871 68 nt |
2 substitutions | 66/68 97% |
| tRNA-His | 11545–11612 68 nt |
11541–11608 68 nt |
5 substitutions | 63/68 93% |
| tRNA-Ser2 | 11613–11671 59 nt |
11609–11667 59 nt |
5 substitutions | 54/59 92% |
| tRNA-Leu2 | 11672–11742 71 nt |
11667–11739 73 nt |
2 insertions 2 substitutions |
69/73 95% |
| tRNA-Glu |
#C14071–14139 69 nt |
C14069–14137 69 nt |
0 changes | 69/69 100% |
| tRNA-Thr | 15289–15355 67 nt |
15287–15353 67 nt |
1 substitution | 66/67 99% |
| tRNA-Pro |
#C15356–15422 67 nt |
C15354–15421 68 nt |
1 insertion 4 substitutions |
63/68 93% |
Gene classification from Mouse Genome Informatics.
Numbering and classification from M. m. domesticus strain C57BL/6J sequence (Bayona-Bafaluy et al., 2003).
C = sequence on complementary (-) strand.
Table 4.
Nucleotide Homology in Noncoding Elements – M. terricolor vs. M. m. domesticus (C57BL/6J; # AY172335).
| Gene | Position in AY172335 (M. m. domesticus) |
±Position in M. terricolor |
Changes in M. terricolor |
% Nucleotide Identity |
|---|---|---|---|---|
| D-loop (overall) | 15423–16299 877 nt |
15422–16310 889 nt |
112 base substitutions 16 insertions, 4 deletions |
85% |
| ETAS 1 | 15445–15504 60 nt |
15445–15504 60 nt |
4 substitutions 7 insertions or deletions |
82% |
| ETAS 2 | 15509–15571 63 nt |
15509–15571 63 nt |
8 substitutions 12 insertions or deletions |
68% |
| CSB I | 16029–16053 25 nt |
16029–16053 25 nt |
0 changes |
100% |
| CSB II | 16084–16100 17 nt |
16084–16100 17 nt |
2 insertions or deletions |
88% |
| CSB III | 16109–16126 18 nt |
16109–16126 18 nt |
0 changes |
100% |
| L-strand Origin of Replication | 5160–5191 32 nt |
0 changes` |
100% |
Abbreviations: ETAS, Extended Termination-Associated Sequences; CSB, Conserved Sequence Block.
Nucleotide homology was 100% in two of the Conserved Sequence Blocks of the D-loop (CSBI and CSBIII), in the L-strand origin of replication, and in the genes for tRNA-Glu and tRNA-Met. The greatest variation appeared in the Extended Termination-Associated Sequences (68% for ETAS 2). All of the tRNA and rRNA genes showed ≥ 91% nucleotide homology. Amino acid homology of 100% was found for subunit II of cytochrome c oxidase (CO2 gene), and homology of 98% or higher was seen for cytochrome c oxidase I, cytochrome c oxidase III, ATPase subunit 6, and cytochrome b. The protein with lowest amino acid homology (91%) was subunit 2 of NADH Dehydrogenase, encoded by ND2. In fact, several of the Complex I subunits showed higher numbers of amino acid changes than other complexes. ND1 had 10 amino acid changes; ND4 had 28; ND5 had 41 and ND6 had 13. By comparison, the cytb subunit of Complex III had only seven amino acid changes; Complex IV subunits had three, zero, and five changes.
Transitions (72%) significantly outnumbered transversions (28%). C-to-T and T-to-C transitions were particularly frequent (968 or 57.5%). The exchange of T → G and C→ and G were the rarest and collectively composed only 37 or 2.2% of the total differences. Transversions tend to increase as species become more divergent (Brown et al., 1982). Due to a correspondingly greater change in shape and structure, transversions represent more dramatic biological effects in vivo, while transitions are more tolerated. Also, with successive generations, the probability of back-mutations and multiple transitions at a single site increases, affecting these ratios.
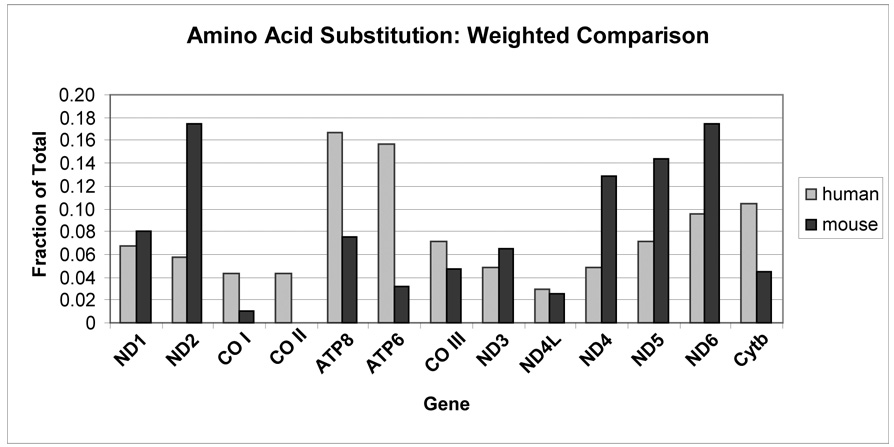
The distribution of amino acid differences among the mtDNA-encoded proteins of the two mouse species was plotted with those observed among humans. This distribution is shown in Fig. 1. Although intraspecific (polymorphic) human differences are compared with interspecific mouse data, such a comparison is useful in gauging trends. Here, there was a greater degree of divergence seen in the mouse complex I subunit genes ND2, ND4, ND5 and ND6, while the CO I, CO II, ATP8, ATP6 and Cytb genes showed fewer differences compared with the level of human polymorphism (Fig. 1).
Figure 1.
Distribution of amino acid substitutions in the 13 mtDNA-encoded proteins in Homo sapiens and between Mus musculus domesticus and Mus terricolor. The graph represents the fraction of total observed polymorphisms for each protein, corrected for the number of amino acids in each protein. The human data are calculated from the non-synonymous substitutions tabulated by Moilanen and Majamaa (2003) from 840 complete human mtDNA sequences. The two mouse species show relatively greater differences for the complex I subunits ND2, ND4, ND5 and ND6, with similar or lower differences in most other proteins, in particular for ATP8 and ATP6.
DISCUSSION
More than 99% of the proteins located in mitochondria are encoded by nuclear DNA; a subset of these interacts with specific mtDNA or mtRNA sequences to carry out various functions. Nuclear-encoded proteins assemble with or bind to mitochondrial-encoded peptides in OXPHOS complexes, and nuclear proteins are involved in mtDNA replication, transcription, and translation. Moreover, mtDNA is believed to mutate 5–10 times faster than nuclear DNA (Ferris et al., 1983). Animal mitochondria therefore represent a unique system of mitochondrial-nuclear genomic cooperation, where co-evolution of interacting proteins occurs (see Rand et al., 2004).
Production of viable xenomitochondrial cybrids correlates with the degree of relatedness between the nuclear and mtDNA donor species. When mtDNA from non-human primates was introduced into human ρ° (mtDNA-depleted) cells, only the transfer of mtDNA from more closely-related primate species resulted in viable cybrids (Kenyon and Moraes, 1997). Likewise, when Mus spretus, Mus terricolor, and Rattus norvegicus mitochondria were introduced into M. m. domesticus embryonic stem (ES) cells, the mouse xenocybrids were viable but the rat cybrids were not (McKenzie et al., 2004). Cells appear to be able to select against foreign mtDNA. In human cell culture studies, cybrids preferentially replicated mtDNA of their own species, even if those mtDNA molecules were replication- or metabolically-deficient (Moraes et al., 1999). Therefore, the mtDNA sequences of M. m. domesticus and M. terricolor were compared here in the context of understanding how sequence differences might contribute to cybrid instability and viability.
First, the origins of replication were considered. A 100% homology in OL was observed; yet, there was a considerable variation in the region assigned to OH. In general, when the mitochondrial origins of replication and promoters were compared between other more divergent species, few conserved sequences were found (Clayton, 1992). The changes in promoters may be highly significant for more diverged xenomitochondrial cybrids, since in vitro studies using human mtDNA show that, mutations in the mitochondrial promoters cause serious defects (Hixon et al, 1986). The D-loop contains three regions known as the Conserved Sequence Blocks (CSBI, II and III). When the L-strand transcript is cleaved to form a primer for H-strand replication, the enzyme RNAse MRP cuts the RNA in these CSB regions (Clayton, 1994). In the mouse species studied here, CSBI and III showed 100% homology; CSBII had two nucleotide changes. CSB II may be involved in the formation of an RNA primer used in replication (Xu and Clayton, 1995). The portion of the genome known as the Extended Termination-Associated Sequences (ETAS) showed only 82% and 68% homology. This region is also known to contain binding sites for the transcription factor mtTFA. Binding sites for mtTFA (Fisher and Clayton, 1988) are highly conserved within the D-loop of human, common chimpanzee, orangutan and gorilla (Kenyon and Moraes, 1997). Overall, the D-loop is the least conserved portion of mtDNA in mammals and is often used for studies of species derivation (Anderson et al., 1982; Saccone et al., 1985). Despite the tolerance for nucleotide differences at some regions in the D-loop, this control region appears to be important in cell viability. OXPHOS defects were reported in human cells that harbored heteroplasmic D-loops (Boles et al., 2003).
Protein-coding sequences appear to be much more stringently controlled than the sequences for replication and transcription. Protein-coding genes are of particular interest with regard to xenomitochondrial mice harboring M. terricolor mtDNA. Several lines of evidence suggest that the most crucial genes for interspecies compatibility are likely to be either nuclear-encoded genes that regulate assembly of the OXPHOS complexes or mtDNA-encoded subunit genes that contribute protein subunits required for normal assembly of these respiration complexes. For example, when Rattus norvegicus mtDNA was introduced to M. m. domesticus ρ° cells, normal transcription and translation occurred, but the activities of Complex I, III, and IV were reduced (Yamaoka et al., 2000; McKenzie and Trounce, 2000). Moreover, in protein complex assembly, human nuclear-encoded OXPHOS subunits appeared to show considerable preference for self-mtDNA encoded subunits over those of foreign species, even when the self-mtDNA was defective and caused respiration defects (Moraes et al., 1999). It seems that the protein-protein interactions between mitochondrial-encoded subunits and nuclear-encoded subunits are a critical contribution to species compatibility.
Our results showed considerable variability in mitochondrial nucleotide sequence between M. terricolor and M. m. domesticus (1802 differences). Interestingly, nucleotide sequence divergence resulted in only 159 amino acid differences. Moreover, 82 of these represented conservative substitutions. For comparison, there are 13 amino acid differences between the most divergent M. m. domesticus strains, the laboratory strain NZB and the common lab mouse strains such as C57BL/6J (Bayona-Bafaluy et al., 2003; Goios et al., 2007). High mtDNA sequence conservation and amino acid identity were also reported for ten inbred and two wild rat strains; with a total of 32 non-synonymous amino acid substitutions identified (Schlick et al., 2006). Moreover, when any two human mtDNAs are compared, a range of a few to perhaps 20 amino acid substitutions are observed. For example, the corrected Cambridge human reference sequence (J01415) and a Ugandan (D38112) sequence showed 17 amino acid differences.
How might the introduced mismatched amino acids be expected to impact oxidative phosphorylation in M. terricolor xenomice? Prior to the creation of M. terricolor : M. m. domesticus cybrids, strategies had focused on cross breeding to produce congenic mice with mtDNA from NZB strain mice or M. spretus on a M. m. domesticus background (these strains are more closely related to M. m. domesticus than is M. terricolor). Robertoux et al. (2003) reported subtle brain anatomical differences and altered behavior in a mouse with NZB strain mtDNA (13 mtDNA encoded amino acid differences) on a M. m. domesticus nuclear background. Furthermore, mice with M. spretus mtDNA on the M. m. domesticus background showed decreased physical performance, without overt pathology (Nagao et al., 1998). While the complete M. spretus mtDNA sequence has not been published, we have previously estimated from limited sequence data that this species will harbor around 70 amino acid substitutions in comparison with M. m. domesticus (Trounce et al., 2004). A similar approach using backcrossing to create conplastic rat strains with different mtDNAs on the same nuclear background suggests that mtDNA effects may have a major influence on diabetic risk (Pravenec et al 2007). Further diverged constructs are not possible using such breeding approaches due to nuclear chromosomal genetic divergence; highlighting the potential of the cybrid xenomitochondrial approach. This suggests that the xenomouse harboring M. terricolor mtDNA on a M. m. domesticus background (McKenzie et al., 2004), with 159 mtDNA-encoded amino acid differences, is more likely to exhibit functional impairment of oxidative phosphorylation, contributing to its usefulness as a model of human mitochondrial disease.
In the present study, complex I subunit genes ND2, ND4, ND5 and ND6 showed the greatest degree of heterogeneity between the two species, with 112 amino acid changes (Table VI and Figure 1). A Complex I deficiency was reported in human-primate xenomitochondrial cybrids (Barrientos et al., 1998). Interestingly, the defect was the same regardless of whether the foreign DNA came from pigmy chimpanzee, common chimpanzee, or gorilla. The Vmax for this protein assembly was reduced in the presence of either of its substrates, NADH or ubiquinone, suggesting that both binding sites were affected. Complex I remains the only unresolved structure of the OXPHOS complexes I to V, although considerable progress has been made with intermediate resolution approaches. All of the mitochondrial-encoded ND subunits form the integral membrane arm of the complex, with ND2, 4 and 5 showing homology to K+/Na+-H+ antiporters. This suggests that these subunits participate in proton-funneling through the inner mitochondrial membrane (Janssen et al., 2006). Due to the mismatched nuclear (M. m. domesticus) and mtDNA (M. terricolor) genomes, we speculate that proton pumping of the hybrid complex may be impaired. This observation may have particular relevance to human disease modeling in which Complex I appears mildly impaired, especially in Parkinson’s disease. It is of interest that the same complex I subunit genes displayed greater polymorphism among rat strains (Schlick et al., 2006) and when marine copepod mtDNAs were sequenced from different populations of the same species (Burton et al., 2007).
As expected, COX proteins in M. m. domesticus and M. terricolor were highly conserved in amino acid sequence. We observed 99, 100, and 98% homology in COI and II and III amino acid sequences, respectively. COX proteins are known to be among the most highly conserved mitochondrial peptides (Barrientos et al., 2000). A comparison of mtDNAs from several primate species as well as rat, mouse, and cattle revealed that the CO I subunit gene of Complex IV was the most conserved among these species (Saccone et al., 1985). Xenocybrid studies have also illustrated the importance of conservation of amino acid sequence in COX genes. Cybrids of orangutan mtDNA against a human nuclear DNA background (supplemented with a limited number of orangutan chromosomes) displayed a dramatic Complex IV deficiency. Although the mtDNA-encoded subunits (CO I and CO II) in these cybrids were translated efficiently, levels of assembled protein were reduced (Barrientos et al., 2000). This defect was attributed to improper association between the nuclear-encoded subunits and the mitochondria-encoded subunits. Yeast mutants have also shown defective association between subunits making up Complex IV (Glerum et al., 1996). Furthermore, in humans, mutations in the mitochondria-encoded CO I and CO II genes also disrupt enzyme assembly (Comi et al., 1998; Rahman et al., 1999). Xenomitochondrial cybrids harboring mtDNA from Mus pahari (intermediate in divergence between M. terricolor and Rattus) against a M. m. domesticus background showed a defect in Complex IV (McKenzie et al., 2003). However, the degree of change in COX genes between M. m. domesticus and M. terricolor appeared to be less than that observed in Mus pahari and M.m. domesticus. The changes between the COX genes in the two species compared in our study were far less than that observed between humans (Fig.1). Thus, COX function would not appear impaired in xenocybrid models created from M. m. domesticus and M. terricolor. As in the M. m. domesticus mtDNA sequence, stop codons were not detected in CO III, ND4, and Cytb genes. Here, terminal “TA” sequences were converted into “TAA” stop codons upon polyadenylation of the mRNA transcript.
The degree of heterogeneity between the ATP8 and ATP6 genes reported here consisted of only eleven amino acid changes, although variation is high at these loci among humans (Moilenen and Majamaa, 2003). The only Complex III gene encoded in mtDNA, cytochrome b, showed a mere seven amino acid changes between the two species. These changes did not include substitutions previously shown to result in a severe Complex III defect in cybrids composed of Rattus norvegicus and M. m. domesticus (McKenzie and Trounce, 2000; McKenzie et al., 2003).
The tRNA genes showed high homology, exclusively above 92%. Interestingly, complete homology was observed in a run of adenine nucleotides in the tRNAArg locus. In contrast, inbred mouse strains of the M. musculus group exhibit polymorphisms at this locus (Bayona-Bafaluy et al., 2003). Such polymorphisms are associated with aging- and environment- related deafness (Johnson et al., 2001). Additionally, position 9821 in tRNA-Arg, which is highly variable in other strains of M. musculus, was the same in the reference and the Mus terricolor sequences.
Finally, differences in rRNA genes were considered. The rRNA genes were 97% (12S rRNA) and 91% conserved (16S rRNA) between the species investigated here. A study of mtDNA of rat, mouse, cattle, man, and non-human primates including chimpanzee, orangutan, gorilla, and gibbon showed that rRNA genes were the most conserved, followed by genes for tRNAs (Saccone et al., 1985). Variations in rRNA genes have been shown to have phenotypic consequences. Changes in the 12S rRNA gene in bovine species were reported to correlate with a significant decrease in fat percentage and milk production, suggesting that the secondary structure of rRNA may be important (Boettcher et al., 1996). However, there are limited data as to the role of RNA genes in xenomitochondrial cybrids.
Knowledge of sequence divergence of mtDNA in various species and subspecies has become necessary as mitochondrial transfer is explored in creating genetically-modified transgenic mouse models. The gap in understanding sequence differences may be one reason why mitochondrial transfer has lagged behind certain other genetic engineering efforts. Despite the prevalence of diseases linked to mtDNA, very few models were created with links to specific human disorders. Accordingly, creation of xenomitochondrial animals appears to be a promising and practical modeling approach. Therefore, complete mtDNA sequence information for the species used in xenomitochondrial modeling is crucial for establishing genotype-phenotype associations.
A large portion of the amino acid substitutions between the species studied here occurred in Complex I genes. While subtle defects in Complexes III, IV and V may also be of interest in this model, we speculate that mild complex I defects may be present. The M. terricolor xenomice appear overtly normal at least until 12 months of age, yet detailed pathological studies are now underway that appear likely to uncover signals of age-related OXPHOS dysfunction. While it is clear that the majority of the introduced mtDNA “polymorphism” does not lead to disease, ongoing studies will seek to identify OXPHOS complex-specific contributions to tissue-specific pathologies.
Supplementary Material
Table 2.
Nucleotide Homology in rRNA Genes – M. terricolor vs. M. m. domesticus (C57BL/6J; #AY172335)
| Gene | *Position in AY172335 (M. m. domesticus) |
±Position in M. terricolor |
Changes in M. terricolor |
Nucleotide Identities (%) |
|---|---|---|---|---|
| 12S rRNA | 70–1024 955 nt |
68–1020 953 nt |
29 substitutions 2 deletions |
922/953 (97%) |
| 16S rRNA | 1094–2675 1582 nt |
1090–2671 1582 nt |
118 substitutions 3 deletions 3 insertions |
1461/1585 (91%) |
Table 3.
Nucleotide Homology in Protein-Coding Genes – M. terricolor vs. M. m. domesticus (C57BL/6J; # AY172335).
| Gene | *Position in AY172335 (M. m. domesticus) |
±Position in M. terricolor |
Changes in M. terricolor |
% Identity |
|---|---|---|---|---|
| ND1 | 2751–3707 957 nt |
2747–3703 957 nt |
110 base substitutions 10 amino acid changes |
89% nucleotide 97% amino acid |
| ND2 | 3914–4951 1038 nt |
3912–4949 1038 nt |
144 base substitutions 30 amino acid changes |
86% nucleotide 91% amino acid |
| CO I | 5328–6872 1545 nt |
5326–6870 1545 nt |
182 base substitutions 3 amino acid changes |
88% nucleotide 99 % amino acid |
| CO II | 7013–7696 684 nt |
7010–7693 684 nt |
86 base substitutions 0 amino acid changes |
87% nucleotide 100% amino acid |
| ATP8 | 7766–7969 204 nt |
7762–7965 204 nt |
21 base substitutions 7 amino acid changes |
90% nucleotide 90% amino acid |
| ATP6 | 7927–8607 681 nt |
7923–8603 681 nt |
79 base substitutions 4 amino acid changes |
88% nucleotide 98% amino acid |
| CO III | 8607–9390 784 nt |
8603–9386 784 nt |
89 substitutions 5 amino acid changes |
88% nucleotide 98% amino acid |
| ND3 | 9459–9806 348 nt |
9455–9802 348 nt |
47 base substitutions 5 amino acid changes |
86% nucleotide 96% amino acid |
| ND4L | 9877–10173 297 nt |
9873–10169 297 nt |
51 base substitutions 6 amino acid changes |
83% nucleotide 94% amino acid |
| ND4 | 10167–11544 1378 nt |
10163–11540 1378 nt |
170 base substitutions 28 amino acid changes |
88% nucleotide 94% amino acid |
| ND5 | 11742–13565 1824 nt |
11740–13572 1833 nt |
257 base substitutions 9 insertions 41 amino acid changes |
86% nucleotide 93% amino acid |
| ND6 | #C13552–14070 519 nt |
C13550–14068 519 nt |
58 base substitutions 13 amino acid changes |
89% nucleotide 92% amino acid |
| mt Cyt b | 14145–15288 1144 nt |
14143–15286 1144 nt |
146 base substitutions 7 amino acid changes |
87% nucleotide 98% amino acid |
Gene Abbreviations: ND, NADH dehydrogenase; CO, cytochrome c oxidase; ATP, ATP Synthase; mt Cyt b, cytochrome b.
ACKNOWLEDGEMENTS
We thank R.L. Howell, M.V. Cannon, C.A. Ingraham, J.L. Littleton, M. McKenzie, S.Vrooman and M.H. Irwin for their helpful comments and assistance over the course of this project. This work was supported in part by funds from NSF (MRI 0420600) and the Drescher Foundation (WKP) and by NIH (RR-16286, DE-12634 and HD-053037), the University of Rochester and Auburn University (CAP). Sequence data were submitted to GenBank for archival purposes and are tentatively available May, 2008.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
LITERATURE CITED
- Anderson S, Bankier AT, Barrell BG, de Bruijn MHL, Coulson AR, et al. Comparison of the human and bovine mitochondrial genomes. In: Slonimski P, Borst P, Attardi G, editors. Mitochondrial Genes. New York: Cold Spring Harbor Press; 1982. pp. 5–43. [Google Scholar]
- Barrientos A, Kenyon L, Moraes CT. Human xenomitochondrial cybrids. Cellular models of mitochondrial complex I deficiency. J. Biol. Chem. 1998;273:14210–14217. doi: 10.1074/jbc.273.23.14210. [DOI] [PubMed] [Google Scholar]
- Barrientos A, Müller S, Dey R, Wienberg J, Moraes CT. Cytochrome c oxidase assembly in primates is sensitive to small evolutionary variations in amino acid sequence. Mol. Biol. Evol. 2000;17:1508–1519. doi: 10.1093/oxfordjournals.molbev.a026250. [DOI] [PubMed] [Google Scholar]
- Barrientos A, Moraes CT. Titrating the effects of mitochondrial complex I impairment in the cell physiology. J. Biol. Chem. 1999;274:16188–16197. doi: 10.1074/jbc.274.23.16188. [DOI] [PubMed] [Google Scholar]
- Bayona-Bafaluy MP, Acín-Pérez R, Mullikin JC, Park JS, Moreno-Loshuertos R, et al. Revisiting the mouse mitochondrial DNA sequence. Nucleic Acids Res. 2003;31:5349–5355. doi: 10.1093/nar/gkg739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck JA, Lloyd S, Hafezparast M, Lennon-Pierce M, Eppig JT, et al. Genealogies of mouse inbred strains. Nature Genet. 2000;24:23–25. doi: 10.1038/71641. [DOI] [PubMed] [Google Scholar]
- Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26:167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Boettcher PJ, Kuh MT, Freeman AE. Impacts of cytoplasmic inheritance on genetic evaluations. J. Dairy Sci. 1996;79:663–675. doi: 10.3168/jds.S0022-0302(96)76412-3. [DOI] [PubMed] [Google Scholar]
- Boles RG, Luna C, Ito M. Severe reversible cardiomyopathy in four unrelated infants associated with mitochondrial DNA D-loop heteroplasmy. Pediatr. Cardiol. 2003;24:484–487. doi: 10.1007/s00246-002-0263-8. [DOI] [PubMed] [Google Scholar]
- Bonhomme F, Guénet J-L. The wild house mouse and its relatives. In: Lyon MF, Searle AG, editors. Genetic Variants and Strains of the Laboratory Mouse. Oxford: Oxford University Press; 1996. pp. 649–662.pp. 1581 [Google Scholar]
- Bonhomme F, Martin S, Taler L. Hybridization between Mus musculus L. and Mus spretus Lataste under laboratory conditions. Experientia. 1978;34:140–141. doi: 10.1007/BF01922917. [DOI] [PubMed] [Google Scholar]
- Brown WM, Prager EM, Wang A, Wilson AC. Mitochondrial DNA Sequences of Primates: Tempo and Mode of Evolution. J. Mol. Evol. 1982;18:225–239. doi: 10.1007/BF01734101. [DOI] [PubMed] [Google Scholar]
- Burton RS, Byrne RJ, Rawson PD. Three divergent mitochondrial genomes from California populations of the copepod Tigriopus californicus. Gene. 2006 doi: 10.1016/j.gene.2007.07.026. epub. [DOI] [PubMed] [Google Scholar]
- Chomyn A, Cleeter MW, Ragan CI, Riley M, Doolittle RF, et al. URF6, last unidentified reading frame of human mtDNA, codes for an NADH dehydrogenase subunit. Science. 1986;234:614–618. doi: 10.1126/science.3764430. [DOI] [PubMed] [Google Scholar]
- Clayton D. Transcription and replication of animal mitochondrial DNAs. Intl. Rev. Cytol. 1992;141:217–232. doi: 10.1016/s0074-7696(08)62067-7. [DOI] [PubMed] [Google Scholar]
- Clayton D. A nuclear function for RNase MRP. Proc. Natl. Acad. Sci (USA) 1994;91:4615–4617. doi: 10.1073/pnas.91.11.4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comi GP, Bordoni A, Salani S, Franceschina L, Sciacco M, et al. Cytochrome c oxidase sununit I microdeletion in a patient with motor neuron disease. Ann. Neurol. 1998;43:110–116. doi: 10.1002/ana.410430119. [DOI] [PubMed] [Google Scholar]
- Crews S, Ojala D, Posakony J, Nishiguchi J, Attardi G. Nucleotide sequence of a region of human mitochondrial DNA containing the precisely identified origin of replication. Nature. 1979;277:192–198. doi: 10.1038/277192a0. [DOI] [PubMed] [Google Scholar]
- Dairaghi DJ, Shadel GS, Clayton DA. Human mitochondrial transcription factor A and promoter spacing integrity are required for transcription initiation. Biochem. Biophys. Acta. 1995;1271:127–134. doi: 10.1016/0925-4439(95)00019-z. [DOI] [PubMed] [Google Scholar]
- Dey R, Barrientos A, Moraes CT. Functional constraints of nuclear-mitochondrial DNA interactions in xenomitochondrial rodent cell lines. J. Biol. Chem. 2000;275:31520–31527. doi: 10.1074/jbc.M004053200. [DOI] [PubMed] [Google Scholar]
- Ferris SD, Sage RD, Wilson AC. Evidence from mtDNA sequences that common laboratory strains of inbred mice are descended from a single female. Nature. 1982;295:163–165. doi: 10.1038/295163a0. [DOI] [PubMed] [Google Scholar]
- Fisher RP, Clayton DA. Purification and characterization of human mitochondrial transcription factor 1. Mol. Cell Biol. 1988;8:3496–3509. doi: 10.1128/mcb.8.8.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glerum DM, Shtanko A, Tzagoloff A. Characterization of COX17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase. J. Biol. Chem. 1996;271:14504–14509. doi: 10.1074/jbc.271.24.14504. [DOI] [PubMed] [Google Scholar]
- Goios A, Pereira L, Bogue M, Macaulay V, Amorim A. mtDNA phylogeny and evolution of laboratory mouse strains. Genome Res. 2007;17:293–298. doi: 10.1101/gr.5941007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guénet J-L, Bonhomme F. Wild mice: an ever-increasing contribution to a popular mammalian model. Trends Genet. 2003;19:24–31. doi: 10.1016/s0168-9525(02)00007-0. [DOI] [PubMed] [Google Scholar]
- Hixson JE, Wong TW, Clayton DA. Both the conserved stem-loop and divergent 5’-flanking sequences are required for initiation at the human mitochondrial origin of light-strand DNA replication. J. Biol. Chem. 1986;261:2384–2390. [PubMed] [Google Scholar]
- Irwin MH, Johnson LW, Pinkert CA. Isolation and microinjection of somatic cell-derived mitochondria and germline heteroplasmy in transmitochondrial mice. Transgenic Res. 1999;8:119–123. doi: 10.1023/a:1008925419758. [DOI] [PubMed] [Google Scholar]
- Irwin MH, Parrino V, Pinkert CA. Construction of a mutated mtDNA genome and transfection into isolated mitochondria by electroporation. Adv. Reprod. 2001;5:59–66. [Google Scholar]
- Kenyon L, Moraes CT. Expanding the functional human mitochondrial DNA database by the establishment of primate xenomitochondrial cybrids. Proc. Natl. Acad. Sci. U S A. 1997;94:9131–9135. doi: 10.1073/pnas.94.17.9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen RJ, Nijtmans LG, van den Heuvel LP, Smeitink JAM. Mitochondrial complex I: Structure, function and pathology. J. Inherit. Metab. Dis. 2006;29:499–515. doi: 10.1007/s10545-006-0362-4. [DOI] [PubMed] [Google Scholar]
- King MP, Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- Larsson N-G, Wang J, Wilhelmsson H, Oldfors A, Rustin P, et al. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nature Genet. 1998;18:199–200. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- McKenzie M, Trounce I. Expression of Rattus norvegicus mtDNA in Mus musculus cells results in multiple respiratory chain defects. J. Biol. Chem. 2000;275:31514–31519. doi: 10.1074/jbc.M004070200. [DOI] [PubMed] [Google Scholar]
- McKenzie M, Chiotis M, Pinkert CA, Trounce IA. Functional respiratory chain analyses in murid xenomitochondrial cybrids expose coevolutionary constraints of cytochrome b and nuclear subunits of complex III. Mol. Biol. Evol. 2003;20:1117–1124. doi: 10.1093/molbev/msg132. [DOI] [PubMed] [Google Scholar]
- McKenzie M, Trounce IA, Cassar CA, Pinkert CA. Production of homoplasmic xenomitochondrial mice. Proc. Natl. Acad. Sci. U S A. 2004;101:1685–1690. doi: 10.1073/pnas.0303184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael NL, Rothbard JB, Shiurba RA, Linke HK, Schoolnik GK, Clayton DA. All eight unassigned reading frames of mouse mitochondrial DNA are expressed. EMBO J. 1984;3:3165–3175. doi: 10.1002/j.1460-2075.1984.tb02275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moilenen JS, Majamaa K. Phylogenetic network and physicochemical properties of nonsynonomous mutations in the protein coding genes of human mitochondrial DNA. Mol. Biol. Evol. 2003;20:1195–1210. doi: 10.1093/molbev/msg121. [DOI] [PubMed] [Google Scholar]
- Moraes CT, Kenyon L, Hao H. Mechanisms of human mitochondrial DNA maintenance: The determining role of primary sequence and length over function. Mol. Biol. Cell. 1999;10:3345–3356. doi: 10.1091/mbc.10.10.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao Y, Totsuka Y, Atomi Y, Kaneda H, Lindahl KF, et al. Decreased physical performance of congenic mice with mismatch between the nuclear and the mitochondrial genome. Genes Genet. Syst. 1998;73:21–27. doi: 10.1266/ggs.73.21. [DOI] [PubMed] [Google Scholar]
- Pinkert CA, Irwin MH, Johnson LW, Moffatt RJ. Mitochondrial transfer into mouse ova by microinjection. Transgenic Res. 1997;6:379–383. doi: 10.1023/a:1018431316831. [DOI] [PubMed] [Google Scholar]
- Pinkert CA, Trounce IA. Production of transmitochondrial mice. Methods. 2002;26:348–357. doi: 10.1016/S1046-2023(02)00041-5. [DOI] [PubMed] [Google Scholar]
- Pravenec M, Hyakukoku M, Houstek J, Zidek V, Landa V, et al. Direct linkage of mitochondrial genome variation to risk factors for type 2 diabetes in conplastic rat strains. Genome Res. 2007;17:1319–1326. doi: 10.1101/gr.6548207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, Taanman JW, Cooper JM, Nelson I, Hargreaves I, et al. A missense mutation of cytochrome oxidase subunit II causes defective assembly and myopathy. Am. J. Hum. Genet. 1999;65:1030–1039. doi: 10.1086/302590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM, Haney RA, Fry AJ. Cytonuclear coevolution: the genomics of cooperation. Trends Ecol. Evol. 2004;19:645–653. doi: 10.1016/j.tree.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Roubertoux PL, Sluyter F, Carlier M, Marcet B, Maarouf-Veray F, et al. Mitochondrial DNA modifies cognition in interaction with the nuclear genome and age in mice. Nature Genet. 2003;35:65–69. doi: 10.1038/ng1230. [DOI] [PubMed] [Google Scholar]
- Saccone C, Attimonelli M, Sbisa E. Primary and higher order structural analysis of animal mitochondrial DNA. In: Quagliariello E, Slater EC, Palmieri F, Saccone C, Kroon AM, editors. Achievements and Perspectives of Mitochondrial Research Volume II: Biogenesis. New York: Elsevier; 1985. pp. 37–47. [Google Scholar]
- Schlick NE, Jensen-Seaman MI, Orlebeke K, Kwitek AE, Jacob HJ, et al. Sequence analysis of the complete mitochondrial DNA in 10 commonly used inbred rat strains. Am. J. Physiol. Cell Physiol. 2006;291:C1183–C1192. doi: 10.1152/ajpcell.00234.2006. [DOI] [PubMed] [Google Scholar]
- Sharma T, Bardhan AM, Bahadur M. Reduced meiotic fitness in hybrids with heterozygosity for heterochromatin in the speciating Mus terricolor complex. J. Biosci. 2003;28:189–198. doi: 10.1007/BF02706218. [DOI] [PubMed] [Google Scholar]
- Silver LM. Mouse Genetics: Concepts and Applications. New York: Oxford University Press; 1995. [Google Scholar]
- Suzuki H, Shimada T, Terashima M, Tsuchiya K, Aplin K. Temporal, spatial, and ecological modes of evolution of Eurasian Mus based on mitochondrial and nuclear gene sequences. Mol. Phylogenet. Evol. 2004;33:626–646. doi: 10.1016/j.ympev.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Trounce IA, McKenzie M, Cassar CA, Ingraham CA, Lerner CA, et al. Development and initial characterization of xenomitochondrial mice. J. Bioenerg. Biomembr. 2004;36:421–427. doi: 10.1023/B:JOBB.0000041778.84464.16. [DOI] [PubMed] [Google Scholar]
- Walberg MW, Clayton DA. Sequence and properties of the human KB cell and mouse L cell D-loop regions of mitochondrial DNA. Nucl. Acids Res. 1981;9:5411–5421. doi: 10.1093/nar/9.20.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mouse models for mitochondrial disease. Am. J. Med. Genet. 2001;106:71–93. doi: 10.1002/ajmg.1393. [DOI] [PubMed] [Google Scholar]
- Wong JF, Ma DP, Wilson RK, Roe BA. DNA sequence of the Xenopus laevis mitochondrial heavy and light strand replication origins and flanking tRNA genes. Nucl. Acids Res. 1983;11:4977–4995. doi: 10.1093/nar/11.14.4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka M, Isobe K, Shitara H, Yonekawa H, Miyabayashi H, et al. Complete repopulation of mouse mitochondrial DNA-less cells with rat mitochondrial DNA restores mitochondrial translation but not mitochondrial respiratory function. Genetics. 2000;155:301–307. doi: 10.1093/genetics/155.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekawa H, Moriwaki K, Gotoh O, Hayashi J-I, Watanabe J, et al. Evolutionary relationships among five subspecies of Mus musculus based on restriction enzyme cleavage patterns of mitochondrial DNA. Genetics. 1981;98:801–816. doi: 10.1093/genetics/98.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.