Abstract
Revealing diversity among extant blood flukes, and the patterns of relationships among them, has been hindered by the difficulty of determining if specimens described from different life cycle stages, hosts, geographic localities, and times represent the same or different species. Persistent collection of all available life cycle stages and provision of exact collection localities, host identification, reference DNA sequences for the parasite, and voucher specimens eventually will provide the framework needed to piece together individual life cycles and facilitate reconciliation with classical taxonomic descriptions, including those based on single life cycle stages. It also provides a means to document unique or rare species that might only ever be recovered from a single life cycle stage. With an emphasis on the value of new information from field collections of any available life cycle stages, here we provide data for several blood fluke cercariae from freshwater snails from Kenya, Uganda, and Australia. Similar data are provided for adult worms of Macrobilharzia macrobilharzia and miracidia of Bivitellobilharzia nairi. Some schistosome and sanguinicolid cercariae that we recovered have peculiar morphological features, and our phylogenetic analyses (18S and 28S rDNA and mtDNA CO1) suggest that 2 of the new schistosome specimens likely represent previously unknown lineages. Our results also provide new insights into 2 of the 4 remaining schistosome genera yet to be extensively characterized with respect to their position in molecular phylogenies, Macrobilharzia and Bivitellobilharzia. The accessibility of each life cycle stage is likely to vary dramatically from one parasite species to the next, and our examples validate the potential usefulness of information gleaned from even one such stage, whatever it might be.
The Schistosomatidae is one of the best known of parasite families, yet even within this group much remains to be learned about global species diversity, biogeography, patterns of vertebrate and snail host usage, and evolutionary relationships. Our knowledge of schistosome origins and diversification also will be improved by better understanding the diversity inherent in closely-related digenean families, including the 2 other families of blood flukes (the Spirorchiidae of turtles and the Sanguinicolidae of fishes), and the Clinostomidae. Blood fluke systematics, particularly of schistosomes, has received considerable attention in the last several years (e.g., Carmichael, 1984; Rollinson et al., 1997; Snyder and Loker, 2000; Attwood et al., 2002; Lockyer et al., 2003; Morgan et al., 2003; Agatsuma et al., 2004). These studies have examined morphology, DNA sequence diversity, and patterns of host use. The molecular studies, in particular, have yielded intriguing new hypotheses that delineate the broad patterns of relationships within the family and have provided an invaluable framework to examine overall schistosome and blood fluke diversity.
One of the long-standing impediments to assessing diversity in these groups (and in other parasite groups with multiple life cycle stages) has been the difficulty of directly relating newly discovered larval ontogenetic stages, usually cercariae released from snails, to the corresponding adult. Often larvae or adults have been recovered from a natural host and have been described and named, but the experimental infection studies that would link the 2 have not been done. This is not surprising as such studies are difficult; the natural hosts are often unknown, many species may have to be tested, and it is often difficult to obtain representatives of the expected natural hosts. Also, even if such experiments have been done, the morphological descriptions may lack precision or be compromised by the worms' developmental plasticity, and the local habitats and ecological conditions may have changed dramatically (Hsu and Hsu, 1957; Kinsella, 1971; Keas and Blankespoor, 1997). Thus, we end up with many tantalizing and often fuzzy dots that remain unconnected. DNA sequences, because of their identity across different life cycle stages within a species, because of their relative stability and reproducibility within a taxon over time, and because they have been shown to differentiate reliably between taxa (e.g., Ross et al., 1978; Dvorak et al., 2002), provide an important tool to overcome these problems. By collecting new specimens of schistosomes and other blood flukes of diverse life cycle stages (cercariae and adults most likely) from across the globe, and providing reference data, such as collection locality, identity of the host species, morphological descriptions, DNA reference sequences (often 18S and 28S rDNA and cytochrome oxidase I, but certainly not exclusively these), and voucher specimens of hosts and parasites if possible, we can provide a series of sharper dots that eventually can be connected into a coherent picture of overall blood fluke diversity. Of course, it is possible that some collections will provide information that never may connect to other life cycle stages, but this information will be valuable in its own right. It may provide the only glimpse we ever get of the existence of some parasites. Inevitably, this combined approach of traditional systematics, classical taxonomy, fieldwork, sequencing, and phylogenetics will provide the touchstones from which both a more accurate and comprehensive nomenclature and understanding of blood fluke biodiversity will emerge.
Although there are about 14 genera and 100 species recognized in Schistosomatidae (Basch, 1991; Khalil, 2002), these counts are based largely on morphological descriptions of adult worms that are often incomplete, potentially duplicative, or difficult to interpret. In aggregate, surprisingly little is known about representatives of genera other than Schistosoma. It is likely that additional study, including acquisition of sequence data, will result in considerable revision of these figures. Once we better understand the full diversity of schistosomes, we will gain a better perspective from which to answer other important questions. For example, what will a more complete and accurate accounting of schistosome diversity tell us about the evolution of dioecy and of patterns of snail or vertebrate host use (e.g., Després and Maurice, 1995; Platt and Brooks, 1997; Morand and Muller-Graf, 2000; Lockyer et al., 2003)? Griphobilharzia amoena from crocodiles (Platt et al., 1991) and Macrobilharzia from cormorants and anhingas (Carmichael, 1984; Morand and Muller-Graf, 2000) are considered to represent ancient lineages (Carmichael, 1984; Platt et al., 1991), yet no molecular information thus far has been available for either genus. More complete sampling of other blood fluke families also will provide further illumination regarding schistosome biology. The Spirorchiidae of turtles are poorly known, but generally considered basal to the Schistosomatidae (Olson et al., 2003; Snyder, 2004) and the Sanguinicolidae of fishes appear to be the basal member of a lineage represented by the blood flukes and the Clinostomidae (Olson et al., 2003).
With an emphasis on the importance of collection of new specimens from the field (often from remote or poorly sampled locations, and with life cycles that are not fully understood) and the fresh insights they bring, the aims of this paper are to highlight the importance of collection of a set of reference data (geographic locality, host of origin, morphological description, and especially DNA reference sequences) from even a single life cycle stage. Herein we provide such data for new blood fluke specimens and compare this new information with the existing blood fluke sequence database (i.e., GenBank) and literature with new phylogenetic analyses. Our results provide new reference points for future studies, report unusual cercariae that alter our concept of the morphology of schistosome and other blood fluke cercariae, and indicate the existence of new lineages of schistosomes that await further characterization. Our results also provide new insights into the phylogenetic positions of 2 of 4 (of a total of 13) schistosome genera that have not been extensively characterized with respect to their position on molecular phylogenetic trees, Macrobilharzia and Bivitellobilharzia. The former is of relevance because of its presumed basal position within the family, and the latter because of its proposed close relationship to species of medically relevant Schistosoma.
MATERIALS AND METHODS
Sample collection, DNA extraction, amplification, and sequencing
Details of the sampling localities and the nature of the specimens obtained and included in this study are provided in Table I. Snails collected from various habitats were isolated in 24-well plates and examined over a 24-hr period for evidence of shedding of furcocercous cercariae. Cercariae were examined, photographed, and provisionally identified to family according to Schell (1985). The specimens then were compared against cercarial descriptions or monographs (see discussion). The venous and arterial systems of various waterfowl were examined for adults of schistosomes such as Macrobilharzia and Trichobilharzia. Elephant dung was sieved and washed using saline and schistosome eggs encouraged to hatch by adding freshwater. Miracidia then were collected using pipettes. All cercariae, adult worms, or miracidia collected were preserved in 95% ethanol for later molecular and morphological analyses. Adult worms were heat killed in alcohol. Worms were deposited in the U.S. National Parasite Collection, Beltsville, Maryland (Table I).
Table I.
List of new taxa and/or new sequences and geographical locations used in this study.
| Taxon Label |
Taxon | Host species | Geographical origin | Lat-Long | Genbank- 28S |
Genbank- 18S |
Genbank- C01 |
USNPC no. |
|---|---|---|---|---|---|---|---|---|
| W133 | Macrobilharzia macrobilharzia, | Anhinga anhinga | Louisiana, Ascension Parish, Sorrento Timberton Hunt Club | 30°10′14″N, 90°52′5″W | AY858885 | AY829260 | AY829248 | |
| W1300 | Bivitellobilharzia nairi | Elephas maximus | Pinnewala Elephant Orphanage, Sri Lanka | AY858888 | AY829261 | AY829249 | ||
| W1285 | Avian schistosome cercaria | Biomphalaria sudanica | Lake Victoria, Kenya Fisheries Association landing site, Kisumu, Kenya | 00°06′S, 34°45′E | AY858886 | AY829258 | AY829246 | |
| W2081 | Avian schistosome cercaria | Ceratophallus natalensis | Lake Victoria, Kenya Fisheries Association landing site, Kisumu, Kenya | 00°06′S, 34°45′E | AY858887 | AY829259 | AY829247 | 96529 |
| W1120 | Spirorchiid cercaria | Biomphalaria sudanica | Kazinga Channel, Jetty no. 2, Mwea Safari Lodge, Queen Elizabeth National Park, Uganda | 00°11′31″S, 29°53′53″E | AY858884 | AY829255 | AY829242 | 96530 |
| W1284 | Sanguinicolid cercaria | Segmentorbis kanisaensis | Lake Victoria, Kenya Fisheries Association landing site, Kisumu, Kenya | 00°06′S, 34°45′E | AY858880 | AY829254 | AY829240 | 96532 |
| W1134 | Sanguinicolid cercaria | Biomphalaria sudanica | Kazinga Channel, Jetty no. 2, Mwea Safari Lodge, Queen Elizabeth National Park, Uganda | 00°11′31″S, 29°53′53″E | AY858881 | AY829253 | AY829239 | 96531 |
| W5003 | Sanguinicolid cercaria | Thiara balannensis | Northern Territory, Victoria River, Coolibah Station, Australia | 15°33′36″S, 130°56′32″E | AY858878 | AY829250 | — | 96533 |
| W5004 | Sanguinicolid cercaria | Glyptophysa gibbosa Amerianna carinata | Northern Territory, Mary River floodplain, Opium Creek Station, Australia | 12°34′37″S, 131°43′29″E | AY858879 | AY829251 | — | 96534 |
| W5005 | Clinostome cercaria | Austropeplea lessoni | Northern Territory, Mary River floodplain, Opium Creek Station, Australia | 12°34′37″S, 131°43′29″E | AY858877 | AY829252 | — | 96535 |
| W118 | Spirorchis scripta* | Chrysemys picta marginata | Corner of Lilac/Johnson Rd. Center Township, St. Joseph Co., South Bend, Indiana | AY858882 | AY829256 | AY829243 | ||
| W119 | Vasotrema robustus* | Apalone spinifera | St. Joseph River, adjacent to 51990 Lilac Rd, St. Joseph Co., South Bend, Indiana | AY858883 | AY829257 | — | ||
| hgrac | Hapalorhynchus gracillis | Chelydra serpentina | Reelfoot Lake, Lake County, Tennessee | AY604718 | AY604710 | AY829241 | ||
| ss17 | Hapalotrema mehrai | Chelonia mydas | Pacific Ocean, Hawaii | AY604716 | AY604708 | AY829244 | ||
| ss22 | Learedius learedi | Chelonia mydas | Pacific Ocean, Hawaii | AY604715 | AY604707 | AY829245 |
Parasites collected and donated by Dr. T. R. Platt, St. Mary's College, Notre Dame, Indiana.
DNA was extracted from fixed whole worms or pools of 10–50 cercariae or 50–100 miracidia with the DNeasy Tissue Kit (Qiagen, Valencia, California) according to manufacturer guidelines. Fragments of DNA were amplified by polymerase chain reaction and sequenced using the primers from Morgan et al. (2003) and those from Snyder (2004) and the Takara Ex Taq kit (Takara Biomedicals, Otsu, Japan). PCR products were purified with Montage Microcon columns (Millipore, Billerica, Maryland). Sequencing reactions were performed with the Applied Biosystems BigDye direct sequencing kit, version 1.1 (Applied Biosystems, Foster City, California).
Sequencing alignment and phylogenetic analysis
Sequences for 18S and 28S rDNA and CO1 mtDNA were assembled and edited using Sequencher ver. 4.2 (Gene Codes, Ann Arbor, Michigan). Alignments were compared to the large alignment generated by Lockyer et al. (2003), subsequently aligned by eye, and edited in Se-Al (Rambaut, A. 1996. Se-Al: Sequence Alignment Editor. http://evolve.zoo.ox.ac.uk). Positions for which alignment was uncertain were removed from the data set (alignments indicating excluded sites are available upon request). New sequences generated by this work were submitted to GenBank (Table I). The remaining taxa used in the study were those used in Lockyer et al. (2003), Morgan et al. (2003, Table I), and Snyder (2004). The ingroup was considered members of Schistosomatidae, and, depending on sequence availability, outgroups were representatives from Spirorchiidae, Sanguinicolidae, Clinostomidae, and other Diplostomida that correspond to those used in Snyder (2004).
Phylogenetic analyses using standard methods of maximum parsimony (MP), maximum likelihood (ML), minimum evolution (ME) were carried out using PAUP* ver. 4.0b10 (Swofford, 2002) and Bayesian inference using the program MrBayes (Huelsenbeck and Ronquist, 2001). Modeltest (Posada and Crandall, 1998) was used to determine the best nucleotide substitution model for the combined data to use for ML and ME analyses. The model GTR+I+G was chosen by Modeltest by best likelihood score, with the following parameters: base frequencies 0.22, 0.18, 0.28; rate matrix 0.46, 5.5, 2.4, 0.37, 3.4, 1.0; proportion of invariable sites 0.56; gamma distribution 0.44. Gaps were treated as missing data information residues. To explore the data for inconsistencies and test the usefulness of the genes used, each gene was analyzed independently and combined, rooted with the spirorchiid, sanguinicolid, and clinostome groups. In some cases, the full length of CO1 was not available, so analyses were carried out using a smaller data (∼1,600 bp 18S and ∼1,200 bp 28S) set to include these taxa. The model TrN+I+G was chosen with the following parameters: base frequencies 0.25, 0.16, 0.26; rate matrix 1.0, 4.1, 1.0, 1.0, 5.6, 1.0; proportion of invariable sites 0.42; gamma distribution 0.69.
Parsimony trees were reconstructed using heuristic searches (500 replicates), random taxon-input order, and tree-bisection and reconnection (TBR) branch swapping. Optimal ME and ML trees were determined from heuristic searches (500 replicates for ME, 5 replicates for ML), random taxon-input order, and TBR. Nodal support was estimated by bootstrap (200 replicates) and was determined for the MP and ME trees using heuristic searches (10 replicates), each with random taxon-input order. For the Bayesian analysis, there were 2 partitions in the data set 18S + 28S and CO1. Each partition was analyzed using its own model and parameter values determined using Modeltest. The model chosen for CO1 was GTR+I+G with rate matrix 0.6, 10.2, 0.9, 2.7, 6.1, 1.0; proportion of invariable sties 0.16; gamma distribution 0.3. For the 18S+28S data set we used the same model as for the ML and ME searches. In all Bayesian analyses, runs were initiated with random starting trees. Four million generations were run with 4 incrementally heated chains sampled at intervals of 100 generations. The first 2,000 trees were discarded (Hall, 2001); the retained trees were used to generate 50% majority-rule consensus trees and posterior probabilities.
RESULTS
Field collections
The following results make available host records, morphological details, and sequence information for 8 furcocercous cercariae: 2 new avian schistosomes, 1 spirorchiid, 3 sanguinicolids, 1 putative sanguinicolid, and 1 clinostome. All cercariae were measured from fixed samples in 95% ethanol (see Fig. 1 for photographs, Table I for localities, and Table II for measurements of the cercariae). Each cercaria below is given a code name that also is used to identify its position on the phylogenetic tree (Figs. 2, 3).
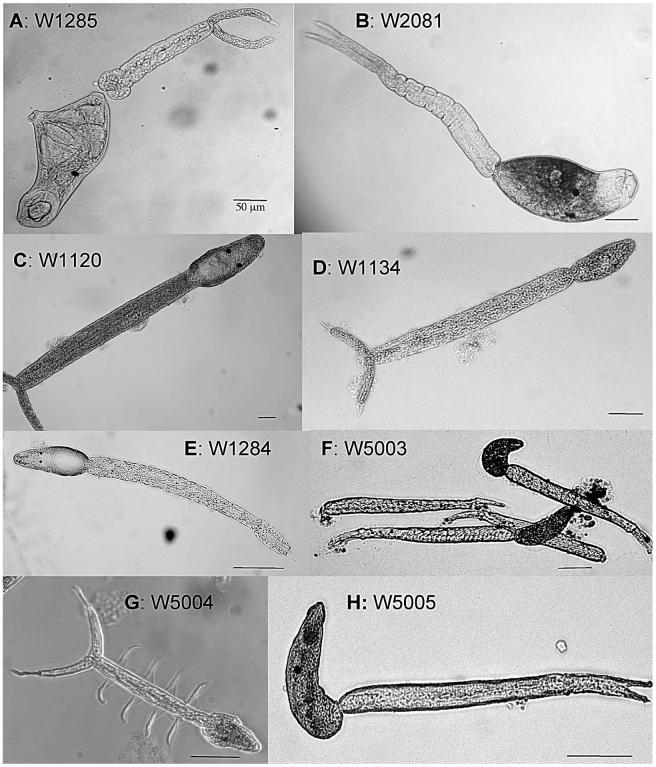
Figure 1.
Light microscope images of the furcocercous cercariae collected from various freshwater snails. Scale bars are all 50 μm.
Table II.
Measurements, in micrometers, of ethanol fixed schistosome cercariae.
| Specimen | n | Total length | Body length | Body width | Tail-stem length | Tail-stem width | Furcal length | Furcal width | Furcal lip length |
Eyespots to anterior |
Eyespots length × width |
Oral sucker/ anterior organ L |
Oral sucker/ anterior organ W |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| W1285 | 4 | 562–665 | 254–305 | 79–111 | 218–245 | 38–48 | 99–125 | 10–18 | 22 | 109 | 7.2 × 7.5 | 72.7 | 65.4 |
| W2081 | 1 | 855.3 | 286.3 | 85.6 | 446.9 | 39.3 | 124.8 | 15 | — | 121.4 | 9.5 × 9 | 81.2 | 60.1 |
| W1120 | 2 | 1,100–1,190.6 | 239.7–278.9 | 119–120 | 655.2–657.7 | 71.6–80.4 | 213.4–291.3 | 29.3–34.6 | 22.8–36 | 113.5–116.8 | 15–16 × 16–20 | 57.2 | 59.6 |
| W1284 | 2 | 635–739 | 143–176.3 | 55–66.6 | 380–440.5 | 35–45.4 | 113–157.5 | 15.8–20 | 23.0–28.5 | 59–62.8 | 5.4 × 4.7 | — | — |
| W1134 | 6 | 473.7–486.5 | 96.9–107.6 | 38.5–42.4 | 280–305.2 | 28–35.4 | 96–102.7 | 12.1–20.6 | 15.5–20 | 40.8–53.6 | 5.4 × 8 | — | — |
| W5003 | 2 | 314.7–348.9 | 69–88.2 | 31.8–33.8 | 175.3–198.2 | 19.9–23.7 | 59–62.5 | 9.1–9.5 | — | — | none | — | — |
| W5004 | 1 | 297.2 | 63.9 | 42.8 | 142.7 | 16.5 | 90.6 | 9.2 | — | 36.4 | 5.5 × 4.6 | — | — |
| W5005 | 1 | 355.2 | 114 | 31.2 | 241.3 | 19.4 | 82.4 | 14.2 | — | 60.8 | 4.8 × 8.7 | 34.2 | 11.6–18.6 |
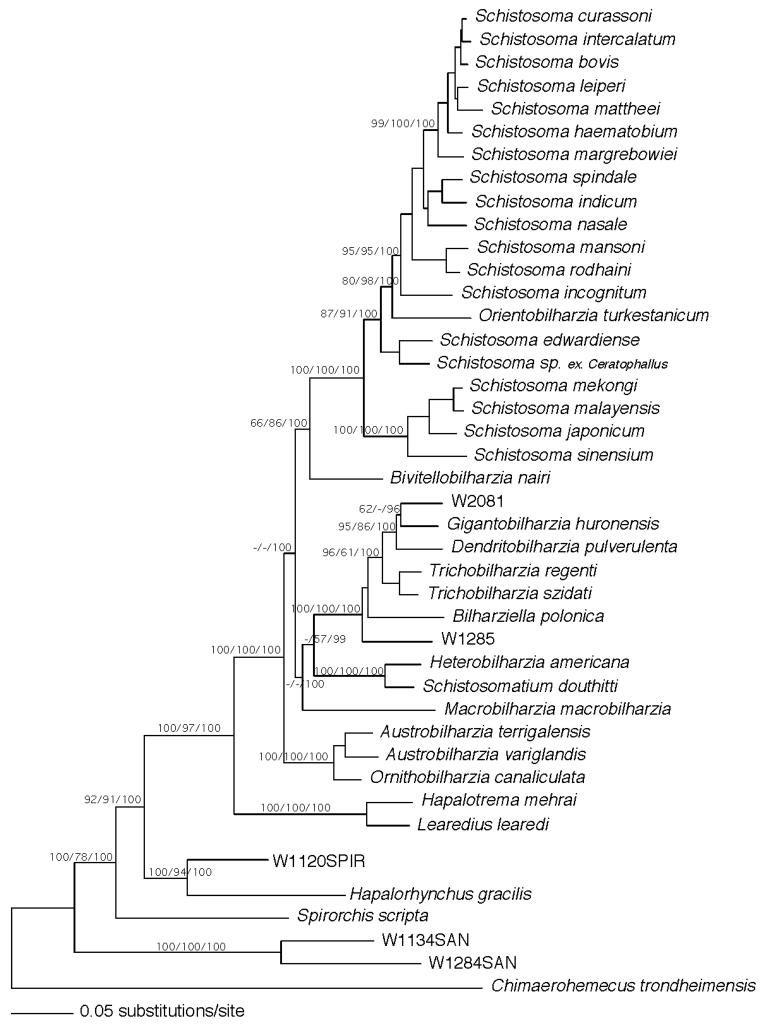
Figure 2.
Maximum likelihood estimated tree from the combined data partitions of 18S, 28S, and partial CO1 genes. Nodal support values are indicated on the branch as bootstrap values for MP/ME/Bayesian posterior probabilities.
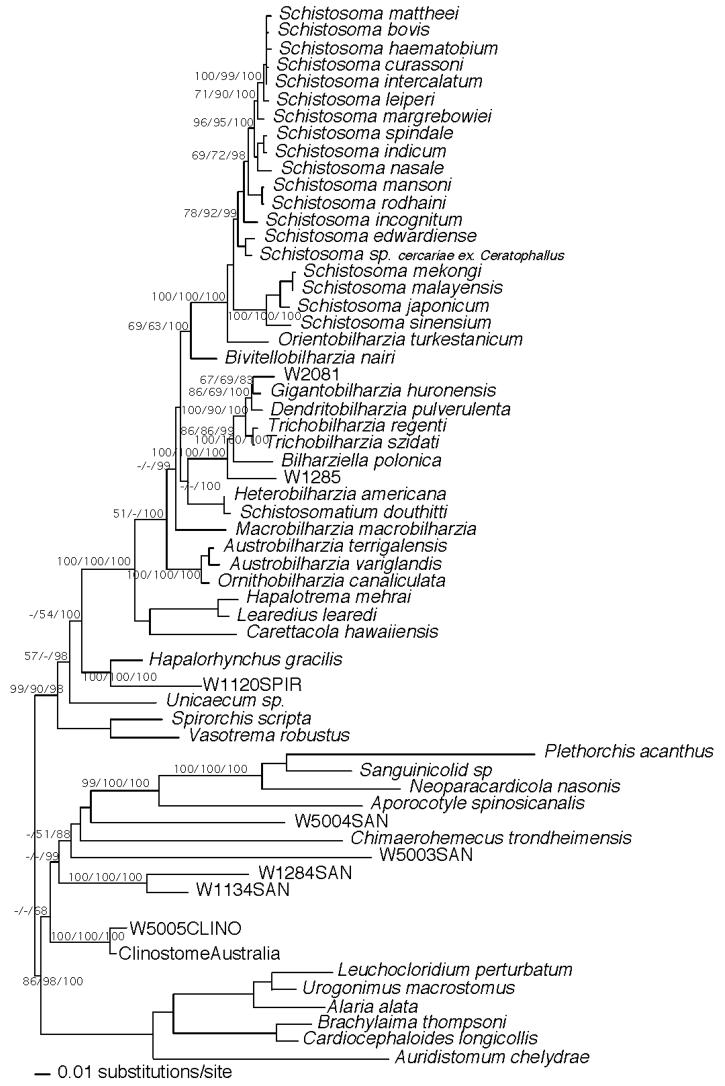
Figure 3.
Maximum likelihood estimated tree from the combined data partitions of 18S, and partial 28S. Nodal support values are indicated on the branch as bootstrap values for MP/ME/Bayesian posterior probabilities.
W1285
These small schistosome cercariae were obtained from a single Biomphalaria sudanica collected from the shore of Lake Victoria, Kisumu, Kenya. The body is widest at the level of the ventral sucker, which occupies the apex of a conical projection of the body. Well-delimited black eyespots are present, and the head organ is distinctive for having a demarcated spherical area within it. The short tail stem has a spherical bulge on either side of its base where it connects to the body. The tail stem and body seem to be easily separable. The furcae have fin-folds, and each furca is extended by a membranous, pointed tip (Fig. 1A).
W2081
These cercariae were derived from a single Ceratophallus natalensis collected from the shore of Lake Victoria, Kisumu, Kenya. They are unlike any other described schistosome cercaria we know of in having a pigmented (brown) body, with the pigment not extending anterior of the head organ (Fig. 1B). They have spherical eyespots and rest on the surface film of the water. It is possible they are released from the snail at night since they were found in the early morning, but this has not been confirmed.
W1120
These spirorchiid cercariae were obtained from a single B. sudanica collected from the Kazinga Channel, Queen Elizabeth National Park, Uganda. The long tail stem is held bent as the cercariae drift in the water. The furcae have fin-folds, but no fin-fold was seen on the body. The body, which is of variable shape, has 2 black eyespots (Fig. 1C).
W1134
These small cercariae were obtained from a single B. sudanica collected at the Kazinga Channel, Queen Elizabeth National Park, Uganda. Indistinct eyespots are present as is a dorsal body fin-fold (Fig. 1D). This cercaria holds its body flexed as it hangs in the water column.
W1284
These cercariae were released from a single Segmentorbis kanisaensis collected from the shore of Lake Victoria, Kisumu, Kenya. They have a relatively small pleomorphic body that has bean-shaped eyespots. No dorsal body fin-fold was observed. This cercaria has a long tail stem. There are no furcal fin-folds, but each furca is continued by a membranous extension that comes to a point (Fig. 1E).
W5003
These were obtained from a single Thiara balannensis collected from a boat from the vertical, vegetated banks of the Victoria River, Northern Territory, Australia. This cercaria has no eyespots, has a dorsal body fold, but lacks furcal fin-folds (Fig. 1F). Its body and tail are held flexed in the water.
W5004
These peculiar cercariae were shed from a single Amerianna carinata collected from the shore of the Mary River floodplain, Northern Territory, Australia. A single specimen of Glyptophysa gibbosa from the same locality also was found to harbor this cercaria. This cercaria has eyespots, is apharyngeate, and the body, which lacks a fin-fold, tapers from its widest part at the level of the eyespots to a blunt point at the anterior end. It is distinctive in that the lateral contours of the body are not smooth, but on each side is raised into a ridge, again at about the level of the eyespots. Most characteristic is the tail stem that has on either side a large membrane that is thrown into 4 folds that give it the appearance of having on either side 4 prominent hairs lying perpendicular to the axis of the tail. The furcae do not bear fins, but each is terminated by a slender extension narrower than the furca and that is about half the length of the furca itself (Fig. 1G).
W5005
These presumed clinostome cercariae were collected from several Austropeplea lessoni from the shore of the Mary River floodplain, Northern Territory, Australia. This cercaria has a body fin-fold, small eyespots, no furcal fin-folds, and accentuated tips to the furcae (Fig. 1H).
Individuals of M. macrobilharzia were found in the mesenteric veins of anhingas collected in a freshwater lake in Sorrento, Louisiana. One bird examined had 2 female worms, and a second had 7 female worms (see Table II for measurements). Miracidia of B. nairi were collected from the feces of Indian elephants from the Pinnewala Elephant Orphanage and the Millennium Elephant Foundation, Rambukkana, Sri Lanka.
Phylogenetic results
In the full data set, 1,796 bp of 18S, 3,827 bp 28S, and 731 bp CO1 of DNA was sequenced for most of the taxa (Fig. 2). We were unable to obtain CO1 sequence or the full complement of 28S for specimens of W5003, W5004, and W5005, so they were used in an analysis without CO1 and only 1,234 bp of 28S sequence and 1,793 bp 18S. This latter analysis of the smaller sequence data set also was undertaken so we could include more spirorchiid and sanguinicolid taxa, avoid biases (Graham et al., 2002), and compare the results with those obtained from the separate analysis based on the larger amount of sequence data (Fig. 3). Branch support obtained from MP, ML, and Bayesian analyses are shown on the ML trees (Figs. 2, 3). Both analyses recovered strong support for the following groups: AO—Austrobilharzia, Ornithobilharzia; HS—Heterobilharzia, Schistosomatium; BTGD—Bilharziella, Trichobilharzia, Gigantobilharzia, Dendritobilharzia; and BSO—Bivitellobilharzia, Schistosoma, Orientobilharzia. The W2081 cercariae aligned with Gigantobilharzia and Dendritobilharzia, W1285 was basal to the BTGD clade, and Bivitellobilharzia consistently grouped basal to SO (Figs. 2, 3). The MP and ME trees on which nodal support was estimated did not find strong support for AO as basal among schistosomes, though in these analyses and ML, AO nonetheless was resolved consistently as basal (Figs. 2, 3). Bayesian results did, however, show strong support for AO as the basal schistosomes, a result found by Olson et al. (2003) and Snyder (2004). In Figure 2, Macrobilharzia is basal to BTGD + HS, an outcome well supported by Bayesian probabilities. However, in Figure 3, Macrobilharzia is also well supported by Bayesian probabilities as basal to BSO, BTGD, and HS. In both data sets, the MP and ME trees tended to place Macrobilharzia in variable positions, usually basal to all other schistosomes, with little support. With the longer sequence data set, Bayesian and ML trees both placed Macrobilharzia as basal to the HS + BTGD clade with strong support in the Bayesian analysis. HS is more likely to group with BTGD, with or without third codon positions, in agreement with some prior analyses (Snyder and Loker, 2000; Snyder, 2004), but in disagreement with the morphologically based analysis of Carmichael (1984) and the molecular study of Lockyer et al. (2003) who noted HS to group with the SO clade. All topologies and both data sets supported the sister relationship of the marine spirorchiids with the schistosomes (Snyder, 2004).
DISCUSSION
Although species determination for the eight furcocercous cercariae discussed here is not presently possible, the collection localities, host records, morphological features, and 18S, 28S, and mtDNA CO1 sequences all will be valuable reference points for future collections of adults and/or larval stages. Additionally, adults of M. macrobilharzia and miracidia of B. nairi were obtained and the phylogenetic positions of these 2 genera explored. Bivitellobilharzia consistently resolved as the sister of SO, but the position of M. macrobilharzia is somewhat uncertain, though it showed no inclination to resolve as the most basal schistosome. Overall, the current tree topology is similar to that from other studies and supports the monophyly of Schistosomatidae, with marine spirorchiids as sister to Schistosomatidae, and freshwater spirorchiids, sanguinicolids, and clinostomes, progressively more divergent (Olson et al., 2003; Snyder, 2004). A persistent topology with strong nodal support is provided for the clades of SO, BTGD, HS, and AO (Figs. 2, 3). In this study, for both data sets all methods of analysis, AO consistently grouped basal in the family, and its position was strongly supported by Bayesian posterior probabilities. Morand and Muller-Graf (2000), Olson et al. (2003), and Snyder (2004) found robust support for the basal position of AO. In agreement with Snyder and Loker (2000), Olson et al. (2003), and Snyder (2004), we found support, albeit weak, for a BTGD + HS clade. HS has been found to be either sister to BTGD (this study, Snyder and Loker, 2000; Olson et al., 2003; Snyder, 2004) or to SO (Carmichael, 1984; Morand and Muller-Graf, 2000; Lockyer et al., 2003).
The variable positions noted in this and other studies of the 4 major schistosome clades may reflect a real evolutionary radiation that has occurred too rapidly and too far in the past to resolve today (Lanyon, 1988). Several lines of evidence suggest that the relationships among schistosomes may be a product of rapid radiation, explaining some of the variation in topologies among studies and lack of statistical difference in the constraint analyses of Lockyer et al. (2003) and Snyder (2004). Here the choice of outgroup is critical and should endeavor to include members of all blood fluke groups and their allies (Graham et al., 2002; Snyder, 2004). First, within the current schistosome tree, the more basal branches are short and have low nodal support. Second, the combined 6353 nucleotides of sequence data (Fig. 2) yield a large number of potentially phylogenetically informative characters from both rapidly (CO1) evolving and slowly (18S, 28S) evolving genes. Third, the data easily recover relationships within clades that are either older or younger than the basal schistosome clades. For example, there is strong branch support for the BSO, AO, HS, and BTGD clades and for other lineages outside of the family (e.g., the marine and freshwater spirorchiids), but, aside from the Bayesian results, there is little overall support for the relationships among the 4 main clades within the Schistosomatidae (Figs. 2, 3).
Given that known schistosome taxon sampling at the generic level in this study is nearly exhaustive, especially in the least resolved areas of the tree, it is unlikely that additional samples will enhance phylogenetic resolution among the major clades of the family. To date, we know that Macrobilharzia, Austrobilharzia, Ornithobilharzia, Heterobilharzia, and Schistosomatium are comprised of 1 or 2 species. Although diversity within Schistosoma, Trichobilharzia, and Gigantobilharzia is greater, additional samples from these groups would not contribute to the resolution of the basal branches. Therefore, in the areas of the tree where relationships are most difficult to determine, taxon sampling likely cannot be improved at this time. Last, there do not seem to be any aberrant sequences or long branches: i.e., each of the 3 genes support the same relative amount of change (Table III). Table III shows that sequence divergences between clades with low node support are similar, thus suggesting rapid or close to simultaneous change. The lack of fossil evidence and usable geologic evidence precludes reasonable and accurate estimates of the timing of cladogenesis in these groups.
Table III.
Sequence divergence between selected taxa and gene.
| Taxon* | 18S | 28S | CO1 |
|---|---|---|---|
| Schistosomatidae-Sangunicolidae | (7.9%) 5.2–10.3% | (11.5%) 9.8–13.7% | (29.91%) 26.6–33.5% |
| Schistosomatidae–Freshwater Spirorchiids | (5.7%) 3.9–7.7% | (9.3%) 6.7–10.1% | (25.9%) 22.4–29.4% |
| Schistosomatidae–Marine Spirorchiids | (5.0%) 1.9–6.7% | (6.5%) 2.9–8.8% | (23.5%) 20.0–26.6% |
| Macrobilharzia–Marine Spirorchiids | (5.3%) 4.9–5.7% | (7.1%) 6.4–7.8% | (25.3%) 24.6–26.0% |
| HS–Marine Spirorchiids | (4.5%) 3.8–5.3% | (6.3%) 4.9–7.7% | (24.9%) 24.4–25.4% |
| BSO–Marine Spirorchiids | (5.5%) 4.2–6.7% | (5.7%) 5.2–8.8% | (23.9%) 20.0–26.6% |
| AO–Marine Spirorchiids | (5.0%) 4.2–5.6% | (5.8%) 4.9–6.6% | (22.7%) 21.7–24.6% |
| BTGD–Marine Spirorchiids | (4.6%) 1.9–7.2% | (5.7%) 2.9–8.2% | (22.7%) 20.2–24.6% |
| Within Schistosomatidae | (3.1%) 0.2–5.6% | (4.6%) 0.5–7.1% | (22.0%) 11.4–28.0% |
| BSO-BTGD | (4.1%) 1.5–5.6% | (6.2%) 4.2–7.0% | (22.3%) 19.1–25.7% |
| BSO-HS | (3.0%) 1.6–3.8% | (6.1%) 4.3–7.1% | (24.7%) 21.4–27.2% |
| BSO-Macrobilharzia | (3.0%) 1.1–3.7% | (5.6%) 4.3–6.3% | (24.8%) 22.6–28.0% |
| AO-BTGD | (3.5%) 2.2–4.6% | (4.7%) 4.5–4.9% | (20.0%) 17.0–23.2% |
| HS-BTGD | (3.1%) 0.2–4.5% | (4.2%) 0.5–4.6% | (23.4%) 21.8–24.5% |
| AO-BSO | (2.9%) 1.3–4.1% | (4.2%) 2.7–6.4% | (22.9%) 19.4–25.2% |
| AO-HS | (2.7%) 2.3–3.3% | (5.1%) 4.7–5.6% | (24.0%) 21.8–26.0% |
| Bivitellobilharzia-SO | (2.3%) 1.7–2.8% | (5.0%) 4.5–5.7% | (23.2%) 20.8–24.6% |
| HS-Macrobilharzia | (1.8%) 1.8–1.9% | (5.8%) 5.5–5.7% | (25.8%) 25.7–25.9% |
| AO-Macrobilharzia | (3.1%) 2.6–3.8% | (5.4%) 5.3–5.6% | (24.0%) 24.0–24.4% |
| Within BSO | (1.9%) 0.4–2.8% | (3.4%) 0.6–5.7% | (21.6%) 13.1–24.8% |
| Within SO | (1.8%) 0.4–2.5% | (2.9%) 0.6–4.3% | (21.1%) 13.1–24.8% |
| Within HS | 0.20% | 0.48% | 19.80% |
| Within BTGD | (1.5%) 0.3–3.1% | (2.2%) 0.6–3.5% | (17.4%) 11.4–20.6% |
| Within AO | (0.8%) 0.5–1.1% | (1.0%) 1.0–1.1% | (16.2%) 15.7–16.9% |
AO = Austrobilharizia, Ornithobilharzia; BSO = Bivitellobilharzia, Schistosoma, Orientobilharzia; HS = Heterobilharzia, Schistosomatium; BTGD = Bilharziella, Trichobilharzia, Gigantobilharzia, Dendritobilharzia.
The 2 schistosome cercariae reported here are distinct from those in previously published reports, by both morphological and genetic features. Cercaria W2081 from C. natalensis from Lake Victoria (Fig. 1b) is unlike any previously described schistosome cercaria in having a pigmented body. In reference to the current sequence database, it has the lowest overall level of sequence divergence (18S, 28S, and CO1 diverge at 0.8%, 1.6%, 16.8%, respectively) with Gigantobilharzia huronensis (see Lockyer et al., 2003). Although our phylogenetic analyses consistently place W2081 closer to Gigantobilharzia, its CO1 sequence is least divergent (15.1%) from that of Trichobilharzia szidati. Soparkar (1921) described a pigmented furcocercous cercaria from Planorbis exustus in India, but it is more like a spirorchiid than a schistosome cercaria. Najim (1956) described the behavior of G. huronensis as follows: “as soon as the cercaria emerges from the snail it moves actively for a short time and then becomes more or less quiet on the surface of the water. The cercaria attaches to the surface film with the body parallel to the surface and the tail hanging downward at various angles.” This behavior is similar to what we observed for W2081.
Species of Gigantobilharzia are typically parasites of passeriform and charadriiform birds (Daniell, 1978). They use planorbid and physid snails as intermediate hosts, although a few authors report Gigantobilharzia from prosobranch and opisthobranch snails (Leigh, 1953, 1955; Fahmy et al., 1976). Interestingly, the planorbids used by species of Gigantobilharzia form a monophyletic clade that includes the tribes Planorbini and Segmentinini (Hubendick, 1955; Morgan et al., 2002), including C. natalensis, the snail from which the pigmented cercariae were shed. Gigantobilharzia has not been studied adequately and as currently conceived includes a wide range of morphological variation, and intermediate and definitive host use, not typical in other schistosome genera (Daniell, 1978). The diversity currently embraced by the genus suggests more detailed study is needed to confirm monophyly.
Cercaria W1285 collected from B. sudanica resembles that of Bilharziella polonica, also from planorbid snails, in having a short tail stem and spherical expansions at the base of the tail stem. Beyond this, W1285 does not closely resemble any other furcocercous cercariae reported from Africa, Europe, or Asia (e.g., Cawston, 1916; Faust, 1921; Sewell, 1922; Porter, 1938; Fain, 1952; Vercammen-Grandjean, 1960; Yamaguti, 1975). Unlike Gigantobilharzia, which parasitizes a wide range of snail families, Bilharziella is reported from only planorbid snails (Khalifa, 1972), although current host records do not suggest a phylogenetic affinity to a particular planorbid clade (Hubendick, 1955; Morgan et al., 2002). With respect to sequence data, W1285 tends to fall basal to the avian clade BTGD, and not sister to Bilharziella (Figs. 2, 3). Additional species of Bilharziella are few. Baugh (1963) described Bilharziella lali and Lal (1937) reported Bilharziella indica from the common teal (Anas crecca) in India. Further identification of cercaria W1285 awaits discovery of the corresponding adults, presumably from East African birds.
Members of Macrobilharzia, first described by Travassos (1922) in Brazil, presently are known as parasites of anhingas in the Americas (Price, 1929; Kohn, 1964) and of cormorants in the Old World (Fain, 1955a,b; Baugh, 1963). Based on morphological characters, the phylogenetic position of Macrobilharzia was equivocal in the preferred tree of Carmichael (1984) and grouped with the SO + HS clade of Morand and Muller-Graf (2000). In our analysis, the first to provide molecular data for this genus, in our larger sequence data set, Macrobilharzia grouped with BTGD + HS in ML and with strong support from the Bayesian analysis, with or without third codon positions (Fig. 2). In the smaller data set, Macrobilharzia was basal to all other schistosomes except the AO clade (Fig. 3).
Bivitellobilharzia was erected based on the discovery of Bivitellobilharzia loxodontae from the African elephant (Vogel and Minning, 1940). These authors also found evidence of schistosome infection in Asian elephants from Burma but did not have sufficient material to describe the species. Mudaliar and Ramanujachary (1945) subsequently named a new species from the Asian elephant in India as B. nairi. Vogel and Minning (1940) attempted to determine the snail host for the Bivitellobilharzia schistosomes from Burmese elephants and were able to obtain experimental infections in Planorbis (=Biomphalaria) pfeifferi from Africa and Planorbis sp. of unknown origin. They concluded that the natural intermediate host must be in Planorbidae. While working in Sri Lanka, we screened over 1,000 snails representing 9 different taxa from freshwater habitats frequented by infected elephants and found no cercariae similar to those described by Vogel and Minning (1940). Snails from these habitats, including Indoplanorbis exustus, an abundant planorbid in Sri Lanka, were exposed to miracidia of B. nairi, but no infections were obtained.
Morphologically, the adults of Bivitellobilharzia resemble those of Schistosoma, and the 2 genera have been considered closely related (Mudaliar and Ramanujachary, 1945; Dutt and Srivastava, 1955; Morand and Muller-Graf, 2000; Agatsuma et al., 2004). Agatsuma et al. (2004) were the first to include Bivitellobilharzia in a molecular phylogeny. They looked at several genes, each analyzed separately with a slightly different complement of schistosome species, and some of their analyses were suggestive of an alliance of Bivitellobilharzia with Schistosoma. Our combined gene analysis provides a solid level of support for placing B. nairi at the base of the SO clade. In no instance, however, did we find B. nairi to nest within Schistosoma as was suggested by Agatsuma et al. (2004). Although the bootstrap support for uniting Bivitellobilharzia with Schistosoma and Orientobilharzia is not as high as noted for some of the other clades, no topology supported B. nairi grouping with the non-Schistosoma mammalian worms ( Schistosomatium, Heterobilharzia) or any of the avian clades.
The large spirorchiid cercaria (W1120) from Uganda is most similar in sequence to Hapalorhynchus gracilis from North American freshwater turtles (Snyder, 2004). Hapalorhynchus is a cosmopolitan spirorchiid genus from freshwater turtles (Platt, 1988; Bourgot, 1990). Goodman (1987) collected Hapalorhynchus beadlei from an African side-necked turtle, Pelusios williamsi, in the Queen Elizabeth National Park in Uganda. Additionally, he described large, furcocercous cercariae belonging to the Spirorchiidae from B. sudanica from Lake Victoria, part of the Nilotic drainage system along with Lake Edward from which W1120 were collected. These findings suggest that W1120 represents a lineage of Hapalorhynchus, or a closely related genus.
The phylogenetic placement of 4 additional cercariae we collected suggests they are likely species of the blood fluke family, Sanguinicolidae. The 2 from Kenya (W1284, W1134) came from different planorbid snails and were dissimilar genetically and morphologically. However, sequence data revealed they were more similar to each other than to the remaining sanguinicolids in the tree (18S: 3.5% vs. ∼10%; 28S: 5.8% vs. ∼13%; CO1: 21.9% vs. ∼30%, respectively). The general lack of sanguinicolid sequences in the database make it premature to speculate as to whether they represent a unique lineage in this complicated digenean family. The 2 putative sanguinicolid cercariae from Australia came from very different snail hosts, W5003 from a thiarid snail (Thiaridae), and W5004 from 2 different Australian planorbid genera (Glyptophysa and Amerianna). Cercaria W5004, although apharyngeate and furcocercous, is unlike any other described cercaria, blood fluke or otherwise, we know of in having extravagant lateral tail membranes and a pointed body shape. In our trees, it clearly groups with the sanguinicolids rather than strigeids or diplostomes, sequences for which were derived from GenBank (Olson et al., 2003; Snyder, 2004). If this placement is correct, the conventional view of blood fluke cercariae must be expanded to accommodate this most peculiar form.
One clinostome cercaria was recovered from a lymnaeid snail in Australia (W5005). The Clinostomidae is of relevance to this study because recent evidence suggests it is the likely sister family to the Sanguinicolidae (Olson et al., 2003). Our sample, which fell outside the blood fluke lineages, was most closely related to a clinostome (AY222094, AY222175) collected as a metacercaria from a freshwater coastal stream fish, Hypseleotris galii, in Queensland (Olson et al., 2003). The relationships of clinostomes with their 3-host life cycles to the 3 families of blood flukes, each with a 2-host life cycle, remain a fascinating area of inquiry that will be better revealed by provision of additional data from more species.
Conclusions and future studies
Our results largely confirm and extend previous analyses of schistosome phylogenetics. There remains consistent, but weak, nodal support among the major groups of schistosomes, suggestive of a rapid radiation among the major clades within Schistosomatidae. This topic is worthy of additional study with larger sequence data sets including protein-coding genes. B. nairi from Sri Lankan elephants was found to be basal to the SO clade. It would be helpful to know more about the molluscan phase of the Bivitellobilharzia life cycle, and about the phylogenetic position of B. loxodontae from African elephants, to our knowledge a parasite not seen nor studied since its original description from a single elephant in 1940. The phylogenetic position of M. macrobilharzia from North American anhingas appears to be basal to the avian clade BTGD plus the mammal clade, HS. Inclusion of additional species of this genus from Old World cormorants may help to clarify its position. Two novel lineages of schistosome cercariae from Kenya were reported, both likely parasites of birds. Very little morphological work, and virtually no molecular work, has yet been done with adult avian schistosomes from Africa, a study that would help to clarify the systematic positions of the novel schistosomes here reported; they may well prove to represent new genera. With the inclusion of Macrobilharzia in this study, the only remaining known schistosome genus that has yet to be incorporated into a molecular phylogenetic study is the enigmatic G. amoena (Brant and Loker, 2005), known only from a single species infecting Australian freshwater crocodiles (Platt et al., 1991).
One spirorchiid cercaria (W1120 ex. Biomphalaria) from Uganda was reported and is closely aligned by sequence with the widespread turtle parasite Hapalorhynchus sp. Data for 4 putative sanguinicolids were provided, but until more samples of sanguinicolids become available, this highly divergent group remains mysterious, and it is likely that major surprises, such as the unusual cercaria (W5004) we found in the Northern Territory, are in store.
Finally, this study demonstrates the value of steadily adding relevant parasitological and sequence data to a growing database for schistosomes and for digeneans in general. An important question that arises is whether a new and blood fluke specific database should be established and kept for this purpose, possibly in the form of an interactive web site, or whether the usual means of data presentation such as publication or sequence databases like GenBank (http://www.ncbi.nlm.nih.gov) are sufficient. GenBank offers a built-in standardized method of data collection and extensive opportunities for annotation of sequence data (e.g., locality, date, host of origin, links to web sites and images), and given its relative permanence as compared to individual-maintained web sites, will remain the gold standard for the accumulation of this type of data. Of course, it always will remain imperative to relate this database to the classical taxonomic literature that has accumulated for the organisms of interest.
No matter which life cycle stage has been obtained or how isolated it might be, from such specimens we will gain an increasingly informed vantage point from which we can connect and clarify life cycles. For some species, descriptions of single life cycle stages may be all we ever get. Although not a substitute for additional studies of individual parasite life cycles or detailed morphological descriptions, such an approach eventually and inevitably will lead to a more complete understanding of the global diversity, host-parasite relationships, evolution, and ecology of these parasites.
ACKNOWLEDGMENTS
We thank Geoffrey Maina, Joseph Kinuthia, Eric Lelo, Diana Karanja, and John Vulule for their assistance in collecting specimens in Kenya. We wish to thank the following people in Louisiana who help us collect ducks: Steve Cardiff and Donna Dittman at the Museum of Natural Science, Louisiana State University; W. Guthrie Perry, Louisiana Department of Wildlife and Fisheries; and Phillip “Scooter” Trosclair, Wildlife Biologist, Rockefeller Refuge; Jim Boyce from the Timberton Hunt Club. In Sri Lanka, we thank R. C. Rajapakse, veterinary surgeon of the Pinnawala Elephant Orphanage, and Dammika Perera and Karunajeewa Kumarawansa for their assistance in collecting snails and elephant fecal samples. From the Northern Territory, Australia, we thank Michael Turner and Nick Robinson for their assistance and watching out for crocodiles, and Jeff Little for permission to collect on his property at Opium Creek Station. In addition, thanks to Owen Pugh, Coolibah Station, for use of his boat for collecting snails and Vicki Simlesa of Berrimah Research Farm, for helping us collect snails on the Victoria River at Coolibah Station. We thank Dr. Coen Adema, Dr. Randy DeJong, and Dr. Ben Hanelt for their invaluable assistance with the project. We thank Dr. Thomas Platt, St. Mary's College, Notre Dame, Indiana, for sending us samples of Vasotrema robustus and Spirorchis scripta. We acknowledge technical support from the University of New Mexico's Molecular Biology Facility, which is supported by NIH Grant Number 1P20RR18754 from the Institute Development Award (IDeA) Program of the National Center for Research Resources.
This study was supported by funds provided by the College of Arts and Sciences at UNM, and NIH grant RO1 AI44913.
LITERATURE CITED
- Agatsuma T, Rajapakse RPVJ, Kuruwita VY, Iwagami M, Rajapakse RC. Molecular taxonomic position of the elephant schistosome, Bivitellobilharzia nairi, newly discovered in Sri Lanka. Parasitology International. 2004;53:69–75. doi: 10.1016/j.parint.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Attwood SW, Upatham ES, Meng XH, Qui D-C, Southgate VR. The phylogeography of Asian Schistosoma (Trematoda: Schistosomatidae) Parasitology. 2002;125:99–112. doi: 10.1017/s0031182002001981. [DOI] [PubMed] [Google Scholar]
- Basch PF. Schistosomes: Development, reproduction and host relations. Oxford University Press; Oxford, U.K.: 1991. p. 248. [Google Scholar]
- Baugh SC. Contributions to our knowledge of digenetic trematodes VI. Zeitschrift für Parasitenkunde. 1963;22:303–315. doi: 10.1007/BF00260191. [DOI] [PubMed] [Google Scholar]
- Bourgot R. Extension taxonomique et biogéographique du genre Hapalorhynchus (Trematoda, Spirorchidae) Bulletin de la Société Française de Parasitologie. 1990;8:289–294. [Google Scholar]
- Brant SV, Loker ES. Can specialized pathogens colonize distantly related hosts? Schistosome evolution as a case study. Public Library of Science Pathogens. 2005;1:e38. doi: 10.1371/journal.ppat.0010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael AC. Phylogeny and historical biogeography of the Schistosomatidae. Michigan State University; East Lansing, Michigan: 1984. p. 246. Ph.D. Thesis. [Google Scholar]
- Cawston FG. The cercariae of the Transvaal. Parasitology. 1916;11:94–97. [Google Scholar]
- Daniell DL. Biology and host-parasite relationships of Gigantobilharzia huronensis (Trematoda: Schistosomatidae) Iowa State University; Ames, Iowa: 1978. p. 166. Ph.D. Thesis. [Google Scholar]
- Després L, Maurice S. The evolution of dimorphism and separate sexes in schistosomes. (series B).Proceedings of the Royal Society of London. 1995;262:175–180. [Google Scholar]
- Dutt SC, Srivastava HD. A revision of the genus Ornithobilharzia Odhner 1912 (Trematoda: Schistosomatidae) Proceedings of the Indian Science Congress. 1955;42:61–73. [Google Scholar]
- Dvorak J, Vanacova S, Hampl V, Flegr J, Horak P. Comparison of European Trichobilharzia species based on ITS1 and ITS2 sequences. Parasitology. 2002;124:307–313. doi: 10.1017/s0031182001001238. [DOI] [PubMed] [Google Scholar]
- Fahmy MAM, Mandour AM, Arafa MS, Omran LAM. Gigantobilharzia sp. adults (Trematoda, Schistosomatidae) recovered from chickens experimentally infected with cercariae from Melania tuberculata in Egypt. Acta Parasitologia Polonica. 1976;24:11–18. [Google Scholar]
- Fain A. Contribution à l'étude des formes larvaires des trématodes au Congo Belge et spécialement de la larve de Schistosoma mansoni. Mémoires Institut Royal Colonial Belge Section des Sciences Naturelles et Médicales. 1952;22:1–312. [Google Scholar]
- Fain A. Un nouveau schistosome du cormoran au Ruanda-Urundi (Congo Belge)—Ornithobilharzia baeri n. sp. Acta Tropica. 1955a;12:356–360. [PubMed] [Google Scholar]
- Fain A. Recherches sur les schistosomes d'oiseaux au Ruanda-Urundi (Congo Belge) Revue de Zoologie Botanique Africaines. 1955b;51:373–387. [Google Scholar]
- Faust EC. Notes on South African larval trematodes. Journal of Parasitology. 1921;8:11–21. [Google Scholar]
- Goodman JD. A new blood fluke, Hapalorhynchus beadlei n. sp. (Spirorchiidae), and a note on Allossostomoides (Paramphistomidae), in Pelusios williamsi lutescens from Uganda. Transactions of the American Microscopical Society. 1987;106:80–84. [Google Scholar]
- Graham SW, Olmstead RG, Barrett CH. Rooting phylogenetic trees with distant outgroups: A case study from the commelinoid monocots. Molecular Biology and Evolution. 2002;19:1769–1781. doi: 10.1093/oxfordjournals.molbev.a003999. [DOI] [PubMed] [Google Scholar]
- Hall BG. Phylogenetic trees made easy. Sinauer Associates; Sunderland, Massachusetts: 2001. p. 179. [Google Scholar]
- Hsu HF, Hsu SYL. On the intraspecific and interstrain variations of the male sexual glands of Schistosoma japonicum. Journal of Parasitology. 1957;43:456–463. [PubMed] [Google Scholar]
- Hubendick B. Phylogeny in the Planorbidae. Transactions of the Zoological Society of London. 1955;28:453–542. [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Keas BE, Blankespoor HD. The prevalence of cercariae from Stagnicola emarginata (Lymnaeidae) over 50 years in northern Michigan. Journal of Parasitology. 1997;83:536–540. [PubMed] [Google Scholar]
- Khalifa R. Studies on Schistosomatidae Looss, 1899 (Trematoda) of aquatic birds of Poland. I. On the life cycle of Bilharziella polonica Kowalewski, 1895, with a discussion of the subfamily Bilharzielliae Price, 1929. Acta Parasitologica Polonica. 1972;20:343–365. [Google Scholar]
- Khalil LF. Family Schistosomatidae Stiles & Hassall, 1898. In: Gibson DI, Jones A, Bray RA, editors. Keys to the Trematoda. CABI Publishing; Wallingford, U.K.: 2002. pp. 419–432. [Google Scholar]
- Kinsella JM. Growth, development, and intraspecific variation of Quinqueserialis quinquerserialis (Trematoda: Notocotylidae) in rodent hosts. Journal of Parasitology. 1971;57:62–70. [Google Scholar]
- Kohn A. Sobre o genero Macrobilharzia Travassos, 1922 (Trematoda, Schistosomatoidea) Memórias do Instituto de Oswaldo Cruz. 1964;62:1–6. doi: 10.1590/s0074-02761964000100001. [DOI] [PubMed] [Google Scholar]
- Lal MB. Studies on the trematode parasites of birds. Morphology and systematic position of some new blood flukes of the family Schistosomidae. (B).Proceedings of the Indian Academy of Science. 1937;6:274–283. [Google Scholar]
- Lanyon SM. The stochastic mode of molecular evolution: What consequences for systematic investigation? Auk. 1988;150:565–573. [Google Scholar]
- Leigh WH. Cercaria huttoni, sp. nov., a dermatitis-producing larva from the marine snail, Haminoea antillarum guadalupensis Sowerby. Journal of Parasitology. 1953;36:625–629. [PubMed] [Google Scholar]
- Leigh WH. The morphology of Gigantobilharzia huttoni (Leigh, 1953) an avian schistosome with marine dermatitis-producing larvae. Journal of Parasitology. 1955;41:262–269. [PubMed] [Google Scholar]
- Lockyer AE, Olson PD, Ostergaard P, Rollinson D, Johnston DA, Attwood SW, Southgate VR, Horak P, Snyder SD, Le TH, Agatsuma T, McManus DP, Carmichael AC, Naem S, Littlewood DTJ. The phylogeny of the Schistosomatidae based on three genes with emphasis on the interrelationships of Schistosoma Weinland, 1858. Parasitology. 2003;126:203–224. doi: 10.1017/s0031182002002792. [DOI] [PubMed] [Google Scholar]
- Morand S, Muller-Graf CDM. Muscles or testes? Comparative evidence for sexual competition among dioecious blood parasites (Schistosomatidae) of vertebrates. Parasitology. 2000;120:45–56. doi: 10.1017/s0031182099005235. [DOI] [PubMed] [Google Scholar]
- Morgan JAT, DeJong RJ, Jung Y, Khallaayoune K, Kock S, Mkoji GM, Loker ES. A phylogeny of planorbi snails, with implications for the evolution of Schistosoma parasites. Molecular Phylogenetics and Evolution. 2002;25:477–488. doi: 10.1016/s1055-7903(02)00280-4. [DOI] [PubMed] [Google Scholar]
- Morgan JAT, DeJong RJ, Kazibwe F, Mkoji GM, Loker ES. A newly identified lineage of Schistosoma. International Journal for Parasitology. 2003;33:977–985. doi: 10.1016/s0020-7519(03)00132-2. [DOI] [PubMed] [Google Scholar]
- Mudaliar SV, Ramanujachary G. Schistosoma nairi n. sp. from an elephant. Indian Veterinary Journal. 1945;22:1–4. [Google Scholar]
- Najim AT. Life history of Gigantobilharzia huronensis Najim, 1950, a dermatitis-producing bird blood-fluke (Trematoda-Schistosomatidae) Parasitology. 1956;46:443–469. doi: 10.1017/s0031182000026597. [DOI] [PubMed] [Google Scholar]
- Olson PD, Cribb TH, Tkach VV, Bray RA, Littlewood DTJ. Phylogeny and classification of the Digenea (Platyhelminthes: Trematoda) Parasitology. 2003;33:733–755. doi: 10.1016/s0020-7519(03)00049-3. [DOI] [PubMed] [Google Scholar]
- Platt TR. Phylogenetic analysis of the North American species of the genus Hapalorhynchus Stunkard, (Trematoda: Spirorchiidae), blood flukes of freshwater turtles. Journal of Parasitology. 1988;74:870–874. [Google Scholar]
- Platt TR, Blair D, Purdie J, Melville L. Griphobilharzia amoena n. gen., n. sp. (Digenea: Schistosomatidae), a parasite of the freshwater crocodile Crocodylus johnstoni (Reptilia: Crocodylia) from Australia, with the erection of a new subfamily, Griphobilharziinae. Journal of Parasitology. 1991;77:65–68. [Google Scholar]
- Platt TR, Brooks DR. Evolution of the schistosomes (Digenea: Schistosomatoidea): The origin of dioecy and colonization of the venous system. Journal of Parasitology. 1997;83:1035–1044. [PubMed] [Google Scholar]
- Porter A. The larval trematodes found in certain South African mollusca with special reference to Schistosomiasis (Bilhariziasis) South African Medical Research. 1938;42:1–492. [Google Scholar]
- Posada D, Crandall KA. Modeltest: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Price EW. A synopsis of the trematode family Schistosomidae with descriptions of new genera and species. Proceedings of the U.S. National Museum. 1929;75:1–39. [Google Scholar]
- Rajapakse J, Iwagami M, Okada T, Agatsuma T. Studies on morphology and surface topography of Bivitellobilharzia nairi in the elephant in Sri Lanka. Journal of Parasitology. in press. [Google Scholar]
- Rollinson D, Kaukas A, Johnston DA, Simpson AJG, Tanaka M. Some molecular insights into schistosome evolution. International Journal for Parasitology. 1997;27:11–28. doi: 10.1016/s0020-7519(96)00169-5. [DOI] [PubMed] [Google Scholar]
- Ross GC, Southgate VR, Knowles RJ. Observations on some isoenzymes of strains of Schistosoma bovis, S. matheei, S. margrebowiei, and S. leiperi. Zeitschrift für Parasitenkunde. 1978;57:49–56. doi: 10.1007/BF00927628. [DOI] [PubMed] [Google Scholar]
- Schell SC. Handbook of trematodes of North America north of Mexico. University of Idaho Press; Moscow, Idaho: 1985. p. 263. [Google Scholar]
- Sewell RBS. Cercariae Indicae. Indian Journal of Medical Research. 1922;10:1–370. (Supplementary number) [Google Scholar]
- Snyder SD. Phylogeny and paraphyly among tetrapod blood flukes (Digenea: Schistosomatidae and Spirorchiidae) International Journal for Parasitology. 2004;34:1385–1392. doi: 10.1016/j.ijpara.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Snyder SD, Loker ES. Evolutionary relationships among the Schistosomatidae (Platyhelminthes: Digenea) and an Asian origin for Schistosoma. Journal of Parasitology. 2000;86:283–288. doi: 10.1645/0022-3395(2000)086[0283:ERATSP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Soparkar MB. Notes on some furcocercous cercariae from Bombay. Indian Journal of Medical Research. 1921;9:23–32. [Google Scholar]
- Swofford DL. Phylogenetic analysis using parsimony* and other methods. Sinauer Associates; New York, New York: 2002. PAUP*. Version 4. [Google Scholar]
- Travassos L. Informações sobre a fauna helmintológica de Mato Grosso. Folha Medica. 1922;3:187–190. [Google Scholar]
- Vercammen-Grandjean PH. Les trématodes du Lac Kivu sud. Zoologische Wetenschappen No. 5. 1960 [Google Scholar]
- Vogel H, Minning W. Bilharziose bei Elefanten. Archiv für Schiffs- und Tropen-Hygiene. 1940;42:562–574. [Google Scholar]
- Yamaguti S. A synoptical review of life histories of digenetic trematodes of vertebrates with special reference to the morphology of their larval forms. Keigaku Shuppan Co.; Tokyo, Japan: 1975. p. 590. [Google Scholar]