Abstract
Jaagsiekte sheep retrovirus (JSRV) causes lung adenocarcinoma in sheep and goats, while the closely-related enzootic nasal tumor virus (ENTV) causes nasal tumors in the same species. The envelope (Env) protein from either virus can transform fibroblasts and epithelial cells in culture, indicating that the Env proteins are responsible for tumorigenesis. However, the primary function of retroviral Env proteins is to mediate virus entry into cells by interacting with specific cell-surface receptors, suggesting that the virus receptor might be a key player in transformation as well. Thus identification of Hyaluronidase-2 (Hyal2) as the cell-entry receptor for both JSRV and ENTV suggested a role for Hyal2 in oncogenesis. Furthermore, Hyal2 is located in a key lung cancer tumor suppressor locus on chromosome 3p21.3, suggesting that Hyal2 might have a tumor suppressor activity that was disrupted by Env thereby leading to tumorigenesis. However, recent experiments showing that expression of the JSRV or ENTV Env protein in mouse lung can induce lung tumors, even though the viral Env proteins cannot bind to or utilize mouse Hyal2 as a receptor for virus entry into cells, indicate that Hyal2 plays no role in cancer induction by these retroviruses. Hyal2 remains an enigmatic member of the hyaluronidase family given its very low hyaluronidase activity in purified form or when expressed in cultured cells, suggesting that it may have evolved to perform some other as yet unknown function.
Keywords: hyaluronidase-2, jaagsiekte sheep retrovirus, enzootic nasal tumor virus, hyaluronidase, lung cancer
1. Oncogenic sheep retroviruses
Jaagsiekte sheep retrovirus (JSRV) causes pulmonary adenocarcinoma (also called sheep pulmonary adenomatosis or jaagsiekte) in sheep and goats [1]. JSRV-induced tumors arise from epithelial cells in the lower airway, and tumor cells express markers of type II alveolar and/or bronchiolar epithelial cells [2]. Two strains of a closely-related retrovirus called enzootic nasal tumor virus (ENTV) have been cloned from sheep (ENTV-1) [3] and goats (ENTV-2) [4] that share ~95% overall amino acid similarity with JSRV. ENTV can be found in the nasal fluid of animals with intranasal tumors, which eventually progress and cause severe cranial deformations and respiratory blockage, resulting in death [5]. JSRV and ENTV can increase production of lung and nasal fluid and can spread by aerosolization of virus present in these secretions. JSRV and ENTV are present in many countries worldwide and have a significant economic and animal health impact. In addition, the disease induced by JSRV exhibits histological features similar to those of many human pulmonary adenocarcinomas, and study of adenocarcinoma induced by JSRV and ENTV may provide insights into the etiology of human lung cancer. While ENTV and JSRV do not appear to cause lung cancer in humans having occupational exposure to these viruses, this possibility has not been definitively excluded.
Until recently, oncogenic retroviruses were divided into those that cause cancer with long latency and do so by insertional activation of host oncogenes, and acutely transforming retroviruses that rapidly induce cancer as a result of virus acquisition and expression of host cell oncogenes. For example, Moloney murine leukemia virus induces leukemia over weeks to months by insertional activation of host cell oncogenes such as lck and myc, while the acutely-transforming Harvey murine sarcoma virus carries a mutant cellular ras oncogene and can acutely transform cells in culture and in animals [6]. JSRV and ENTV are examples of a small but growing new class of retroviruses that are acutely transforming and induce cancer as a direct result of expression of viral genes that show no relation to host cell genes. In the case of JSRV, cancer induction can occur in as little as 10 days in newborn sheep [7], showing that it is acutely transforming, yet JSRV does not contain sequences related to mammalian genes. JSRV and ENTV are simple retroviruses (Fig. 1) that carry genes required for viral replication and that lack accessory genes typical of complex retroviruses or cell-derived genes typical of most acutely-transforming retroviruses. Analysis of the transforming activity of JSRV and ENTV in cell culture has revealed that the env genes of these viruses are necessary and sufficient to induce transformation [8–10]. The primary role of Env in viral replication is to promote virus entry into cells following binding to specific cell-surface receptors, and it seemed likely that these receptors might play a key role in transformation as well.
Fig. 1. Genetic structure of JSRV and ENTV.
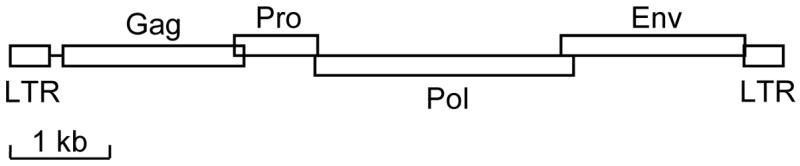
The structure of the integrated DNA form of the retroviruses is shown. Long terminal repeat (LTR) sequences that function to initiate and terminate mRNA transcription are shown flanking the protein coding regions. Protein coding regions are: Gag, virion core polypeptide; Pro, protease; Pol, reverse transcriptase (polymerase) and integrase; Env, viral coat protein (envelope) required for cell entry. The reading frames of the protein coding regions are indicated by the elevation of the boxes, for example, the Pro and Env coding regions are in the same reading frame but Pol and Gag are in different reading frames. kb, distance in kilobases.
2. Identification of Hyal2 as the cell-entry receptor for JSRV and ENTV
Retrovirus entry into cells depends on the presence of specific proteins that bind the viral Env protein and help trigger conformational changes in Env that lead to fusion of the virus and cell membranes and entry of the virus core into the cell. A wide variety of proteins have been found to serve as receptors for different retroviruses, based primarily on their ability to promote virus entry after expression in cells that are not naturally permissive for virus entry (Table 1). In most cases, a single protein suffices to render otherwise nonpermissive cells susceptible to virus entry. Typically, these proteins promote virus binding, and may also promote virus fusion with the cell membrane. For other viruses (for example, HIV) there are distinct binding and fusion receptors that are required for virus entry. Retrovirus receptors are key determinants of the species and cell types that a retrovirus can infect, and thus are primary determinants of the host range and the type of disease induced by the virus.
Table 1.
Retrovirus Receptors
| Retrovirus | Receptor | Type a | Function |
|---|---|---|---|
| Human immunodeficiency virus, simian immunodeficiency virus | CD4 and
CXCR4, CCR5, others |
TM1
TM7 |
Immune function
G protein-coupled chemokine receptors |
| Feline immunodeficiency virus | CD134 and
CXCR4 |
TM1
TM7 |
Immune function
G protein-coupled chemokine receptor |
| Human T-cell leukemia virus | GLUT-1 | TM12 | Glucose transporter |
| Ecotropic murine leukemia virus | CAT-1 (SLC7A1) | TM14 | Basic amino acid transport |
| Gibbon ape leukemia virus, amphotropic murine leukemia virus, 10A1 murine leukemia virus, feline leukemia virus type B, woolly monkey virus | Pit1 (SLC20A1) or Pit2 (SLC20A2) | TM10–13 | Phosphate transport |
| RD114, type D simian retroviruses, baboon endogenous virus, human endogenous retrovirus type W | RDR (SLC1A5) or
RDR2 (SLC1A4) |
TM9–10
TM9–10 |
Neutral amino acid transport
Glutamate and neutral amino acid transport |
| Xenotropic and polytropic murine leukemia viruses | XPR1 | TM8 | G protein-coupled signaling? Transport? |
| Feline leukemia virus type A | Thtr1 | TM12 | Thiamine transport |
| Feline leukemia virus type C | Flvcr | TM12 | Heme export |
| Feline leukemia virus type T | Felix and
Pit1 (SLC20A1) |
soluble
TM10–13 |
Env-like protein
Phosphate transport |
| Pig endogenous retrovirus type A | Par-1 (GPR172A) or Par-2 (GPR172B) | TM10–11 | G protein-coupled receptors |
| M813 murine leukemia virus | Smit1 (SLC5A3) | TM14 | myo-inositol transport |
| Avian leukosis virus type A | Tva | TM1 | LDL receptor-like protein |
| Avian leukosis virus types B, D, E | Tvb | TM1 | Fas/TNFR-like receptor |
| Avian leukosis virus type C | Tvc | TM1 | Butyrophilin-like (immunoglobulin superfamily) |
| Avian leukosis virus type J | NHE1 (SLC9A1) | TM12 | Na+/H+ antiporter |
| Mouse mammary tumor virus | Tfr1 | TM1 | Transferrin receptor |
| Jaagsiekte sheep retrovirus, Enzootic nasal tumor virus | HYAL2 | GPI-anchored | Hyaluronidase (weak) |
TM indicates a transmembrane protein and the number after TM indicates the number of times the protein is predicted to span the membrane. GPI-anchored indicates a glycosylphosphatidylinositol-anchored membrane protein.
To identify the cell-entry receptor for JSRV we used a retroviral vector that encodes human placental alkaline phosphatase (AP) and that was packaged into virions bearing the JSRV Env protein on the virion surface [11]. In early experiments we found that this vector could transfer and express (transduce) the AP marker protein gene to sheep and human cells, but not to rodent cells including those from mice, rats, and hamsters. This allowed us to develop a genetic screen to identify the human gene that when expressed in rodent cells would allow vector transduction. As target cells we used a set of 80 hamster cell lines carrying different fragments of DNA that had been produced by fusing hamster cells with irradiated human cells. This allowed us to identify the chromosomal location of the receptor within a few hundred kilobase pairs of DNA [11]. We were lucky to find that this region had been cloned as a set of overlapping cosmid clones, and it was relatively straightforward to identify the gene encoding the receptor by testing hamster cells for JSRV vector susceptibility following transfection of individual cosmids into the cells [8]. This genetic analysis indicated that only one gene served as a receptor for JSRV, but to reinforce this conclusion, we tested all of the human hyaluronidase family members for receptor activity, and found no activity associated with human Hyal1, Hyal3, Hyal4, or Spam1 [8]. These data indicate that Hyal2 is the only protein in the human genome that functions as a JSRV receptor.
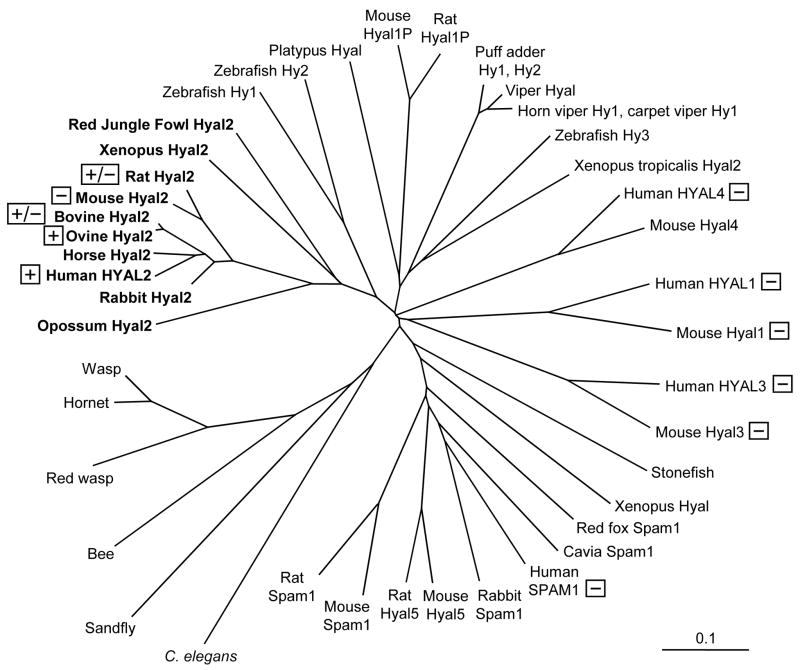
We also tested hyaluronidase family members from other species for receptor function to determine if receptor function correlates with the ability of the JSRV vector to transduce cells from different species (Fig. 2). Indeed, mouse Hyal2 functioned poorly as a receptor for JSRV vector cell-entry when expressed in mouse or hamster cells, consistent with the inability of the JSRV vector to transduce mouse cells [8]. In contrast, sheep Hyal2 functioned well as a receptor for JSRV vector cell-entry when expressed in mouse or hamster cells, consistent with the high susceptibility of sheep cells to JSRV vector transduction [10]. An intermediate result was obtained for rat Hyal2, where overexpression of rat Hyal2 in mouse, hamster, or rat cells rendered the cells susceptible to JSRV vector transduction, but rat cells are normally resistant to vector transduction [12]. Additional experiments showed that JSRV Env binds rat Hyal2 less well than it does human Hyal2, supporting the interpretation that higher levels of rat Hyal2 are required to promote efficient JSRV vector transduction than are normally expressed on rat cells [12]. In conclusion, these experiments show that expression of a functional Hyal2 protein is the primary determinant of JSRV Env-mediated virion entry into cells.
Fig. 2. Receptor activity of Hyal2 orthologs (bold) and paralogs.
Proteins related to Hyal2 obtained by BLAST search of GenBank are shown. Proteins that exhibit high receptor activity when expressed in cells that are normally nonpermissive for JSRV and ENTV vector transduction are indicated by “+”, those that exhibit moderate receptor activity are indicted by “+/−“, and those that exhibit very low to no activity are indicated by “−“ [8,10,13,16].
Using a retroviral vector encoding AP packaged into virions bearing either the ENTV or JSRV Env proteins on the virion surface, we found that ENTV Env promotes infection of a more restricted range of cell types than does the JSRV Env [10]. Given the similarity in ENTV and JSRV Env amino-acid sequences, we first tested whether ENTV Env might use Hyal2 for cell entry as does JSRV Env. Indeed, both sheep and human Hyal2 can serve as cell-entry receptors for virus bearing the ENTV Env. Interestingly, Hyal2 expression was not sufficient to promote entry of the ENTV vector into all cells, indicating that other factors are important for ENTV Env-mediated entry into cells [13].
3. Hyal2 location and enzymatic activity
Hyal2 was initially identified as a lysosomal hyaluronidase by addition of a green fluorescent protein (GFP) tag to the carboxy terminus of Hyal2 and by showing that GFP fluorescence localized to lysosomes after expression of the hybrid protein in a rat glioma cell line [14]. Hyal2 exhibited low but detectable hyaluronidase activity with an acidic pH optimum in these experiments. However, later studies have conclusively shown that Hyal2 is actually a glycosylphosphatidylinositol (GPI)-anchored cell-surface protein, consistent with its role as a cell-surface receptor for sheep retroviruses [8,12,13,15–17]. GPI-anchored proteins have an endoplasmic reticulum (ER) signal sequence at the amino terminus, which directs the proteins to the ER and is removed during protein translocation into the ER, and have hydrophobic carboxy termini that are replaced with a GPI anchor that tethers the proteins to the cell surface. The latter feature likely explains the original result indicating that Hyal2 was a lysosomal protein -the GFP tag added to the carboxy end of Hyal2 in those experiments was likely removed during GPI anchor addition and was sent to lysosomes for degradation, while the processed Hyal2 was exported to the cell surface.
Hyal2 exhibits very low hyaluronidase activity in comparison to Hyal1 or Spam1 under all conditions analyzed to date. While the hyaluronidase activity of Hyal2 could be detected in cells engineered to greatly overexpress Hyal2 by infection with a vaccinia virus vector encoding human Hyal2 [14], we have had difficulty detecting Hyal2 activity in cells transduced with a retroviral vector that encodes human Hyal2 [8]. Therefore we studied a soluble preparation of Hyal2 made by inserting a stop codon into the human Hyal2 cDNA at the position of the GPI-anchor cleavage site, and by expressing this truncated protein in insect cells using a baculovirus vector [18,19]. The endoplasmic reticulum signal sequence is properly removed from the translated protein, and without the hydrophobic tail and GPI-anchor signal sequence, the protein is secreted from the cells. This secreted form of Hyal2 (sHyal2) corresponds exactly to native Hyal2 expressed on the cell surface except that it lacks the GPI anchor. The sHyal2 protein appears to be a properly-folded monomeric protein by size-exclusion chromatography and is stable in solution at 4°C for months. Initially, the hyaluronidase activity of purified sHyal2 appeared to have a neutral pH optimum [18], but we later showed that this was due to the presence of a very small amount of a highly active baculoviral hyaluronan lyase [19], and that the hyaluronidase activity of sHyal2 actually has an acidic pH optimum consistent with previous analysis of Hyal2 activity in cultured cells. This activity could be greatly reduced by mutation of amino acid residues that correspond to the active site residues common to the bee venom and Spam1 hyaluronidases, indicating that the active site for hyaluronan digestion in Hyal2 is similar to those of other hyaluronidases [19]. However, the hyaluronidase activity of sHyal2 is ~400-fold lower than that of Spam1 [19].
The availability of purified sHyal2 allowed us to further analyze the kinetics of hyaluronan degradation by Hyal2 [18,19]. Others have noted a 20 kDa intermediate of hyaluronan degradation that appeared to be uniquely associated with hyaluronan degradation by Hyal2, and that this intermediate did not disappear even after prolonged incubation with Hyal2 [14]. We also find this intermediate following digestion of hyaluronan with sHyal2, but in contrast, this intermediate can be completely digested following prolonged incubation with sHyal2. Indeed, similar kinetics of hyaluronan digestion are observed for Hyal1 and Spam1, with initial rapid digestion of hyaluronan to a 20 kDa intermediate followed by a 25-fold slower digestion of the 20 kDa form to smaller products. Thus digestion of hyaluronan by Hyal1, Hyal2 and Spam1 appears to follow similar biphasic kinetics involving a relatively stable 20 kDa intermediate corresponding to 50 – 60 disaccharide units.
Although purified Hyal2 has low hyaluronidase activity, it is possible that other cellular proteins or cofactors might modulate Hyal2 activity and/or be required for conversion of Hyal2 into a more active enzyme. Indeed, a requirement for CD44 to promote acidification of the extracellular environment and activate the hyaluronidase activity of Hyal2 has been described [20,21]. Given the long incubation times used for detection of hyaluronidase activity in these experiments it still appears that Hyal2 is a relatively weak hyaluronidase, although a direct comparison of Hyal2 activities to a highly active hyaluronidase such as Spam1 was not performed. Perhaps in the local space adjacent to the cell only a small amount of hyaluronidase is required to mediate biologically-relevant changes in hyaluronan properties and production of a highly active enzyme would be deleterious.
4. Hyal2 role in sheep retrovirus oncogenesis?
Interaction of JSRV and ENTV Env proteins with human Hyal2, location of the human Hyal2 gene in the 3p21.3 lung cancer tumor suppressor locus, and the presumed role of Hyal2 in metabolism of the extracellular matrix all pointed to a potential role of Hyal2 in transformation by the sheep retrovirus Env proteins. Support for this hypothesis was provided by studies in the human bronchial epithelial cell line BEAS-2B [22]. In these cells, Hyal2 can bind to the RON receptor tyrosine kinase rendering it inactive. JSRV Env can transform the cells, and in cells expressing Env, Env associated with Hyal2 and caused its degradation, releasing RON from suppression by Hyal2 and activating the Akt and mitogen-activated protein kinase oncogenic pathways. Most importantly, expression of a dominant-negative RON protein blocked Env transformation of the cells indicating that RON played a critical role in Env transformation.
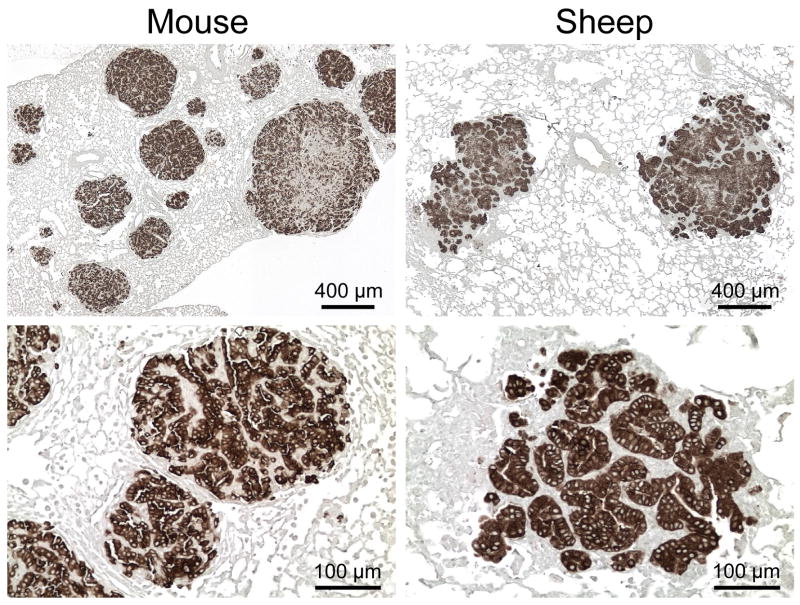
On the other hand, JSRV and ENTV Env proteins cannot mediate virus entry into mouse cells and do not bind mouse Hyal2 [12], yet both Env proteins can transform cultured NIH 3T3 mouse fibroblasts [9,12,23]. Furthermore, expression of either JSRV or ENTV Env in mouse lung can induce lung adenocarcinoma similar to that seen in sheep infected with replication-competent JSRV (Fig. 3) [24–26]. These results indicate that mouse Hyal2 plays no role in oncogenic transformation by either Env protein in mice. Whether Hyal2 plays some role in sheep tumorigenesis is uncertain but an interaction of Env with Hyal2 seems unlikely to be required based on the results in mice.
Fig. 3. Lung tumors induced by Env expression in mice and JSRV infection in sheep.
Fixed paraffin-embedded lung sections were stained for JSRV Env expression using a monoclonal antibody against Env which stains transformed lung cells in tumors. For more details see Wootton et al. [26].
So how do the JSRV and ENTV Env proteins transform cells if not by interaction with Hyal2? Several studies have shown that sequences in the cytoplasmic domain of the Env proteins are critical for transformation, and that oncogenic signaling occurs through the phosphoinositide 3-kinase (PI3K)/Akt and mitogen-activated protein kinase (MAPK) pathways [27]. The mechanisms by which the Env proteins activate these pathways are currently unknown.
5. Hyal2 role in sheep placental morphogenesis
Perhaps one of the most interesting findings relating to the interaction of sheep retrovirus Env proteins with Hyal2 is the role of Env proteins synthesized from endogenous sheep retroviruses and Hyal2 in placental morphogenesis in sheep. Mammals carry many copies of retroviruses in their genomes. Sheep carry ~20 copies of endogenous retroviruses related to JSRV and ENTV, but the Env proteins synthesized from these viruses are either nonfunctional or contain mutations that render the Env proteins nontransforming. However, some of these Env proteins can still interact with Hyal2, and in this case, can mediate fusion not between virions and cells but between cells. It has been hypothesized that endogenous retroviruses play a role in mammalian reproduction, particularly in placental morphogenesis, because intact retroviral Env genes are expressed in the syncytiotrophoblasts of human and mouse placenta and can elicit fusion of cells in culture. The importance of endogenous sheep retrovirus Env expression during pregnancy in sheep was confirmed by administration of morpholino antisense oligonucleotides to block Env expression in utero, which resulted in termination of pregnancy [28]. This study dramatically confirms that retroviruses are not simply pathogens but can contribute in a positive way to mammalian evolution, and shows that Hyal2 plays a critical role in sheep reproduction. Further work is necessary to decipher other potential functions of Hyal2 in mammals.
Acknowledgments
This work was supported by grants from the Fred Hutchinson Cancer Research Center and the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Palmarini M, Sharp JM, De las Heras M, Fan H. Jaagsiekte sheep retrovirus is necessary and sufficient to induce a contagious lung cancer in sheep. J Virol. 1999;73:6964–72. doi: 10.1128/jvi.73.8.6964-6972.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmarini M, Dewar P, De las Heras M, Inglis NF, Dalziel RG, Sharp JM. Epithelial tumour cells in the lungs of sheep with pulmonary adenomatosis are major sites of replication for Jaagsiekte retrovirus. J Gen Virol. 1995;76:2731–7. doi: 10.1099/0022-1317-76-11-2731. [DOI] [PubMed] [Google Scholar]
- 3.Cousens C, Minguijon E, Dalziel RG, Ortin A, Garcia M, Park J, et al. Complete sequence of enzootic nasal tumor virus, a retrovirus associated with transmissible intranasal tumors of sheep. J Virol. 1999;73:3986–93. doi: 10.1128/jvi.73.5.3986-3993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ortin A, Cousens C, Minguijon E, Pascual Z, Villarreal MP, Sharp JM, et al. Characterization of enzootic nasal tumour virus of goats: complete sequence and tissue distribution. J Gen Virol. 2003;84:2245–52. doi: 10.1099/vir.0.19125-0. [DOI] [PubMed] [Google Scholar]
- 5.Vitellozzi G, Mughetti L, Palmarini M, Mandara MT, Mechelli L, Sharp JM, et al. Enzootic intranasal tumour of goats in Italy. J Vet Med. 1993;40:459–68. doi: 10.1111/j.1439-0450.1993.tb00164.x. [DOI] [PubMed] [Google Scholar]
- 6.Rosenberg N, Jolicoeur P. Retroviral pathogenesis: oncogenesis. Retroviruses, Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1997. pp. 478–523. http://web.ncbi.nlm.nih.gov/books/bv.fcgi?rid=rv.section.4717. [PubMed] [Google Scholar]
- 7.Sharp JM, Angus KW, Gray EW, Scott FM. Rapid transmission of sheep pulmonary adenomatosis (jaagsiekte) in young lambs. Arch Virol. 1983;78:89–95. doi: 10.1007/BF01310861. [DOI] [PubMed] [Google Scholar]
- 8.Rai SK, Duh F-M, Vigdorovich V, Danilkovitch-Miagkova A, Lerman MI, Miller AD. Candidate tumor suppressor HYAL2 is a glycosylphosphatidylinositol (GPI)-anchored cell-surface receptor for jaagsiekte sheep retrovirus, the envelope protein of which mediates oncogenic transformation. Proc Natl Acad Sci USA. 2001;98:4443–8. doi: 10.1073/pnas.071572898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maeda N, Palmarini M, Murgia C, Fan H. Direct transformation of rodent fibroblasts by jaagsiekte sheep retrovirus DNA. Proc Natl Acad Sci USA. 2001;98:4449–54. doi: 10.1073/pnas.071547598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dirks C, Duh F-M, Rai SK, Lerman MI, Miller AD. Mechanism of cell entry and transformation by enzootic nasal tumor virus. J Virol. 2002;76:2141–9. doi: 10.1128/jvi.76.5.2141-2149.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rai SK, DeMartini JC, Miller AD. Retrovirus vectors bearing jaagsiekte sheep retrovirus Env transduce human cells by using a new receptor localized to chromosome 3p21.3. J Virol. 2000;74:4698–704. doi: 10.1128/jvi.74.10.4698-4704.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu S-L, Duh F-M, Lerman MI, Miller AD. Role of virus receptor Hyal2 in oncogenic transformation of rodent fibroblasts by sheep betaretrovirus env proteins. J Virol. 2003;77:2850–8. doi: 10.1128/JVI.77.5.2850-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Hoeven NS, Miller AD. Improved enzootic nasal tumor virus pseudotype packaging cell lines reveal virus entry requirements in addition to the primary receptor Hyal2. J Virol. 2005;79:87–94. doi: 10.1128/JVI.79.1.87-94.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lepperdinger G, Strobl B, Kreil G. HYAL2, a human gene expressed in many cells, encodes a lysosomal hyaluronidase with a novel type of specificity. J Biol Chem. 1998;273:22466–70. doi: 10.1074/jbc.273.35.22466. [DOI] [PubMed] [Google Scholar]
- 15.Liu S-L, Halbert CL, Miller AD. Jaagsiekte sheep retrovirus envelope efficiently pseudotypes human immunodeficiency virus type 1-based lentiviral vectors. J Virol. 2004;78:2642–7. doi: 10.1128/JVI.78.5.2642-2647.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duh FM, Dirks C, Lerman MI, Miller AD. Amino acid residues that are important for Hyal2 function as a receptor for jaagsiekte sheep retrovirus. Retrovirology. 2005;2:59. doi: 10.1186/1742-4690-2-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller AD, Vigdorovich V, Strong RK, Fernandes RJ, Lerman MI. Letter to the Editor: Hyal2, where are you? Osteoarthritis Cartilage. 2006;14:1315–7. doi: 10.1016/j.joca.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Vigdorovich V, Strong RK, Miller AD. Expression and characterization of a soluble, active form of the jaagsiekte sheep retrovirus receptor, Hyal2. J Virol. 2005;79:79–86. doi: 10.1128/JVI.79.1.79-86.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vigdorovich V, Miller AD, Strong RK. Ability of hyaluronidase 2 to degrade extracellular hyaluronan is not required for its function as a receptor for jaagsiekte sheep retrovirus. J Virol. 2007;81:3124–9. doi: 10.1128/JVI.02177-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bourguignon LY, Singleton PA, Diedrich F, Stern R, Gilad E. CD44 interaction with Na+-H+ exchanger (NHE1) creates acidic microenvironments leading to hyaluronidase-2 and cathepsin B activation and breast tumor cell invasion. J Biol Chem. 2004;279:26991–7007. doi: 10.1074/jbc.M311838200. [DOI] [PubMed] [Google Scholar]
- 21.Harada H, Takahashi M. CD44-dependent intracellular and extracellular catabolism of hyaluronic acid by hyaluronidase-1 and -2. J Biol Chem. 2007;282:5597–607. doi: 10.1074/jbc.M608358200. [DOI] [PubMed] [Google Scholar]
- 22.Danilkovitch-Miagkova A, Duh F-M, Kuzmin I, Angeloni D, Liu S-L, Miller AD, et al. Hyaluronidase 2 negatively regulates RON receptor tyrosine kinase and mediates transformation of epithelial cells by jaagsiekte sheep retrovirus. Proc Natl Acad Sci USA. 2003;100:4580–5. doi: 10.1073/pnas.0837136100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu S-L, Lerman MI, Miller AD. Putative phosphatidylinositol 3-kinase (PI3K) binding motifs in ovine betaretrovirus Env proteins are not essential for rodent fibroblast transformation and PI3K/Akt activation. J Virol. 2003;77:7924–35. doi: 10.1128/JVI.77.14.7924-7935.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wootton SK, Halbert CL, Miller AD. Sheep retrovirus structural protein induces lung tumours. Nature. 2005;434:904–7. doi: 10.1038/nature03492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wootton SK, Halbert CL, Miller AD. Envelope proteins of jaagsiekte sheep retrovirus and enzootic nasal tumor virus induce similar bronchioalveolar tumors in lungs of mice. J Virol. 2006;80:9322–5. doi: 10.1128/JVI.00865-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wootton SK, Metzger MJ, Hudkins KL, Alpers CE, York D, Demartini JC, et al. Lung cancer induced in mice by the envelope protein of jaagsiekte sheep retrovirus (JSRV) closely resembles lung cancer in sheep infected with JSRV. Retrovirology. 2006;3:94. doi: 10.1186/1742-4690-3-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu SL, Miller AD. Oncogenic transformation by the jaagsiekte sheep retrovirus envelope protein. Oncogene. 2007;26:789–801. doi: 10.1038/sj.onc.1209850. [DOI] [PubMed] [Google Scholar]
- 28.Dunlap KA, Palmarini M, Varela M, Burghardt RC, Hayashi K, Farmer JL, et al. Endogenous retroviruses regulate periimplantation placental growth and differentiation. Proc Natl Acad Sci USA. 2006;103:14390–5. doi: 10.1073/pnas.0603836103. [DOI] [PMC free article] [PubMed] [Google Scholar]