Abstract
Caspases are essential components of the apoptotic machinery in both vertebrates and invertebrates. Here, we report the isolation of a mutant allele of the Drosophila effector caspase drICE as a strong suppressor of hid- (head involution defective-) induced apoptosis. This mutant was used to determine the apoptotic role of drICE. Our data are consistent with an important function of drICE for developmental and irradiation-induced cell death. Epistatic analysis suggests that drICE acts genetically downstream of Drosophila inhibitor of apoptosis protein 1 (Diap1). However, although cell death is significantly reduced in drICE mutants in all assays, it is not completely blocked. A double-mutant analysis between drICE and death caspase-1 (dcp-1), another effector caspase, reveals that some cells (type I) strictly require drICE for apoptosis, whereas other cells (type II) require either drICE or dcp-1. Thus, these data demonstrate a barely appreciated complexity in the apoptotic pathway, and are consistent with current models about effector caspase regulation in both vertebrates and invertebrates.
Keywords: DrICE, Dcp-1, Drosophila, programmed cell death, Diap1, Dronc
Introduction
Programmed cell death or apoptosis is an essential physiological process required for normal development of metazoan organisms and tissue homeostasis. The key mediators of cell death are caspases, a highly specialized class of Cysproteases. Caspases are produced as inactive zymogens. During apoptosis, activation of caspases involves proteolytic processing, cleaving off an N-terminal prodomain, and generating the large and the small catalytic subunits (reviewed in Ref.1).
Two classes of caspases have been defined based on the length of the prodomain. Initiator caspases contain long prodomains that harbor regulatory motifs such as the caspase activation and recruitment domain (CARD) in the prodomain of Caspase-9.1 These regulatory motifs serve as binding sites for upstream apoptotic signaling factors. For example, through homotypic interactions of the CARD motif of Caspase-9 with the CARD motif of Apaf-1, Caspase-9 is recruited into the apoptosome, a large multisubunit complex, where it undergoes autoprocessing and activation.1 Once activated, Caspase-9 cleaves and activates the effector Caspase-3, which is characterized by the presence of a short prodomain. Effector caspases execute the cell death process by cleaving a large number of cellular proteins, triggering the morphological events leading to apoptotic cell death.
Caspases are negatively regulated by inhibitor of apoptosis proteins (IAPs). IAPs bind to processed caspases and inhibit them (recently reviewed in Ref.2). Thus, IAPs provide the last line of defense against inappropriate caspase activity. In apoptotic cells, IAP antagonists such as the Drosophila Reaper, Hid (Head involution defective), and Grim (RHG) proteins (recently reviewed in ref.3) displace IAPs directly from caspases4,5 which are then released from IAP inhibition, and induce apoptosis. In addition, Reaper, Hid and Grim also promote proteolytic degradation of Drosophila IAP1 (Diap1).3
The Drosophila genome contains a total of seven caspase genes, three of which encode putative initiator caspases (Drosophila Nedd-2 like Caspase (Dronc), Dredd and Strica), whereas the remaining four are putative effector caspases (Drosophila ICE (DrICE), death caspase-1 (Dcp-1), Decay and Damm) (reviewed in Ref.1,6). Mutations in these caspase genes that would allow their genetic characterization have been described for dredd, dcp-1 and dronc. However, the available evidence suggests that dredd is not an apoptotic caspase, but instead appears to have a fundamental role in innate immunity (reviewed in Ref.1). Homozygous dcp-1 mutants are viable and fertile.7 The only cell death phenotype reported for dcp-1 is lack of germline cell death during mid-oogenesis in response to nutrient deprivation.7 dronc is the only caspase gene described so far whose mutations display a clear apoptotic phenotype. Genetic inactivation of dronc blocks most developmental cell death during embryogenesis, imaginal disc development and metamorphosis.8-11 Dronc is functionally similar to human Caspase-9 because it contains a CARD motif in the prodomain,12 and interacts with Drosophila Apaf-1-related killer (Ark), also known as Dark, Hac-1 and D-Apaf-1 (reviewed in Ref.3).
It has not been genetically determined whether any of the effector caspases in Drosophila are apoptotic (with the exception of dcp-1 during oogenesis).7 However, several observations suggest that drICE is an important component of the apoptotic machinery in Drosophila. First, overexpression of drICE sensitizes Drosophila S2 cells to apoptosis.13 Immunodepletion of DrICE in S2 cells reduces the ability of cycloheximide and reaper expression to induce apoptotic morphology.14 Furthermore, silencing of DrICE by RNA interference (RNAi) blocks S2 cell apoptosis.15 These findings suggest that DrICE is required for apoptosis in S2 cells. Second, drICE expression is induced by the insect hormone ecdysone, which stimulates apoptosis during metamorphosis.16 Active DrICE was also found during cell death in both mid- and late oogenesis.17 Third, the initiator caspase Dronc can cleave and activate DrICE in vitro.18,19 DrICE also cleaves Dronc,15 and it has been proposed that this cleavage constitutes a caspase amplification loop.16 Fourth, Diap1, which is essential for cellular survival,2,3 can inhibit DrICE through direct physical interactions.4,5,20,21 These observations imply, but do not prove, that drICE encodes an important component of the apoptotic machinery in Drosophila. Thus, to clarify the role of drICE for developmental apoptosis, analysis of mutations in the endogenous gene are necessary.
In this study, we describe the isolation and genetic characterization of an EMS-induced allele of the effector caspase drICE. This mutant is characterized by reduced levels of developmental and irradiation-induced cell death, and contains additional cells. We show that the strong apoptotic phenotype of diap1 mutants is partially suppressed by drICE inactivation, suggesting that inappropriate activation of DrICE contributes to the diap1 mutant phenotype, and that drICE acts genetically downstream of diap1. However, even though the drICE mutant reduces developmental apoptosis, it does not completely block it. We show that drICE and dcp-1 share a partially overlapping function in such a way that some cells (type I) strictly require drICE for apoptosis, whereas other cells (type II) are more flexible and die either through drICE or dcp-1.
Results
Isolation and identification of a drICE mutant
Recently, we described GheF (GMR-hid ey-FLP) screening method which allows to identify mutants in genes required for hid-induced cell death.10 The GheF method takes advantage of the eye-ablation phenotype caused by expression of hid under eye-specific GMR-enhancer control (GMR-hid; Figure 1b). During GheF screening, suppressors of GMR-hid (su(GMR-hid)) are identified in homozygous mutant eye clones obtained by ey-FLP/FRT-mediated recombination in otherwise heterozygous animals (for further technical details see Xu et al., 2005).10
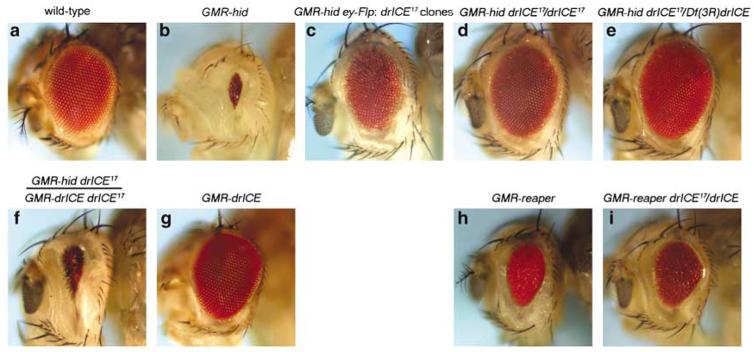
Figure 1.
Isolation of drICE17 as strong suppressor of GMR-hid. (a) Wild-type eye. (b) GMR-hid-induced eye-ablation phenotype. (c) Suppression of GMR-hid in ey-Flp-induced drICE17 clones. Exact genotype: GheF; FRT82B drICE17/FRT82B P[w+]. (d) Suppression of GMR-hid in homozygous drICE17 background. Exact genotype: GMR-hid drICE17/drICE17. (e) Suppression of GMR-hid in trans-heterozygous drICE17 over Df(3R)drICE mutant background. Exact genotype: GMR-hid drICE17/Df(3R)drICE.(f) Transgenic rescue of the GMR-hid-induced eye-ablation phenotype in drICE17 mutant background by a GMR-drICE transgene. Exact genotype: GMR-hid drICE17/GMR-drICE drICE17. (g) GMR-drICE does not cause an eye-ablation phenotype on its own. (h) Eye-ablation phenotype caused by GMR-reaper. Exact genotype: CyO, 2x GMR-reaper/ + . (i) Weak suppression of GMR-reaper by drICE17. Exact genotype: CyO, 2x GMR-reaper; drICE17/drICE17
The drICE locus maps to cytological position 99C1 of the polytene map on the right arm of chromosome 3 (3R). Thus, to isolate mutants in drICE, we carried out an EMS mutagenesis screen using the GheF method for 3R (see Materials and Methods). The strongest suppressor of GMR-hid obtained in the screen, su(GMR-hid)17, was chosen for further characterization. In ey-FLP/FRT clones, su(GMR-hid)17 suppresses GMR-hid strongly (Figure 1c). su(GMR-hid)17 is homozygous viable (see also below), and homozygously suppresses GMR-hid even stronger (Figure 1d), restoring the eye back to wild-type size (Figure 1a).
To identify the gene mutant in su(GMR-hid)17, we mapped the mutation to the distal tip of 3R by P-element mapping. In parallel to the above described EMS screen, a small deficiency deleting cytological range 99B3/B8–99C2/C4 on the polytene map was isolated by X-ray irradiation (see Materials and Methods). This deficiency, referred to as Df(3R)drICE, over su(GMR-hid)17 strongly suppresses GMR-hid (Figure 1e). Thus, this analysis suggests that su(GMR-hid)17 maps to the cytological range 99B3/B8–99C2/C4. None of the genes residing in this cytological range have been implicated in apoptosis, with the exception of drICE which maps to 99C1. Thus, we tested whether a GMR-drICE transgene22 can restore the small eye phenotype of GMR-hid in homozygous su(GMR-hid)17 animals. This was found to be the case (Figure 1f). GMR-drICE does not display a small eye phenotype on its own22 (Figure 1g). These findings establish that the suppression of GMR-hid by su(GMR-hid)17 is caused by genetic inactivation of drICE. This is also confirmed by DNA sequencing analysis and immunoblotting (see next section). Therefore, we refer to su(GMR-hid)17 from now on as drICE17. The rescue of GMR-hid by drICE17/Df(3R)drICE is slightly better than the one of homozygous drICE17 animals (Figure 1d and e) suggesting that drICE17 is a very strong hypomorphic allele, but not a null allele.
We also determined whether drICE17 could suppress the GMR-reaper-induced small eye phenotype. The GMR-reaper eye-ablation phenotype is weaker compared to GMR-hid (Figure 1h). Surprisingly, although homozygous drICE17 animals do suppress GMR-reaper (Figure 1i), the suppression is significantly weaker compared to the suppression of GMR-hid (Figure 1d). The weak suppression of GMR-reaper could reflect an unanticipated complexity of the apoptotic process, or could be allele specific for drICE17. Additional drICE mutants are necessary to distinguish between these possibilities.
In summary, this analysis identifies a mutation in the drICE gene and provides evidence that drICE+ is genetically required for GMR-hid-induced cell death. Homozygous drICE17 adults are viable, and carry wings that appear less transparent compared to wild type (data not shown). This wing phenotype which is difficult to illustrate in photographs, appears to be characteristic for mutants of cell death genes as it has been previously observed in hid, ark and dronc mutants.10 Although drICE17 mutant animals are homozygous viable, they were not obtained at mendelian ratios. Only about 1/3 of the expected progeny (∼500 offspring scored) was found relative to controls (drICE17/TM3). A similar semilethality was observed for drICE17/Df(drICE) animals. The lethal phase of those homozygous individuals which die occurs during embryogenesis without detectable phenotype. In addition, it is difficult to keep drICE17 flies in a homozygous condition. They appear to be semisterile, especially the males, but we have not characterized this phenotype in detail.
drICE17 encodes for an unstable protein
DNA sequencing reveals one single base pair change in the drICE17 open reading frame changing Asn116 to Tyr. Asn116 lies in a well conserved domain of the large subunit of DrICE (Figure 2a). This residue is conserved in all invertebrate caspases and in some mammalian caspases including Caspase-6 from mouse and even human Caspase-9, an initiator caspase (Figure 2a). In mouse and human Caspase-3 and Caspase-7, this position is occupied by a semiconserved Asp residue (Figure 2a). Immunoblot analysis showed that drICE17 encodes for an unstable protein (Figure 2b). We quantified that <5% of the wild-type levels of DrICE protein are detectable in immunoblots of drICE17 mutant embryos.
Figure 2.
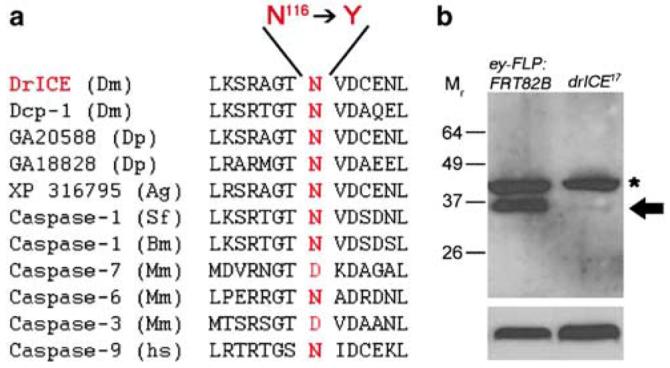
drICE17 encodes for an unstable protein. (a) Alignment of the amino-acid residues surrounding Asn116 in DrICE and various effector caspases in insects and mammals, and human Caspase-9. drICE17 carries a mutation in Asn116, changing it to Tyr. Dm, Drosophila melanogaster; Dp, Drosophila pseudoobscura; Ag, Anopheles gambiae; Sf, Spodoptera frugiperda; Bm, Bombyx mori; Mm, Mus musculus; Hs, Homo sapiens. (b) Embryonic extract obtained from homozygous ey-Flp; FRT82B (the stock used for mutagenesis) and drICE17 mutant flies were analyzed by immunoblotting using an antibody raised against the prodomain of DrICE (upper panel). Lower panel is the same blot probed with antiactin antibody as loading control. The arrow indicates full-length DrICE, the asterisk an unspecific protein
drICE17 mutants exhibit reduced developmental cell death
The drICE17 mutant was isolated as a strong suppressor of GMR-hid (Figure 1). Furthermore, immunoblot analysis showed that drICE17 encodes for an unstable protein. These observations suggest that drICE17 represents a loss-of-function allele. To determine the genetic requirement of drICE for normal developmental cell death, we analyzed drICE17 mutant embryos by acridine orange (AO), terminal deoxynucleotidyl transferase-mediated biotinylated UTP nick end labeling (TUNEL) and anticleaved Caspase-3 (referred to as Caspase-3*) labelings. AO stains specifically dying cells. Compared to wild-type embryos, AO-positive cell death is significantly reduced in drICE17 mutants (Figure 3a and b). Similar observations were made using TUNEL, an alternative method to label dying cells (Figure 3c and d). Thus, consistent with its postulated role as effector caspase, drICE is required for developmental cell death. However, even though cell death is reduced in drICE17 mutants, it is not completely blocked (Figure 3b and d). Because drICE17 is not a defined null allele, the incomplete block of developmental apoptosis could be due to partial activity of drICE17; or, drICE is functionally redundant with another effector caspase. One candidate for an alternative effector caspase is dcp-1 (see Song et al.23) which is most similar to drICE at the sequence level (57% identical).22 Deletion of dcp-1, using dcp-1Prev1 a protein null allele,7 does not or only weakly affect the global embryonic cell death pattern (Figure 3e), consistent with previous reports.7 However, dcp-1Prev1 drICE17 double-mutant embryos (see Materials and Methods) display significantly reduced levels of embryonic apoptosis compared to drICE17 single mutants, and contain only a few dying cells (Figure 3f and b). This residual apoptosis might be the result of weak activity of drICE17, or may imply that a third effector caspase (Damm or Decay) is required for embryonic cell death. In any case, this analysis establishes that drICE and dcp-1 have overlapping functions in the apoptotic pathway in Drosophila embryos (see Discussion).
Figure 3.
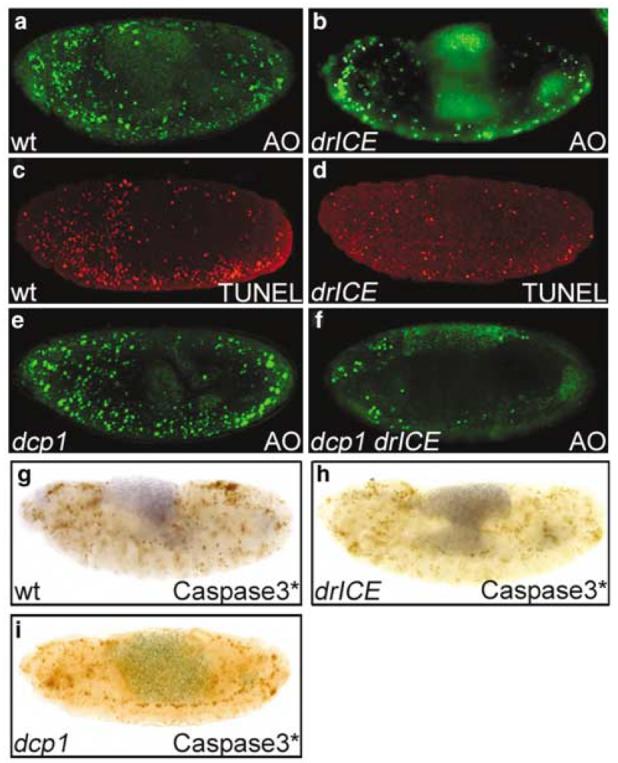
Acridine orange (AO), TUNEL and anticleaved caspase-3 labelings. (a) Wild-type embryo (stage 13) labeled with AO. (b) Homozygous drICE17 mutant embryo (stage 13) labeled with AO. This embryo was obtained from a cross of homozygous drICE17 males and females to remove the maternal contribution. (c) Wild-type embryo (stage 13) labeled with TUNEL. (d) Homozygous drICE17 mutant embryo (stage 13) labeled for TUNEL. This embryo was obtained from a cross of homozygous drICE17 males and females to remove the maternal contribution. (e) Homozygous dcp-1Prev1 mutant embryo (stage 13) labeled for AO. This embryo was obtained from a cross of homozygous males and females to remove the maternal contribution. (f) dcp-1Prev1drICE17 double-mutant embryo (stage 13) labeled for AO. This embryo was obtained by induction of germline clones as described in Materials and Methods. (g) Wild-type embryo (stage 13) labeled with anticleaved caspase-3 antibody. (h) Homozygous drICE17 embryo (stage 13) labeled with anticleaved caspase-3 (Caspase-3*) antibody. The labeling pattern is not appreciately altered compared to wild-type (g). (i) Homozygous dcp-1Prev1 mutant embryo (stage 13) labeled with Caspase-3* antibody. This embryo was obtained from a cross of homozygous males and females to remove the maternal contribution
The anticleaved Caspase-3 antibody (Caspase-3*; Cell Signaling Technology) was raised against a peptide N-terminal to Asp175 in the large subunit of human Caspase-3. It is thought that this antibody recognizes its epitope only after cleavage and activation of Caspase-3. In uncleaved Caspase-3 the epitope is buried inside the protein inaccessible to the antibody. Interestingly, immunolabeling of Drosophila embryos using this antibody mimics the known AO and TUNEL pattern (Figure 3g). Furthermore, labeling with this antibody is dependent on the RHG genes reaper, hid and grim, as homozygous embryos deficient for the RHG genes (Df(3L)H99) fail to give a staining signal (data not shown). diap1 mutant embryos which are characterized by a strong apoptotic phenotype3,24 (Figure 7a) show increased labeling (data not shown). These observations suggest that the Caspase-3* antibody crossreacts with a Drosophila protein that becomes activated during apoptotic cell death. Such an apoptotic protein might be the initiator caspase Dronc or an effector caspase acting downstream of Dronc, because the Caspase-3* antibody shows reduced immunoreactivity in dronc mutants.10 Another antibody, termed CM1, was raised against the same peptide and was shown to crossreact with cleaved DrICE in immunoblots.25 It is not clear whether Caspase-3* antibody also crossreacts with cleaved DrICE. To address this question, we analyzed drICE17 mutant embryos with Caspase-3* antibody. However, compared to wild type, the level of immunoreactivity is not significantly altered in drICE17 embryos (Figure 3h), and surprisingly does not match the AO- and TUNEL patterns of drICE17 mutants (Figure 3b and d). As we showed in Figure 2b, that drICE17 produces an unstable protein with <5% of the total amount of DrICE protein left, it is unlikely that the Caspase-3* antibody specifically recognizes cleaved DrICE. We also tested the Caspase-3* antibody on homozygous dcp-1Prev1 embryos which lack Dcp-1 protein.7 However, the immunolabeling is indistinguishable from that of wild-type embryos (Figure 3i). Thus, this antibody does not appear to bind to either DrICE or Dcp-1, or it may recognize multiple epitopes (see Discussion).
Figure 7.
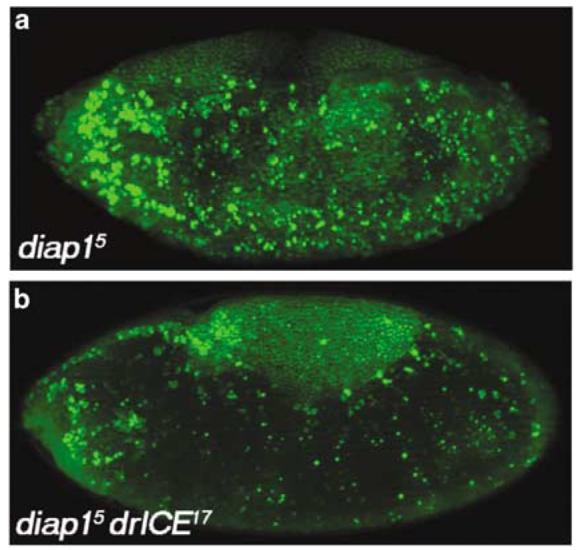
drICE17 suppresses the diap1 mutant apoptotic phenotype. (a) diap15 mutant embryos labeled with AO display a strong apoptotic phenotype. (b) diap15 drICE17 double-mutant embryos strongly suppress the diap1 phenotype
The data presented in Figure 3 provide strong evidence that drICE+ is genetically required for developmental cell death. We therefore determined the consequences of reduced rates of cell death at the cellular level in drICE17 mutants. The best-characterized apoptotic model during Drosophila embryogenesis is the development of the midline glia (MG) in the central nervous system.26 The MG are transient cells during embryogenesis and are required for the separation and ensheathment of commissural axon tracts.26 At stage 13 of embryogenesis, about 10 MG cells per segment have been generated. Subsequent to the establishment of commissure morphology, a subset of the MG cells undergo apoptosis, leaving about three ensheathing MG cells per segment by the end of embryogenesis at stage 17 (Figure 4a and c).26 The reduction in the number of MG cells is dependent on reaper, hid, grim, ark and dronc. In homozygous H99 (deleting reaper, hid and grim),27 ark28 and dronc8,10 mutant embryos, the MG cells fail to die by apoptosis. We determined the fate of MG cells in drICE17 mutants. At stage 17, drICE17 mutant embryos contain on average more than twice the number of MG cells (∼80–100; n = 5) compared to wild-type (∼40; n = 5) (Figure 4b and d). This number is similar to hid mutant embryos,27 but is significantly less compared to H99 and dronc mutant embryos which contain on average 140–160 MG cells.10,27 Thus, drICE is required for some, but not all, MG cell death.
Figure 4.
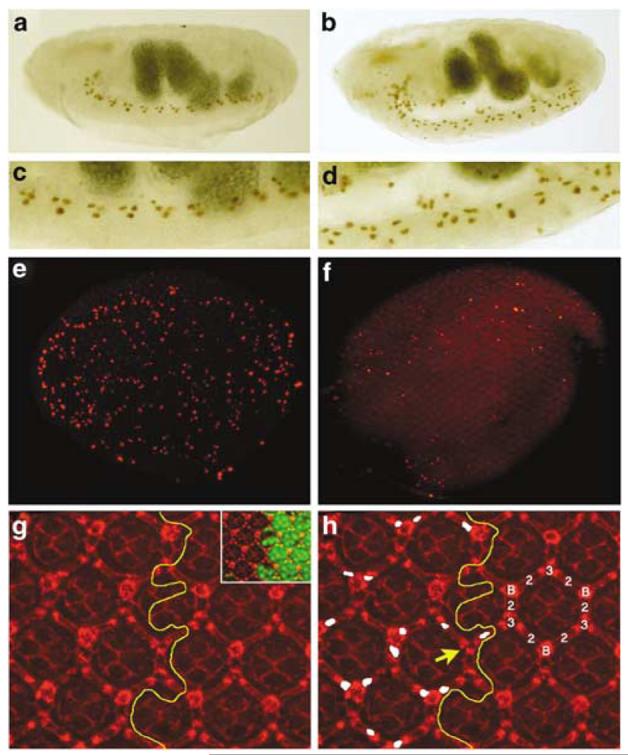
drICE17 mutants contain additional cells. (a) The midline glia (MG) of a stage 17 wild-type embryos visualized by a P[sli-1.0]lacZ reporter transgene. (b) The MG of stage 17 drICE17 embryos contains additional cells. (c) Enlargement of the ventral nerve cord of the wild-type embryo in (a). (d) Enlargement of the ventral nerve cord of the drICE17 mutant in (b). (e) Whole mount of a wild-type pupal eye disc 26 h APF labeled for TUNEL. (f) Whole mount of a drICE17 mutant eye disc 26 h APF labeled for TUNEL. The global cell death pattern is reduced compared to (e). (g and h) IOC survival in drICE17 mutant clones. (g) Overview of a Dlg-labeled (red) drICE17 mosaic eye disc 42 h APF to visualize the weak disorganization of the mutant lattice. The yellow line marks the clonal boundary. The inset shows the drICE17 clone marked by absence of GFP (green). (h) Shows the same field as (g). The interommatidial cell (IOC) cluster is composed of six secondary (2), three tertiary (3) and three bristle cells (B). The yellow line marks the clonal boundary. Extra IOCs in the drICE17 clone are marked in white. The yellow arrow points to a rare patterning defect in which a bristle cell is replaced by a tertiary pigment cell
Another very well-characterized cellular model system to study developmental cell death in Drosophila is the fate of interommatidial cells (IOCs) in the developing compound eye which is composed of approximately 750 individual unit eyes, called ommatidia.29 Individual ommatidia are positioned within a hexagonal lattice of shared pigment cells and mechanosensory bristles.29 Formation of the ommatidial core, composed of eight photoreceptors, four cone cells and two primary pigment cells, is completed 24 h after pupariation formation (APF), leaving an excess of undifferentiated cells in the interommatidial space.29 The final step of ommatidial development is the differentiation of the IOCs into secondary (2°) and tertiary (3°) pigment cells (Figure 4h), and the elimination of excess IOCs by hid-dependent cell death in order to refine the hexagonal pigment cell lattice.25,29,30 By 42 h APF, IOC differentiation and cell death is completed bringing individual ommatidia into register within the lattice.
IOC apoptosis throughout the eye disc is highest between 26 and 28 h APF29 (Figure 4e). Thus, we tested whether drICE17 mutants change the global pattern of cell death by TUNEL labeling. Similar to drICE17 mutant embryos, the global pattern of TUNEL-positive cell death is significantly reduced, but not completely blocked (Figure 4f). Although we have been unable to test whether dcp-1 accounts for the remaining death in drICE17 discs, it is likely that this is the case based on our findings in embryos.
We also determined the number of IOCs in drICE17 mutant clones in discs 42 h APF. Using an antibody against the Discs-large (Dlg) protein to visualize membranes and thus cell outline, we found that ommatidia in drICE17 mutant clones 42 h APF contained on average 1.6±0.75 S.E.M. additional IOCs (Figure 4g and h; marked in white; see Materials and Methods) which is approximately half the number of additional IOCs reported for dronc mutant ommatidia.10 Thus, this analysis provides further support for a redundant function of effector caspases. Nevertheless, these data suggest that drICE+ is genetically required for some IOC apoptosis in the developing retina. Interestingly, occasionally we also observe a patterning defect in which a bristle cell is replaced by a 3° pigment cell (Figure 4h, yellow arrow).
drICE17 partially protects against irradiation-induced cell death
Ionizing radiation induces apoptosis in both mammalian cells and in Drosophila embryos. Radiation-induced apoptosis requires the RHG genes, ark, and dronc. Thus, we determined whether drICE is required for irradiation-induced cell death. Wild-type and drICE17 mutant embryos were irradiated with 4000 rad. In wild-type embryos this treatment induces a strong apoptotic response (Figure 5a). However, compared to irradiated wild-type embryos, fewer apoptotic cells were consistently observed in drICE17 mutants (Figure 5b). Therefore, drICE17 partially protects against radiation-induced cell death.
Figure 5.
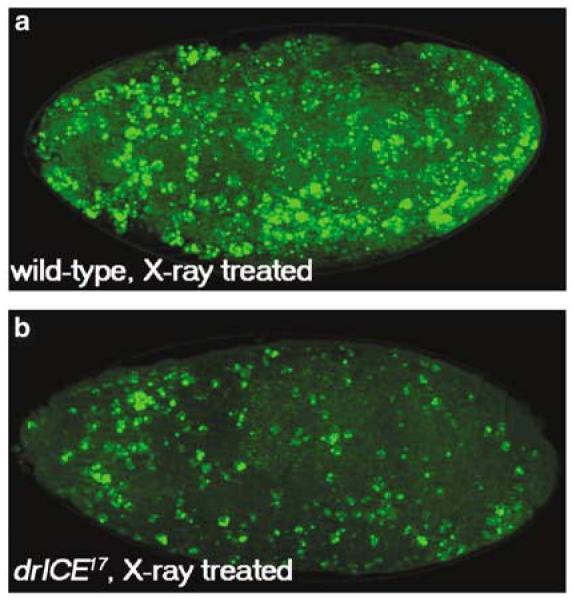
drICE17 partially protects against irradiation-induced cell death. Wild-type (a) and drICE17 (b) were exposed to X-ray irradiation (4000 rad), aged for 1.5 h and labeled with acridine orange. These embryos were X-ray treated at stage 9
drICE17 fails to suppress GMR-dronc- and GMR-dcp-1-induced eye phenotypes
Based on its domain structure, DrICE has been classified as an effector caspase.6,13 Consistent with this notion is the observation that the initiator caspase Dronc cleaves and activates DrICE in vitro.18,19 However, DrICE can also cleave Dronc in vitro,15 and it was proposed that DrICE is required for amplification of Dronc processing in a caspase activation loop following the initial activation of Dronc.15,16 We attempted to determine the genetic relationship between dronc and drICE by epistasis analysis. Expression of the full-length form of Dronc in third instar larval eye discs using the GMR enhancer (GMR-pro-dronc) causes induction of apoptosis18,19,31 (Figure 6a). To determine the genetic relationship between dronc and drICE, we analyzed the apoptotic phenotype of GMR-pro-dronc in a drICE17 mutant background. Surprisingly, in homozygous drICE17 animals the apoptotic phenotype of GMR-pro-dronc in larval eye discs is not suppressed (Figure 6b). Similarly, the strong apoptotic phenotype caused by GMR-induced expression of a dominant active allele of Dronc which deletes the prodomain (GMR-ΔN-dronc)18 is not significantly suppressed by drICE17 (Figure 6c and d). Consistently, the adult eye phenotype of GMR-pro-dronc and GMR-ΔN-dronc are not significantly rescued by drICE17 (data not shown). These observations suggest that overexpressed Dronc can induce apoptosis independently of drICE either through activation of other effector caspases, or it behaves as an effector caspase itself (see Discussion).
Figure 6.
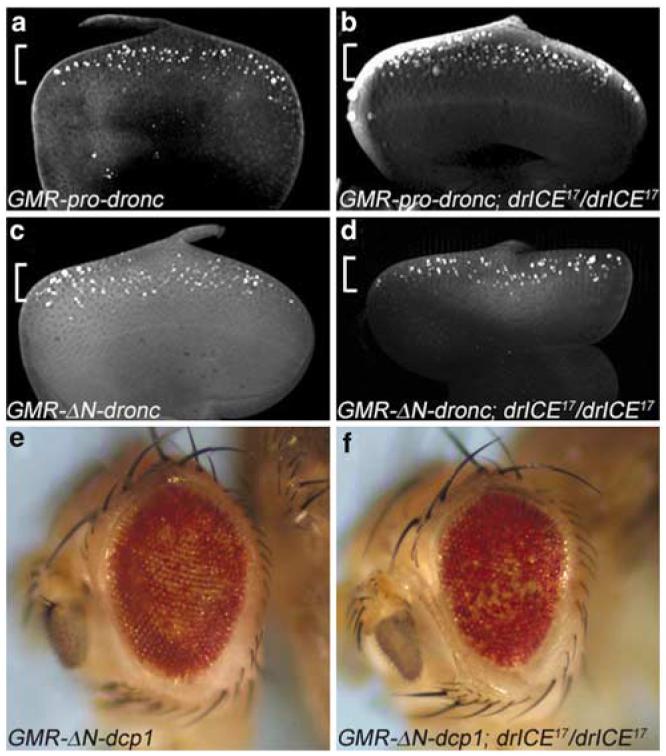
drICE17 fails to suppress GMR-dronc- and GMR-dcp-1-induced eye phenotypes. (a) Third instar GMR-pro-Dronc larval eye disc labeled by TUNEL. The brackets in a–d indicate the expression domain of the GMR-dronc transgenes, overlapping with TUNEL-positive apoptosis. (b) Third instar GMR-pro-dronc larval eye disc mutant for drICE17 labeled by TUNEL. There is no significant difference compared to (a). (c) Third instar GMR-ΔN-dronc larval eye disc labeled by TUNEL. (d) Third instar GMR-ΔN-dronc larval eye disc mutant for drICE17 labeled by TUNEL. There is no significant difference compared to (c). (e) The ‘spotted’ eye phenotype caused by GMR-ΔN-dcp-1. (f) Homozygous drICE17 flies do not suppress GMR-ΔN-dcp-1
Dcp-1 is a candidate effector caspase activated by Dronc in parallel to DrICE.7,22,23 GMR-ΔN-dcp-1 which lacks the N-terminal prodomain gives rise to a ‘spotted’ eye phenotype22 (Figure 6e). As expected, drICE17 mutants are unable to suppress this eye phenotype (Figure 6f), implying that dcp-1 either acts downstream of drICE, or in parallel. Thus, this observation suggests that dcp-1 can induce cell death independently of drICE.
drICE17 partially suppresses the apoptotic phenotype of diap1 mutants
IAPs, most notably Diap1, are important regulators of apoptosis.1,2 The diap15 allele is a strong loss-of-function allele characterized by a dramatic apoptotic phenotype. Essentially every cell is TUNEL positive in these embryos24 (Figure 7a) suggesting an essential function of Diap1 for cellular survival. There is overwhelming biochemical evidence that IAPs regulate apoptosis through inhibition of caspases.1,2,5,18,21 Diap1 has been shown to be able to directly bind to and inhibit DrICE in vitro.4,5,20,21 However, the functional significance of this interaction has never been genetically demonstrated. Thus, we determined whether Diap1 inhibits DrICE in vivo by double-mutant analysis. diap15 drICE17 double-mutant embryos contain fewer apoptotic cells compared to diap15 single-mutant embryos (Figure 7b). This finding suggests that Diap1 indeed regulates the apoptotic activity of DrICE. Furthermore, this analysis places drICE genetically downstream of diap1, consistent with the expectation. However, we note that the diap1 mutant phenotype is only partially suppressed by drICE17. The partial suppression can be explained either by the hypomorphic nature of drICE17 or by the activity of other effector caspases such as Dcp-1 which is also target of Diap1 inhibition.
Discussion
The importance of caspases for programmed cell death was first revealed in genetic studies in Caenorhabditis elegans,32 and later confirmed by targeted gene disruptions in mice.33-35 In Drosophila, the first report implicating caspases as important mediators of programmed cell death took advantage of the universal caspase inhibitor P35. In P35-overexpressing animals, cell death is significantly reduced. More recently, dominant-negative constructs of cloned caspases and RNAi experiments further supported the involvement of caspases in the cell death response in Drosophila.18,31 Finally, the availability of mutations in the initiator caspase dronc confirmed an essential role of caspases for developmental cell death.8-11 Here, we report the isolation and characterization of a mutant in the effector caspase drICE. The phenotypic characterization of this mutant is consistent with a role of drICE for developmental and irradiation-induced cell death.
The su(GMR-hid)17 mutant was isolated as a strong recessive suppressor of GMR-hid by GheF screening. We demonstrated in four ways that su(GMR-hid)17 corresponds to a mutation in drICE. First, su(GMR-hid)17 in trans to the deficiency Df(3R)drICE, which deletes 99B3/B8–99C2/C4 including drICE, strongly suppressed GMR-hid (Figure 1d) suggesting that su(GMR-hid)17 maps to this cytological range. Second, expression of a GMR-drICE transgene restored the eye-ablation phenotype of GMR-hid in homozygous su(GMR-hid)17 mutants (Figure 1f). Third, by DNA sequencing we identified a missense mutation in the drICE gene of su(GMR-hid)17 changing the conserved Asn116 to Tyr. Finally, immunoblot analysis using an anti-DrICE antibody suggests that su(GMR-hid)17 encodes for an unstable DrICE protein. Thus, these observations suggest that su(GMR-hid)17 carries a mutation in drICE, and we referred to this mutant as drICE17.
drICE17 carries a missense mutation in a conserved residue, changing Asn116 to Tyr, and thus, is not a defined null mutant. However, we have reasons to believe that drICE17 is a very strong hypomorphic allele. It is a strong suppressor of GMR-hid and produces <5% of the wild-type levels of DrICE protein. However, drICE17/Df(3R)drICE mutants suppress the GMR-hid eye-ablation phenotype slightly better than homozygous drICE17 animals (Figure 1d and e) suggesting that drICE17 is a very strong, but not a null allele.
Partial redundancy between drICE and dcp-1
The phenotypic analysis of drICE17 in embryos and imaginal eye discs establishes that drICE+ functions in developmental cell death. It is also partially required for irradiation-induced cell death and for establishment of the diap1 mutant phenotype. However, the cell death phenotypes observed for drICE17 are weaker compared to the phenotypes reported for mutations of the initiator caspase dronc (see Daish et al.,8 Chew et al.,9 Xu et al.10 and Waldhuber et al.11) suggesting that at least one additional effector caspase is required for apoptosis in Drosophila. One potential effector caspase which can compensate for the loss of drICE may be dcp-1.23 Consistently, we showed by double-mutant analysis with dcp-1, that drICE and dcp-1 have overlapping functions in the apoptotic pathway. The double-mutant phenotype is similar, although slightly weaker compared to the dronc null phenotype. 10 This slightly weaker phenotype could be caused by the hypomorphic nature of drICE17, or alternatively by a third effector caspase such as Damm or Decay which may also be activated by Dronc.6 In any case, this study demonstrates that effector caspases in Drosophila have overlapping functions.
However, it is interesting to note that dcp-1 mutants display an apoptotic phenotype only in a double mutant with drICE (Figure 3f). In contrast, the drICE mutant has an apoptotic phenotype on its own. These observations suggest that Dcp-1 is not sufficient to induce apoptosis in those cells which survive in drICE17 mutants, but would otherwise die in wild-type embryos, implying that some cells strictly require DrICE for apoptosis independently of Dcp-1 (we refer to these cells as type I cells), and these cells survive in drICE mutants, whereas other cells (type II) require either DrICE or Dcp-1, and these cells still die in either drICE or dcp-1 mutants, but survive in the double mutant. This model also explains why drICE17 mutants fail to suppress the GMR-ΔN-dcp-1 eye phenotype. One example of a class I cell type are S2 cells. Immunodepletion of DrICE and gene silencing by RNAi results in block of apoptosis14,15 suggesting that DrICE is the only effector caspase required for S2 apoptosis. How this partial redundancy is regulated and why type I cells can tolerate Dcp-1 is unclear. However, it reveals an unanticipated complexity in the apoptotic pathway in Drosophila. Similarly, analysis of genetic knockouts in mouse has revealed that Caspase-3 is essential in some cell types for apoptosis, but not in others, and that additional effector caspases can compensate for the loss of Caspase-3.36
Interestingly, labeling of drICE17 mutant embryos with cleaved caspase-3 (Caspase-3*) antibody produced a wild-type pattern. This is puzzling because the Caspase-3* pattern does not match the AO- and TUNEL patterns of drICE17 mutants (Figure 3). This observation suggests that Caspase-3* recognizes an epitope which is produced upstream of drICE such as the initiator caspase Dronc. We have previously shown that the immunoreactivity of the Caspase-3* antibody is strongly reduced in dronc mutants.10 This observation implies, but does not prove, that the Caspase-3* antibody recognizes cleaved Dronc. However, it is also possible that this antibody recognizes multiple apoptotic proteins including Dronc, effector caspases and even caspase substrates.
The genetic relationship between diap1, dronc and drICE
Mutations in diap1 cause a dramatic apoptotic phenotype in early embryos.3,24 We showed that drICE17 partially suppresses the apoptotic phenotype of diap1 mutants suggesting that DrICE is regulated by Diap1 and acts genetically downstream of Diap1. However, it is unclear from this genetic analysis whether Diap1 directly inhibits DrICE, or whether the diap1 mutant phenotype is caused by loss of inhibition of Dronc, which then activates DrICE. The available biochemical evidence suggests a combination of both. Binding studies in vitro have shown that Diap1 can be a negative regulator of DrICE.4,5,20,21 However, Diap1 cannot inhibit DrICE until DrICE becomes activated and proteolytically removes the 20 N-terminal residues of Diap1,5,37,38 which constitute an autoinhibitory domain for the function of Diap1.5,37 These findings suggest that Dronc has to cleave and activate DrICE first before Diap1 can inhibit it. Dronc is also target of negative regulation by Diap1,18 and we have previously shown that dronc suppresses the diap1 mutant phenotype in the ovary, placing dronc downstream of diap1.10 Thus, consistent with a previously proposed model,37 Diap1 appears to inhibit primarily Dronc, whereas the inhibition of activated DrICE constitutes a minor activity of Diap1, which might be necessary to protect the cell against weak apoptotic signals or against inappropriately activated DrICE. Nevertheless, consistent with the role of DrICE as effector caspase, this analysis establishes that drICE acts genetically downstream of diap1.
The genetic relationship between dronc and drICE is less clear. drICE17 is unable to suppress apoptosis induced by GMR-pro-dronc and GMR-ΔN-dronc. This observation can be explained in several ways. First, dronc and drICE might act in independent pathways. However, this possibility is unlikely as both dronc and drICE mutants suppress GMR-hid, suggesting that they indeed do act in the same pathway. Second, Dronc might activate several effector caspases. Our data suggest that at least in embryos Dcp-1 is an alternative effector caspase which under the unphysiologically high concentration of pro-Dronc and ΔN-Dronc may be sufficiently active to compensate for the loss of drICE. Consistent with this scenario is our observation that GMR-ΔN-dcp-1 is unaffected by drICE17 suggesting that dcp-1 acts in parallel or downstream of drICE.
However, there is another possibility. The GMR-pro-dronc and GMR-ΔN-dronc eye phenotypes are insensitive to expression of the caspase inhibitor P35.18,19 In contrast, Dcp-1 and DrICE can be inhibited by P35.22 Thus, these observations suggest that the GMR-dronc-induced eye phenotypes are independent of Dcp-1 and DrICE. It is unclear how GMR-pro-dronc and GMR-ΔN-dronc induce apoptosis independently of Dcp-1 and DrICE. It is possible that the dronc transgenes induce the activation of another P35-insensitive effector caspase, or that overexpressed Dronc can also act as effector caspase. More experiments are needed to clarify these observations.
In summary, we have isolated a strong loss-of-function mutant in drICE. The phenotypic analysis is consistent with a role of drICE as an effector caspase. Our data establish that drICE and dcp-1 function redundantly in some cells, whereas other cells strictly require drICE for apoptosis. Future studies will reveal how specificity is conferred in these paradigms.
Materials and Methods
Isolation and identification of drICE17
ey-Flp; FRT82B males were starved for 12 h, followed by treatment with 25 mm EMS in 5% sucrose solution for 24 h. After recovery for 3 h, the mutagenized males were mated to GheF; FRT82B w+ females and incubated at 25°C. 40 000 F1 progeny were screened for suppression of the GMR-hid-induced small eye phenotype. The strongest suppressor, su(GMR-hid)17, was selected for further analysis. su(GMR-hid)17 was identified as a drICE allele by genetic tests described in the Results section and by DNA sequencing.
X-ray mutagenesis
Df(3R)drICE was obtained in the following manner: males carrying the P-element l(3)05884 inserted in 99C1–2 were treated with X-ray, crossed to TM2,ry/TM6B,ry females, and F1 progeny was screened for loss of the eye color marker (ry+) of l(3)05884. l(3)05884 is a P-element insertion in the ncd gene, approximately 6 kb distal from the drICE locus. Loss of the eye color marker indicates that the P-element along with flanking genomic sequences has been deleted from the genome. Loss of the P-element was confirmed by PCR analysis. Df(3R)drICE has a proximal breakpoint between 99B3 and 99B8, because it complements Dr at 99B3, and fails to complement ca at 99B8. Its distal breakpoint lies between 99C2 (because it lacks the original P-element) and 99C4 (because it complements the lethality of CG18041EY04131 which maps to 99C4).
Fly stocks and genetics
The following mutant and transgenic fly stocks were used: drICE17 and Df(3R)drICE (this study); dcp-1Prev1 (see Laundrie et al.7); diap15 (see Lisi et al.24); UAS-pro-dronc and UAS-ΔN-dronc;18 GMR-ΔN-dcp-1 and GMR-drICE;22 GheF.10 The wild-type stock used for comparison was the ey-Flp; FRT82B stock used for the mutagenesis.
The following stocks were obtained by meiotic recombination:
GMR-hid drICE17
GMR-drICE drICE17
GMR-ΔN-dcp-1 drICE17
GMR-Gal4 drICE17
UAS-pro-dronc drICE17
GMR-Gal4 UAS-ΔN-dronc drICE17
th5drICE17
For embryonic analysis, homozygous drICE17 males and females (unless otherwise noted) were crossed with each other to remove maternal and zygotic drICE. Double-mutant diap15 drICE17 embryos were obtained by crossing males and females of diap15 drICE17/drICE17 genotypes. All embryos in these collections are phenotypically similar to drICE17 single mutants, suggesting that the diap1 mutant phenotype is effectively suppressed.
Generation of dcp-1 drICE double-mutant embryos in germline clones (GLCs): dcp-1 drICE double mutants are almost completely lethal. Only four double homozygous adult flies were recovered in <1000 control flies. Such a small number of flies is impractical for embryonic analysis. However, homozygous dcp-1Prev1 flies in a heterozygous drICE17 mutant background are viable which enabled us to remove the maternal contribution of both dcp-1 (see Song et al.23) and drICE (see Fraser and Evan13) by GLC analysis39 to increase the number of double homozygous embryos lacking both maternal and zygotic dcp-1 and drICE. Double-mutant GLC were obtained by crossing females of genotype hs-FLP; dcp-1Prev1/dcp-1Prev1; FRT82B drICE17/FRT82B P[ovoD] with males of genotype dcp-1Prev1/dcp-1Prev1; drICE17/TM6B,lacZ. To induce GLCs, first instar larvae were heat shocked at 37°C for one hour.
To visualize the MG, males of the genotype P[sli-1.0-lacZ]; drICE17/TM6B, ubx-lacZ were crossed to drICE17/drICE17 females, and labeled by β-Gal immunohistochemistry.
For Dlg labelings, drICE17 mosaic pupal eye discs 42 h after pupariation were dissected and labeled with anti-Dlg antibody and GFP to mark the clones. Cell counting was done using criteria established by Cordero et al.29 Ten hexagons corresponding to 20 ommatidia from four individuals each were analyzed.
Fly crosses were carried out under standard conditions at 25°C.
X-ray treatment of embryos
Embryos were treated with 4000 rad in a Nasatron X-ray machine with a Caesium137 source. After recovery the embryos were fixed and prepared for AO labeling.
Immunohistochemistry
TUNEL, AO and immunohistochemistry were carried out as described.40 Anticleaved Caspase-3 antibody (Cell Signaling Technology) was used at a dilution of 1 : 50, β-Gal antibody (Promega) at dilution of 1 : 500, and anti-Dlg antibody (a kind gift of G Halder) at 1 : 2000. The MG was visualized by β-Gal immunohistochemistry. Fluorescent photography was carried out using a Zeiss Axio Imaginer Z1 with ApoTome technology.
Immunoblotting
Embryos were collected, decorionated and snap frozen in liquid nitrogen. Embryos were sonicated in Laemmli SDS loading buffer while being frozen. The equivalent of 20 lysed embryos was loaded per lane. Immunoblots were carried out using standard procedures and were probed with anti-DrICE antibodies (diluted 1 : 1000) raised against the prodomain of DrICE (provided by Andy Fraser). This antibody recognizes only the full-length form of DrICE.
Acknowledgements
We apologize to all our colleagues whose work could not be cited due to space constraints. We thank Masayuki Miura and Bruce Hay for sharing unpublished information; Marvette Hobbs for use of the Nasatron X-ray machine; Kim McCall, Pascal Meier, Hermann Steller, Kristin White, Georg Halder, Andy Fraser and the Bloomington stock center for fly stocks and reagents; Pascal Meier for stimulating discussions; the MD Anderson DNA Analysis Core Facility for sequencing (supported by Core Grant #CA16672 from the National Cancer Institute); and Mary Ellen Lane and Pierrette Lo for critical discussions about the project and the manuscript. AB is a fellow of the MD Anderson Research Trust. This work was supported by grants from the NIH (GM068016) and the Robert A Welch Foundation (G1496) to AB.
Abbreviations
- AO
acridine orange
- APF
after pupariation formation
- Ark
Apaf-1-related killer
- CARD
caspase activation and recruitment domain
- Dcp-1
death caspase-1
- Diap1
Drosophila inhibitor of apoptosis protein 1
- DrICE
Drosophila ICE
- Dronc
Drosophila Nedd-2-like caspase
- ey
eyeless
- FLP
Flippase
- FRT
Flippase recombination target
- GheF
GMR-hid ey-Flp
- GMR
glassmultimer reporter
- hid
head involution defective
- IAP
inhibitor of apoptosis proteins
- MG
midline glia
- RHG
Reaper Hid Grim
- RNAi
RNA interference
- su
suppressor
- TUNEL
terminal deoxynucleotidyl transferase-mediated biotinylated UTP nick end labeling
References
- 1.Salvesen GS, Abrams JM. Caspase activation – stepping on the gas or releasing the brakes? Lessons from humans and flies. Oncogene. 2004;23:2774–2784. doi: 10.1038/sj.onc.1207522. [DOI] [PubMed] [Google Scholar]
- 2.Vaux DL, Silke J. IAPs, RINGs and ubiquitylation. Nat. Rev. Mol. Cell. Biol. 2005;6:287–297. doi: 10.1038/nrm1621. [DOI] [PubMed] [Google Scholar]
- 3.Cashio P, Lee TV, Bergmann A. Genetic control of programmed cell death in Drosophila melanogaster. Semin. Cell Dev. Biol. 2005;16:225–235. doi: 10.1016/j.semcdb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Zachariou A, Tenev T, Goyal L, Agapite J, Steller H, Meier P. IAP-antagonists exhibit non-redundant modes of action through differential DIAP1 binding. EMBO J. 2003;22:6642–6652. doi: 10.1093/emboj/cdg617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan N, Wu JW, Chai J, Li W, Shi Y. Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim. Nat. Struct. Mol. Biol. 2004;11:420–428. doi: 10.1038/nsmb764. [DOI] [PubMed] [Google Scholar]
- 6.Kumar S, Doumanis J. The fly caspases. Cell Death Differ. 2000;7:1039–1044. doi: 10.1038/sj.cdd.4400756. [DOI] [PubMed] [Google Scholar]
- 7.Laundrie B, Peterson JS, Baum JS, Chang JC, Fileppo D, Thompson SR, McCall K. Germline cell death is inhibited by P-element insertions disrupting the dcp-1/pita nested gene pair in Drosophila. Genetics. 2003;165:1881–1888. doi: 10.1093/genetics/165.4.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daish TJ, Mills K, Kumar S. Drosophila caspase DRONC is required for specific developmental cell death pathways and stress-induced apoptosis. Dev. Cell. 2004;7:909–915. doi: 10.1016/j.devcel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 9.Chew SK, Akdemir F, Chen P, Lu WJ, Mills K, Daish T, Kumar S, Rodriguez A, Abrams JM. The apical caspase dronc governs programmed and unprogrammed cell death in Drosophila. Dev. Cell. 2004;7:897–907. doi: 10.1016/j.devcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 10.Xu D, Li Y, Arcaro M, Lackey M, Bergmann A. The CARD-carrying caspase Dronc is essential for most, but not all, developmental cell death in Drosophila. Development. 2005;132:2125–2134. doi: 10.1242/dev.01790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waldhuber M, Emoto K, Petritsch C. The Drosophila caspase DRONC is required for metamorphosis and cell death in response to irradiation and developmental signals. Mech. Dev. 2005;122:914–927. doi: 10.1016/j.mod.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Dorstyn L, Colussi PA, Quinn LM, Richardson H, Kumar S. DRONC, an ecdysone-inducible Drosophila caspase. Proc. Natl. Acad. Sci. U.S.A. 1999;96:4307–4312. doi: 10.1073/pnas.96.8.4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fraser AG, Evan GI. Identification of a Drosophila melanogaster ICE/ CED-3-related protease, drICE. EMBO J. 1997;16:2805–2813. doi: 10.1093/emboj/16.10.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fraser AG, McCarthy NJ, Evan GI. drICE is an essential caspase required for apoptotic activity in Drosophila cells. EMBO J. 1997;16:6192–6199. doi: 10.1093/emboj/16.20.6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muro I, Monser K, Clem RJ. Mechanism of Dronc activation in Drosophila cells. J. Cell Sci. 2004;117:5035–5041. doi: 10.1242/jcs.01376. [DOI] [PubMed] [Google Scholar]
- 16.Kilpatrick ZE, Cakouros D, Kumar S. Ecdysone-mediated up-regulation of the effector caspase DRICE is required for hormone-dependent apoptosis in Drosophila cells. J. Biol. Chem. 2005;280:11981–11986. doi: 10.1074/jbc.M413971200. [DOI] [PubMed] [Google Scholar]
- 17.Peterson JS, Barkett M, McCall K. Stage-specific regulation of caspase activity in Drosophila oogenesis. Dev. Biol. 2003;260:113–123. doi: 10.1016/s0012-1606(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 18.Meier P, Silke J, Leevers SJ, Evan GI. The Drosophila caspase DRONC is regulated by DIAP1. EMBO J. 2000;19:598–611. doi: 10.1093/emboj/19.4.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hawkins CJ, Yoo SJ, Peterson EP, Wang SL, Vernooy SY, Hay BA. The Drosophila caspase DRONC cleaves following glutamate or aspartate and is regulated by DIAP1, HID, and GRIM. J. Biol. Chem. 2000;275:27084–27093. doi: 10.1074/jbc.M000869200. [DOI] [PubMed] [Google Scholar]
- 20.Kaiser WJ, Vucic D, Miller LK. The Drosophila inhibitor of apoptosis D-IAP1 suppresses cell death induced by the caspase drICE. FEBS Lett. 1998;440:243–248. doi: 10.1016/s0014-5793(98)01465-3. [DOI] [PubMed] [Google Scholar]
- 21.Tenev T, Zachariou A, Wilson R, Ditzel M, Meier P. IAPs are functionally non-equivalent and regulate effector caspases through distinct mechanisms. Nat. Cell Biol. 2005;7:70–77. doi: 10.1038/ncb1204. [DOI] [PubMed] [Google Scholar]
- 22.Song Z, Guan B, Bergman A, Nicholson DW, Thornberry NA, Peterson EP, Steller H. Biochemical and genetic interactions between Drosophila caspases and the proapoptotic genes rpr, hid, and grim. Mol. Cell. Biol. 2000;20:2907–2914. doi: 10.1128/mcb.20.8.2907-2914.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song Z, McCall K, Steller H. DCP-1, a Drosophila cell death protease essential for development. Science. 1997;275:536–540. doi: 10.1126/science.275.5299.536. [DOI] [PubMed] [Google Scholar]
- 24.Lisi S, Mazzon I, White K. Diverse domains of THREAD/DIAP1 are required to inhibit apoptosis induced by REAPER and HID in Drosophila. Genetics. 2000;154:669–678. doi: 10.1093/genetics/154.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu SY, Yoo SJ, Yang L, Zapata C, Srinivasan A, Hay BA, Baker NE. A pathway of signals regulating effector and initiator caspases in the developing Drosophila eye. Development. 2002;129:3269–3278. doi: 10.1242/dev.129.13.3269. [DOI] [PubMed] [Google Scholar]
- 26.Klambt C, Jacobs JR, Goodman CS. The midline of the Drosophila central nervous system: a model for the genetic analysis of cell fate, cell migration, and growth cone guidance. Cell. 1991;64:801–815. doi: 10.1016/0092-8674(91)90509-w. [DOI] [PubMed] [Google Scholar]
- 27.Zhou L, Schnitzler A, Agapite J, Schwartz LM, Steller H, Nambu JR. Cooperative functions of the reaper and head involution defective genes in the programmed cell death of Drosophila central nervous system midline cells. Proc. Natl. Acad. Sci. U.S.A. 1997;94:5131–5136. doi: 10.1073/pnas.94.10.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez A, Chen P, Oliver H, Abrams JM. Unrestrained caspase-dependent cell death caused by loss of Diap1 function requires the Drosophila Apaf-1 homolog, Dark. EMBO J. 2002;21:2189–2197. doi: 10.1093/emboj/21.9.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cordero J, Jassim O, Bao S, Cagan R. A role for wingless in an early pupal cell death event that contributes to patterning the Drosophila eye. Mech. Dev. 2004;121:1523–1530. doi: 10.1016/j.mod.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 30.Wolff T, Ready DF. Cell death in normal and rough eye mutants of Drosophila. Development. 1991;113:825–839. doi: 10.1242/dev.113.3.825. [DOI] [PubMed] [Google Scholar]
- 31.Quinn LM, Dorstyn L, Mills K, Colussi PA, Chen P, Coombe M, Abrams J, Kumar S, Richardson H. An essential role for the caspase dronc in developmentally programmed cell death in Drosophila. J. Biol. Chem. 2000;275:40416–40424. doi: 10.1074/jbc.M002935200. [DOI] [PubMed] [Google Scholar]
- 32.Yuan J, Shaham S, Ledoux S, Ellis HM, Horvitz HR. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 beta-converting enzyme. Cell. 1993;75:641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- 33.Woo M, Hakem R, Soengas MS, Duncan GS, Shahinian A, Kagi D, Hakem A, McCurrach M, Khoo W, Kaufman SA, Senaldi G, Howard T, Lowe SW, Mak TW. Essential contribution of caspase 3/CPP32 to apoptosis and its associated nuclear changes. Genes Dev. 1998;12:806–819. doi: 10.1101/gad.12.6.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuida K, Haydar TF, Kuan CY, Gu Y, Taya C, Karasuyama H, Su MS, Rakic P, Flavell RA. Reduced apoptosis and cytochrome c-mediated caspase activation in mice lacking caspase 9. Cell. 1998;94:325–337. doi: 10.1016/s0092-8674(00)81476-2. [DOI] [PubMed] [Google Scholar]
- 35.Kuida K, Zheng TS, Na S, Kuan C, Yang D, Karasuyama H, Rakic P, Flavell RA. Decreased apoptosis in the brain and premature lethality in CPP32-deficient mice. Nature. 1996;384:368–372. doi: 10.1038/384368a0. [DOI] [PubMed] [Google Scholar]
- 36.Zheng TS, Hunot S, Kuida K, Momoi T, Srinivasan A, Nicholson DW, Lazebnik Y, Flavell RA. Deficiency in caspase-9 or caspase-3 induces compensatory caspase activation. Nat. Med. 2000;6:1241–1247. doi: 10.1038/81343. [DOI] [PubMed] [Google Scholar]
- 37.Yokokura T, Dresnek D, Huseinovic N, Lisi S, Abdelwahid E, Bangs P, White K. Dissection of DIAP1 functional domains via a mutant replacement strategy. J. Biol. Chem. 2004;279:52603–52612. doi: 10.1074/jbc.M409691200. [DOI] [PubMed] [Google Scholar]
- 38.Ditzel M, Wilson R, Tenev T, Zachariou A, Paul A, Deas E, Meier P. Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nat. Cell Biol. 2003;5:467–473. doi: 10.1038/ncb984. [DOI] [PubMed] [Google Scholar]
- 39.Chou TB, Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 1996;144:1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCall K, Peterson JS. Detection of apoptosis in Drosophila. Methods Mol. Biol. 2004;282:191–205. doi: 10.1385/1-59259-812-9:191. [DOI] [PubMed] [Google Scholar]