Abstract
Although much has been learned about short noncoding RNAs, long noncoding transcripts are largely uncharacterized. Here, we describe Caenorhabditis elegans rncs-1, a highly base-paired, 800-nucleotide noncoding RNA expressed in hypodermis and intestine. Transcription of rncs-1 is modulated in response to food supply. Although highly double-stranded, we show that rncs-1 RNA is not a substrate for Dicer because of branched structures at its termini. However, rncs-1 RNA inhibits Dicer cleavage of a second dsRNA in vitro, presumably by competition. We validate this observation in vivo by demonstrating that mRNA levels of several Dicer-regulated genes vary with changes in rncs-1 expression. Certain viruses express dsRNA to compete with cellular dsRNA-mediated pathways, and our data suggest that rncs-1 provides a cellular correlate of this phenomenon.
Keywords: adenosine deaminase that acts on RNA (ADAR), Caenorhabditis elegans, double-stranded RNA, miRNA, siRNA
With a growing number of genomes sequenced, it is clear that an increase in organismal complexity is not explained by an increase in protein-coding genes. The genome of the ≈1,000-cell nematode Caenorhabditis elegans contains ≈20,000 protein-coding genes, an only slightly smaller number than the 22,000 genes predicted for humans (1). Certainly, posttranscriptional processes such as RNA splicing and editing account for some of this discrepancy. Another solution to this paradox lies in the nonprotein-coding transcripts produced from genomes. Although protein-coding sequences encompass 26 Mb of the C. elegans genome, an additional 33 Mb of transcripts are noncoding. In contrast, the 34 Mb of human coding sequence are dwarfed by the 1,600 Mb of noncoding transcription products (2). Noncoding RNA (ncRNA) is implicated in a variety of functions that include dosage compensation, chromatin remodeling and epigenetic imprinting, modulation of transcription and translation, regulation of RNA splicing and editing, and stress response (3). However, in light of the immense repertoire of noncoding transcripts in complex organisms, our knowledge of ncRNA is merely anecdotal.
Recent large-scale approaches have caused the number of ncRNAs to surge from a handful of known transcripts to thousands of candidates (4, 5). Yet, experimental design usually restricted searches to a size range or to binding partners of known proteins. In addition, reverse transcription techniques used for cloning noncoding transcripts are biased against long, highly structured ncRNAs, and such transcripts are likely underrepresented in the current catalog of ncRNAs.
Previous studies of our laboratory used a biochemical approach to identify polyadenylated C. elegans and human transcripts that are substrates of adenosine deaminases that act on RNA (ADARs), editing enzymes that introduce adenosine to inosine changes in double-stranded regions of RNA (6, 7). This work revealed extended double-stranded regions in noncoding sequences of mRNAs (UTRs and introns) and in completely noncoding transcripts. One of the C. elegans ADAR substrates is the product of the F55A4.9 gene (Fig. 1 A and B). This highly structured 800-nucleotide (nt) transcript, referred to as rncs-1 (RNA noncoding, starvation up-regulated), is spliced and polyadenylated but does not contain a functional ORF and is not found on polysomes (7).
Fig. 1.
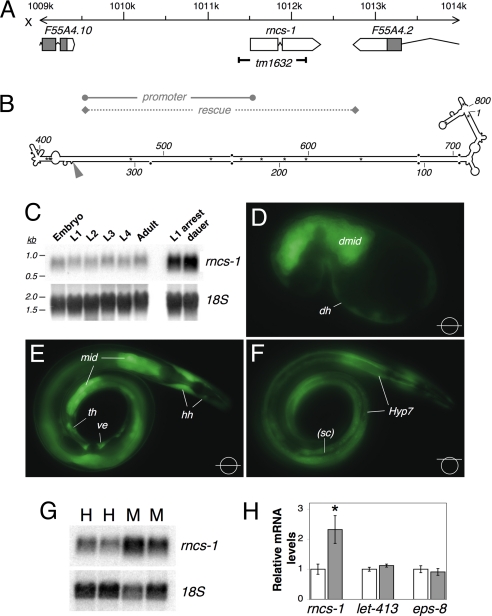
Genomic location, structure, and expression of rncs-1. (A) The X chromosome region that encodes the 847-nt rncs-1 gene is shown. Gray boxes, coding exons; white boxes and arrows, noncoding exons; connecting lines, introns. The 815-bp deletion in rncs-1(tm1632) is indicated. Regions used as promoter in Prncs-1::GFP and as rescue fragment in rncs-1 rescue and overexpressing lines are marked. (B) Secondary structure of mature rncs-1 RNA as predicted by mfold and biochemical methods (nuclease probing; data not shown). Asterisks, GU pairs; dots, mismatches and bulges; gray arrowhead, exon–exon junction; numbering, nucleotide relative to transcription start. (C) Northern blot of rncs-1 in total RNA from wild-type embryos, larval stages (L1–L4), young adults, arrested L1s, and dauer larvae purified from exhausted liquid culture. Blot was rehybridized with probe for 18S ribosomal RNA (rRNA) as a loading control. Data are representative of multiple blots (n = 2–4, depending on stage). (D–F) GFP expression from putative rncs-1 upstream regulatory and promoter sequences. (D) Comma stage embryo with fluorescence in the developing midgut (dmid) and cells of developing hypodermis (dh). (E and F) Adult with Prncs-1::GFP expression in the midgut (mid), cells of the head hypodermis (hh), tail hypodermis (th), Hyp 7 syncytium (Hyp 7), and vulval epithelium (ve). GFP expression is absent in seam cells (sc). Focal planes are shown at Bottom Right. Because transgenes are often silenced in the germ line, the lack of GFP expression in the germ line is not necessarily indicative of lack of expression. Multiple transgenic lines showed the same expression pattern. (G) Northern blot of rncs-1 in total RNA of wild-type hermaphrodites (H) and males (M); 18S rRNA, loading control. (H) Quantification of relative RNA levels in four independent samples of males and hermaphrodites. rncs-1 levels were determined by Northern blotting; let-413 and eps-8 levels were quantified by qRT-PCR. Error bars, SEM; *, P < 0.05, t test.
Intrigued by the unique nature of the rncs-1 transcript, we studied its expression, regulation, and function. We find that rncs-1 transcription is regulated by environmental signals. Importantly, our data suggest that the double-stranded structure of rncs-1 allows it to modulate expression of Dicer-regulated genes.
Results
rncs-1 Is Expressed in Intestine and Hypodermis and Enriched in Males.
C. elegans develop from embryo through four larval stages (L1–L4) into adults. Postembryonic development depends on the presence of food, and worms arrest at the beginning of L1 under starved conditions (L1 arrest). Further, lack of food and high culture density in early development prompt entry into an alternative larval form called dauer. To study rncs-1 expression, we isolated total RNA from synchronized cultures at each developmental stage and from arrested L1s and dauer larvae from starved and crowded liquid culture. Northern blot analysis revealed an RNA species of ≈800 nt (Fig. 1C). The transcript was present at constant levels in embryos, larvae, and adults but increased in arrested L1 worms and dauers.
To investigate the spatial expression of rncs-1, we injected Prncs-1::GFP, a reporter with ≈2 kb of putative rncs-1 regulatory and promoter sequences driving expression of green fluorescent protein (Fig. 1A), into wild-type (N2) worms. GFP expression initiated in the early gastrula (data not shown). We observed robust expression of Prncs-1::GFP in the midgut (E cell lineage) starting at the 28-cell stage and continuing into adulthood (Fig. 1 D–F). By the comma stage (Fig. 1D), fluorescence was also visible in the embryo periphery in cells that give rise to hypodermis. In L1 larva and subsequent stages, we observed strong expression of GFP in hypodermal cells, including Hyp 7 syncytium and head and tail hypodermis. The expression pattern was identical in hermaphrodites and males, but adult hermaphrodites displayed fluorescence in vulval epithelium. Expression was absent in seam cells, nervous system, and pharynx. As described below, like rncs-1, the Prncs-1::GFP reporter showed increased expression during starvation. Although fluorescence intensity was enhanced under starved conditions, the spatial expression pattern was unchanged.
Expression of the Prncs-1::GFP transgene was also enhanced in males. We observed an ≈2.5-fold increase in rncs-1 expression in total RNA prepared from wild-type, well fed males, compared with hermaphrodites (Fig. 1 G and H). In contrast, differences in mRNA levels were not observed for mRNAs of let-413 and eps-8, intestinal genes involved in midgut formation and maintenance (8, 9) (Fig. 1H).
rncs-1 Transcription Is Regulated by Food Supply.
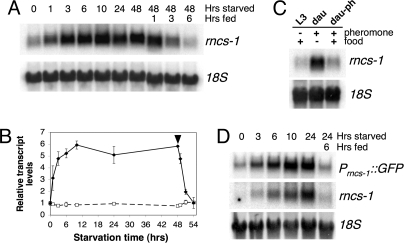
To determine whether the increase in rncs-1 levels in arrested L1 and dauer larvae was a response to lack of food, we removed food from cultures of L4 worms and harvested worms for RNA isolation over 2 days. We observed an ≈3-fold induction of rncs-1 levels over unstarved controls in as little as 1 h of starvation [Fig. 2 A and B and supporting information (SI) Fig. S1]. After 10 h, rncs-1 RNA reached a maximum of ≈6-fold enrichment, and this level was maintained for the 48-h starvation period. After 48 h of starvation, food was reintroduced, and within 6 h rncs-1 levels decreased to baseline. Unlike rncs-1, the general population of polyadenylated RNA did not increase in the absence of food (Fig. 2B, dashed line).
Fig. 2.
Regulation of rncs-1 transcription in response to food supply. (A) Northern blot for rncs-1 in RNA of wild-type L4 larvae starved for indicated times and then reintroduced to food for several hours (48-h starved, 1- to 6-h fed); 18S rRNA, loading control. (B) Quantified Northern blot data for two independent cohorts of worms. Black diamonds/solid line, rncs-1 RNA; white squares/dashed line, total polyadenylated RNA detected by an oligo(dT) probe; black arrowhead, time of food addition; error bars, SEM. Transcript levels were normalized to 18S rRNA and are shown relative to unstarved 0-h sample. The possibility that the increase in rncs-1 levels compared with rRNA was caused by reduction of total RNA during starvation was excluded by measuring total RNA yield per worm in a starvation time course (Fig. S1). (C) Comparison of rncs-1 levels in well fed L3 larvae (L3), dauer larvae isolated from starved and overcrowded liquid culture (dau), and dauer larvae grown with food on dauer pheromone-rich plates (dau-ph). (D) Northern blot of GFP mRNA and rncs-1 in RNA from wild-type worms carrying the Prncs-1::GFP transgene that were subjected to a starvation/feeding time course; 18S rRNA, loading control.
The C. elegans dauer is a nonfeeding stage, and entry into dauer is mediated by sensory integration of two environmental cues: lack of food and abundance of dauer pheromone (daumone). High levels of daumone in the medium, indicative of high population density, prompt entry into dauer despite sufficient food (10). We took advantage of the different pathways to dauer to investigate whether rncs-1 was elevated because of absence of food in the medium or lack of food in the digestive tract. When RNA was isolated from dauer larvae purified from exhausted liquid culture, where starvation conditions and high pheromone levels existed, we observed increased levels of rncs-1 compared with L3 worms (Fig. 2C). However, in samples where dauer formation was initiated on agar plates containing high levels of daumone but also sufficient food, no increase was detected. The oral orifices of dauer animals are plugged so they cannot feed. Thus, regulation of rncs-1 occurs in response to the presence or absence of food in the environment but does not require actual food uptake.
To investigate whether rncs-1 RNA increased during starvation because of posttranscriptional stabilization of the transcript, or transcriptional up-regulation, wild-type animals carrying the Prncs-1::GFP reporter were subjected to a starvation time course. Using RNA isolated at various times of starvation and after refeeding, we compared GFP mRNA levels with endogenous rncs-1 levels by Northern blot analyses. Consistent with the enhanced fluorescence observed in starved worms carrying the Prncs-1::GFP transgene, induction of GFP under control of the putative rncs-1 promoter paralleled the regulation of genomic rncs-1 (Fig. 2D).
Although these experiments do not rule out a component of regulation at the level of RNA stability, they indicate that upstream regulatory and promoter sequences likely contribute to regulation of rncs-1 in response to food supply. In fact, the upstream regulatory sequence of rncs-1 contains seven motifs with the (A/T)GATA(A/G) consensus recognized by GATA transcription factors, including three extended GATA consensus sites often found in C. elegans intestinal genes (Fig. S2A) (11). Indeed, starved animals in which the mRNA coding for the intestinal GATA factor ELT-2 was targeted by RNAi showed a significantly weaker induction of rncs-1 (≈2.5-fold compared with ≈7-fold) (Fig. S2 B and C).
Terminal Branched Structures Protect rncs-1 RNA from Processing by Dicer.
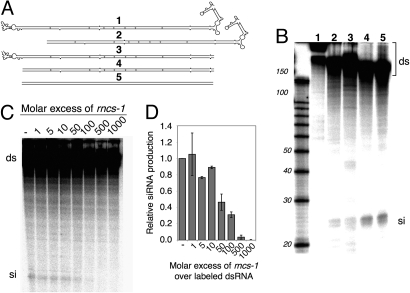
dsRNA, when introduced into C. elegans, is cleaved by the RNase III enzyme Dicer into ≈23-nt small interfering RNAs (siRNAs). The rncs-1 secondary structure contains an almost perfectly double-stranded helix of ≈300 bp (Fig. 1B), suggesting that rncs-1 RNA may be a substrate for Dicer. Although C. elegans Dicer has not been purified in an active form, Dicer activity can be assayed by incubating 32P-labeled dsRNA in embryo extracts (12). We used this assay to test whether siRNAs are generated from rncs-1 RNA in vitro (Fig. 3). When 32P-labeled rncs-1 RNA was incubated in extract, no small RNAs were observed (Fig. 3B, lane 1); the absence of detectable siRNAs persisted over multiple reaction times and RNA and extract concentrations (Fig. S3A; data not shown).
Fig. 3.
rncs-1 is not cleaved by Dicer but inhibits Dicer activity in vitro. (A) Schematic of full-length rncs-1 (1) and derivatives (2–5) synthesized by in vitro transcription. Substrate 5 is identical to 4 except GU base pairs and mismatches were repaired. (B) Autoradiogram showing reaction products of 1-h incubations of 32P-labeled rncs-1 and derivatives in wild-type embryo extract as analyzed by denaturing gel electrophoresis. Processing of dsRNA (ds) by Dicer is evidenced by the appearance of siRNA (si). Size standards, RNA Decade Markers (Ambion). (C) Inhibition of Dicer cleavage of a 32P-labeled 300-bp dsRNA (20 nM; unc-22; see Materials and Methods) by unlabeled rncs-1. Autoradiogram shows products of a representative experiment. (D) Quantification of siRNA production from [32P]dsRNA relative to reaction in absence of rncs-1. The average of two sets of experiments with independent preparations of embryo extract is shown. Error bars, SEM.
Human Dicer cleaves dsRNA more efficiently from its termini (13), and we speculated that the branched structures flanking the central RNA helix of rncs-1 hindered processing. We synthesized rncs-1 derivatives lacking one or both terminal structures (Fig. 3A, substrates 2–4) and a substrate consisting of the rncs-1 helix with all mismatches repaired, which served as a positive control (Fig. 3A, substrate 5). Incubation of rncs-1 derivatives in extract produced siRNAs when at least one helix terminus was blunt-ended (Fig. 3B, lanes 2–5). This indicated that the terminal branched structures of rncs-1 protect it from cleavage by Dicer in vitro; because both termini provided protection, yet were different sequences, protection was not caused by a specific sequence. Northern blot analyses of RNA isolated from adult C. elegans showed similar levels of rncs-1 in wild-type, dcr-1(ok247), and rde-4(ne299) animals (Fig. S3B). These mutant strains are incapable of siRNA production (14, 15), and thus, consistent with our in vitro studies, under these conditions rncs-1 is not a substrate for Dicer in vivo.
rncs-1 RNA Inhibits Dicer Cleavage in trans.
C. elegans siRNA production involves at least two dsRNA-binding proteins (dsRBPs), Dicer (15) and RDE-4 (16, 17). dsRBPs are not sequence-specific, and although rncs-1 was not cleaved by Dicer, we wondered whether it could bind Dicer and/or RDE-4 to sequester these factors and inhibit cleavage of another dsRNA, in trans. We added increasing amounts of unlabeled rncs-1 to extract containing a 32P-labeled 300-bp dsRNA substrate. As expected, the [32P]dsRNA gave rise to siRNAs (Fig. 3C, − lane). However, when extract was preincubated with rncs-1, [32P]siRNA decreased, beginning at 5- to 10-fold molar excess (Fig. 3 C and D). Thus, rncs-1 RNA inhibits production of siRNAs in vitro, presumably by competing with dsRNA substrate for binding to Dicer or RDE-4; indeed, as proof of principle, we confirmed that rncs-1 can bind RDE-4 in vitro (Fig. S3C). Inhibition of Dicer activity depended on the presence of the duplex region of rncs-1 but was independent of its branched terminal structures (Fig. S3D). Although the in vivo role of the terminal structures is unclear, possibly they evolved to prevent spurious siRNA formation.
Analysis of rncs-1 Deletion and Overexpressing Lines.
In hopes of understanding the in vivo role of rncs-1 we analyzed a deletion strain [rncs-1(tm1632); Fig. 1A], and transgenic lines overexpressing rncs-1 in a rncs-1(tm1632) or wild-type background. All animals appeared healthy. Because rncs-1 is induced during starvation, we assayed survival during starvation and dauer formation (data not shown). rncs-1(tm1632) and overexpressing lines were indistinguishable from wild-type animals in these assays.
We noticed that overexpressing lines had an increased frequency of males among hermaphrodite self-progeny. To quantify this defect, for each strain tested, 500–1,000 L4 hermaphrodites were transferred to a separate plate and grown to day 1 of adulthood. Progeny were isolated by hypochlorite treatment and synchronized by hatching without food overnight, and hermaphrodites and males were counted at the adult stage. When rncs-1 was overexpressed in the mutant or wild-type background, male frequency was significantly increased (8- to 15-fold; P < 0.0001; Table 1). Occurrence of males (XO genotype) in offspring of virgin hermaphrodites (XX) results from X chromosome nondisjunction; males in overexpressing lines had XO genotypes, producing ≈50% male offspring when mated to XX hermaphrodites (data not shown). Male frequency in rncs-1(tm1632) and wild-type worms was similar (0.041 and 0.042%, respectively), but in this case counting the number of animals (500,000) necessary to confirm a statistically significant difference was infeasible. Similarly, we reasoned that up-regulation of rncs-1 during starvation might lead to an increased frequency of males, but the low levels of progeny in starved worms hindered our ability to obtain statistically significant numbers.
Table 1.
Frequency of spontaneous male offspring
| Strain* | H/M† | % Males | P value, χ2 |
|---|---|---|---|
| N2 | 11,802/5 | 0.042 | |
| N2 + rncs-1 + GFP | 6,846/43 | 0.624 | <0.0001‡ |
| N2 + GFP | 8,312/11 | 0.132 | <0.0001§, 0.026‡ |
| rncs-1(tm1632) | 9,654/4 | 0.041 | |
| rncs-1(tm1632) + rncs-1 + GFP | 9,646/33 | 0.341 | <0.0001¶ |
| rncs-1(tm1632) + GFP | 4,370/5 | 0.114 | 0.017‖, 0.114¶ |
| rncs-1(tm1632) + rncs-1 + RFP | 3,101/6 | 0.193 | 0.009¶ |
*GFP under rncs-1 promoter, RFP under myo-3 promoter.
†Hermaphrodite (H) and male (M) counts (two or more experiments).
‡Compared with N2.
§Compared with N2 + rncs-1 + GFP.
¶Compared with rncs-1(tm1632).
‖Compared with rncs-1(tm1632) + rncs-1 + GFP.
Expression of Dicer-Regulated Genes Depends on rncs-1 Levels.
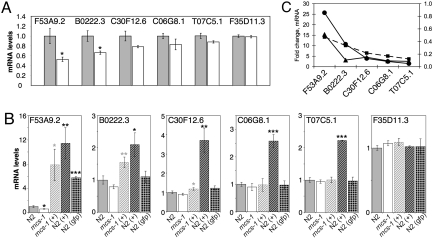
Because rncs-1 inhibits Dicer in vitro, we wondered whether it could exert this function in vivo, and we used our rncs-1 mutant and overexpressing lines to test this idea. We first searched for Dicer-regulated genes with altered expression in rncs-1(tm1632) animals. Our laboratory recently used microarray analyses to identify genes whose mRNA levels are Dicer-dependent (18). We chose 18 genes from these datasets with a predicted or confirmed expression pattern similar to that of rncs-1 and used quantitative RT-PCR (qRT-PCR) to compare their expression in wild-type and rncs-1(tm1632) animals. If rncs-1 antagonizes Dicer activity in vivo, genes normally silenced by Dicer would show a further decrease in expression in an rncs-1-deficient background, and we identified five genes with this property (Fig. 4A). Reduction in mRNA levels varied from ≈50% (F53A9.2), to ≈10% (T07C5.1). Dicer-dependent silencing of F35D11.3 was not further enhanced in the rncs-1(tm1632) mutant, and this gene was included as a negative control.
Fig. 4.
Dicer-regulated genes respond to rncs-1 levels. (A) Relative expression of Dicer-regulated genes in wild-type (gray) and rncs-1(tm1632) (white) adult hermaphrodites as quantified by qRT-PCR and normalized to gpd-3 (n = 6; error bars, SEM; *, P < 0.05, t test). (B) Relative expression of genes in A for the strains indicated. +, strains expressing the rncs-1 transgene; gfp, strain expressing Prncs-1::GFP. The mean value for three to six independent samples is plotted, normalized to let-413 (error bars, SEM). *, P < 0.05; **, P < 0.01; ***, P < 0.001, t test. Gray asterisks, comparison with rncs-1(tm1632); black asterisks, comparison with N2. (C) Fold change in mRNA level compared with wild-type is plotted for dcr-1(ok247) (circles), rncs-1(tm1632) (squares), and wild-type animals overexpressing rncs-1 (triangles). Data for dcr-1(ok247) are the average of qRT-PCR values for three or four independent samples. P < 0.001 for all genes (compared with wild type) except T07C5.1 (P < 0.05) and F35D11.3 (P < 0.01).
There were many possible explanations for the decreased expression of Dicer-regulated genes in rncs-1(tm1632) animals. However, if rncs-1 was truly competing with endogenous dsRNA [i.e., precursors of siRNA or microRNA (miRNA)] for binding to Dicer, animals overexpressing rncs-1 should show increased expression of Dicer-regulated genes. Indeed, genes with reduced mRNA levels in the rncs-1(tm1632) mutant had increased mRNA levels when rncs-1 was overexpressed (+, Fig. 4B). qRT-PCR analyses verified that Dicer mRNA levels were unchanged in these strains (data not shown).
For each gene, we also quantified mRNA levels in dcr-1(ok247) mutant animals. We then compared how much wild-type mRNA levels were altered in dcr-1(ok247) animals with how much they were altered in rncs-1(tm1632) and overexpressing strains (Fig. 4C). For the five Dicer-regulated genes we observed a broad range in regard to how much the wild-type expression level was altered in dcr-1(ok247) knockout animals (circles), in rncs-1(tm1632) knockout animals (squares), and in rncs-1-overexpressing lines (triangles). Interestingly, the magnitude of response for a given gene in one strain correlated with its response in other strains (Fig. 4C). For example, in each strain, F53A9.2 showed the greatest change, C30F12.6 showed an intermediate response, and T07C5.1 was least affected. The characteristic way in which each gene responded in the various strains suggested that the same property of the gene was being interrogated in each case. It seems likely that this property is the relative amount of mRNA associated with each gene, compared with the dsRNA involved in its silencing (see Discussion).
F53A9.2 expression was most sensitive to changes in Dicer and rncs-1 (Fig. 4C), and for this gene, but not others, mRNA levels were also increased by the Prncs-1::GFP transgene. We found that this transgene, as often observed in repetitive, trangenic arrays (19), produces both sense and antisense transcripts (Fig. S4). Analysis of additional transgenic strains showed a correlation with F53A9.2 expression and the amount of dsRNA synthesized by the transgenic array and verified that the effect of dsRNA on F53A9.2 was independent of sequence (Fig. S5).
Northern blot analyses revealed a 16- to 19-fold increase in rncs-1 in overexpressing lines compared with wild type (Fig. S4A), the same order of magnitude as the 6-fold increase in rncs-1 that occurs during starvation (Fig. 2B). Further, from qRT-PCR assays of mRNA from the Dicer-regulated genes we estimate rncs-1 is 14- to 125-fold more abundant in unstarved animals (SI Materials and Methods). If dsRNA involved in silencing these genes was equimolar to the mRNA, rncs-1 would be present at an excess similar to that which showed competition in vitro (Fig. 3D). However, in repeated assays, we did not observe a simple up-regulation of Dicer-regulated genes under starved conditions. Instead, expression of rncs-1-sensitive genes showed a large degree of fluctuation during starved conditions. Starvation initiates many events, in addition to up-regulation of rncs-1, that presumably are responding to subtle differences between experiments, despite our careful attempts to maintain constancy.
siRNAs Are Reduced by rncs-1 Overexpression.
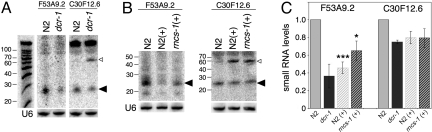
If the change in expression of Dicer-regulated genes in response to changes in rncs-1 levels was caused by inhibition of Dicer by rncs-1, we should also see changes in levels of small RNAs that were Dicer products. Indeed, using sense-oriented DNA oligonucleotide probes (to detect antisense RNA), we detected small RNAs complementary to F53A9.2 and C30F12.6 (black arrowheads, Fig. 5 A and B). dcr-1(ok247) animals are sterile, and because homozygous animals are isolated from F1 progeny of heterozygous mothers, they contain some maternal Dicer. Thus, as expected, small RNAs were reduced but not eliminated in this strain (Fig. 5A). Previous assays of miRNAs in dcr-1(ok247) show that reduction in the ≈23-nt miRNA correlates with an accumulation of the pre-miRNA (20). C30F12.6 reduction of small RNAs in dcr-1(ok247) coincided with appearance of a ≈65-nt band reminiscent of a pre-miRNA (white arrowheads, Fig. 5 A and B). Although future studies will be necessary to delineate the role of Dicer in regulating the C30F12.6 and F53A9.2 genes, possibly the former is regulated by a miRNA and the latter an siRNA.
Fig. 5.
rncs-1 alters small RNA production. (A) Northern blot of 3 μg of size-selected RNA from the indicated strains, probed for sequences complementary to F53A9.2 and C30F12.6. U6 RNA, loading control; black arrowhead, small RNA; white arrowhead, possible small RNA precursor. Putative precursor band in dcr-1 sample comigrates with that in (+) samples (data not shown). (B) As in A with strain designation as in Fig. 4. (C) Multiple analyses as in A and B were quantified to show small RNA levels in various strains relative to wild type. Results were normalized to U6 (F53A9.2: n = 2 for dcr-1, n = 4 for other strains; C30F12.6: n = 2 for all strains). Error bars, SEM. *, P < 0.05; ***, P < 0.01, t test.
Consistent with our competition model, F53A9.2 and C30F12.6 small RNAs were reduced in strains overexpressing rncs-1 (Fig. 5 B and C). Reduction in small RNAs correlated with the increase in mRNA (Fig. 4B and Fig. S6 A and B). Previous studies suggest that small RNAs act catalytically, and this finding agrees with our data. In the N2(+) strain, F53A9.2 mRNA levels increased ≈12-fold in response to a 2-fold decrease in small RNA levels (Figs. 4B and 5C; Fig. S6A). Thus, it is not surprising that small RNAs of C30F12.6 changed only slightly upon rncs-1 overexpression (≈20%, Fig. 5 B and C) because a very moderate up-regulation of its mRNA occurred (1.5- to 3.5-fold; Fig. 4B). We did not detect changes in small RNAs in the rncs-1(tm1632) mutant (data not shown), likely because of limited sensitivity of the Northern blot analysis.
Discussion
rncs-1 is a ncRNA whose double-stranded structure is unprecedented among characterized ncRNAs. Changes in rncs-1 expression lead to changes in expression of several Dicer-regulated genes, suggesting that rncs-1 binds to Dicer or accessory dsRBPs to compete with endogenous dsRNA involved in silencing. Although rncs-1 now provides an example of a cellular RNA that modulates activity of a dsRBP, viruses have long been known to employ such strategies (21–23).
What Is Important for Modulating Expression of Dicer-Regulated Genes?
Both C. elegans and Homo sapiens encode only a single Dicer, but the enzyme is responsible for regulating expression of numerous genes. How Dicer coordinates precise expression of many genes is entirely unexplored, and our data provide clues in this regard. The Dicer-regulated genes we analyzed showed a broad range in sensitivity to changes in rncs-1 expression or the deletion of the dcr-1 gene [dcr-1(ok247); Fig. 4]. Yet, the magnitude of response observed for a given gene under one condition correlated with the magnitude of its response in other conditions (Fig. 4C). Genes showing a large change in expression in dcr-1(ok247) animals showed large changes when the levels of rncs-1 were altered. Our data suggest that Dicer is limiting compared with its dsRNA substrates (siRNA or miRNA precursors), and for a given gene, the amount of dsRNA substrate relative to the cognate mRNA differs. High mRNA levels that correlate with equally high levels of dsRNA would be very sensitive to the loss of DCR-1 but relatively insensitive to rncs-1 levels because it would be hard to compete with the high levels of dsRNA substrate. However, high mRNA levels associated with much less than equimolar dsRNA would show only a small increase in dcr-1(ok247) animals, but the low level of silencing would be very sensitive to rncs-1 levels. A gene that gives rise to low levels of mRNA but equimolar amounts of dsRNA would be very sensitive to the loss of Dicer, and sensitive to changes in rncs-1. This latter scenario correlates with the F53A9.2 gene we analyzed, and in fact, this gene is associated with very low levels of mRNA (SI Materials and Methods). These considerations may explain why only a subset of the Dicer-regulated genes we assayed were affected by rncs-1 levels and why processing of the abundant lin-4 miRNA is not altered by rncs-1 overexpression (Fig. S6C).
What Is the Biologic Role of rncs-1?
We observed a decrease in mRNA levels of several Dicer-regulated genes in the rncs-1(tm1632) mutant compared with wild type. Thus, rncs-1 modulates expression of these genes in vivo. However, as yet, we have not observed a physiological phenotype in rncs-1(tm1632) animals that correlates with this molecular phenotype. We did, however, observe an increased frequency of males in rncs-1-overexpressing lines. The low numbers of progeny in starved animals precluded our efforts to determine whether increased levels of rncs-1 during starvation also leads to increased male progeny. However, initiation of genetic exchange by switching from asexual to sexual reproduction is a common strategy for adaptation to challenging environments (24), and it remains possible that rncs-1 is involved in such a strategy. In this regard, rncs-1 may reiterate a theme of ncRNAs, many of which relate to stress (3).
rncs-1 functions are mediated by its structure rather than its sequence, so it is not surprising that sequence searches have not revealed homologs. Here, rncs-1 is similar to other long ncRNAs, including essential transcripts like mammalian Xist, that for unknown reasons are poorly conserved in sequence, even in closely related species (25). Although the sequence of rncs-1 is unique and not repeat-derived, inverted DNA repeats with extensively base-paired stems are recombinogenic and cause genome instability (26). This could explain the absence of an identifiable rncs-1 homolog in Caenorhabditis briggsae and Caenorhabditis remanei, species closely related to C. elegans. Indeed, in C. briggsae and C. remanei the region of the X chromosome syntenic with the rncs-1 neighborhood underwent an inversion, with a putative breakpoint close to rncs-1. The fact that C. elegans has retained an unstable element in its genome hints at a selective advantage provided by rncs-1.
Materials and Methods
Strain designation, cloning of rncs-1, and construction of transcription templates are described in SI Materials and Methods.
Strains and Culture.
C. elegans strains were grown under standard conditions (27): rncs-1(tm1632) was backcrossed six times to generate BB15 (SI Materials and Methods). Arrested L1 were harvested after hatching embryos in M9 for 16–24 h. Males were picked to ≈99% purity from progeny of male-fertilized hermaphrodites.
For starvation time courses, L4 from well fed liquid culture (Fig. 2A) or plates (Fig. 2D) were starved in M9.
Dauer larvae were harvested from liquid culture grown to high density/food exhaustion, by treatment with 1% SDS (28). Pheromone plates were seeded with Escherichia coli OP50 and contained sufficient partially purified daumone to induce ≈100% dauer formation in N2 at 20°C. Adults placed on each plate laid eggs for 8 h before removal. After 3 days, progeny that entered dauer were harvested.
Transgenics.
For the Prncs-1::GFP reporter, ≈2,000 bp of upstream sequences of genomic rncs-1 (Fig. 1A, promoter) were inserted into the multiple cloning site of pPD49.26. Plasmid was injected into N2 worms. Lines overexpressing rncs-1 were produced by injecting a ≈3,400-bp PCR product (Fig. 1A, rescue) into rncs-1(tm1632) along with Prncs-1::GFP reporter to generate BB17 or with pRF4 into N2 to generate BB18 (Figs. 4 and 5). Excess nonspecific DNA (1-kb ladder; Invitrogen) was coinjected.
Northern Blot Analysis and RT-PCR.
For nonintegrated lines, ≈99% pure transgenic populations were handpicked. dcr-1(ok247);unc-32(e189) homozygotes were sorted based on coiling phenotype (18).
Total RNA was isolated by using TRIzol (Invitrogen) and used directly for Northern blotting. For RT-PCR, RNA was treated with TURBO DNase (Ambion) followed by RNeasy chromatography (Qiagen). For Northern blot analyses to detect siRNA and miRNA, RNA was size-selected (mirVana miRNA isolation kit; Ambion).
Northern blot analyses were performed with standard protocols for 1% formaldehyde–agarose gel electrophoresis and nylon membrane blotting. rncs-1 probe (nucleotides 323–442 of cDNA, not edited by ADAR) was prepared by in vitro transcription (Strip-EZ T7 kit; Ambion). ULTRAhyb buffer (Ambion) was used for Northern blot analyses of rncs-1 or 18S. Polyadenylated RNA quantified in Fig. 2B was detected with 32P-labeled (dT)30 (29).
Northern blot analyses of small RNA were as published in ref. 30. To increase sensitivity, some Northern blotting was performed with carbodiimide-mediated cross-linking (31). Probes used to detect F53A9.2 and C30F12.6 siRNAs are shown in Table S1.
For qRT-PCR, 1 μg of total RNA was reverse-transcribed with SuperScript II reverse transcriptase and oligo(dT) primers (Invitrogen). Samples were treated with RNase H (New England Biolabs) and diluted 4-fold with ddH2O. Five microliters of cDNA was used per PCR in a LightCycler 2.0 instrument with the LightCycler FastStart DNA MasterPLUS SYBR Green I kit (Roche). Primer pairs spanned at least one exon–exon junction and produced products of 150–300 bp.
In Vitro Transcription.
Internally 32P-labeled RNA was transcribed with T7 RNA polymerase (32) and purified from a 4% denaturing polyacrylamide gel. Duplex RNAs were annealed as described in ref. 17 and purified from a 5% native gel. Unlabeled rncs-1 RNA was prepared similarly, with modifications to improve yield.
In Vitro Dicer Activity Assay.
Extract preparation and Dicer activity assays were as described in ref. 17, but for competition assays glycerol was 20%, and extract was cleared by centrifugation (100,000 × g; 60 min). Dicer assays (20 μl) contained 50 μg of total protein and 10 nM labeled RNA. Reactions with rncs-1 as inhibitor were preincubated (5 min; 20°C) with unlabeled rncs-1 before adding labeled 300-bp unc-22 dsRNA.
Supplementary Material
Acknowledgments.
We thank J. Habig and N. Welker for helpful discussions and access to unpublished data, D. Morse for information on rncs-1, G. Parker for assays with RDE-4, and Susan Mango and her laboratory for advice and assistance. We thank the National Bioresource Project for the Nematode, Japan, for rncs-1(tm1632) and the Caenorhabditis Genetics Center, funded by the National Institutes of Health/National Center for Research Resources (NCRR). This work was supported by National Institutes of Health Grant GM044073 (to B.L.B.). B.L.B. is a Howard Hughes Medical Institute Investigator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805118105/DCSupplemental.
References
- 1.Hubbard TJP, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frith MC, Pheasant M, Mattick JS. The amazing complexity of the human transcriptome. Eur J Hum Genet. 2005;13:894–897. doi: 10.1038/sj.ejhg.5201459. [DOI] [PubMed] [Google Scholar]
- 3.Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: An answer to the “genome complexity” conundrum. Genes Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- 4.Huttenhofer A, Vogel J. Experimental approaches to identify noncoding RNAs. Nucleic Acids Res. 2006;34:635–646. doi: 10.1093/nar/gkj469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mattick JS, Makunin IV. Noncoding RNA. Hum Mol Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 6.Morse DP, Aruscavage PJ, Bass BL. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc Natl Acad Sci USA. 2002;99:7906–7911. doi: 10.1073/pnas.112704299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morse DP, Bass BL. Long RNA hairpins that contain inosine are present in Caenorhabditis elegans poly(A)+ RNA. Proc Natl Acad Sci USA. 1999;96:6048–6053. doi: 10.1073/pnas.96.11.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bossinger O, Fukushige T, Claeys M, Borgonie G, McGhee JD. The apical disposition of the Caenorhabditis elegans intestinal terminal web is maintained by LET-413. Dev Biol. 2004;268:448–456. doi: 10.1016/j.ydbio.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Croce A, et al. A novel actin barbed-end-capping activity in EPS-8 regulates apical morphogenesis in intestinal cells of Caenorhabditis elegans. Nat Cell Biol. 2004;6:1173–1179. doi: 10.1038/ncb1198. [DOI] [PubMed] [Google Scholar]
- 10.Golden JW, Riddle DL. A pheromone influences larval development in the nematode Caenorhabditis elegans. Science. 1982;218:578–580. doi: 10.1126/science.6896933. [DOI] [PubMed] [Google Scholar]
- 11.Pauli F, Liu Y, Kim YA, Chen PJ, Kim SK. Chromosomal clustering and GATA transcriptional regulation of intestine-expressed genes in C. elegans. Development. 2006;133:287–295. doi: 10.1242/dev.02185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ketting RF, et al. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parrish S, Fire A. Distinct roles for RDE-1 and RDE-4 during RNA interference in Caenorhabditis elegans. RNA. 2001;7:1397–1402. [PMC free article] [PubMed] [Google Scholar]
- 15.Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ-line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA-binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 17.Parker GS, Eckert DM, Bass BL. RDE-4 preferentially binds long dsRNA, and its dimerization is necessary for cleavage of dsRNA to siRNA. RNA. 2006;12:807–818. doi: 10.1261/rna.2338706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welker NC, Habig JW, Bass BL. Genes misregulated in C. elegans deficient in Dicer, RDE-4, or RDE-1 are enriched for innate immunity genes. RNA. 2007;13:1090–1102. doi: 10.1261/rna.542107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knight SW, Bass BL. The role of RNA editing by ADARs in RNAi. Mol Cell. 2002;10:809–817. doi: 10.1016/s1097-2765(02)00649-4. [DOI] [PubMed] [Google Scholar]
- 20.Grishok A, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 21.Andersson MG, et al. Suppression of RNA interference by adenovirus virus-associated RNA. J Virol. 2005;79:9556–9565. doi: 10.1128/JVI.79.15.9556-9565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKenna SA, Kim I, Liu CW, Puglisi JD. Uncoupling of RNA binding and PKR kinase activation by viral inhibitor RNAs. J Mol Biol. 2006;358:1270–1285. doi: 10.1016/j.jmb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 23.Tian B, Bevilacqua PC, Diegelman-Parente A, Mathews MB. The double-stranded RNA-binding-motif: Interference and much more. Nat Rev Mol Cell Biol. 2004;5:1013–1023. doi: 10.1038/nrm1528. [DOI] [PubMed] [Google Scholar]
- 24.de Visser JA, Elena SF. The evolution of sex: Empirical insights into the roles of epistasis and drift. Nat Rev Genet. 2007;8:139–149. doi: 10.1038/nrg1985. [DOI] [PubMed] [Google Scholar]
- 25.Pang KC, Frith MC, Mattick JS. Rapid evolution of noncoding RNAs: Lack of conservation does not mean lack of function. Trends Genet. 2006;22:1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 26.Gordenin DA, et al. Inverted DNA repeats: A source of eukaryotic genomic instability. Mol Cell Biol. 1993;13:5315–5322. doi: 10.1128/mcb.13.9.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Kim SK. Global analysis of dauer gene expression in Caenorhabditis elegans. Development. 2003;130:1621–1634. doi: 10.1242/dev.00363. [DOI] [PubMed] [Google Scholar]
- 29.Harley CB. Hybridization of oligo(dT) to RNA on nitrocellulose. Gene Anal Tech. 1987;4:17–22. doi: 10.1016/0735-0651(87)90013-6. [DOI] [PubMed] [Google Scholar]
- 30.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 31.Pall GS, Codony-Servat C, Byrne J, Ritchie L, Hamilton A. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA, and piRNA by northern blot. Nucleic Acids Res. 2007;35:e60. doi: 10.1093/nar/gkm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bass BL, Weintraub H. A developmentally regulated activity that unwinds RNA duplexes. Cell. 1987;48:607–613. doi: 10.1016/0092-8674(87)90239-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.