Abstract
The eukaryotic MCM2–7 complex is recruited at origins of replication during the G1 phase and acts as the main helicase at the replication fork during the S phase of the cell cycle. To characterize the interplay between the MCM helicase and DNA prior to the melting of the double helix, we determined the structure of an archaeal MCM orthologue bound to a 5.6-kb double-stranded DNA segment, using cryo-electron microscopy. DNA wraps around the N-terminal face of a single hexameric ring. This interaction requires a conformational change within the outer belt of the MCM N-terminal domain, exposing a previously unrecognized helix-turn-helix DNA-binding motif. Our findings provide novel insights into the role of the MCM complex during the initiation step of DNA replication.
Keywords: AAA+ ATPase, archaea, DNA topology, electron microscopy, MCM2–7
Introduction
The eukaryotic MCM2–7 complex is essential during the S phase of the cell cycle and is believed to act as the replicative helicase (Forsburg, 2004). It is recruited onto origins of replication in the G1 phase and, together with the origin recognition complex (ORC), Cdc6 and Cdt1, forms the pre-replicative complex (Arias and Walter, 2007). In yeast, Cdc6 and ORC catalyse sequential ATP hydrolysis events, responsible for the multiple loading of the MCM complex (Randell et al, 2006). The MCM proteins are found at the replication fork together with Cdc45 and GINS (Pacek et al, 2006) and display helicase activity (Moyer et al, 2006). Most archaeal genomes contain only one copy of an MCM orthologue and their gene product forms a homo-oligomeric assembly, which is able to unwind fork-shaped DNA substrates (Kelman et al, 1999; Chong et al, 2000; Shechter et al, 2000; Carpentieri et al, 2002; Grainge et al, 2003; Haugland et al, 2006; Barry et al, 2007).
Up to 40 MCM protein complexes per origin of replication have been found bound to chromatin (Edwards et al, 2002). This abundance is hard to reconcile with a simple model of a canonical helicase, acting at the replication fork (Dimitrova et al, 1999; Takahashi et al, 2005). The MCM2–7 complex is required both for replication initiation and fork progression (Labib et al, 2001): it is possible that this protein complex may possess a dual role, being actively involved in origin melting during the initiation step and in the unwinding of the fork during the elongation step of DNA replication (Costa and Onesti, 2008). In this context, the former step could require multiple copies of the MCM complex, whereas the latter step would only require one active helicase assembly per fork.
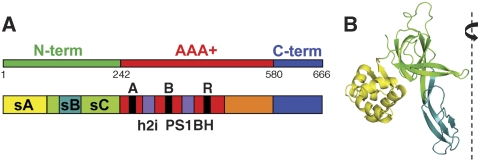
The MCM proteins belong to the AAA+ superfamily of ATPases and include a N-terminal domain, a AAA+ domain and a C-terminal region that was predicted to fold into a winged-helix (WH) domain. Crystallographic studies have shown that the N-terminal domain from Methanothermobacter thermautotrophicus MCM (MthMCM) can form a double hexameric ring with a head-to-head configuration (Fletcher et al, 2003). Each chain folds into three subdomains (sA, sB and sC; Figure 1), with subdomain A located in the peripheral belt of the MCM ring (Fletcher et al, 2003; Kasiviswanathan et al, 2004). The atomic structure of the MCM N-terminal domain from the archaeon Sulfolobus solfataricus (SsoMCM) has also been recently published, which crystallized as a single planar hexamer (Liu et al, 2008).
Figure 1.
Domain structure of MthMCM. (A) Schematic representation of the primary structure of MthMCM, comprising a N-terminal domain (green) a AAA+ domain (red) and a C-terminal domain (blue). The N-terminal domain can be further subdivided into 3 subdomains (sA, sB, sC, in yellow, teal and green, respectively). The AAA+ domain folds into an α/β-subdomain (red) and a α-domain (orange); the former contains the canonical Walker A, Walker B and R finger motifs found in AAA+ protein, as well as the unique insertions (h2i and PS1BH, in violet), which characterize the MCM clade. The C-terminal domain is predicted to fold into a winged-helix motif. (B) The crystal structure of a monomer of the MthMCM N-terminal domain (Fletcher et al, 2003; PDB entry 1LTL), with subdomains sA, sB and sC colour-coded as in (A). A dashed line indicates the position of the six-fold axis around which the hexameric complex assembles.
Initial electron microscopy studies carried out on negatively stained MthMCM samples revealed heptameric (Yu et al, 2002) as well as hexameric (Pape et al, 2003) single rings. Subsequent work showed the presence of hexameric and heptameric (head-to-head) double-ring configurations (Gomez-Llorente et al, 2005; Costa et al, 2006a, 2006b), as well as head-to-tail helical arrangements (Chen et al, 2005).
We have used electron microscopy (EM) to analyse the MthMCM complex bound to a 5.6-kb segment of double-stranded DNA. We show that multiple copies of a single hexameric complex bind to the same segment of DNA and bend it, and that MthMCM filaments can also be found interacting with DNA. Cryo-EM single-particle reconstruction reveals that the dsDNA wraps around single-hexameric MthMCM assemblies, interacting with a previously unrecognized helix-turn-helix fold located in the outer belt of the N-terminal domain. We suggest that this represents an initial site of interaction between MCM and double-stranded DNA, distinct from the canonical one in which the protein ring encircles single-stranded DNA, prior to the loading and activation of the complex to function as a helicase at the fork.
Results
EM analysis of various MthMCM mutants bound to long segments of dsDNA
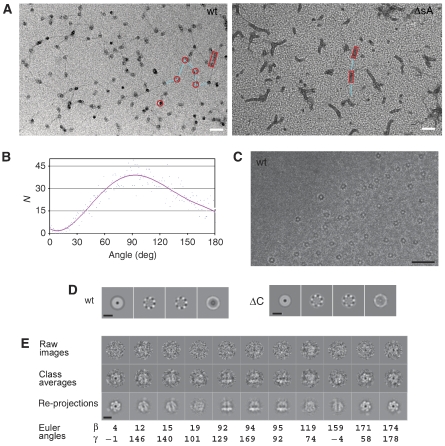
In a previous study, we used EM to characterize the MthMCM protein bound to short stretches (∼100 bp) of dsDNA (Costa et al, 2006a). The corresponding three-dimensional 3D reconstruction is consistent with the DNA segment partially threading through the central hole of the MCM double hexameric ring. We have now characterized the protein bound to longer segments of dsDNA (∼5600 bp). We found conditions, in negative stain EM, which allowed us to visualize both the protein particles and the DNA (Figure 2A and Supplementary Figure 1), working at 5 μm defocus and × 27 500 magnification, to maximize contrast. The majority of the protein particles appear to interact with dsDNA as single rings. Whenever dsDNA is interacting with an MCM particle it undergoes a sharp kink, with a very different topology from the dsDNA visualized in the absence of protein (Supplementary Figure 1). A statistical analysis of the DNA kinks on a set of 5000 angles shows that they follow a broad distribution roughly centred around 90° (Figure 2B). It should be noted that a possible interpretation of this result is that preferential orientations of the MCM protein particles on a carbon support could influence the angular distribution of dsDNA kinks.
Figure 2.
Electron microscopy of MthMCM bound to long dsDNA molecules. (A) Micrographs of a positively stained complex between dsDNA (5600 bp) and either wt MthMCM (left panel) or the protein lacking subdomain A (ΔsA, right panel). Images were taken at a magnification of × 27 500 and 5 μm defocus. A few features have been highlighted: MCM complexes are shown as red rings, rectangular boxes indicate the presence of fibres, whereas DNA segments are highlighted in cyan. Scale bar 500 Å. (B) Statistical analysis of angle distribution performed on a sample of 5000 protein-induced dsDNA kinks carried out on the wt MthMCM-negative stain data. (C) Micrograph of MthMCM bound to long dsDNA molecules embedded in vitreous ice. Scale bar 500 Å. (D) Eigenimages obtained from multivariate statistical analysis performed on translationally aligned cryo-EM particles, both for the wt (left panel) and a mutant lacking the C-terminal domain (ΔC, right panel). Scale bar 100 Å. (E) Overview of the 3D reconstruction procedure. Scale bar 100 Å.
A few higher order oligomers are also visible on the micrographs, compatible with the formation of double rings and short fibres (Supplementary Figure 2). These fibres look similar to the filaments that can be seen in samples that are highly contaminated by endogenous dsDNA (Supplementary Figure 2) and disappear when the protein is further purified.
Cryo-EM single-particle analysis of the same nucleoprotein preparation (Figure 2C) highlights a strong hexameric symmetry component, detected by inspection of the eigenimages after multivariate statistical analysis (MSA; Figure 2D) on translationally aligned particles (Dube et al, 1993). This confirms that, independent of the length of the dsDNA segment, a hexameric configuration is stabilized upon DNA binding, consistent with our previous studies (Costa et al, 2006a). A further implication of this analysis is that a large portion of the particle data set is showing an end-on view. This suggests a model in which an MCM complex sits across the dsDNA rather than encircling it, as a ring encircling DNA would mainly produce side views, and no hexameric pattern would be detected. Interestingly, the fourth eigenimage (Figure 2D) shows a high variance of the electron density distribution around the MCM ring, a feature which was absent in previous studies of the MCM protein (Costa et al, 2006a). These observations are compatible with a model in which MCM is interacting with dsDNA using one face of the ring.
To assess whether the N- or the C-terminal face of the ring is mediating this interaction, we characterized by EM various deletion mutants complexed to the same long dsDNA molecule (Supplementary Figure 1C). A very different behaviour was observed for the subdomain A deletion mutant (ΔsA; Figure 2A), where most of the protein formed fibrous assemblies, with DNA filaments emerging at the ends. This suggests that subdomain A has a critical role in the primary interaction between the protein and long dsDNA stretches.
Cryo-EM structure of the wild-type MthMCM protein bound to long segments of dsDNA
We carried out a cryo-EM single-particle reconstruction of the wt MthMCM protein treated with 5600 bp of dsDNA. A 3D structure was obtained, based on 10 010 particles and refined with no symmetry imposition (Supplementary Figure 2). As clearly visible from side-view class averages and re-projections (Figure 2E), one of the two faces of the MCM ring displays higher electron density than the other, which is consistent with dsDNA interacting with one face of the ring.
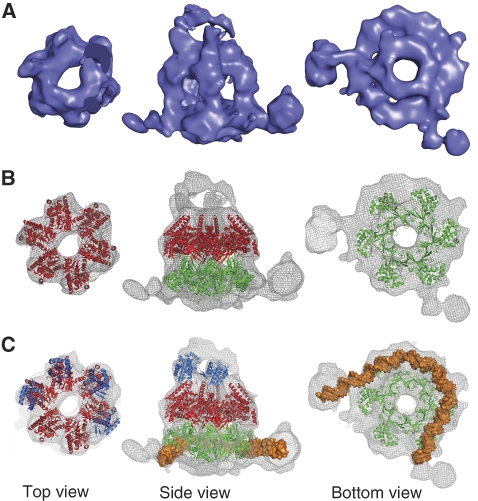
The structure consists of a hexameric hollow single ring, made of two tiers stacked upon each other (Figure 3A). The main body of the molecule has a width of 130 Å and a height of 100 Å. One face of the ring (top view) displays six-fold symmetry, whereas the other face (bottom view) appears to be rounded, so that no clear hexameric pattern can be detected. Furthermore, two filamentous protrusions depart radially from the bottom face of the ring. The side view of the molecule shows electron density features budding from the top tier of the ring.
Figure 3.
Cryo-EM structure of wt MthMCM bound to long dsDNA molecules. (A) Surface rendering of the molecule (displayed at 2.5 σ) showing a slab through the top, side and bottom views, respectively. The structure consists of a single ring with a large central cavity. Whereas the top face of the ring displays six-fold symmetry, the bottom face is rounded, with two filamentous protrusions departing radially. (B) Fitting of the AAA+ domain from MkaMCM (red; Bae et al, submitted) and the N-terminal domain from MthMCM (green; Fletcher et al, 2003). The AAA+ hexamer fits well to the dome-shaped top face, whereas additional electron density surrounds the atomic model in the bottom tier. (C) Four copies of the WH domain of Cdc6 (blue; Liu et al, 2000) have been fitted to the protrusions from the AAA+ domain (Supplementary Figure 4). Roughly 80 bp of dsDNA has been fitted to the additional electron density at the bottom of the ring (Supplementary Figure 5); a re-orientation of subdomain A is required to avoid steric clashes with the modelled dsDNA.
Assignment of the MthMCM domains
The atomic coordinates for the MthMCM N-terminal domain are available (Fletcher et al, 2003), whereas for the AAA+ domain we used the recently determined crystal structure of a Methanopyrus kandleri MCM (MkaMCM; Bae et al, submitted). MkaMCM was crystallized as a monomer and lacks both the N-terminal B subdomain and the C-terminal WH domain. A hexameric hybrid model was obtained by superposing six MkaMCM monomers onto a MthMCM hexamer. When placed into the EM map, the AAA+ tier fits well to the dome-shaped hexameric top face of the ring (Figure 3B).
Additional electron density budding from the AAA+ tier can be assigned to the C-terminal domain. Only four out of six electron density features could be visualized (Figure 3B): this is compatible with the notion that the C-terminal WH domain is only partially ordered in this configuration. A similar positioning of the C-terminal domain has been previously suggested, based on EM reconstructions (Costa et al, 2006a). The assignment of the C-terminal domain has been controversial and two other putative locations had been proposed (Pape et al, 2003; Gomez-Llorente et al, 2005). To establish with confidence the location of the C-terminal domain, we determined the 3D structure of the ΔC mutant treated with the same long dsDNA and generated a map, based on 5000 particles. A comparison between the two maps, filtered to the same resolution, shows that the overall body of the molecule is similar (Supplementary Figure 4), but the buds on the top of the AAA+ tier are only present in the wt reconstruction, suggesting that these represent partially disordered C-terminal domains. A similar localization for the C-terminal WH domain can be modelled by overlaying the atomic structure of the C-terminal fragment of Pyrobaculum aerophilum Cdc6 (Liu et al, 2000; PDB entry 1FNN) onto the alpha domain of MkaMCM (Supplementary Figure 4). The atomic coordinates of the Cdc6 WH domain were therefore used for fitting (Figure 3C).
Double-stranded DNA interacts with subdomain A and encircles the MCM ring
Once the three domains of the MthMCM complex have been located, unassigned electron density can be found surrounding the outer belt of the N-terminal domain and in the form of two radially departing filaments (Figure 3B), forming an angle of roughly 90°. This angle is in agreement with the statistical analysis carried out on the DNA kinks (Figure 2B). When displaying the electron density map at higher threshold, most of the features in the structure disappear, with the exception of a rod of electron density partially surrounding the N-terminal domain (Supplementary Figure 5). We can fit roughly 80 bp of dsDNA in the map displayed at high threshold (Figure 3C). Although additional electron density can be found all around the bottom ring, in one sector of the ring the density features are weaker and the N-terminal subunits appear better defined, as exemplified by the presence of holes in between subunits. This suggests that the DNA may only partially run along the ring and then depart from the complex.
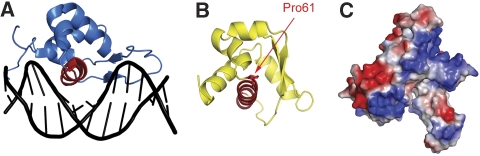
This preliminary modelling implies that the lower face of the outer belt of the N-terminal domain tier is involved in the interaction with dsDNA. A cluster of positively charged residues can be found in the same region, corresponding to the bottom of subdomain A (Figure 4C). A database search for homologous structures with the program SSM (Krissinel and Henrick, 2004) detects a similarity between subdomain A and a variety of proteins containing HTH motifs, including a number of basic HTH domains, as well as WH domains, such as in the E2F and the Genesis transcription factors. All of these proteins contact dsDNA through the recognition helix (Figure 4A). The same cluster of helices forming an HTH domain can be found within subdomain A (Figure 4B).
Figure 4.
Subdomain A contains a helix-turn-helix. (A) The crystal structure of the Genesis transcription factor bound to dsDNA (Jin et al, 1999; PDB entry 2HDC) with the recognition helix is highlighted in red. (B) The structure of subdomain A in the same orientation: the atomic model of proline 61 is shown in red, respectively. (C) Surface electrostatic potential of the N-terminal domain, in the same orientation as in Figure 1B, with negative charges shown in red and positive charges in blue.
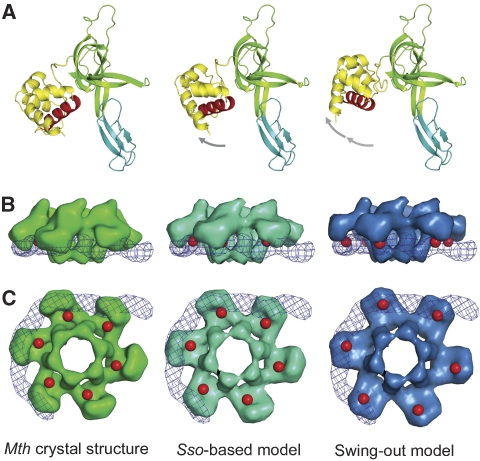
When analysed in the context of the entire N-terminal domain structure, the putative recognition helix is packed against subdomain C, so that a conformational change would be needed to allow for a protein–DNA interaction similar to Genesis (Figure 5). We have modelled this swing-out movement in four out of the six A subdomains, to avoid steric clashes and to allow productive interaction with the fitted dsDNA (Figure 5 and Supplementary Figure 5). The modelling took into account the more ‘open' conformation of subdomain A observed in the crystal structure of SsoMCM (Liu et al, 2008). This re-orientation allows for a better fit to the EM map at the interface between the N-terminal and AAA+ domains (Supplementary Figure 6). A swing-out movement of subdomain A had already been characterized both by crystallography as well as EM studies (Fletcher et al, 2003; Chen et al, 2005).
Figure 5.
Modelling of the swing-out movement of subdomain A. The left panel shows the crystal structure of the N-terminal domain of MthMCM (Fletcher et al, 2003), the central panel the same structure with subdomain A remodelled based on the crystal structure of the S. solfataricus MCM N-terminal domain (Liu et al, 2008), and the right panel the structure modelled to fit the EM data. The modelling was carried out by extrapolating the movement derived from the differences between the MthMCM and SsoMCM structures. (A) A ribbon diagram representation of the N-terminal domain monomer, colour-coded as in Figure 1B, with the helix beginning with proline 61 highlighted in red. The left panel shows that the putative recognition helix packs against subdomain C, whereas in the conformation shown on the right panel is available for interaction with dsDNA. (B) Side view surface representation of the hexameric N-terminal domain obtained by filtering to 20-Å resolution the electron density calculated from the models shown above, overlaid with a surface representation of the modelled dsDNA (depicted as a blue mesh). The location of the recognition helix is highlighted in red. (C) The same surface representation viewed from the bottom.
It has to be stressed that multiple conformations may be present in the sample, probably including small subsets of complexes where the dsDNA interacts with a different number of subunits. However, due to technical limitations, we choose not to separate these sub-populations, and to carry out a single 3D reconstruction. This may have caused a small degree of averaging and some additional noise. However, the higher intensity of electron density around three-quarter of the ring, the better definition of two N-terminal subunits, as well as the presence of the protrusions, suggest that the model presented here is likely to be representative of a majority of the existing conformations.
Biochemical characterization of the interaction with DNA
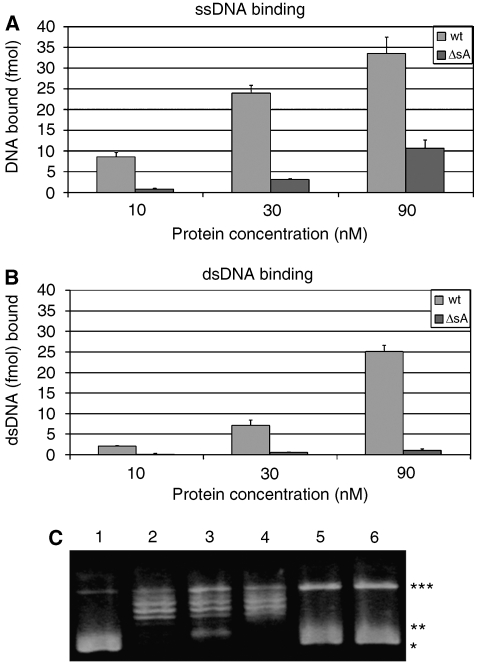
A biochemical study of the functional properties of various N-terminal deletion mutants of MthMCM, including the ΔsA mutant, has been already published (Kasiviswanathan et al, 2004). However, no data were available on dsDNA binding. We have therefore performed a detailed analysis of the ssDNA- and dsDNA-binding properties of the ΔsA mutant (Figure 6). The mutant shows a decreased ability to bind both ssDNA and dsDNA. Although the impairment in ssDNA binding can be partly counteracted by increasing the amount of protein, no binding to short stretches of dsDNA (50 bp) is observed even at high protein concentration. It is possible to detect some dsDNA binding for the ΔsA mutant when using longer dsDNA segments (250 bp, data not shown).
Figure 6.
Biochemical characterization of the MthMCM–DNA interaction. DNA-binding properties of the ΔsA mutant: filter-binding assays were performed using both the wt and ΔsA mutant proteins, in the presence of (A) a ssDNA substrate of 50 nucleotides and (B) a 50 bp dsDNA substrate. The ΔsA mutant shows a decreased binding to both ssDNA and dsDNA. Whereas the binding of ssDNA increases with the amount of protein, binding to short stretches of dsDNA (50 bp) remains virtually undetectable. (C) Topology footprint assay. Plasmid DNA relaxation at saturating concentration of Topoisomerase I and in the presence of MthMCM demonstrates a role of the MCM complex in the modulation of plasmid topological state and confirms the involvement of subdomain A. Lane 1: 5.7 nM pUC18. Lane 2: pUC18 incubated with 20 U of wheat germ Topoisomerase I. Lane 3: pUC18 incubated with 1 μM of wt MthMCM (hexamer), for 15 min, followed by a 60 min incubation with Topoisomerase I. Lane 4: the same experiment as in lane 3, where the ΔsA mutant is used instead of wt MthMCM. Lanes 5 and 6: pUC18 incubated with 1 μM of MthMCM or ΔsA, respectively. Different topological states can be visualized: *negatively supercoiled DNA; **partially relaxed DNA; ***totally relaxed DNA.
If dsDNA wraps around a protein ring one would expect a detectable change in the nucleic acid topology. To assess whether the interaction between the outer belt of the MthMCM N-terminal tier and nucleic acid affects the topology of dsDNA, we have carried out a topology footprint assay (Figure 6C), which is usually employed for DNA replication initiator proteins (Erzberger et al, 2006). The naturally negatively supercoiled pUC18 was incubated with MthMCM and then treated with saturating concentrations of Topoisomerase I. Reactions were stopped by the addition of a buffer containing Proteinase K, to degrade all DNA-bound proteins. Upon incubation with MthMCM, a band corresponding to a partially relaxed plasmid appeared. This band is not present when using the MthMCM mutant that lacks subdomain A. The observed behaviour is compatible with the dsDNA wrapping around the protein and suggests a possible role of MthMCM in modulating the topological state of the DNA substrate.
Discussion
In the present study, we report the cryo-EM structure of MthMCM bound to long dsDNA molecules, which shows a single hexameric ring sitting across dsDNA and bending it. We suggest that this is an initial site of interaction between protein and DNA, prior to the loading of the active helicase onto the replication fork. Our proposal for two different modes of protein–nucleic acid interaction is reminiscent of the model presented for the Rho termination factor, another hexameric AAA+ ATPase helicase (Skordalakes and Berger, 2003, 2006). As in the case of Rho, after an initial recognition step where the nucleic acid binds to the periphery of the hexamer, we envisage a second step in which the helicase loads onto the DNA, ready for translocation. A study on the S. cerevisiae pre-RC in vitro assembly describes two types of interaction between the MCM2–7 complex and DNA at origins, distinguishing between ‘association' and ‘loading' interactions, where the ‘associated' protein is sensitive to high salt washes, unlike the ‘loaded' protein (Bowers et al, 2004). The interaction between dsDNA and MCM described in this paper may provide a model for the ‘association' mode.
According to our single-particle analysis, a high variance in the electron density distribution is detected around the hexameric particles both in the wt and ΔC mutants (Figure 2D). Indeed, our EM structure shows density for dsDNA interacting with the lower face of the outer belt of the MthMCM N-terminal domain (corresponding to the N-terminal subdomain A; Figure 3). Secondary structure comparison allowed us to identify a similarity between subdomain A and a number of dsDNA-binding proteins containing HTH motifs (Figure 4). Although the topology of subdomain A does not entirely conform to a simple HTH or classical WH fold, a very large number of variations on the HTH theme have been described (Aravind et al, 2005). We have chosen the Genesis transcription factor as an example, as not only the three canonical helices of the helix-turn-helix fold match with corresponding helices in subdomain A, but an additional small helix inserted just before the recognition helix in Genesis (Gajiwala and Burley, 2000) is also present in subdomain A.
Although the recognition helix is partly buried at the interface between subdomains A and C in the crystal structure of the N-terminal domain, a swing-out movement of subdomain A, which would unmask the functional elements essential for dsDNA binding, has been previously proposed (Fletcher et al, 2003; Chen et al, 2005; Fletcher and Chen, 2006; Hoang et al, 2007). This conformational change has been correlated with a molecular switch, which initiates DNA replication in eukaryotes. The switch is triggered by a Cdc7-mediated phosphorylation event, which can be by-passed by a P83L substitution in yeast Mcm5, causing mutant cells to enter S phase prematurely (Hardy et al, 1997). The crystal structure of MthMCM harbouring the equivalent mutation (P61L) shows a small but significant opening of subdomain A (Fletcher et al, 2003). A similar but far more dramatic movement has been visualized in the EM structure of the MthMCM N-terminal domain (Chen et al, 2005), suggesting that subdomain A can undergo large conformational changes. Interestingly, the structural changes coupled to the Cdc7 phosphorylation event involve both the opening of a replication bubble and small but origin-dependent changes in DNA topology (Geraghty et al, 2000). Intriguingly, the same Pro → Leu mutation in yeast MCM3 is defective for MCM2–7 recruitment to replication origins (Lei et al, 2002).
Very recently, the structure of the MCM N-terminal domain from S. solfataricus was published (Liu et al, 2008). Although the overall fold and topology of the domain is very similar to MthMCM, there are significant differences in the relative orientation of the subdomains. In particular, S. solfataricus subdomain A is rotated by about 17° around the hinge connecting subdomains A and C. This causes subdomain A to open up in such a way as to partially expose the putative recognition helix. We therefore modelled the swing-out movement of subdomain A in MthMCM by further extrapolating from the position of the subdomain in the SsoMCM structure and rotating by roughly 50° in the same direction.
We also observed the formation of fibres, which seem to be correlated with the presence of endogenous or exogenous nucleic acid: fibres are visualized for the wt MthMCM purified by Ni-affinity chromatography (Supplementary Figure 2A), disappear when the protein is subjected to heparin chromatography to eliminate co-purified DNA contaminants, and can be reconstituted upon addition of long segments of dsDNA (Supplementary Figure 2B). Filaments of dsDNA can indeed be observed departing from the MCM fibres (Supplementary Figure 2B). Helical arrangements have been previously described for MthMCM (Chen et al, 2005), although the physiological role of the MCM helices was unclear. Although we have not demonstrated that the fibres we observe have a helical nature, the similarity to the ones in the publication mentioned above, is striking. We therefore observe that in the case of the wt protein, the formation of helices is favoured by the presence of dsDNA. Deletion of subdomain A makes the protein far more prone to the formation of helices, with or without DNA (Chen et al, 2005). A possible explanation is that the presence of subdomain A in the ‘closed' conformation somehow hinders the formation of the helical fibres: either the deletion of the subdomain or the structural change triggered by dsDNA allow the protein to adopt a conformation more compatible with the helical pitch.
The EM and modelling studies are supported by biochemical results confirming that the deletion of subdomain A strongly impairs binding of MthMCM to dsDNA, whereas it has a milder effect on ssDNA (Figure 6). It has to be stated that the possibility of multiple sites of DNA binding on the MCM complex (probably with different relative specificities and/or affinities) makes it difficult to design a definitive biochemical experiment that clearly targets one site rather than the other. A previous biochemical study showed that the ΔsA mutant still displays a weak helicase activity (Kasiviswanathan et al, 2004), suggesting that the impairment in the initial DNA binding does not hinder the canonical mode of interaction through the central channel, but somehow weakens the activity of the protein.
A model in which dsDNA encircles the MCM ring has consequences for the topology of DNA, modifying the degree of supercoiling. We have carried out topology footprint assays (Figure 6C). Although we cannot rule out that the effect may be simply due to the MCM protein protecting the dsDNA and limiting topoisomerase access, the result of the assay, coupled with our EM work, suggests that MthMCM has indeed an effect on the degree of writhing of a negatively supercoiled plasmid. The effect is not present with the ΔsA mutant, suggesting that subdomain A is involved in the topological change. Notably, a role of the MCM2–7 protein in stabilizing negative supercoils upon loading onto the origin of replication has been previously reported in an in vivo study in yeast (Chiani et al, 2006).
An intriguing set of structural similarities can be found between the MCM protein and a number of DNA replication initiator proteins, which all contain a AAA+ domain and an HTH or WH dsDNA-binding domain. In the past couple of years, the structural characterization of various initiator proteins has enhanced our understanding of the nucleoprotein architecture at the origin of replication (Erzberger et al, 2006; O'Donnell and Jeruzalmi, 2006; Dueber et al, 2007; Gaudier et al, 2007). Unlike other classical AAA+ complexes that form planar rings, the initiator proteins can form non-planar open-ring configurations or helical assemblies (Clarey et al, 2006; Erzberger et al, 2006). A general model has been proposed for initiator proteins, involving the regulation of the degree of DNA supercoiling at the origin of replication, and hence facilitating its melting (Erzberger et al, 2006; Grainge et al, 2006). In particular, the bacterial initiator DnaA forms an ATP-dependent helical structure around which dsDNA is wrapped and has a direct influence on the topology of the DNA, facilitating the opening of the origin of replication (Erzberger et al, 2006). In addition to the helical configuration observed for the ATP-bound form (Erzberger et al, 2006), DnaA has also been shown to form globular assemblies, around which dsDNA is wrapped (Funnell et al, 1987). This interaction stabilizes the formation of DNA kinks, in a configuration very much reminiscent of the one we characterize for MthMCM. Moreover, DnaA-like helical arrangements can be observed for MthMCM (Chen et al, 2005).
We find it tempting to speculate that the primary mode of DNA binding that we characterized in the context of a single ring can be extrapolated to a helical architecture, with a similar pattern to that proposed for the ATP-bound form of DnaA (Erzberger et al, 2006), supporting the notion that MCM has a role in origin melting, prior to being loaded as an active helicase onto the replication fork. Although the type of DNA binding we see in our EM map, with the dsDNA encircling MCM, may account for the MCM-induced changes in DNA topology, a more robust effect would be generated by the DNA wrapping around an MCM helical fibre. Our results may therefore confirm and extend the suggestion that MCM may bind DNA in a helical form in vivo (Chen et al, 2005), thus explaining the relative abundance of MCM proteins in the cell.
The novel helix-turn-helix motif in the N-terminal domain of MCM is in addition to the WH motif predicted within the C-terminal domain (Koonin, 1993). Whether the C-terminal WH motif can bind DNA is still controversial (Jenkinson and Chong, 2006; Barry et al, 2007; Pucci et al, 2007). By carrying out a 3D reconstruction for both the wt and ΔC mutants, we confirm the localization of the C-terminal domain on top of the AAA+ module (Supplementary Figure 4). Some unassigned electron density adjacent to the WH domain remains in the cryo-EM map (Figure 3C): although the region is partially disordered, we cannot rule out the possibility that the unassigned density represents dsDNA interacting with the C-terminal WH domain.
Taken together, the observations presented in this study provide new insights into the initial stages of interaction between MCM and DNA, describing the role for the MCM complex during the initiation step of DNA replication. A crucial question remains unanswered, as to what triggers the transition between the two modes of DNA interaction (wrapping around the N-terminal domain or threading through the central channel).
Materials and methods
EM sample preparation
Expression and purification of the wt and mutant MthMCM proteins were performed as previously reported (Kasiviswanathan et al, 2004; Costa et al, 2006b; Jenkinson and Chong, 2006) and all samples were buffer exchanged in buffer A (30 mM Tris–HCl pH 7.5, 150 mM NaCl, 5 mM MgCl2 and 5 mM β-mercaptoethanol). The vector pET15b (Novagen) was digested using BamHI and XbaI restriction enzymes, to generate a 5599-bp dsDNA fragment, which was purified from a 2% agarose gel and eluted in buffer A. The samples were prepared by incubating for 10 min at room temperature a solution containing both protein and dsDNA at a final concentration of 12.5 μg/ml in buffer B (30 mM Tris–HCl pH 7.5, 50 mM NaCl, 5 mM MgCl2, 5 mM β-mercaptoethanol). Negative-stain preparations were performed as follows: a sample of 4 μl was incubated for 1 min on glow-discharged continuous carbon-coated formvar copper or formvar nickel grids. The grid was washed with a 40 μl drop of 2% uranyl-acetate solution, blotted and air-dried. For cryo preparations, a sample of 4 μl was applied on a glow discharged holy-carbon copper grid (Quantifoil R 1.2/1.3), blotted for 2.5 s at an off-set of −1 and flash frozen in liquid ethane, using an automated plunger (FEI Vitrobot), yielding MthMCM nucleoprotein complexes embedded in a thin film of vitrified ice.
EM data collection
Negative-stain EM of the wt and mutant nucleoprotein preparations was performed on either a Tecnai12 electron microscope (FEI) at an accelerating voltage of 120 kV, or a CM200 (FEI) field-emission gun EM at an accelerating voltage of 200 kV. To enhance low-frequency components typical of the long stretches of dsDNA, a magnification of × 11 500–27 500 and 5–10 μm nominal defocus were used. For fibre diffraction studies, defocal pair micrographs were imaged on a CM200 EM under low-dose conditions, at a magnification of × 50 000 and at 0.5 and 5 μm nominal defocus. Cryo-EM was performed on a CM200 EM under low-dose conditions, at a magnification of × 50 000 and recording defocal pairs at 3.5 and 2.5–0.7 μm nominal defocus. Micrographs were digitized using a Nikon Super Coolscan 8000 and coarsened by a factor of 2, leading to a pixel size of 2.6 Å on the specimen scale.
2D single-particle analysis
Statistical analysis of the angular distribution of DNA kinks was carried out by selecting 2D coordinates of three neighbouring MthMCM particles contacting DNA (using the program Boxer; Ludtke et al, 1999) and deriving the comprised angle.
Cryo-EM particles were selected automatically using the program Boxer (Ludtke et al, 1999) from 3.5 μm defocus micrographs, using a few manually selected particles as a reference. Images were cut from the corresponding low defocus micrograph for each defocal pair. A data set of 12 000 particles were selected from micrographs of the wt–DNA complex, and 7000 particles were selected from the ΔC–DNA complex. Contrast-transfer-function correction was applied on 512 × 512 pixel images using the findctf2d program (Grant et al, manuscript in preparation) and the size of the image frame was subsequently reduced to 256 × 256 pixels and band-pass filtered with a low-frequency cutoff of 200 Å and a high-frequency cutoff of 6 Å. Images were iteratively centred translationally to their rotationally averaged total sum and their symmetry components were determined by MSA symmetry analysis (Dube et al, 1993).
3D single-particle reconstruction
An initial six-fold symmetric reference was used for angular assignment to MSA-based class averages. After symmetry relaxation, non-symmetric electron density features surrounding the bottom face of the molecule started to appear. To further investigate the statistical relevance of these features, six-fold symmetry was imposed again on the structure, and the map was binarized at a threshold, which allowed visualization of six rod-shaped electron densities departing from the outer belt of the bottom face of the MthMCM ring. Upon MSA classification of images aligned to the re-projections of this symmetry-imposed reference structure, class averages were obtained, showing only two and not six filamentous electron density features projecting radially from the body of the molecule. This departure from six-fold symmetry (Boekema et al, 1986) was in agreement with highly defocused images of the wt nucleoprotein complex, confirming the significance of the rod-like electron density features. Further refinement of the nucleoprotein complex was performed without any symmetry imposition, resulting in a structure based on ∼10 000 particles with a resolution of 19 and 25 Å, according to the 1/2 bit and 0.5 FSC criteria, respectively (Supplementary Figure 2). To minimize anisotropy-derived artefacts, only a limited portion of the abundant end-on-view-oriented particles was included in the final reconstruction (Supplementary Figure 2 and Supplementary data; Boisset et al, 1998). The reconstruction of the ΔC MthMCM mutant was performed as described for the wt protein, based on 5000 particles at a resolution of 25 and 30 Å according to the 1/2 bit and 0.5 FSC criteria, respectively. The electron density maps of the wt and ΔC proteins were displayed at a threshold corresponding to 130% of their molecular weight (to account for the flexible nature of the C-terminal domain and the interacting dsDNA). The wt and ΔC mutant structures were superposed automatically using Chimera (Pettersen et al, 2004).
Atomic modelling and map fitting
The atomic model of a hexameric MCM complex was obtained by superposing six monomers of the MkaMCM protein (Bae et al, submitted) to a planar single hexamer of the MthMCM N-terminal domain structure (Fletcher et al, 2003; PDB entry 1LTL), resulting in a hybrid model consisting of the MthMCM N-terminal domains and the MkaMCM AAA+ ATPase domains. The model was fitted automatically into the EM map using Chimera (Pettersen et al, 2004). dsDNA was manually fitted to the circular belt of electron density obtained by displaying the EM map at a threshold of 5.5σ.
A switchblade movement of subdomain A (residues 1–92) within the MthMCM N-terminal domain was modelled based on the crystal structure of the N-terminal domain of SsoMCM (Liu et al, 2008). The atomic coordinates of the N-terminal MthMCM hexamer (PDB entry 1LTL; Fletcher et al, 2003) were superposed to the N-terminal SsoMCM hexamer (PDB entry 2VL6; Liu et al, 2008) using EBI SSM (Krissinel and Henrick, 2004). In particular, the two hexamers formed by sC alone were used to perform the superposition, as this represents the statistically most significant secondary structure match (with a Q score of 0.21 and a Z score of 10, whereas superposition of the full-length hexamers yields a Q score of 0.11 and a Z score of 2.03). It should be noted that this modelling approach differs drastically from the one performed by Liu and colleagues where monomeric full-length N-terminal domains were superposed. The swing-out movement was modelled by superposing the subdomain A of MthMCM onto the corresponding positions in SsoMCM (Figure 5, central panels), and then further extrapolated (Figure 5, right panel).
The structure of subdomain A (Fletcher et al, 2003; PDB entry 1LTL) was submitted to the SSM server (Krissinel and Henrick, 2004) to be compared with the entire Protein Data Bank. When subdomain A from monomer C or D in the hexamer was used, the program detected a number of matches to proteins containing HTH or WH motifs (PDB entries: 1K61, 1X2N, 1CF7, 1B72, 1PUF, 2CQX, 1APL, 1P7J, 2H13, 1IG7, 2HFH). No matches with a significant Q score were found when subdomain A from monomers A, B, E and F was used, although the monomers in the N-terminal hexamer show no obvious differences.
Atomic modelling of the C-terminal domain was performed using SSM (Krissinel and Henrick, 2004) to superpose the α-domain (residues 190–276) of Cdc6 from P. aerophilum (Liu et al, 2000; PDB entry 1FNN) onto the α domain of MkaMCM (residues 410–492). The resulting position of the C-terminal WH domain of Cdc6 was manually adjusted to fit the MCM electron density map and optimized by automatic fitting using Chimera (Pettersen et al, 2004).
Handling of atomic coordinate files was performed using the CCP4 package (CCP4, 1994) and figures were generated using Pymol (DeLano, 2002).
Nitrocellulose filter DNA-binding assay
Single- and double-stranded DNA substrates were prepared by labelling a 49 mer oligonucleotide (5′-GCAGATAACAGTTGTCCTGGAGAACGACCTGGTTGACACCCTCACACCC-3′) using [γ-32P]ATP and T4 polynucleotide kinase. To generate the duplex substrate, the oligonucleotide was annealed to the complementary strand and unincorporated [γ-32P]ATP was removed from the DNA substrate by extraction from polyacrylamide gel as previously described (Shin et al, 2003).
Filter binding assays were carried out in a reaction mixture (20 μl) containing 25 mM Hepes-NaOH (pH 7.5), 2 mM dithiothreitol (DTT), 10 mM MgCl2, 100 μg/ml bovine serum albumin, 50 fmol of 32P-labelled DNA substrates, and 10, 30 and 90 nM of MCM proteins (as monomers). After incubation at 60°C for 10 min the mixture was filtered through an alkaline-washed nitrocellulose filter (Millipore HA 0.45 μm), which was subsequently washed with 20 mM Tris–HCl (pH 7.5; McEntee et al, 1980). The radioactivity adsorbed to the filter was measured by liquid scintillation counting. All DNA-binding experiments were repeated three times and their averages with standard deviations are shown in Figure 6.
Topology footprint assay
Topology footprint assays were carried out in 15 μl of reaction buffer (50 mM Tris, pH 7.5, 1 mM DTT, 5 mM MgCl2, 30% (v/v) glycerol) containing 150 ng of DNA (5.7 nM). The DNA was incubated for 15 min at 37°C with 1 μM of wt MthMCM or with the same amount of ΔsA mutant, followed by the addition of 20 U of wheat germ Topoisomerase I. Reactions were stopped by incubating the reaction mixture with 5 μl of Stop buffer (1 mg/ml Proteinase K, 100 mM EDTA, 1% SDS and 50% glycerol) for 15 min at 37°C. The samples were loaded on a 1.2% agarose gel in TBE 1 × and electrophoresis was performed for 90 min at 11 V/cm. Results were visualized after ethidium bromide staining.
Accession numbers
The electron density map has been deposited in the EMDB database (accession code EMD-1526).
Supplementary Material
Supplementary Data
Acknowledgments
We thank Elizabeth R Jenkinson and Rajesh Kasiviswanathan for the purification of the ΔC and ΔsA deletion mutants, Marta Carroni for the assistance in nucleoprotein sample preparation, Cecilia Bebeacua for helpful discussion and assistance with the software, Peter Brick for assistance with the software and critical reading of the paper. A special thank goes to Timothy Grant, for sharing image analysis programs in the phase of development. AC was supported by a studentship from the Department of Life Science, Imperial College. BM was supported by a studentship from ELETTRA. Research in the Kelman lab is supported by a Grant from the National Science Foundation (MCB-0815646).
References
- Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM (2005) The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev 29: 231–262 [DOI] [PubMed] [Google Scholar]
- Arias EE, Walter JC (2007) Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev 21: 497–518 [DOI] [PubMed] [Google Scholar]
- Barry ER, McGeoch AT, Kelman Z, Bell SD (2007) Archaeal MCM has separable processivity, substrate choice and helicase domains. Nucleic Acids Res 35: 988–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekema EJ, Berden JA, van Heel MG (1986) Structure of mitochondrial F1-ATPase studied by electron microscopy and image processing. Biochim Biophys Acta 851: 353–360 [DOI] [PubMed] [Google Scholar]
- Boisset N, Penczek PA, Taveay JC, You V, de Haas F, Lamy J (1998) Overabundant single-particle electron microscope views induce a three-dimensional reconstruction artifact. Ultramicroscopy 74: 201–207 [Google Scholar]
- Bowers JL, Randell JC, Chen S, Bell SP (2004) ATP hydrolysis by ORC catalyzes reiterative Mcm2–7 assembly at a defined origin of replication. Mol Cell 16: 967–978 [DOI] [PubMed] [Google Scholar]
- Carpentieri F, De Felice M, De Falco M, Rossi M, Pisani FM (2002) Physical and functional interaction between the mini-chromosome maintenance-like DNA helicase and the single-stranded DNA binding protein from the crenarchaeon Sulfolobus solfataricus. J Biol Chem 277: 12118–12127 [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Chen YJ, Yu X, Kasiviswanathan R, Shin JH, Kelman Z, Egelman EH (2005) Structural polymorphism of Methanothermobacter thermautotrophicus MCM. J Mol Biol 346: 389–394 [DOI] [PubMed] [Google Scholar]
- Chiani F, Di Felice F, Camilloni G (2006) SIR2 modifies histone H4-K16 acetylation and affects superhelicity in the ARS region of plasmid chromatin in Saccharomyces cerevisiae. Nucleic Acids Res 34: 5426–5437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JP, Hayashi MK, Simon MN, Xu RM, Stillman B (2000) A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase. Proc Natl Acad Sci USA 97: 1530–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarey MG, Erzberger JP, Grob P, Leschziner AE, Berger JM, Nogales E, Botchan M (2006) Nucleotide-dependent conformational changes in the DnaA-like core of the origin recognition complex. Nat Struct Mol Biol 13: 684–690 [DOI] [PubMed] [Google Scholar]
- Costa A, Onesti S (2008) The MCM complex: (just) a replicative helicase? Biochem Soc Trans 36: 136–140 [DOI] [PubMed] [Google Scholar]
- Costa A, Pape T, van Heel M, Brick P, Patwardhan A, Onesti S (2006a) Structural basis of the Methanothermobacter thermautotrophicus MCM helicase activity. Nucleic Acids Res 34: 5829–5838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Pape T, van Heel M, Brick P, Patwardhan A, Onesti S (2006b) Structural studies of the archaeal MCM complex in different functional states. J Struct Biol 156: 210–219 [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002) The Pymol Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific [Google Scholar]
- Dimitrova DS, Todorov IT, Melendy T, Gilbert DM (1999) Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J Cell Biol 146: 709–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube P, Tavares P, Lurz R, van Heel M (1993) The portal protein of bacteriophage SPP1: a DNA pump with 13-fold symmetry. EMBO J 12: 1303–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueber EL, Corn JE, Bell SD, Berger JM (2007) Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science 317: 1210–1213 [DOI] [PubMed] [Google Scholar]
- Edwards MC, Tutter AV, Cvetic C, Gilbert CH, Prokhorova TA, Walter JC (2002) MCM2–7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J Biol Chem 277: 33049–33057 [DOI] [PubMed] [Google Scholar]
- Erzberger JP, Mott ML, Berger JM (2006) Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat Struct Mol Biol 13: 676–683 [DOI] [PubMed] [Google Scholar]
- Fletcher RJ, Bishop BE, Leon RP, Sclafani RA, Ogata CM, Chen XS (2003) The structure and function of MCM from archaeal M. thermoautotrophicum. Nat Struct Biol 10: 160–167 [DOI] [PubMed] [Google Scholar]
- Fletcher RJ, Chen XS (2006) Biochemical activities of the BOB1 mutant in Methanobacterium thermoautotrophicum MCM. Biochemistry 45: 462–467 [DOI] [PubMed] [Google Scholar]
- Forsburg SL (2004) Eukaryotic MCM proteins: beyond replication initiation. Microbiol Mol Biol Rev 68: 109–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funnell BE, Baker TA, Kornberg A (1987) In vitro assembly of a prepriming complex at the origin of the Escherichia coli chromosome. J Biol Chem 262: 10327–10334 [PubMed] [Google Scholar]
- Gajiwala KS, Burley SK (2000) Winged helix proteins. Curr Opin Struct Biol 10: 110–116 [DOI] [PubMed] [Google Scholar]
- Gaudier M, Schuwirth BS, Westcott SL, Wigley DB (2007) Structural basis of DNA replication origin recognition by an ORC protein. Science 317: 1213–1216 [DOI] [PubMed] [Google Scholar]
- Geraghty DS, Ding M, Heintz NH, Pederson DS (2000) Premature structural changes at replication origins in a yeast minichromosome maintenance (MCM) mutant. J Biol Chem 275: 18011–18021 [DOI] [PubMed] [Google Scholar]
- Gomez-Llorente Y, Fletcher RJ, Chen XS, Carazo JM, San Martin C (2005) Polymorphism and double hexamer structure in the archaeal minichromosome maintenance (MCM) helicase from Methanobacterium thermoautotrophicum. J Biol Chem 280: 40909–40915 [DOI] [PubMed] [Google Scholar]
- Grainge I, Gaudier M, Schuwirth BS, Westcott SL, Sandall J, Atanassova N, Wigley DB (2006) Biochemical analysis of a DNA replication origin in the archaeon Aeropyrum pernix. J Mol Biol 363: 355–369 [DOI] [PubMed] [Google Scholar]
- Grainge I, Scaife S, Wigley DB (2003) Biochemical analysis of components of the pre-replication complex of Archaeoglobus fulgidus. Nucleic Acids Res 31: 4888–4898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy CF, Dryga O, Seematter S, Pahl PM, Sclafani RA (1997) mcm5/cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc Natl Acad Sci USA 94: 3151–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugland GT, Shin JH, Birkeland NK, Kelman Z (2006) Stimulation of MCM helicase activity by a Cdc6 protein in the archaeon Thermoplasma acidophilum. Nucleic Acids Res 34: 6337–6344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang ML, Leon RP, Pessoa-Brandao L, Hunt S, Raghuraman MK, Fangman WL, Brewer BJ, Sclafani RA (2007) Structural changes in Mcm5 protein bypass Cdc7–Dbf4 function and reduce replication origin efficiency in S. cerevisiae. Mol Cell Biol 27: 7594–7602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson ER, Chong JP (2006) Minichromosome maintenance helicase activity is controlled by N- and C-terminal motifs and requires the ATPase domain helix-2 insert. Proc Natl Acad Sci USA 103: 7613–7618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Marsden I, Chen X, Liao X (1999) Dynamic DNA contacts observed in the NMR structure of winged helix protein–DNA complex. J Mol Biol 289: 683–690 [DOI] [PubMed] [Google Scholar]
- Kasiviswanathan R, Shin JH, Melamud E, Kelman Z (2004) Biochemical characterization of the Methanothermobacter thermautotrophicus minichromosome maintenance (MCM) helicase N-terminal domains. J Biol Chem 279: 28358–28366 [DOI] [PubMed] [Google Scholar]
- Kelman Z, Lee JK, Hurwitz J (1999) The single minichromosome maintenance protein of Methanobacterium thermoautotrophicum DeltaH contains DNA helicase activity. Proc Natl Acad Sci USA 96: 14783–14788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV (1993) A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res 21: 2541–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D 60: 2256–2268 [DOI] [PubMed] [Google Scholar]
- Labib K, Kearsey SE, Diffley JF (2001) MCM2–7 proteins are essential components of prereplicative complexes that accumulate cooperatively in the nucleus during G1-phase and are required to establish, but not maintain, the S-phase checkpoint. Mol Biol Cell 12: 3658–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Cheng IH, Roberts LA, McAlear MA, Tye BK (2002) Two mcm3 mutations affect different steps in the initiation of DNA replication. J Biol Chem 277: 30824–30831 [DOI] [PubMed] [Google Scholar]
- Liu J, Smith CL, DeRyckere D, DeAngelis K, Martin GS, Berger JM (2000) Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control. Mol Cell 6: 637–648 [DOI] [PubMed] [Google Scholar]
- Liu W, Pucci B, Rossi M, Pisani FM, Ladenstein R (2008) Structural analysis of the Sulfolobus solfataricus MCM protein N-terminal domain{dagger}. Nucleic Acids Res 36: 3235–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W (1999) EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128: 82–97 [DOI] [PubMed] [Google Scholar]
- McEntee K, Weinstock GM, Lehman IR (1980) recA protein-catalyzed strand assimilation: stimulation by Escherichia coli single-stranded DNA-binding protein. Proc Natl Acad Sci USA 77: 857–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer SE, Lewis PW, Botchan MR (2006) Isolation of the Cdc45/Mcm2–7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci USA 103: 10236–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell M, Jeruzalmi D (2006) Helical proteins initiate replication of DNA helices. Nat Struct Mol Biol 13: 665–667 [DOI] [PubMed] [Google Scholar]
- Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC (2006) Localization of MCM2–7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell 21: 581–587 [DOI] [PubMed] [Google Scholar]
- Pape T, Meka H, Chen S, Vicentini G, van Heel M, Onesti S (2003) Hexameric ring structure of the full-length archaeal MCM protein complex. EMBO Rep 4: 1079–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Pucci B, De Felice M, Rocco M, Esposito F, De Falco M, Esposito L, Rossi M, Pisani FM (2007) Modular organization of the Sulfolobus solfataricus mini-chromosome maintenance protein. J Biol Chem 282: 12574–12582 [DOI] [PubMed] [Google Scholar]
- Randell JC, Bowers JL, Rodriguez HK, Bell SP (2006) Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2–7 helicase. Mol Cell 21: 29–39 [DOI] [PubMed] [Google Scholar]
- Shechter DF, Ying CY, Gautier J (2000) The intrinsic DNA helicase activity of Methanobacterium thermoautotrophicum delta H minichromosome maintenance protein. J Biol Chem 275: 15049–15059 [DOI] [PubMed] [Google Scholar]
- Shin JH, Jiang Y, Grabowski B, Hurwitz J, Kelman Z (2003) Substrate requirements for duplex DNA translocation by the eukaryal and archaeal minichromosome maintenance helicases. J Biol Chem 278: 49053–49062 [DOI] [PubMed] [Google Scholar]
- Skordalakes E, Berger JM (2003) Structure of the Rho transcription terminator: mechanism of mRNA recognition and helicase loading. Cell 114: 135–146 [DOI] [PubMed] [Google Scholar]
- Skordalakes E, Berger JM (2006) Structural insights into RNA-dependent ring closure and ATPase activation by the Rho termination factor. Cell 127: 553–564 [DOI] [PubMed] [Google Scholar]
- Takahashi TS, Wigley DB, Walter JC (2005) Pumps, paradoxes and ploughshares: mechanism of the MCM2–7 DNA helicase. Trends Biochem Sci 30: 437–444 [DOI] [PubMed] [Google Scholar]
- Yu X, VanLoock MS, Poplawski A, Kelman Z, Xiang T, Tye BK, Egelman EH (2002) The Methanobacterium thermoautotrophicum MCM protein can form heptameric rings. EMBO Rep 3: 792–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data