Abstract
Ras of complex proteins (Roc) belongs to the superfamily of Ras-related small G-proteins that always occurs in tandem with the C-terminal of Roc (COR) domain. This Roc–COR tandem is found in the bacterial and eukaryotic world. Its most prominent member is the leucine-rich repeat kinase LRRK2, which is mutated and activated in Parkinson patients. Here, we investigated biochemically and structurally the Roco protein from Chlorobium tepidum. We show that Roc is highly homologous to Ras, whereas the COR domain is a dimerisation device. The juxtaposition of the G-domains and mutational analysis suggest that the Roc GTPase reaction is stimulated and/or regulated by dimerisation in a nucleotide-dependent manner. The region most conserved between bacteria and man is the interface between Roc and COR, where single-point Parkinson mutations of the Roc and COR domains are in close proximity. The analogous mutations in C. tepidum Roc–COR decrease the GTPase reaction rate, most likely due to a modification of the interaction between the Roc and COR domains.
Keywords: dimerisation device, LRRK2, Parkinson, Roc–COR, structure
Introduction
G-proteins are molecular switches that cycle between a GTP-bound active and a GDP-bound inactive form. In the case of the superfamily of Ras-like proteins, which are 20–25 kDa proteins, nucleotides are bound very tightly such that the exchange of GDP for GTP in the activation process requires guanine nucleotide exchange factors (GEFs), which increase the otherwise slow intrinsic dissociation reaction by orders of magnitude. The GTPase reaction of these proteins is slow such that GTPase-activating proteins (GAPs) cognate for each class of Ras proteins are required for stimulation of the phosphoryl transfer reaction by orders of magnitude (Vetter and Wittinghofer, 2001).
A second group of G-proteins use a different mode of reaction for the switch cycle. They have a much lower affinity for nucleotide such that GDP dissociation is fast and does not require the action of a GEF. Their GTPase reaction is also slow but stimulation of the reaction requires the dimerisation of the G-domains. A particular well-studied example is the human guanylate binding protein hGBP1, an interferon-γ-induced G-protein of the dynamin superfamily (Martens and Howard, 2006). Here, the dimerisation through the G-domains optimally rearranges the catalytic machinery for catalysis (Ghosh et al, 2006). In the case of the tRNA-modifying enzyme MnmE and the regulator of Ni insertion into hydrogenases HypB, the G-domain homo-dimerisation rearranges the active site and/or supplements that of one protomer by residues from the other (Egea et al, 2004; Focia et al, 2004; Shan et al, 2004; Scrima et al, 2005; Gasper et al, 2006; Scrima and Wittinghofer, 2006). A special case of this type of mechanism has been documented for the pseudo-dimerisation of signal recognition particle and its receptor, where the two highly homologous proteins stimulate the GTPase of each other by complementing the active site of each other (Egea et al, 2004; Focia et al, 2004, 2006).
Ras of complex proteins (Roc) is a Ras-like GTP-binding domain, which is 20–25 kDa in size and forms a separate group of the Ras superfamily (Bosgraaf and Van Haastert, 2003). Its name stems from its presence in very large multidomain proteins that occur in bacteria, plants and animals. It is always found in tandem with the C-terminal of Roc (COR) domain. It was first discovered in a very large protein in Dictyostelium, GbpC, which was identified as a candidate cyclic GMP target protein (Goldberg et al, 2002). Dictyostelium contains a large number of these proteins, whereas mammals contain four members of the Roco family (Bosgraaf and Van Haastert, 2003). Two of these were originally called Roco1 and Roco2 and consist of a leucine-rich repeat (LRR) domain N-terminal and a protein kinase homologous MAPKKK domain C-terminal to the Roc–COR tandem. They are commonly called LRRK1 and LRRK2. The two other human orthologues are MASL1 with LRR and Roc–COR domains and DAPk with a kinase domain N-terminal and death domain C-terminal to Roc–COR (Bosgraaf and Van Haastert, 2003).
Parkinson disease (PD) (OMIM no. 168600) is the second most common neurodegenerative disease, the hallmarks of which are loss of dopaminergic neurons and deposition of protein aggregates termed Lewis bodies in the substantia nigra (Farrer, 2006). Although it occurs spontaneously with an incidence of 2% in the general population later in life, an estimated number of nine genes responsible for the rare inherited forms of the disease have been identified. Mutations in PARK8 on chrosomome 12 were identified in several families with PD as the LRRK2 gene coding for a protein that is also called dardarin (basque for tremor) (Paisan-Ruiz et al, 2004; Zimprich et al, 2004). Many more families with mutations in LRRK2/dardarin (LRRK2 from now) have been identified since then, with mutations localised to one apparent hot spot each in the LRR, Roc and COR domains, and several mutations in the kinase domain (Zimprich et al, 2004; Gilks et al, 2005; Nichols et al, 2005; West et al, 2007).
The molecular mechanism of how mutations of LRRK2 lead to loss of dopaminergic neurons is not well known (Smith et al, 2005; West et al, 2007). It has been speculated that mutations result in enhanced kinase activity due to an activation of the Roc domain. For the Dictyostelium protein GbpC, it was demonstrated that the protein combines elements of signal receiver, modulator and signal output in the same polypeptide (van Egmond et al, unpublished data). In the case of LRRK2, it could be argued that the Roc–COR domain tandem is regulated by LRR, a frequently used protein–protein or protein–ligand interaction domain, whereas the G-domain switch regulates protein kinase activity. Most of previous studies have indeed shown that the mutations associated with PD are gain-of-function mutations that increase the signal output as measured by increased kinase activity towards a non-specific substrate MBP, auto-phosphorylation and/or cellular toxicity (Smith et al, 2005, 2006; Greggio et al, 2006; Korr et al, 2006; Guo et al, 2007; Lewis et al, 2007; Li et al, 2007; Luzon-Toro et al, 2007; West et al, 2007).
The function of Roc–COR domains as the putative regulator of kinase activity is not well understood. However, it is known that it functions as a bona GTP-binding protein with a low GTPase activity that somehow stimulates the kinase activity (Korr et al, 2006; Greggio et al, 2007; Guo et al, 2007; Ito et al, 2007; West et al, 2007). Various scenarios have been proposed whereby the mutations in the Roc or COR domains lead to a increase or an decrease in GTP binding (unlikely) and/or the GTPase activity as the most likely regulatory module of LRRK2 (Korr et al, 2006; Greggio et al, 2007; Guo et al, 2007; Ito et al, 2007; Li et al, 2007; West et al, 2007). To clarify the role of the conserved Roc–COR unit, we have solved the structure of a bacterial Roc–COR unit and studied this Roco protein biochemically. The structure obtained is fundamentally different from a recently described Roc domain structure and the conclusions drawn from it (Deng et al, 2008). In analogy to similar G-domain proteins, we would postulate that COR is a constitutive dimerisation motif that is required for GTP-dependent juxtaposition of Roc, and that the dimerisation-dependent GTPase might regulate protein kinase activity.
Results
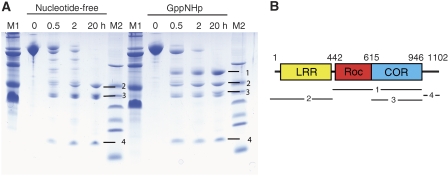
Chlorobium tepidum Roco protein is a 124 kDa protein of 1102 residues consisting of LRRs, Roc and COR domains (Figure 1). In contrast to the human LRRK2 or any fragments thereof, it can be expressed in Escherichia coli and purified as a soluble and stable protein (see below). As the full-length protein could not be crystallised, we searched for smaller fragments that might be more amenable to structural investigation. Mild digestion with trypsin gave a distinct pattern of fragments (Figure 1), and the same pattern was obtained with elastase. The bands observed in PAGE were identified by ESI mass spectrometry and N-terminal sequencing. Band 2 has a molecular mass of 48.8 kDa and contains the LRR, from residue −6 (from the tag) to Arg441. Band 3 has a molecular mass of 38.3 kDa and is created by trypsin-cutting N-terminal (at Ser615) and C-terminal (at Arg946) to the COR domain. Band 4 is obtained by proteolysis of the C-terminal end behind the COR domain. No band for the Roc domain can be identified after proteolysis of the nucleotide-free protein, which indicates that either the Roc domain alone is completely digested by trypsin even under conditions of limited proteolysis or insoluble. We favour the latter explanation as the Roc domain alone cannot be expressed in our hands (see below). The proteolysis experiments also show that the connection between the Roc and COR domains is flexible and very susceptible to cleavage. GTP induces a major conformational change of the Ct Roco protein as demonstrated by performing the proteolysis experiment in the presence of saturating concentrations of GppNHp. While the protein is just as easily digested C-terminal to the LRR domain, the Roc–COR tandem is now considerably more stable. Roc–COR corresponds to band 1 in PAGE, comprising residues 442–946, and is only obtained in the presence of GppNHp (Figure 1). This indicates that the Roc–COR connection is less flexible in the presence of nucleotide (this can also be shown by using the separately expressed Roc–COR tandem, data not shown). We conclude from this that Roc is stabilised in the presence of the COR domain and the Roc–COR connection is stabilised by binding to nucleotide.
Figure 1.
Tryptic digestion of the C. tepidum Roco protein in the nucleotide-free (left lanes) and the GppNHp-bound state (right lanes) as described in Materials and methods. At the indicated time points of 0, 0.5, 2 and 20 h, samples were taken and analysed by PAGE, as indicated. M, molecular mass markers. (B) Scheme of the domain structure of C. tepidum Roco and the boundaries of fragments 1–4 obtained as indicated to the right of the gel in (A). Fragment 1 includes the Roc–COR tandem (442–946), 2 the LRR-domain (−6-441), 3 the COR domain (615–946) and 4 the C terminus (947–1090).
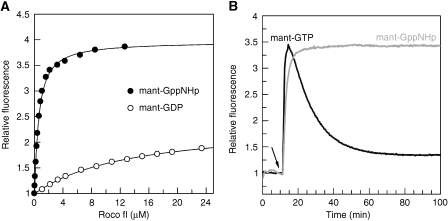
In contrast to Ras-like proteins, the purified protein was only partially bound with nucleotide. Titration of the nucleotide-free protein with fluorescent-labelled mant-GDP shows that binding with nucleotide can be saturated, with an equilibrium dissociation constant of 13.4 μM (Figure 2A), which indicates that the protein has a fast dissociation rate and would not require any GEF for nucleotide exchange. The affinity to the GTP analogue mant-GppNHp is considerably higher, with a KD of 0.5 μM. Whereas the twofold fluorescence increase is typical for the binding of mant-labelled nucleotides, the binding of mant-GppNHp nucleotide induces a much larger fluorescence amplitude change, indicating a significant conformational change around the nucleotide-binding site when going from the GDP- to the GTP-bound state. The high affinity of the nucleoside triphosphate is not due to the label, as unlabelled nucleotide has a similar affinity.
Figure 2.
Biochemical analysis of C. tepidum Roco. (A) Affinities of fluorescent nucleotides measured by the fluorescence increase obtained by adding increasing concentrations of protein to 0.2 μM mant-GDP (empty circles) or mant-GppNHp (filled circles). The equilibrium constant KD was obtained by fitting the data to a quadratic equation. (B) Time-resolved fluorescence obtained by addition of 0.2 μM mant-GTP (black) or mant-GppNHp (grey) to 2 μM Roco protein. The addition of mant-nucleotide is indicated with a black arrow.
The single-turnover GTPase of Ct Roco protein can be measured by fluorescence spectroscopy using mant-GTP. By adding mant-GTP to Roco, there is an increase of fluorescence on binding and a decrease due to GTP hydrolysis, which can be fitted to a rate constant of 0.063 min−1, approximately twofold faster than the intrinsic hydrolysis of wild-type Ras (Figure 2B). The amplitude of the fluorescence change shows a large increase in fluorescence due to mant-GTP binding, but also a large difference in fluorescence emission between GDP and GTP, confirming the equilibrium titration experiments. After addition of the non-hydrolysable GTP-analog mant-GppNHp to Roco, an increase of fluorescence also occurs, which remains at a constant level. This indicates that the observed fluorescence decrease in the presence of mant-GTP is due to hydrolysis. The experiment has also been performed with fluorescent tamra-GTP (Eberth et al, 2005) and gives the same rate.
The structure of the COR domain
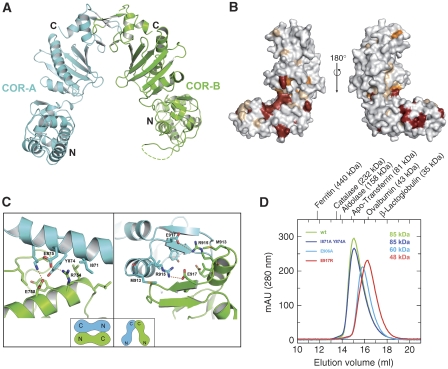
Having determined the boundaries of stable fragments, the LRR, the COR and the Roc–COR fragments were expressed individually. The LRR (1–441) and COR (615–946) domains were soluble and could easily be purified and concentrated. The 38.8 kDa COR domain crystallised in space group P3(2)21 and the structure was solved by phasing with Se-Met-SAD (Supplementary Table 1). The COR domain consists of two subdomains connected by a long, partially flexible linker. The N-terminal half is an (mainly) α-helical domain with a short three-stranded antiparallel β-sheet, and the C-terminal subdomain contains a central seven-stranded antiparallel β-sheet flanked by four helices and a β-hairpin motif near the C terminus (Figure 3A, Supplementary Figure 1). The two subdomains are linked by a single polypeptide chain connection and the interface shows hydrophobic interactions involving main chain and side chains. If residues conserved between bacterial Roco proteins and LRRK from various organisms (Supplementary Figure 1) are painted onto the surface of the COR domain, most of the invariant and highly conserved residues are located in or near the interface between the two subdomains of COR (Figure 3B).
Figure 3.
Structural analysis of the COR domain dimer. (A) Ribbon diagram of one (physiological dimer) of the COR dimers found in the crystal, with different protomers shown in green and cyan. Loops, that are not visible in the structure are indicated as dashed lines. (B) Surface representation of the COR monomer in two different orientations separated by 180°, with residues totally invariant between bacteria and man in red and those highly conserved in orange. (C) Details of the two dimer interfaces found in the crystal, including residues mutated for gel permeation chromatography analysis. Schematic drawings of the two types of dimers are shown in the insets. The interface on the right corresponds to the dimer in A and is the solution dimer. (D) Gel permeation analysis of wild-type and mutant COR proteins, with apparent molecular masses as obtained from equilibration with the indicated marker proteins.
Within the COR crystal, two different types of dimers can be identified. In the dimer forming the asymmetric unit, the N-terminal subdomain of one protomer interacts with the C-terminal of the other and vice versa (Figure 3C, see also Supplementary Figure 2). This interaction creates a solvent-inaccessible surface of 2610 Å2. The other dimer in the crystal, where the protomers are separated by a twofold crystallographic axis, involves only the C-terminal subdomains with a buried surface of 1290 Å2 (Figure 3A, C). When the COR protein is chromatographed on an S200 Sepharose gel filtration column equilibrated with various molecular mass markers, it elutes with an apparent molecular mass of 85 kDa, which is very close to twice the calculated mass of 38.8 kDa. This shows that COR is also a dimer in solution, under various conditions of ionic strength (data not shown).
To determine which of the two different dimers (N–C, C–C) is the one formed in solution, we introduced mutations into the corresponding interfaces (Figure 3C), purified the mutant proteins and analysed their behaviour by gel permeation chromatography. The I871A/Y874A (Figure 3D) and more drastic Y847R/E875R (data not shown) double mutants of the N–C interaction interface have similar gel filtration behaviour as the wild-type protein with molecular mass of ∼85 kDa. However, the elution volume of the mutants E917R or E906A (Figure 3D) is larger and corresponds to a calculated molecular mass of 48 and 60 kDa, respectively. The mutations of the C–C interaction thus shift the monomer–dimer equilibrium either partially (E906A) or largely (E917R) towards monomer. This is clear evidence that the dimer is formed through the C-terminal subdomains, and that the one with the smaller interface is the dimer in solution.
The Roc–COR tandem
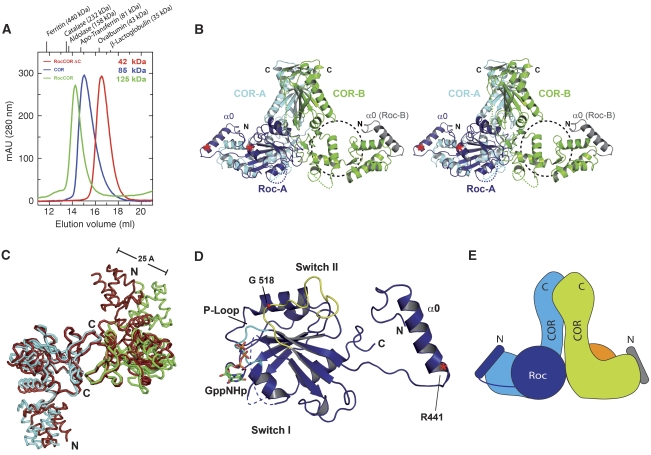
The Roc–COR (residues 442–946) tandem is the product of trypsin digestion in the presence of GppNHp. This same fragment could, however, not be expressed in a soluble form in E. coli. However, an N-terminal extended construct (residues 412–946), which incorporates another element of sequence conserved between bacteria and man (α0, Supplementary Figure 1), is soluble and can be purified. As expected, owing to the presence of the COR domain, the protein runs as a dimer on gel permeation chromatography with an apparent molecular mass of 125 kDa, very close to the calculated mass of 124 kDa (Figure 4A). To show that dimerisation is due to the same dimer interface as found for the COR domain and not due to domain swapping as recently reported for the Roc domain (Deng et al, 2008), we created a Roc–COR–ΔC fragment where the C-terminal subdomain of COR is missing. The protein with a calculated molecular mass of 42.2 kDa runs with an apparent molecular mass of 42.3 on an S200 column, showing that the C-terminal subdomain of COR is responsible for the constitutive dimerisation (Figure 4A–C).
Figure 4.
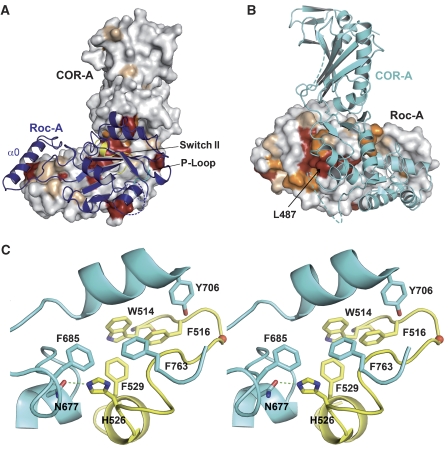
Structure and properties of the Roc–COR tandem. (A) Gel permeation analysis of the COR domain, the Roc–COR tandem and Roc–COR-ΔC on an S200 gel permeation chromatography column, highlighting the apparent molecular masses obtained from equilibration with marker proteins. (B) Stereo-ribbon diagram of the Roc–COR dimer with colours for COR domains A and B as in Figure 3, and for Roc domain A and parts of Roc domain B in different shades of blue. The putative position of the non-visible G-domain is indicated with a dashed circle. Positions of the tryptic cleavages sites are marked with red asterisks. (C) Superimposition of the main chain worm plots of COR domains A and B from the free COR (red) and the Roc–COR (blue, cyan) structures, using the C-terminal domains for the overlay. (D) Ribbon plot of Roc domain A and helix α0, highlighting particular features as discussed in the text. The tryptic cleavage site (R441) is indicated with a red asterisk. (E) Schematic view of the complete Roc–COR dimer including the full Roc-B subunit. Disordered regions, not visible in the structure, are indicated with dashed lines.
We also tested nucleotide binding and the intrinsic GTPase activity of Roc–COR. The affinity for GDP is 30.2 μM, slightly lower as compared with the full-length protein. The affinity to GppNHp is very similar with a 0.33 μM KD for Roc–COR. Furthermore, the single-turnover GTPase activity, as measured with the fluorescent GTP derivative, gives a kcat of 0.057 min−1 and similarly shows that the LRR domain has no significant influence on these biochemical properties of the Roc domain (data not shown).
The structure of the Roc–COR tandem
When testing conditions for crystallisation, Roc–COR (E917R) turned out to be the most suitable protein. This mutant, which in COR prevents dimerisation, is in fast equilibrium between monomer and dimer (data not shown) in the context of the Roc–COR tandem, for reasons that became apparent in the structure (see below). The mutant, Roc–COR from now, crystallised in space group P2(1)2(1)2(1) and the structure could be solved by molecular replacement, using the structure of the COR domain as model (Supplementary Table 1).
Surprisingly, we found that there were two COR domains, which dimerise as before through their C-terminal subdomains, but only one Roc domain (Roc-A) visible in the asymmetric unit. Only the N-terminal helix α0 from the second Roc (Roc-B), which is bound by the N-terminal subdomain of COR, is visible (Figure 4B). As the protein was full length after purification and the α0 helix of Roc-B is present, we exclude the possibility that one of the Roc subunits is proteolytically removed during crystallisation. We favour a second explanation that the other G-domain is present but highly mobile. This is supported by residual density in the area where we would expect the second Roc domain, especially in the area of the Roc–COR interface, and that there is enough space in the crystal lattice to accommodate it. Further support comes from the observation that the Roc-A domain is stabilised by crystal contacts to an adjacent molecule (Supplementary Figure 3), strengthening the notion that the Roc domains are generally highly mobile, and that RocA is only visible due to stabilising crystal contacts.
The COR domains in the Roc–COR structure were superimposed on the free COR dimer structure. Comparison of the C-terminal subdomains shows similar structures with an r.m.s. deviation of 0.9 and 1.4 Å for COR-A and COR-B, respectively. However, when the C-terminal subdomains of COR are superimposed, the N-terminal subdomains for the A protomers show an r.m.s. deviation of 3.7 Å using 133 Cα atoms (Figure 4C). The difference is even larger for the B protomer, which apparently does not interact with Roc. It deviates from the B protomer in the COR structure by an r.m.s. of 10.8 Å for 120 Cα atoms, whereas the largest distance between particular atoms is 25 Å. Although it is not possible to assign which of the structural changes are at least in part mediated by crystal contacts, these data nevertheless show that N- and C-terminal subdomains, which are connected by a single pass of polypeptide chain, are very mobile with respect to each other.
The structure (Figure 4D) of the Roc domain is that of a canonical G-domain with six β-strands and helices on either side of the sheet. The structure overlays well with that of Ras (Supplementary Figure 4) except for a few minor deviations such as a partially disordered α2 between β3 and β4 and a shorter α4 helix, indicating that, as in other low-affinity G-proteins, the absence of nucleotide does not grossly perturb the structure (Prakash et al, 2000; Scrima et al, 2005). The comparison gives an r.m.s.d. of 2.4 Å for 126 of the 166 residues from RasGppNHp (Pai et al, 1989). For a better comparison, we keep the canonical G-domain nomenclature for secondary structure elements. The N-terminal extra helix α0 of Roc-B is located in exactly the same place as α0 from the visible subunit, strengthening the assumption that the second Roc domain should occupy a similar position in the Roc–COR dimer. As α0 has no direct interface with the G-domain and as the linking polypeptide is unstructured and proteolytically cleaved behind Arg441 (Figure 4B, D), we consider it to be a helical connector between the LRR and the Roc–COR tandem. In the proteolysis experiments (Figure 1), the connector was part of the LRR domain and can be recombinantly expressed as part of the LRR. As the protein was crystallised in the absence of excess GDP/GTP and the affinity to nucleotide is low, the nucleotide-binding site is empty. However, from the overlay with Ras, and as the nucleotide binding site is open, nucleotide could be fitted into the structure without any severe backbone clashes.
Although the exact boundaries of the putative switch regions can only be defined after solving the structure of Roc in various nucleotide binding states, the switch regions are conventionally defined as the sequence motifs around the invariant threonine (Thr35 in Ras) and glycine (Gly60 in Ras), which connect these motifs to the γ-phosphate. In Roc, structural overlay with Ras identifies Thr484 as the canonical Thr residue, but the polypeptide chain around switch I is not well defined in the nucleotide-free state, which is not unexpected considering that switch regions are usually not well defined even in the presence of GDP (Vetter and Wittinghofer, 2001). Leu487, three residues C-terminal to Thr484, is, however, visible and is part of a hydrophobic pocket involving Phe516, Ile703 and Tyr706.
Sequence alignment of Roco proteins shows a great variation in the residues and length of polypeptide between α1 and β2, the presumed site of switch I. The sequence element around the canonical switch II DxxG motif, WDFGG in C. tepidum Roco, WDFAG in LRRK2, is conserved and the structure is well defined, indicating an important function for the region. The sequence in the switch II area between β3 and β4, which is only partially folded into an helix (α2) as in the canonical G-domain fold, is in fact the most highly conserved element of Roco proteins between bacteria and man (Supplementary Figure 1). This part of the polypeptide is folded into an extended chain and a loop that is situated at the side of the G-domain away from the nucleotide. Gly518 from the DxxG motif is 12 Å away from the modelled γ-phosphate (Figure 4D, see below).
The Roc/COR interface and human LRRK2 mutations
Roc sits between the two legs of the triangular-shaped COR domain (Figure 4B, E, Supplementary Figure 3). It makes contact with both COR protomers, where the buried surface between Roc-A and COR-A is 3867 Å2 and to COR-B 998 Å2. The interaction of Roc-A with COR-B apparently contributes to dimer formation and could explain the finding that the E917R mutation does not lead to complete dissociation of the dimer as in COR alone. Incidentally, the double mutation in the COR interface, E917R/E906A, is sufficient to create a Roc–COR monomer.
The N-terminal helix α0, which was necessary for expression of a soluble construct, extends from the G-domain and makes contact with the N-terminal COR subdomain. Figure 5A shows that all the invariant and most of the highly conserved residues on one face of COR are involved in forming the interface with Roc. Similarly, most of the invariant and highly conserved residues on Roc are in or very close to the interface with COR (Figure 5B), demonstrating that the function of this interface is conserved between bacteria and man. A major part of the Roc-A/COR-A interaction occurs between the switch II region of Roc and the most highly conserved patch on COR in the bend between the N- and C-terminal subdomains. Figure 5C shows that the interface between Switch II and COR is mediated by mostly hydrophobic residues and one hydrogen bond between Asn677 and His526 (see also Supplementary Figure 6).
Figure 5.
The Roc–COR interface. (A) Ribbon model of Roc-A on the surface representation of COR-A, with invariant and conserved residues of COR as in Figure 3B. (B) Ribbon model of COR-A on the surface of Roc-A (180° orientation relative to (A)), with invariant and conserved residues taken from the sequence alignment (Supplementary Figure 1). (C) Detailed stereo view of the interface between Roc and COR interface, highlighting residues from switch II (yellow) of Roc and contacting residues from COR (cyan).
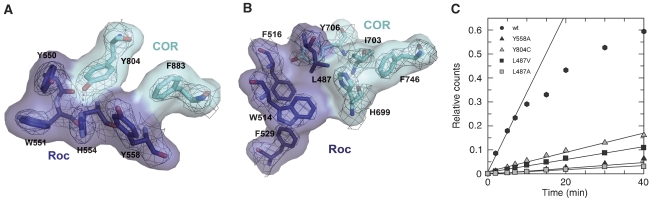
Parkinson patients carry mutations in the LRRK2 Roc and COR domain. These are point mutations of Arg1441 of Roc to Cys or Gly, Ile 1371 to Val in Roc and Tyr1699 to Cys in COR. Tyr1699 in the COR domain is almost fully conserved as an aromatic residue from bacterial Roco to LRRK2 and is Tyr804 in Ct Roco (Figure 6A). It is situated in the major contact site between the Roc and COR involving helix α3 of Roc, which is in close proximity to Switch II (Supplementary Figure 5). It points into a hydrophobic interface and forms a hydrogen bond to His554. At the equivalent position in the human LRRK2, there is an Asn1437 instead of a His and could thus involve a similar hydrogen. Its mutation to Cys would be expected to disrupt the hydrogen bond and weaken the interaction.
Figure 6.
GTPase activity and Parkinson mutations. (A, B) Atomic model and corresponding 2fo-fc electron density (contoured at 1σ) of residues analogous to PD-related LRRK2 mutants and their location in the C. tepidum Roc/COR interface. Residues and surface of COR are coloured in cyan and of Roc in blue. (C) Multi-turnover GTP hydrolysis of wild-type and mutant Roc–COR proteins, measured with charcoal assay using 1 μM protein and 10 μM GTP at 15 °C. The following initial rates could be determined: wt, 3.3 × 10−2; Y558A, 1.2 × 10−3; Y804C, 4.0 × 10−3; L487V, 2.7 × 10−3; and L487A, 8.0 × 10−4 min−1.
Arg1441 in LRRK2 is not conserved between bacterial and metazoan proteins (Supplementary Figure 1). As the proteins can nevertheless be aligned with high confidence in this area without having to use a gap or insertion, the corresponding residue in Ct Roco would be Tyr558. Surprisingly, it is very close to and points into the same hydrophobic interaction area as Tyr804. This suggests that its mutation would have a similar effect on the Roc/COR interaction. We propose that it is the aliphatic side chain of Arg1441 in human LRRK2 that is important rather than the positive charge and that the mutation to Cys or Gly found in patients would weaken the interface as well. Superimposition of Ct Roc with one G-domain of the human Roc structure, neglecting the idea of domain swapping (Deng et al, 2008) and treating it as a single polypeptide, gives an r.m.s.d. of 1.7 Å for 120 of the 177 residues from hs RocGDP. In such a superimposition, Tyr558 aligns well with Arg1441, supporting the analogy of the two residues. Leu487, which would correspond to Ile1371 in LRRK2, is also located in the interface between Roc and COR (Figures 5B and 6B), although at a different hydrophobic contact site involving residues Tyr706, Ile703 and Phe746. The I1371V mutation would be expected to weaken this hydrophobic pocket.
We created the mutants Y804C, Y558A, L487V and L487A of the Ct Roc–COR protein corresponding to the mutations found in Parkinson patients and determined their properties. They behave similar to wild type on gel permeation chromatography, indicating that these mutations do not affect the ability of Ct Roc–COR to dimerise. For the Y804C mutation, it was also shown that the affinity to mGDP is similar to that of the wild-type protein, with a KD of 6 μM. The other mutants have similar nucleotide-binding properties compared with wild-type Roc–COR. The most prominent feature of the mutation is the strongly reduced GTPase activity. While the wild-type Roc–COR has an initial rate of 3.3 × 10−2 min−1 in a multi-turnover assay under these conditions, it is reduced eightfold in the Y804C mutation (Figure 6C). Similarly, the Y558A mutation has a GTPase rate of 1.2 × 10−3 min−1, and L487V shows 12-fold reduction (2.7 × 10−3 min−1). This indicates that an efficient GTPase is dependent on the integrity and/or stability of the Roc/COR interface, and that even an apparently minor reduction of the aliphatic side chain from Leu to Val disturbs the GTPase reaction. Accordingly, the more drastic L487A mutation has an even slower GTPase reaction as compared with L487V with a rate of 8 × 10−4 min−1. The results of the charcoal assay were additionally confirmed with HPLC measurements.
Dimerisation and GTPase activity
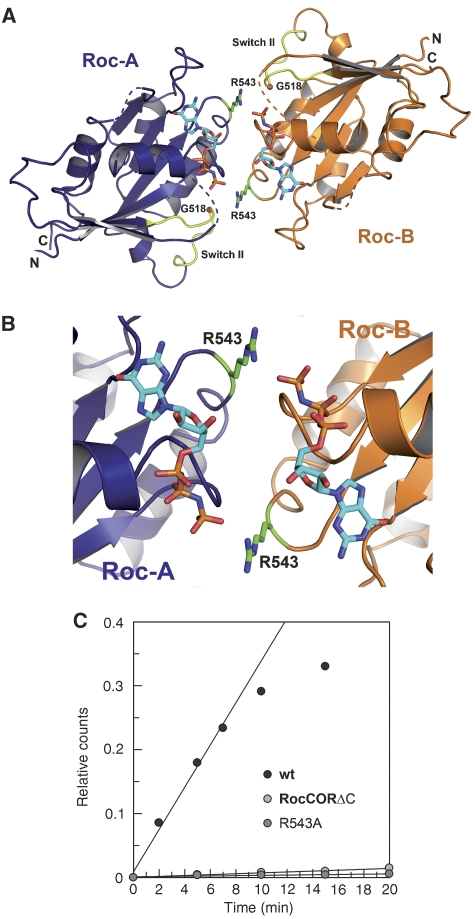
When we model the second Roc domain into the structure by positioning Roc-B into a position analogous to Roc-A, the two Roc domains face each other with their nucleotide-binding region (Figure 7, Supplementary Figure 6). The presumed close juxtaposition of the G-domains in the dimeric Roc–COR is reminiscent of many other G-proteins that use homo-dimerisation to complement the active site of each other for efficient catalysis, such as MnmE, hGBP1, Septin 2 and HypB. If we use the overlay with Ras to position the GppNHp nucleotide into the structural model, which is possible without any major backbone clashes as noted above, the nucleotides in the interface between the two G-domains face each other in a head-to-tail fashion, where the ribose of one GppNHp is pointing towards the γ-phosphate of the other, although the distance of 9 Å is too far in the current model to suspect any influence on the GTPase activity (Figure 7A, B). This relative location of nucleotides very much resembles the head-to-tail orientation of nucleotides in the SRP-SR pseudo-dimer (Egea et al, 2004; Focia et al, 2004); but is unlike the situation in the hGBP1 (Ghosh et al, 2006), HypB (Gasper et al, 2006), Septin2 (Sirajuddin et al, 2007) and MnmE dimers (Scrima and Wittinghofer, 2006), which dimerise in a head-to-head fashion. Assuming a conformational transition where the Roc domains change their relative orientation in the presence of GTP, it can be envisioned that dimerisation induces a catalytically active conformation (see below) similar to the SRP-SR interaction.
Figure 7.
Dimerisation and GTPase activity. (A, B) Ribbon diagram of the full Roc–COR model, highlighting juxtaposition of the G-domains, in particular the position of Arg543 close to γ-phosphate of the neighbouring protomer. GppNHp was modelled into the structure by superimposition with Ras-GppNHp (PDB 5P21). Owing to the absence of side-chain density, Arg543 residue was modelled into the most favourable rotamer conformation. (C) The GTPase activities of wt Roc–COR, the R543A and Roc–COR-ΔC mutants, measured as described in Figure 6C.
To analyse whether dimerisation is required for efficient GTP hydrolysis, we made use of the Roc–COR-ΔC construct where the C-terminal subdomain of COR is deleted. This protein is monomeric, as shown by the gel permeation chromatography (Figure 4A). It does not dimerise in the presence of GDP or GTP or any other conditions analysed. Even though the Roc domain should not be perturbed by this construct and actually has bound nucleotide after purification, it has no measurable GTPase activity in the GTPase (charcoal) assay even at higher concentrations (Figure 7C). On closer inspection of the dimer model, we find an arginine residue, Arg543, from Roc-B pointing into the active site of Roc-A and vice versa (Figure 7B). This is reminiscent of the arginine finger that is supplied into the active site of Ras proteins by their cognate GAPs, as shown for RasGAP (Scheffzek et al, 1996), RhoGAP (Rittinger et al, 1997), RabGAP (Pan et al, 2006) and the Arl3GAP RP2 (Veltel et al, 2008). An arginine-like finger is also found in the dimerising G-protein hGBP1 where it is also involved in stabilising the transition state of the GTPase reaction (Ghosh et al, 2006). When we mutate the arginine and measure the GTPase activity of the R543A protein, it shows no measurable GTPase activity, whereas its ability to bind nucleotides is unchanged (Figure 7C).
Discussion
The structural analysis has shown that the Roc domain of C. tepidum is a normal G-domain protein, unlike the previously determined structure of the human Roc domain where it was found as domain-swapped dimer, in which the N-terminal part of one G-domain interacts with the C-terminal of the other, thus forming a constitutive dimer (Deng et al, 2008). Although we cannot exclude that the human Roc protein has a different structure than the bacterial Roco protein, it seems rather unlikely. First of all, domain swapping has previously been observed as a crystallographic artefact for many proteins (for a review, see Liu and Eisenberg, 2002) as well as for the small GTP-binding protein Rab27b. In the latter case, many different biochemical experiments did not show dimer formation for Rab27b in solution (Chavas et al, 2007). This is in contrast to the report from Deng et al (2008), which did not conclusively show dimerisation in solution. In our hands, Ct Roc–COR is indeed a dimer, but dimerisation is clearly mediated by the C-terminal half of the COR domain, and its mutation or deletion causes the protein to be monomeric in solution, indicating that the COR-free Roc domains do not form a constitutive dimer either in the presence or absence of nucleotide, nor under any other condition that we have analysed (data not shown).
The Roc/COR interface is the most highly conserved area of both domains, lending credibility to its existence in all Roc–COR tandem proteins. Using the swapped dimer structure from the human Roc dimer for a superimposition of the Roc domain from C. tepidum and LRRK2, we would obtain serious clashes between the second G-domain and the N-terminal subdomain of COR, such that the proposed swapped dimer would not be consistent with that interface (Supplementary Figure 7).
We show here that COR is a constitutive dimerisation device and that all Roc G-domain proteins seem to belong to the class of molecular switches whose GTPase switch-off reaction is regulated by transient dimer formation. This is supported by the inability of the Roc–COR-ΔC mutant to hydrolyse GTP. Although the structure presented here is that of a nucleotide-free protein, GTP can be easily modelled into the canonical nucleotide-binding site. Although not all the features of a GTP-competent active site can be inferred from our model, we can nevertheless show that the active site of one protomer is complemented by an arginine finger from the second. The inability of the R543A mutant to hydrolyse GTP proves that this is relevant for the GTPase reaction. Complementation of the active site of each other is also found for the dimers of hGBP1 and MmnE, as described above. This structural setup is also reminiscent of many ATP-binding proteins such as ABC transporters where the ATP-dependent dimerisation drives transport of a large variety of transport substrates across the membrane (Hollenstein et al, 2007).
We would postulate that the G-domains are mobile and that dimerisation is triggered by some input signal, which may be derived from the LRR domain, resulting in GTP loading and/or mutual stimulation of the GTPase reaction by the protomers. The mobility of the G-domain is evident from the proteolytic digestion and from the structure where one of the two G-domains is too mobile to be visible in the crystal and where the other is only visible because it is fixed by crystal–crystal contacts. The proteolysis experiments have also shown that the Roc–COR tandem is more compact in the presence of nucleotide, and that the Roc domain, once cleaved from COR, is not soluble anymore. As the invariant Gly from the DxxG motif in the Roc G-domain is likely to contact the γ-phosphate, as in any other G-domain, this would lead to a major rearrangement of switch II and in turn to a change in the Roc/COR interface. The large difference in fluorescence amplitude between the GDP and GTP-bound states indicates that the presence of the γ-phosphate would induce a large conformational change compared with GDP, at least in the nucleotide-binding site.
The function and mechanism of LRRK2 and its activation in Parkinson patients are not immediately obvious from the present structure. However, the output signal of LRRK2 is the kinase activity that in many protein kinases is regulated by transphosphorylation of the activation loop. That native LRRK2 is a dimer in solution and that autophosphorylation is required for kinase activity have recently been shown (Jaleel et al, 2007; Greggio et al, 2008). Thus, bringing the G-domains into close proximity in a GTP-dependent manner could be a device for bringing the activation loops of the two protomers of the LRRK2 dimer close to each other for trans-phosphorylation. As the arginine finger observed in the C. tepidum Roco protein is not present in LRRK2, other ways for complementation of the LRRK2 active site would have to be considered. In any way, the GTPase reaction would then determine the level of kinase activity. We have shown here that the Y804C Parkinson mutations in the COR domain, and the two mutations L487V and Y558A in the Roc domain, analogous to the Roc Parkinson mutations I1371V and R1441G/C, have a much lower GTPase activity. Assuming a similar effect of the mutations in LRRK2, this reduced GTPase activity would then lead to a higher kinase activity, consistent with reports that the R1441C or R1441G mutations reduce the GTPase activity (Guo et al, 2007; Lewis et al, 2007; Li et al, 2007) and with the reported increased kinase activity of LRRK2 (Y1699C) and all other kinases derived from patients with mutations in Dardarin/LRRK2 (Smith et al, 2005; Korr et al, 2006; Luzon-Toro et al, 2007; West et al, 2007). This is analogous to the small G-protein Ras in the Raf-MEK-Erk Map kinase module, where activation due to a reduced GTPase activity leads to a higher Erk kinase activity in trans (Barbacid, 1987) as compared to cis in LRRK2. The kinase activity of LRRK2 is thus a valuable drug target for treating PD. Our data on the structure and function of the C. tepidum Roc–COR tandem should contribute to a better understanding of its function in the regulation of this important protein kinase.
Materials and methods
Protein expression and purification
For expression of Roco fl (M1-T1102) and Roc–COR (D412-R946) from C. tepidum (Swiss-Prot accession number: Q8KC98), encoding DNA fragments were amplified from genomic PCR and cloned into a pProEx plasmid (Invitrogen). N-terminally His6-tagged Roco fl and Roc–COR were expressed in E. coli strain BL21DE3 (RIL) in TB medium. At an OD600 of 0.7, the expression was induced by addition of 0.1 mM IPTG. After overnight induction, cells were harvested by centrifugation and resuspended in buffer A (30 mM Tris pH 7.5, 150 mM NaCl, 5 mM MgCl2, 3 mM β-mercaptoethanol, 0.1 mM GDP) with 2.5% glycerol. The cells were lysed in a microfluidiser. After centrifugation, the supernatant was applied to a Ni-NTA column, washed with lysis buffer and an additional washing step with buffer A containing 300 mM KCl and 1 mM ATP. Samples were eluted with buffer A containing 300 mM imidazol. Peak fractions were concentrated and purified by gel filtration on a Superdex S200 26/60 equilibrated in buffer B (30 mM Tris pH 7.5, 50 mM NaCl, 5 mM MgCl2, 3 mM dithioerythritol (DTE)). Fractions corresponding to a molecular weight of a dimer were concentrated, flash-frozen in liquid nitrogen and stored at −80°C.
COR (S615-R946) from C. tepidum was cloned into a pGEX4T1 NTEV plasmid, and GST–COR in BL21DE3 pLysS was expressed in TB medium. After induction and harvesting the cells as described for Roco fl, cells were resuspended in buffer C (30 mM Tris pH 7.5, 150 mM NaCl, 3 mM DTE), lysed by microfluidiser. The supernatant was applied to a GSH-Sepharose column, followed by two washing steps with buffer C as well as with buffer C containing 300 mM KCl and 1 mM ATP. After elution with buffer C containing 20 mM glutathione, GST–COR was concentrated and GST was removed by tobacco etch virus protease. Ct COR was purified by gel filtration on a Superdex S200 26/60 equilibrated with buffer B, and residual GST was removed by applying the eluate to a GSH-column using the gel filtration buffer. Overexpression and purification of Roc–COR-ΔC (D412-L780) was analogous to the purification of COR with buffer C containing 5 mM MgCl2 and 0.1 mM GDP.
Mutagenesis was performed using the QuickChange method by Stratagene. Overexpression and purification of the COR and Roc–COR mutants were analogous to purification of the corresponding wild-type proteins.
Overexpression of COR for selenomethionine incorporation was performed in minimal medium containing 50 mg l−1 selenomethionine. Purification was analogous to purification of native COR except for using 10 mM DTE in all buffers.
Protein methods
Fluorimetry/equilibrium titration. Roco fl and mutant protein were titrated against 200 nM 3′-O-(N-methyl)anthranoyl (mant)-nucleotides until saturation was reached. The mant fluorophore was excited at 360 nm, and emission was monitored at 450 nm (Fluoromax4, Horiba Jobin Yvon). The increase in fluorescence upon addition of protein was integrated over at least 10 min and determination of the equilibrium dissociation constant, Kd, was calculated by fitting a quadratic function to the data. Experiments were performed at 25°C in buffer B.
For determination of the single-turnover hydrolysis rate, 200 nM mant-GTP and mant-GppNHp as control was mixed with 2 μM protein and fluorescence was measured for 100 min at 25°C.
For tryptic proteolysis, a 5 mg ml−1 Roco fl protein solution was incubated with 10 μg ml−1 trypsin or elastase protease at 25°C in buffer B. Aliquots were taken at various time points and analysed by SDS–PAGE. N-terminal sequencing by Edman degradation was carried out by the Center for Molecular Medicine, University of Cologne. The mass of the SDS molecular-weight markers used are for M1: 200, 116, 97, 66, 45 and 29 kDa (SDS6H2, Sigma); for M2: 66, 45, 36, 29, 24, 20 and 14 kDa (SDS7, Sigma).
All analytical gel permeation experiments were performed with 300 μg protein in buffer B on a Superdex 200 10/30 equilibrated previously with various marker proteins.
Measurement of GTP hydrolysis by charcoal assay. For GTP hydrolysis measurements, 1 μM of Roc–COR and corresponding mutants were incubated in buffer B at 15 °C. The GTP-hydrolysis reaction was started by adding 10 μM GTP including 30 nM [32-P-γ]GTP (15 μCi). Aliquots of 10 μl were taken at certain time points and mixed with 400 μl of charcoal solution (50 g l−1 charcoal in 20 mM phosphoric acid) to stop the reaction. The charcoal was pelleted and the amount of free 32Pi in 180 μl of the supernatant was determined by scintillation counting. Relative counts were plotted against reaction time and initial reaction rates were determined by linear regression.
Crystallography
Optimised crystals of C. tepidum COR were obtained using the hanging-drop/vapour diffusion method. In all, 1 μl drops of 10 mg ml−1 COR solution were mixed with 1 μl of reservoir solution (600 mM Succinat pH 6.5, 5% 2-methyl-2, 4-pentan-diol). After 3–5 days, crystals grew at 20 °C. For data collection, crystals were cryoprotected in reservoir solution containing 20% PEG400 as cryoprotectant. A SAD data set was collected on beam line X10SA at the Swiss Light Source (SLS) (Paul Scherer Institut, Villingen, Switzerland).
Data were indexed, integrated and scaled with the XDS package (Kabsch, 1993). Initial heavy atom sites for SAD phasing were identified with SOLVE (Terwilliger, 2002) and initial phases were calculated with RESOLVE (Terwilliger, 2002). The model was built in COOT (Emsley and Cowtan, 2004) and refined with REFMAC5 using TLS-refinement (CCP4 suite) (Murshudov et al, 1997).
Crystals of C. tepidum Roc–COR E917R were only obtained in 96-well plates using the sitting-drop/vapour diffusion method. A total of 100 nl of 10 mg ml−1 Roc–COR E917R were mixed with 100 nl of reservoir solution (0.1 M HEPES pH 7.0, 1 M NaCitrate). Crystals were cryoprotected by adding 25% glycerol to the reservoir solution and a data set was collected at SLS X10SA.
Molecular replacement was performed with Phaser from the CCP4 package using the COR domain coordinates as searching model. The additional electron density was clearly identified as a G-domain fold and the Roc-domain was build from scratch using COOT. Refinement was carried out with REFMAC5 using TLS refinement.
Figures were generated using PYMOL (DeLano Scientific LLC).
Atomic coordinates and structural factors have been deposited within the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB) under the accession code 3PDT (COR) and 3PDU (Roc–COR).
Supplementary Material
Supplementary Figures and Table
Acknowledgments
We thank Wulf Blankenfeldt, Shen Yu, Aymelt Itzen, Nils Schrader and Antje Schulte for data collection. The data sets were collected at the Swiss Light Source, beamline X10SA, Paul Scherrer Institut, Villigen, Switzerland. We are grateful for the technical help of the beamline staff. This research project has been supported by the European Commission under the 6th Framework Programme: Strengthening the European Research Area, Research Infrastructures. Contract no: RII3-CT-2004-506008′. We also thank Lothar Gremer, Andrea Scrima and Ingrid R Vetter for help in crystallography, Patricia Stege for technical and Rita Schebaum for secretarial assistance.
References
- Barbacid M (1987) ras Genes. Ann Rev Biochem 56: 779–827 [DOI] [PubMed] [Google Scholar]
- Bosgraaf L, Van Haastert PJM (2003) Roc, a Ras/GTPase domain in complex proteins. Biochim Biophys Acta—Mol Cell Res 1643: 5–10 [DOI] [PubMed] [Google Scholar]
- Chavas LMG, Torii S, Kamikubo H, Kawasaki M, Ihara K, Kato R, Kataoka M, Izumi T, Wakatsuki S (2007) Structure of the small GTPase Rab27b shows an unexpected swapped dimer. Acta Crystallogr Sec D BiolCrystallogr 63: 769–779 [DOI] [PubMed] [Google Scholar]
- Deng J, Lewis PA, Greggio E, Sluch E, Beilina A, Cookson MR (2008) Structure of the ROC domain from the Parkinson's disease-associated leucine-rich repeat kinase 2 reveals a dimeric GTPase. PNAS 105: 1499–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberth A, Dvorsky R, Becker CFW, Beste A, Goody RS, Ahmadian MR (2005) Monitoring the real-time kinetics of the hydrolysis reaction of guanine nucleotide-binding proteins. Biol Chem 386: 1105–1114 [DOI] [PubMed] [Google Scholar]
- Egea PF, Shan SO, Napetschnig J, Savage DF, Walter P, Stroud RM (2004) Substrate twinning activates the signal recognition particle and its receptor. Nature 427: 215–221 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr Sec D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Farrer MJ (2006) Genetics of Parkinson disease: paradigm shifts and future prospects [Review]. Nat Rev Genet 7: 306–318 [DOI] [PubMed] [Google Scholar]
- Focia PJ, Gawronski-Salerno J, Coon JS, Freymann DM (2006) Structure of a GDP: AlF4 complex of the SRP GTPases Ffh and FtsY, and identification of peripheral nucleotide interaction site. J Mol Biol 360: 631–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focia PJ, Shepotinovskaya IV, Seidler JA, Freymann DM (2004) Heterodimeric GTPase core of the SRP targeting complex. Science 303: 373–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasper R, Scrima A, Wittinghofer A (2006) Structural insights into HypB, a GTP-binding protein that regulates metal binding. J Biol Chem 281: 27492–27502 [DOI] [PubMed] [Google Scholar]
- Ghosh A, Praefcke GJK, Renault L, Wittinghofer A, Herrmann C (2006) How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP. Nature 440: 101–104 [DOI] [PubMed] [Google Scholar]
- Gilks WP, Abou-Sleiman PM, Gandhi S, Jain S, Singleton A, Lees AJ, Shaw K, Bhatia KP, Bonifati V, Quinn NP, Lynch J, Healy DG, Holton JL, Revesz T, Wood NW (2005) Common LRRK2 mutation in idiopathic Parkinson's disease. Lancet 365: 415–416 [DOI] [PubMed] [Google Scholar]
- Goldberg JM, Bosgraaf L, Van Haastert PJM, Smith JL (2002) Identification of four candidate cGMP targets in Dictylostelium. Proc Natl Acad Sci USA 99: 6749–6754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greggio E, Jain S, Kingsbury A, Bandopadhyay R, Lewis P, Kaganovich A, van der Brug MP, Beilina A, Blackinton J, Thomas KJ, Ahmad R, Miller DW, Kesavapany S, Singleton A, Lees A, Harvey RJ, Harvey K, Cookson MR (2006) Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol Dis 23: 329–341 [DOI] [PubMed] [Google Scholar]
- Greggio E, Lewis PA, van der Brug MP, Ahmad R, Kaganovich A, Ding JH, Beilina A, Baker AK, Cookson MR (2007) Mutations in LRRK2/dardarin associated with Parkinson disease are more toxic than equivalent mutations in the homologous kinase LRRK1. J Neurochem 102: 93–102 [DOI] [PubMed] [Google Scholar]
- Greggio E, Zambrano I, Kaganovich A, Beilina A, Taymans JM, Daniels V, Lewis P, Jain S, Ding J, Syed A, Thomas KJ, Baekelandt V, Cookson MR (2008) The Parkinson's disease associated Leucine rich repeat kinase 2 (LRRK2) is a dimer that undergoes intra-molecular autophosphorylation. J Biol Chem 283: 16906–16914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo LX, Gandhi PN, Wang W, Petersen RB, Wilson-Delfosse AL, Chen SG (2007) The Parkinson's disease-associated protein, leucine-rich repeat kinase 2 (LRRK2), is an authentic GTPase that stimulates kinase activity. Exp Cell Res 313: 3658–3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenstein K, Frei DC, Locher KP (2007) Structure of an ABC transporter in complex with its binding protein. Nature 446: 213–216 [DOI] [PubMed] [Google Scholar]
- Ito G, Okai T, Fujino G, Takeda K, Ichijo H, Katada T, Iwatsubo T (2007) GTP binding is essential to the protein kinase activity of LRRK2, a causative gene product for familial Parkinson's disease. Biochemistry 46: 1380–1388 [DOI] [PubMed] [Google Scholar]
- Jaleel M, Nichols RJ, Deak M, Campbell DG, Gillardon F, Knebel A, Alessi DR (2007) LRRK2 phosphorylates moesin at threonine-558: characterization of how Parkinson's disease mutants affect kinase activity. Biochem J 405: 307–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr 26: 795–800 [Google Scholar]
- Korr D, Toschi L, Donner P, Pohlenz HD, Kreft B, Weiss B (2006) LRRK1 protein kinase activity is stimulated upon binding of GTP to its Roc domain. Cell Signal 18: 910–920 [DOI] [PubMed] [Google Scholar]
- Lewis PA, Greggio E, Beilina A, Jain S, Baker A, Cookson MR (2007) The R1441C mutation of LRRK2 disrupts GTP hydrolysis. Biochem Biophys Res Commun 357: 668–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XT, Tan YC, Poulose S, Olanow CW, Huang XY, Yue ZY (2007) Leucine-rich repeat kinase 2 (LRRK2)/PARK8 possesses GTPase activity that is altered in familial Parkinson's disease R1441C/G mutants. J Neurochem 103: 238–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Eisenberg D (2002) 3D domain swapping: as domains continue to swap. Protein Sci 11: 1285–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzon-Toro B, de la Torre ER, Delgado A, Perez-Tur J, Hilfiker S (2007) Mechanistic insight into the dominant mode of the Parkinson's disease-associated G2019S LRRK2 mutation. Hum Mol Genet 16: 2031–2039 [DOI] [PubMed] [Google Scholar]
- Martens S, Howard J (2006) The interferon-inducible GTPases [Review]. Ann Rev Cell Develop Biol 22: 559–589 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson J (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallograph Sec D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Nichols WC, Pankratz N, Hernandez D, Paisan-Ruiz C, Jain S, Halter CA, Michaels VE, Reed T, Rudolph A, Shults CW, Singleton A, Foroud T (2005) Genetic screening for a single common LRRK2 mutation in familial Parkinson's disease. Lancet 365: 410–412 [DOI] [PubMed] [Google Scholar]
- Pai EF, Kabsch W, Krengel U, Holmes KC, John J, Wittinghofer A (1989) Structure of the guanine-nucleotide-binding domain of the Ha-ras oncogene product p21 in the triphosphate conformation. Nature 341: 209–214 [DOI] [PubMed] [Google Scholar]
- Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der BM, de Munain AL, Aparicio S, Gil AM, Khan N, Johnson J, Martinez JR, Nicholl D, Carrera IM, Pena AS, de Silva R, Lees A, Marti-Masso JF, Perez-Tur J, Wood NW et al. (2004) Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron 44: 595–600 [DOI] [PubMed] [Google Scholar]
- Pan XJ, Eathiraj S, Munson M, Lambright DG (2006) TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature 442: 303–306 [DOI] [PubMed] [Google Scholar]
- Prakash B, Renault L, Praefcke GJK, Herrmann C, Wittinghofer A (2000) Triphosphate structure of guanylate-binding protein 1 and implications for nucleotide binding and GTPase mechanism. EMBO J 19: 4555–4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittinger K, Walker PA, Eccleston JF, Nurmahomed K, Owen D, Laue E, Gamblin SJ, Smerdon SJ (1997) Crystal structure of a small G protein in complex with the GTPase-activating protein RhoGAP. Nature 388: 693–697 [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Lautwein A, Kabsch W, Ahmadian MR, Wittinghofer A (1996) Crystal structure of the GTPase-activating domain of human p120GAP and implications for the interaction with Ras. Nature 384: 591–596 [DOI] [PubMed] [Google Scholar]
- Scrima A, Vetter IR, Armengod ME, Wittinghofer A (2005) The structure of the TrmE GTP-binding protein and its implications for tRNA modification. EMBO J 24: 23–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrima A, Wittinghofer A (2006) Dimerisation-dependent GTPase reaction of MnmE: how potassium acts as GTPase-activating element. EMBO J 25: 2940–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan SO, Stroud RM, Walter M (2004) Mechanism of association and reciprocal activation of two GTPases. PLOS Biol 2: 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirajuddin M, Farkasovsky M, Hauer F, Kuhlmann D, Macara IG, Weyand M, Stark H, Wittinghofer A (2007) Structural insight into filament formation by mammalian septins. Nature 449: 311. [DOI] [PubMed] [Google Scholar]
- Smith WLW, Pei Z, Jiang HB, Dawson VL, Dawson TM, Ross CA (2006) Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nature Neuroscience 9: 1231–1233 [DOI] [PubMed] [Google Scholar]
- Smith WW, Pei Z, Jiang HB, Moore DJ, Liang YD, West AB, Dawson VL, Dawson TM, Ross CA (2005) Leucine-rich repeat kinase 2 (LRRK2) interacts with parkin, and mutant LRRK2 induces neuronal degeneration. Proc Natl Acad Sci USA 102: 18676–18681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC (2002) Automated structure solution, density modification and model building. Acta Crystallograph Sec D Biol Crystallogr 58: 1937–1940 [DOI] [PubMed] [Google Scholar]
- Veltel S, Gasper R, Eisenacher E, Wittinghofer A (2008) The retinitis pigmentosa 2 gene product is a GTPase-activating protein for Arf-like 3. Nat Struct Mol Biol 15: 373–380 [DOI] [PubMed] [Google Scholar]
- Vetter IR, Wittinghofer A (2001) Signal transduction—the guanine nucleotide-binding switch in three dimensions. Science 294: 1299–1304 [DOI] [PubMed] [Google Scholar]
- West AB, Moore DJ, Choi C, Andrabi SA, Li XJ, Dikeman D, Biskup S, Zhang Z, Lim KL, Dawson VL, Dawson TM (2007) Parkinson's disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum Mol Genet 16: 223–232 [DOI] [PubMed] [Google Scholar]
- Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, Kachergus J, Hulihan M, Uitti RJ, Calne DB, Stoessl AJ, Pfeiffer RF, Patenge N, Carbajal IC, Vieregge P, Asmus F, Muller-Myhsok B, Dickson DW, Meitinger T, Strom TM et al. (2004) Mutations in LRRK2 cause autosomal-dominant Parkinsonism with pleomorphic pathology. Neuron 44: 601–607 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures and Table