Abstract
A novel, high-throughput method for determining deacetylase substrate specificity was developed using a one-bead, one-compound (OBOC) acetyl-peptide library with a quantum dot tagging strategy and automated bead-sorting. A 5-mer OBOC peptide library of 104,907 unique sequences was constructed around a central ε-amino acetylated lysine. The library was screened using the human NAD+-dependent deacetylase SIRT1 for the most efficiently deacetylated peptide sequences. Beads preferentially deacetylated by SIRT1 were biotinylated and labeled with streptavidin-coated quantum dots. After fluorescent bead-sorting, the top thirty-nine brightest beads were sequenced by mass spectrometry. In-solution deacetylase assays on randomly chosen hit and non-hit sequences revealed that hits correlated with increased catalytic activity by as much as 20-fold. We found that SIRT1 can discriminate peptide substrates in a context dependent fashion.
Keywords: Sir2, sirtuin, NAD, peptide library, one-bead, one-compound, deacetylase, quantum dot
The Silent information regulator 2 (Sir2) family of NAD+ dependent protein deacetylases has been extensively documented in recent years (1) (2) (3). This burgeoning interest can be attributed to the crucial roles of Sir2 enzymes (sirtuins) in regulating chromatin architecture (4), promoting transcriptional silencing and longevity (5) and in fatty acid metabolism (6). NAD+ dependent lysyl deacetylation is characterized by the stoichiometric release of nicotinamide and a novel metabolite, O-acetyl-ADP-ribose (OAADPr) (7) (8). The Sir2 family of deacetylases is highly conversed among all forms of life (9) with seven known human homologs (SIRT1−7) (10) (11). The most studied mammalian homolog, SIRT1, is a nuclear enzyme (12) that has been found to deacetylate a number of proteins (3). Histones H3 and H4 (5), p53 (12), p300 (13), TAFI68 (14), PCAF/MyoD (15), PGC-1α (16), FOXO1(17) and 4 (18), NF-κB (19), and Tat (20) are examples reported to be either biological targets and/or in vitro substrates of SIRT1. Collectively, the variety of proposed physiological targets reflects the functional diversity of SIRT1.
Identifying biological substrates is a necessary step in understanding the molecular basis for sirtuin phenotypes. However, in many of the cases cited above, a certain degree of logical bias was used to link the target protein and SIRT1, as unbiased global substrate screening procedures were not used. Varying conclusions have been reached in regard to sirtuin substrate specificity and recognition. Most striking are the conclusions that sirtuins display minimal side-chain recognition (21, 22) and that SIRT1 displays no substrate sequence specificity (23). In contrasting reports, clear substrate preferences were noted for yeast Sir2 and HST2 (24), and human SIRT2 (24) (25). To date, only one study has attempted to probe sirtuin substrate specificity using an acetyl-peptide library approach (23). Curiously, the study reported that SIRT1 displayed no substrate specificity in vitro, a conclusion based on an oriented peptide library. With this method, only globally preferred amino acids could be resolved, and the actual sequence of individual peptides was not elucidated. Although this technique has been successful for examining protein kinase substrate specificity (26), its usefulness toward protein deacetylases remains uncertain. To resolve the issue of SIRT1 specificity, we have generated a one-bead one-compound (OBOC) (27) (28) combinatorial acetylated peptide library to examine the sequence preferences (if any) of SIRT1.
OBOC libraries offer a solution to the problem of SIRT1 substrate specificity because they allow the isolation and sequence characterization of the most catalytically efficient peptide substrates, as opposed to “consensus sequences”. In these OBOC libraries, all possible peptide sequences from a group of selected amino-acid building blocks are represented in an on-bead format in which many copies of only one sequence exist on each bead. OBOC libraries permit sifting through a list of privileged peptide substrate sequences to correlate top hits with protein sequence databases. This strategy has been used successfully to determine the optimal peptide substrates of peptide deformylase, a Fe2+ metalloenzyme that catalyzes N-terminal deformylation of nascent polypeptides in eubacteria (29). In addition to probing substrate specificity, OBOC libraries provide starting points for the development of peptidomimetics for use as potential therapeutics (27) (30, 31).
Using the OBOC library approach, we found that SIRT1 displays substrate preferences and have generated a list of the top peptide substrate sequences. The legitimacy of the library approach for determining optimal SIRT1 substrates was established by the resynthesis and in-solution assay of select hits and non-hits. In our efforts to elucidate SIRT1 substrate specificity, we have developed a general, high-throughput methodology for the evaluation of protein deacetylase substrates. Our approach utilized on-bead deacetylation assays with subsequent quantum dot tagging and fluorescent bead-sorting instrumentation. Quantum dots are nanoparticles that exhibit exquisite photochemical properties owing to their semiconductor cores and are emerging as ideal fluorophores for screening OBOC libraries (32). These properties include robust photostability (33), a high quantum yield and a sharp emission with a broad range of excitation wavelengths (34). Coupled with a bead-sorting instrument, quantum dots allow the screening of hundreds of thousands of peptide sequences for deacetylase activity in a single day.
Experimental Procedures
General methods
All amino acid derivatives and resins were purchased from Peptides International (Louisville, KY) or from Bachem (Bubendorf, Switzerland). Peptides used in the solution deacetylase assays were obtained from the University of Wisconsin Biotechnology Core Facility. All other chemical reagents were obtained from Sigma-Aldrich, Acros (Geel, Belgium), Novabiochem (San Diego, CA), Amersham Biosciences (Buckinghampshire, England) or Quantum Dot (Hayward, CA). Reaction vessels for peptide library synthesis were purchased from Alltech Chromatography (Deerfield, IL). Analytical gradient HPLC was conducted on a Shimadzu series 2010C HPLC with a Vydac C18 column (10 μm, 4.6 × 250 mm). All runs used linear gradients of 0.05% aqueous TFA and 0.02% TFA in acetonitrile. MALDI-TOF MS was performed on a Bruker REFLEX II using α-cyano-4-hydroxy-cinnamic acid as matrix. Fluorescent bead sorting was carried out on a COPAS Select (Union Biometrica; Holliston, MA) instrument and fluorescent microscopy was done on an Olympus IX81 instrument (Tokyo, Japan) equipped with a Hamamatsu digital camera (Hamamatsu-City, Japan).
Enzyme preparation
SIRT1, SIRT2 and ySir2 were expressed and purified as previously described (24) (49). Trypanasome Sir2 was cloned, expressed and purified using a similar strategy (unpublished data, T.M. Kowieski, J.M. Denu).
Determination of peptide concentrations
Prior to kinetic analysis, peptide concentrations were established by amino acid analysis (AAA) or by with a coupled assay in which NAD+ leftover from exhaustive deacetylation reactions (acetylated peptide was typically incubated with 5−10 μM Sir2 and 80 μM NAD+ for 20 min.) was quantitatively converted to NADH with alcohol dehydrogenase and monitored spectrophotometrically in real-time at 340 nm. Peptide concentrations were obtained by subtracting the amount of NADH formed from the original amount of NAD+ used in the reaction.
Solution deacetylation assays
All solution phase Sir2 assays were carried out at 25 ° C in 50 mM Tris•HCl at a pH of 7.5. Reactions were done in 50−100 μL with 0.1 to 1.5 μM enzyme, 0.1−1.2 mM NAD+, 0.5−1000 μM peptide and 1 mM DTT. Reaction mixtures were quenched with TFA to a final concentration of 1% after 5−10 minutes and nicotinamide levels were quantitated by HPLC at 264 nm. Alternatively, [32P]-NAD+ (10 mCi/mL) was used in assays and quenched reaction mixtures were spotted on a silica TLC plate and run in a chamber containing 60 % ethanol and 40 % 2.5 mM ammonium acetate for 3−4 hours. Levels of 32P-OAADPr and 32P-NAD+ were then quantitated by phosphorimaging and the fraction turnover was calculated. Saturation curves were done at varying concentrations of peptide while holding that of NAD+ constant. Time points were chosen such that product formation never exceeded 20 % conversion and data were plotted as rate (s−1) vs. peptide concentration. Plots were fitted to the Michaelis-Menten equation, v = [(kcat/Km)[S]]/(1 + [S]/Km) using Kaleidagraph software (Reading, PA) to extract Km and kcat/Km.
Library construction
The acetylated peptide library was constructed on TentaGel Macrobead NH2 resin (280− 320 μm, 0.21 mmol/g loading, 65,550 beads/g) using the split-pool approach (27) (28). Fmoc/tBu methodology (50) was used to carry out the library synthesis on 4.80 g of resin. Prior to randomization, a four amino acid linker, BBRM (B = β-alanine) was synthesized. After deprotecting the N-terminus with 20 % (v/v) piperidine in DMF for fifteen minutes, the resin was split equally into eighteen separate reaction vessels (one for each amino acid in the library). To each vessel was added four equivalents of amino acid and coupling reagent in addition to 5% (mol/mol) capping reagent for later sequencing. Capping reagents included phenylacetic acid and 4-pentenoic acid. Phenylacetic acid was used in conjunction with norleucine, while 4-pentenoic acid was used with all other amino acids. Equimolar ratios of both capping reagents were used for isoleucine, asparagine, glutamine and histidine. After a second coupling, the resin from all vessels was washed three times each with DCM and DMF, pooled and deprotected. Next, the resin was redistributed into the reaction vessels for coupling of the second randomized residue. The process was repeated and after pooling, N-ε-acetyl lysine was installed as the third residue with no capping. The split-pool technique was repeated for the fourth and fifth randomized residues. Finally, after the final N-terminal deprotection, the N-termini of all the peptides were acetylated (70% DCM, 25% acetic anhydride, 5% triethylamine) and washed with DCM. Reagent K (TFA/EDT/thioanisole/water/phenol: 82.5%, 2.5%, 5%, 5%, 5%) (51) was used as the global deprotection cocktail. The resin was washed thoroughly with DCM and stored at −20° C until use.
Determining the viability of quantitative quantum dot analysis
Five 10 mg portions of TentaGel S NH2 resin (90 μm, 0.26 mmol/g loading, 2.86 × 106 beads/g) were divided out and swollen in DCM. After washing with DCM (3 × 1mL) and DMF (3 × 1 mL), the beads were labeled with 1, 0.5, 0.01 and 0.001 molar equivalents of N-hydroxysuccinimidobiotin in 200 μL portions of DMF. After an hour of rocking at room temperature, the solutions were drained and washed with DMF (3 × 1 mL). Approximately 5 mg of resin from each of the above reactions were combined and incubated with 1 mL BSA (1 mg/mL) in TBST buffer (25 mM Tris•HCl, pH 8.0, 150 mM NaCl, and 0.1% Tween 20) for 1 hr. Next, the resin was washed with TBST buffer (3 × 1 mL) and drained to the level of the resin bed. At this point, 500 μL of 75 nM streptavidin coated Q-dot 605 in TBST buffer was poured over the resin and rocked for two hours, after which the solution was drained to the resin bed before washing with TBST (10 × 1 mL). Beads were then photographed using a fluorescence microscope with a FITC filter and sorted on the basis of fluorescence (λex = 488 nm, λem = 610 nm) with the COPAS Select sorting instrument. Sorting data were evaluated with FCS Express (De Novo Software; Thornhill, Ontario) in histogram and dot plot form.
On-bead peptide library deacetylation by SIRT1
The entire library was assayed in a 75-mL column equipped with a filter. Prior to the assay, the resin was sequentially washed with DCM (3 × 50 mL), DMF (3 × 50 mL) and deacetylation assay buffer (50 mM Tris, pH 7.5) (1 × 50 mL). The enzymatic reaction was initiated upon addition of 50 mL of deacetylation cocktail (0.35 μM SIRT1, 1.5 mM β-NAD+, 1 mM DTT). The reaction mixture was allowed to rock gently for 12 minutes. After draining, the resin was washed with doubly distilled water (5 × 50 mL) and DMF (5 × 50 mL). Afterwards, the resin was rocked with biotin N-hydroxy-succinimide ester in DMF (3.5 mM, 50 mL) for 20 minutes. At this point, the solution was drained and the resin was washed with DMF (6 × 50 mL) and TBST buffer (2 × 50 mL). To reduce nonspecific binding, the beads were incubated with 50 mL of BSA (2 mg/mL) in TBST buffer for 1.5 hours. After draining and washing with TBST buffer (1 × 50 mL), 50 mL of 4 nM streptavidin coated Q-Dot 605 in TBST buffer was added and the mixture was allowed to rock for two hours. Again, the solution was drained and washed with TBST buffer (10 × 50 mL). The resin was then suspended in a minimal amount of TBST buffer and refrigerated at 4° C overnight.
Library screening
Beads were sorted on the basis of fluorescence (λex = 488 nm, λem = 610 nm) using the COPAS instrument. First, an initial sort was conducted such that the 300 beads exhibiting the highest fluorescence readings were collected. These beads were then subjected to a more stringent cutoff in which the 45 most fluorescent beads (from the pool of 300) were collected in a 96-well plate to generate an enriched sample.
Hit sequencing with MALDI-MS
Beads from the enriched sample were pooled and washed with 8 M guanidinium hydrochloride (2 × 1mL), doubly distilled water (10 × 1 mL) and DMF (3 × 1 mL). Individual beads were then deposited into separate microcentrifuge tubes containing 20 μL of cleavage cocktail (70% TFA, 30% doubly distilled water and 20% cyanogen bromide by weight) (29). After incubation overnight in the dark, the samples were dried and resuspended in 5 μL of 0.1% TFA. Each sample (1 μL) was combined with saturated matrix solution (1 μL) and dried on the target for MALDI-TOF MS analysis (positive ion mode).
Results
Library Design: Considerations of Peptide Length
Prior to library construction, it was essential to determine whether relatively short acetyl-peptides would function as efficient substrates of Sir2 enzymes. To evaluate peptide length requirements, ten acetyl-lysine containing peptides corresponding to the histone H3 sequence surrounding Lys-14 and of varying length were assayed using SIRT1 and a variety of other sirtuins (SIRT2; yeast Sir2, ySir2; and Trypanosoma brucei Sir2, TbSir2). We chose this sequence based on our experience that Lys-14 of H3is generally a good substrate for sirtuins. Deacetylation assays were conducted at fixed NAD+ concentrations while peptide concentrations were varied to produce saturation curves. The resulting data were fitted to the Michaelis-Menten equation to yield catalytic efficiencies, as defined by the apparent second order rate constant (kcat/Km), which takes into consideration both binding and catalysis. All peptides used in these studies were N-terminally acetylated, but the N-terminus was not deacetylated by sirtuins in control assays (data not shown). The results (Table 1) are represented as relative kcat/Km values, with the longest peptide AcTGG(AcK)APRK given a value of one. In these studies, all sirtuins surveyed showed no more than a two to three-fold difference in kcat/Km for the various peptide substrates. Thus, the shortest peptide, a 5-mer, was similar in catalytic efficiency to the longest peptides in this preliminary set, regardless of the enzyme assayed. These observations suggest amino acids beyond the −2 and +2 positions are not necessary for efficient binding and catalysis by sirtuins. For library construction, balancing minimal peptide length with practical limitations of library complexity were important considerations. Consequently, we elected to construct a 5-mer library with an acetylated lysine residue in the central position.
Table 1.
A summary of the relative catalytic efficiencies (kcat/Km) of various Sir2 homologs with ten peptide substrates.
| Relative kcat/Km | ||||
|---|---|---|---|---|
| Peptide | SIRT1 | SIRT2 | ySir2 | TbSir2 |
| AcGG(AcK)AP | 1.7 ± 0.5 | 0.6 ± 0.1 | 1.5 ± 0.7 | 1.3 ± 0.2 |
| AcTGG(AcK)AP | 0.5 ± 0.1 | 0.6 ± 0.1 | 1.6 ± 0.6 | 0.7 ± 0.2 |
| AcSTGG(AcK)AP | 1.6 ± 0.5 | 0.8 ± 0.1 | 1.8 ± 0.8 | 0.6 ± 0.1 |
| AcGG(AcK)APR | 0.7 ± 0.1 | 0.6 ± 0.1 | 2.5 ± 1.3 | 1.2 ± 0.2 |
| AcTGG(AcK)APR | 0.6 ± 0.1 | 0.7 ± 0.1 | 1.7 ± 0.8 | 0.9 ± 0.2 |
| AcSTGG(AcK)APR | 0.7 ± 0.1 | 0.9 ± 0.2 | 1.6 ± 0.8 | 1.4 ± 0.2 |
| AcGG(AcK)APRK | 1.0 ± 0.2 | 0.7 ± 0.1 | 2.3 ± 0.9 | 1.8 ± 0.2 |
| AcGG(AcK)APRKQ | 0.8 ± 0.2 | 0.7 ± 0.1 | 2.1 ± 0.8 | 1.4 ± 0.2 |
| AcKSTGG(AcK)AP | 0.7 ± 0.1 | 0.6 ± 0.1 | 2.1 ± 0.8 | 1.8 ± 0.7 |
| AcTGG(AcK)APRK | 1.0 ± 0.1 | 1.0 ± 0.1 | 1.0 ± 0.4 | 1.0 ± 0.1 |
Strategy for Sequencing Peptides on Beads
In order to extract peptide sequences from individual beads in the library, we devised a variation on the capping method developed by Youngquist, in which sequence decoding is done by reading a mass spectral peptide ladder (35). Instead of using the acetyl group for capping during peptide synthesis, we chose to use two carboxylic acids: phenylacetic acid and 4-pentenoic acid. First, a four amino acid linker was synthesized onto TentaGel beads to extend the bound peptide into solution and to bring the peptide mass out of the MALDI matrix region. This linker was composed of methionine (for a cyanogen bromide cleavage point), arginine (for improved mass spectral analysis) and two β-alanines (for added flexibility). In each coupling step of a randomized residue, a small amount of capping reagent was added to terminate chain growth for later sequencing. In each capping step, either one or both of the capping reagents were used. The use of two reagents assisted in deciphering amino acids of similar or identical masses. In cases, where both caps were used, a signature doublet would appear on the mass spectrum. By HPLC analysis, we found that 5 mol % capping at each step in the synthesis of a prototypical 5-mer yielded ∼ 79 % full-length peptide. This amount of capping reagent provided a more than adequate amount for on-bead assay, yet produced enough capped material to produce quality peptide ladders in the mass spectra. An acetyl group served as the N-terminal cap.
Screening Methodology
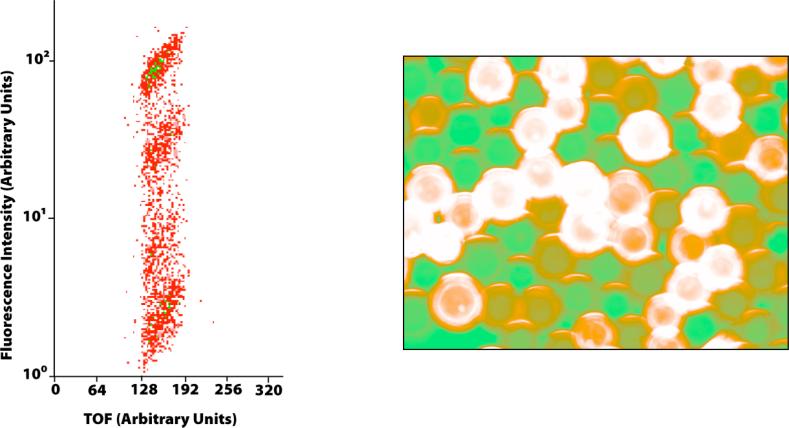
Our screening strategy relies on the reaction of biotin N-hydroxy-succinimide ester with the newly generated ε-amino group formed upon SIRT1 deacetylation. Subsequent binding of the streptavidin conjugated quantum dots provides the fluorescent tag for screening. Initially, it was important to establish that quantum dot labeling was proportional to the molar abundance reacted biotin. Resin bearing free amino groups was aliquoted into five reaction vessels and labeled with 1, 0.5, 0.01, 0.001 and 0 equivalents of biotin N-hydroxy-succinimide ester. After differential labeling, the resin was pooled into reaction vessel and a streptavidin conjugated quantum dot (λem = 605 nm) solution was added. After draining the quantum dot solution and washing the resin, the resulting pooled beads displayed differential levels of associated quantum dots, correlating with the amount of covalently linked biotin. Indeed, viewing the labeled beads under a fluorescence microscope showed the predicted dichromic color scheme (Fig. 1). Quantum dot labeled beads had an orange/red color while the background autofluorescence of the TentaGel resin appeared green. The varying shades of orange correlated qualitatively to the amount of bound quantum dot.
Figure 1.
Differntial biotinylation experiments with quantum dot tagging. Left: Log scale plot of fluorescence intensity vs. TOF (bead size) for quantum dot labeled beads with biotinylation levels of 1, 0.5, 0.01, 0.001 and 0 equivalents as analyzed by the COPAS beadsorter at 610 nm (individual beads are red and positions with ten or more beads are green). Right: A microscopic representation of the differentially biontinylated TentaGel beads after incubation with streptavidin coated quantum dots. Quantum dot labeled beads appear orange while TentaGel autofluorescence is green.
To provide a more quantitative assessment of fluorescent quantum dot labeling, a complex object parametric analyzer and sorter (COPAS) instrument was utilized. COPAS sorts beads based on fluorescence intensity while also gathering data on bead size (time of flight). Using this instrument, beads labeled in the previously mentioned experiment were sorted with an excitation of 488 nm and an emission of 610 nm. The fluorescence distribution was plotted and distinct populations could be visualized (Fig. 1). These populations resided in a fluorescence regime that encompassed more than two orders of magnitude. Moreover, the bead groupings corresponded to the differential levels of biotinylation (although the 0.001 and 0 biotin equivalents coalesced into a single cluster). It is important to note that when the emission wavelength was set to green light ((λex = 510 nm) corresponding to the intrinsic TentaGel autofluorescence, only a single population was observed. From these studies, we concluded that quantum dot labeling is quantitative in sub-stoichiometric amounts and that it can be coupled to the COPAS instrument for sorting beads on the basis of fluorescence intensity.
Library Construction and Screening
After validating that quantitative quantum dot labeling could be used in conjunction with fluorescence-based bead sorting, an OBOC peptide library was constructed using the split-pool method (27) (28). Eighteen variable amino acids were used at four positions centered around an acetylated lysine. All common natural amino acids excluding cysteine, lysine, methionine and arginine were used. To mimic charged residues, dimethyl arginine was substituted for lysine and arginine. To avoid unwanted cyanogen bromide cleavage points, isosteric norleucine was used in place of methionine. Lysine and cysteine were not included in the library because both residues would produce false hits (in addition to the problems posed by disulfide formation in the latter case) because the nucleophilicity of the amine and sulfhydryl groups respectively would result in their biotinylation and subsequent quantum dot labeling (Fig. 2). In preliminary studies, incorporation of arginine residues beyond the linker position gave false positive signals in the on-bead assays, due to reaction with the biotin ester during the labeling step. This was an unfortunate result, as it precluded incorporation of arginine in the library. The same problems posed by the reactivity of arginine have prevented its incorporation in a previous library (29). In order to mimic positively charged residues: lysine and arginine, we used symmetrical dimethyl arginine. Thus, 184 = 104,907 sequences were represented in the library. A threefold excess of beads was used to give 95 % probability that all sequences were represented (36).
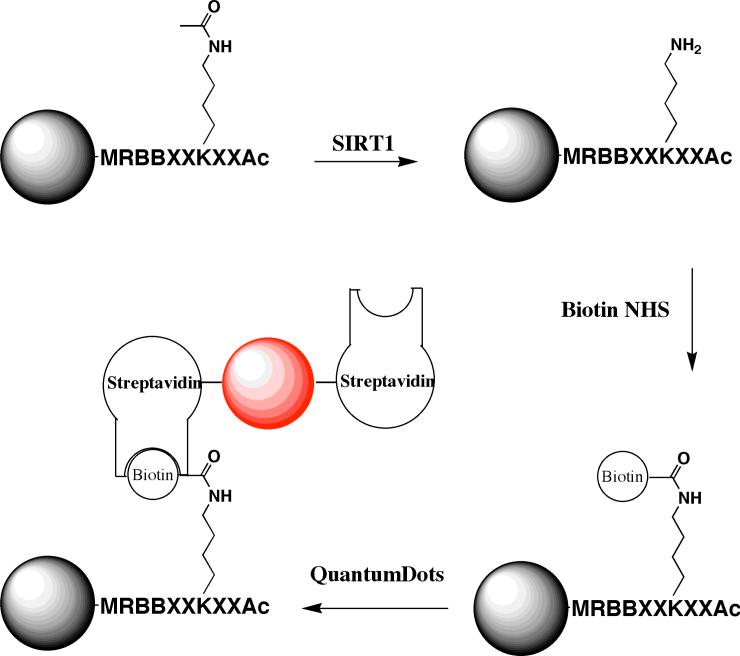
Figure 2.
Quantum dot bead-based assay. After deacetylation with SIRT1, beads are washed and free lysyl amines are biotinylated. Next, the beads are blocked from nonspecific binding with a BSA solution and incubated with quantum dots (red sphere). Finally, the beads are sorted with a COPAS instrument.
After library synthesis, the on-bead SIRT1 deacetylation assay was carried out (Fig. 2). In this assay, all beads were simultaneously subjected to deacetylation conditions (0.35 μM SIRT1, 12 min. at 25 °C), allowing competition of all peptide sequences for reaction with SIRT1. Afterwards, the beads were washed and subjected to biotinylation conditions in DMF. Excess reagent was removed prior to blocking non-specific protein binding sites with BSA and subsequent quantum dot labeling. Lastly, beads were washed a final time and sorted using the COPAS instrument (Fig. 3).
Figure 3.
Example of the fluorescence distribution of library members and an example of the mass spectrum obtained for microsequencing of a top hit sequence. Left: A histogram displaying the number of beads versus fluorescence intensity of a portion of the library (the sharp peak on the left corresponds to bubbles trapped in the instrument). Right: A representative mass spectrum of the cleavage products of one of the top forty most fluorescent beads. The amino acids corresponding various mass differences are annotated. Signature doublets are obtained for asparagine and histidine as result of the use of both capping reagents (1 and 2) during those coupling reactions.
Top 40 Hits
Initially, the three-hundred most intensely fluorescent beads (0.1 %) were collected, pooled and then sorted a second time to generate an enriched sample of the forty-five brightest beads. After washing in a guanidinium hydrochloride solution, single beads were placed in separate microcentrifuge tubes and treated overnight with a cyanogen bromide cleavage cocktail. The cleavage products were subsequently subjected to MALDI-TOF MS for sequence analysis (Fig. 3). Of those forty-five beads, thirty-three were sequenced successfully from their mass spectra (Table 2), six were found to be damaged and were not sequenced, while the remaining six yielded spectra that were not interpretable. BLAST (37) searches of the mammalian proteome were performed in the short, nearly exact mode for the thirty-three sequences obtained from the library (see discussion and supplemental data).
Table 2.
Peptide sequences of hits from the SIRT1 combinatorial library screen. Position -2 is the N-terminal end and Position +2 is the C-terminal end. Uncertainty in the order of N-terminal (and adjacent) amino acids is signified by /.
| Position -2 | Position -1 | Position 0 | Position +1 | Position +2 |
|---|---|---|---|---|
| Leu | Asn | AcLys | Asp | Gln |
| Trp | His | AcLys | Phe | Gln |
| Trp | His | AcLys | Phe | Glu |
| Ser | Tyr | AcLys | Gln | Trp |
| Gln | Pro | AcLys | Gln | Ile |
| Val | Gln | AcLys | Ile | Ile |
| His | Me2Arg | AcLys | Nle | Pro |
| Ala | Val | AcLys | Phe | Nle |
| Asn | His | AcLys | Leu | Leu |
| Me2Arg | Phe | AcLys | Pro | Glu |
| Nle | Nle | AcLys | Gln | Gln |
| Trp | Gly | AcLys | Ser | Pro |
| Phe | Glu | AcLys | Tyr | Me2Arg |
| Trp | Pro | AcLys | Trp | Gln |
| Me2Arg | Ala | AcLys | Nle | Asp |
| Gly | Thr | AcLys | Thr | Gly |
| Gly | Tyr | AcLys | Pro | Thr |
| Ile | Phe | AcLys | Thr | Phe |
| Thr | Glu | AcLys | Gln | Glu |
| His | Trp | AcLys | Thr | His |
| Asp | Ser | AcLys | Gly | Ala |
| Ser | Asp | AcLys | Tyr | His |
| Asn | His | AcLys | Ile | Ile |
| Trp | Trp | AcLys | His | Gly |
| Pro | Ile | AcLys | Glu | Gln |
| Me2Arg | Pro | AcLys | Gln | Phe |
| Asp | Val | AcLys | Nle | His |
| Ile | Tyr | AcLys | Asn | Asp |
| Thr | Pro | AcLys | Asn | Ala |
| Pro | Gly | AcLys | Leu | Tyr |
| Me2Arg/Trp | Me2Arg/Trp | AcLys | Ile | Thr |
| Pro/Trp | Pro/Trp | AcLys | Ile | Thr |
| Me2Arg/Pro | Me2Arg/Pro | AcLys | Ser | Ile |
Library Validation
To validate the results of our library screen, select hits and non-hits were resynthesized and subjected to in-solution kinetic analysis (Table 3). A radioactive TLC-based assay was employed with subsaturating levels of [32P]-NAD+ to determine the relative catalytic efficiencies (38). In addition, we analyzed two “consensus” peptides containing residues occurring with the highest and lowest frequency at each position, independent of context. For comparison, a 5-mer comprised of a sequence corresponding to a known site for p53 deacetylation by SIRT1 was assayed (Table 3). We found that hit sequences had significantly higher catalytic activity than non-hits. In fact, some hits were near or greater than an order of magnitude more catalytically active than their non-hit counterparts. We also noted that most hit sequences assayed in solution had significantly higher activity than the peptide sequence relevant to in vivo p53 deacetylation. One non-hit sequence AY(AcK)EV however, had a catalytic activity comparable to those of a few of the hits. It is important note that although the apparent second order rate constant (kcat/Km) varied widely among the peptides tested, the turnover number (kcat) was relatively constant at ∼0.1 s−1, suggesting that differences in kcat/Km reflect differences in peptide binding affinity.
Table 3.
Peptide sequences and catalytic efficiencies of resynthesized select hits and non-hits from the SIRT1 peptide library screen.
| Peptide | kcat/Km (× 10-3 M-1s-1) |
|---|---|
| Select Hits | |
| QP(AcK)QI | 27.2 ± 4.2 |
| Me2RP(AcK)QF | 14.7 ± 3.2 |
| Me2RP(AcK)SI | 8.4 ± 0.6 |
| NH(AcK)II | 3.6 ± 0.8 |
| WH(AcK)FQ | 3.3 ± 0.4x |
| VQ(AcK)IIa | ≥ 2.5 ± 1.3x |
| Select Non-hits | |
| AY(AcK)EV | 5.3 ± 0.6 |
| QNIe(AcK)GF | 2.3 ± 0.1 |
| LNIe(AcK)VG | 1.6 ± 0.5x |
| For Comparison | |
| WH(AcK)QQb | 7.2 ± 1.1 |
| WP(AcK)QQb | 1.5 ± 1.1 |
| EL(AcK)ASc | 1.4 ± 0.1 |
| HK(AcK)LMd | 3.1 ± 05 |
Efficiencies (x = average) were obtained by fitting the data from [32P]-NAD+ assays to the modified Michaelis-Menten equation, v = [(kcat/Km)[S]]/(1 + [S]/Km).
We were not able to determine a definite catalytic efficiency for VQ(AcK)II due to problems with insolubility but were able to establish a lower limit. Catalytic efficiencies of peptides containing the residues of the highest
lowest
frequency at each position and the sequence relevant to p53 deacetylation in vivo
are shown for comparison.
Discussion
We have developed a new high throughput method for constructing, screening and identifying novel SIRT1 substrates that can be applied to all protein deacetylases. This method takes advantage of the split-pool approach to synthesizing all possible peptide permutations from a diverse set of amino-acid building blocks and utilizes a screening methodology that incorporates fluorescent quantum dots and a bead-sorting instrument. In this approach, substrate identification is achieved by a capping strategy that permits the facile differentiation of isomeric amino acids in randomized peptide sequences. Although this approach is not without a few caveats (e.g., not all 20 amino acids are used in the library and the length of peptide sequences were restricted), here, we demonstrate its usefulness in probing sirtuin substrate recognition.
Data obtained from the combinatorial peptide library suggests that SIRT1 shows considerable preference for certain substrate sequences over others. For example, QP(AcK)QI is over an order of magnitude more reactive than most non-hits assayed against SIRT1. Other sequences that showed especially high activity in the validation studies are those that contained N-terminal dimethyl arginine residues. In addition, the catalytic activity of all peptide hits assayed in the library validation were superior to or in the range of that determined for a 5-mer sequence corresponding to the reported deacetylation site of p53, an in vivo substrate of SIRT1 (12). This data suggests that the hit sequences isolated from the library are preferred peptide substrates of SIRT1.
One of the main advantages of the OBOC library is its context specific nature. In other words, there is no implicit assumption that residues in substrate sequences function independently of one another. While oriented peptide libraries, on the other hand, can be useful in resolving globally-preferred “consensus” sequences, (26) (23) they do not provide contextual information. It is interesting to note that our so-called “consensus” peptides WH(AcK)QQ and WP(AcK)QQ show a seven-fold difference in catalytic activity in favor of WH(AcK)QQ (Table 3). Thus, in the context of WX(AcK)QQ, a histidine is greatly preferred at position −1. Within the XP(AcK)QX context, QP(AcK)QI is favored over WP(AcK)QQ by 18-fold. This suggests that SIRT1 mediated deacetylation is stringently context dependent and that there is no best “average sequence”. Further support comes from the fact that although proline residues (at −1) are not well tolerated when adjacent to a tryptophan at −2, they appear to function well when adjacent to dimethyl arginine at −2. These observations imply that there are synergistic/antagonistic relationships among certain residues and that this plays a significant role in substrate recognition by SIRT1.
The top 40 hits represents 0.04% of the total unique sequences of the library. These acetylated peptides were deacetylated ∼10-fold faster than randomly selected sequences from non-hits beads. Because the non-hit sequences were randomly chosen from 99.7 % of the total library, the catalytic efficiency of these sequences likely represents an average peptide deacetylation rate. Thus, a peptide displaying an average deacetylation rate is likely to be up to 10-fold lower than the top 40 peptides identified here. Accordingly, the worst peptide substrates (far left of the symmetrical Gaussian curve, Fig 3) would be predicted to be deacetylated ∼100-fold slower than the top 40. Given this context-dependent sequence preference of SIRT1, it is not surprising that Blander et al (23) observed no significant consensus. With SIRT1 capable of deacetylating a variety of peptide sequences, identification of the very best and the very worst peptide sequences would be masked in an oriented-peptide library analysis (23).
Although there is a strong correlation between the on-bead and the in-solution deacetylation assays, there was one exception, AY(AcK)EV, which was a randomly selected non-hit that displayed a relatively high kcat/Km value. It is quite possible that this sequence falls on the far right side of the Gaussian-type distribution (shown in Fig. 3), but narrowly failed to make our stringent selection of Top 40 sequences. Moreover, substrate recognition and binding on the solid phase may differ slightly from that in the solution phase for certain peptide sequences. Nonetheless, a general correlation between peptide hits and increased catalytic efficiency has been established.
BLAST (37) searches of the SIRT1 hits (Table 2) within the mammalian proteome reveal correspondence to a number of proteins (see supplementary data), some of which are known to be acetylated in vivo. A few proteins that have exact-match sequences with our peptide hits include moesin (functions in cell survival) (39), p33 (negatively regulates cell proliferation through acetylation of p53) (40), RNA guanylyltransferase (mRNA processing) (41), ROS1 (an oncogene highly-expressed in various tumor cell lines) (42), amyloid precursor-like protein (APPs have been linked to Alzhheimer's disease and transcriptional activation) (43) and PARPs 6 and 8 (DNA damage repair and aging) (44). Perhaps, one of the more intriguing matches was that for the Werner syndrome protein, a RecQ helicase known to be acetylated in vivo and thought to be involved in the repair of double strand DNA breaks (45) (46). Mutations of the Werner gene trigger a rare autosomal disorder that results in pre-mature aging (45).
Our findings suggest that substrate sequence recognition may play an important role in controlling acetyl-protein selectivity of SIRT1 activity in vivo. However, in a complex cellular environment, additional factors likely contribute to SIRT1 specificity. A recent study suggests that there may be conformational requirements for SIRT1 catalyzed deacetylation (47). Other parameters that would influence SIRT1 activity are the availability of NAD+, the levels of the inhibitor nicotinamide, and subcellular localization. For example, it is thought that SIRT1 interacts and localizes with PML nuclear bodies (48) and that this may partially dictate its deacetylase activity on localized targets. Also, substrate specificity may be influenced through associations with other protein factors that can either tether potential targets or alter the intrinsic binding affinity for certain substrates. Although we have established that SIRT1 does indeed harbor intrinsic substrate preferences, further studies will be needed to determine how these others factors contribute to substrate selection.
Various applications of libraries of this type can be envisioned. For example, the sequence information obtained from this library could be used to generate acetyl-peptide specific antibodies for Western blot analysis. This would provide in vivo validation of acetylation at protein sites discovered in BLAST searches. Such antibodies could also be employed in cell extract immunoprecipitation studies. Mass spectral analysis could then be performed to identify the acetylated proteins. Other potential uses of the library include the creation of super-substrates for the in vivo generation of OAADPr to elucidate its cellular roles. In another application, co-crystal studies could be executed to uncover how Sir2 interacts with these optimal substrate sequences. As noted earlier, limited peptide substrates co-crystallized with Sir2 have shown interactions primarily with the peptide backbone (21) (22). Here, we have demonstrated that side-chain interactions are important factors in the ability of sirtuins to bind and efficiently catalyzed protein deacetylation.
In addition to applications noted above, hits from libraries of this type could serve as starting points for the design of peptidomimetics. Our data suggests that optimized substrates reflect higher binding affinity to SIRT1. Modification of the peptide to prohibit enzymatic turnover and protease degradation could be implemented to generate a specific, tight-binding in vivo inhibitor. The original peptide sequence could be further modified to confer altered chemical and biological properties (30, 31). This strategy has been used to tailor-make peptides into therapeutics that avoid the pitfalls of proteolytic cleavage, rapid clearance from the circulatory system, inability to pass through the blood brain barrier and lack of oral activity (30).
Supplementary Material
Acknowledgments
We thank Matt Hanson and Bo Wang for assistance with the COPAS, and Dehua Pei and Thomas Kodadek for helpful discussions.
This work was supported by NIH Grant GM065386.
The abbreviations used are
- Sir2
silent information regulator 2
- NAD+
Nicotinamide Adenine Dinucleotide
- OAADPr
O-Acetyl-ADP-ribose
- OBOC
one bead- one compound
- COPAS
complex object parametric analyzer and sorter
- MALDI-TOF
matrix-assisted laser desorption/ionization time of flight mass spectrometry
- BLAST
basic local alignment search tool
- AAA
amino acid analysis
References
- 1.North BJ, Verdin E. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 2004;5:224. doi: 10.1186/gb-2004-5-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Denu JM. The Sir2 family of protein deacetylases. Curr Opin Chem Biol. 2005 doi: 10.1016/j.cbpa.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 4.Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 5.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 6.Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science. 2002;298:2390–2392. doi: 10.1126/science.1077650. [DOI] [PubMed] [Google Scholar]
- 7.Jackson MD, Denu JM. Structural identification of 2'- and 3'-O-acetyl-ADP-ribose as novel metabolites derived from the Sir2 family of beta -NAD+-dependent histone/protein deacetylases. J Biol Chem. 2002;277:18535–18544. doi: 10.1074/jbc.M200671200. [DOI] [PubMed] [Google Scholar]
- 8.Sauve AA, Celic I, Avalos J, Deng H, Boeke JD, Schramm VL. Chemistry of gene silencing: the mechanism of NAD+-dependent deacetylation reactions. Biochemistry. 2001;40:15456–15463. doi: 10.1021/bi011858j. [DOI] [PubMed] [Google Scholar]
- 9.Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 10.Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- 11.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 12.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 13.Bouras T, Fu M, Sauve AA, Wang F, Quong AA, Perkins ND, Hay RT, Gu W, Pestell RG. SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. J Biol Chem. 2005;280:10264–10276. doi: 10.1074/jbc.M408748200. [DOI] [PubMed] [Google Scholar]
- 14.Muth V, Nadaud S, Grummt I, Voit R. Acetylation of TAF(I)68, a subunit of TIF-IB/SL1, activates RNA polymerase I transcription. Embo J. 2001;20:1353–1362. doi: 10.1093/emboj/20.6.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, Hoffman E, Veech RL, Sartorelli V. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 16.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 17.Yang Y, Hou H, Haller EM, Nicosia SV, Bai W. Suppression of FOXO1 activity by FHL2 through SIRT1-mediated deacetylation. Embo J. 2005;24:1021–1032. doi: 10.1038/sj.emboj.7600570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Horst A, Tertoolen LG, de Vries-Smits LM, Frye RA, Medema RH, Burgering BM. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1) J Biol Chem. 2004;279:28873–28879. doi: 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- 19.Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. Embo J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pagans S, Pedal A, North BJ, Kaehlcke K, Marshall BL, Dorr A, Hetzer-Egger C, Henklein P, Frye R, McBurney MW, Hruby H, Jung M, Verdin E, Ott M. SIRT1 regulates HIV transcription via Tat deacetylation. PLoS Biol. 2005;3:e41. doi: 10.1371/journal.pbio.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Avalos JL, Celic I, Muhammad S, Cosgrove MS, Boeke JD, Wolberger C. Structure of a Sir2 enzyme bound to an acetylated p53 peptide. Mol Cell. 2002;10:523–535. doi: 10.1016/s1097-2765(02)00628-7. [DOI] [PubMed] [Google Scholar]
- 22.Zhao K, Chai X, Marmorstein R. Structure of the yeast Hst2 protein deacetylase in ternary complex with 2'-O-acetyl ADP ribose and histone peptide. Structure (Camb) 2003;11:1403–1411. doi: 10.1016/j.str.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 23.Blander G, Olejnik J, Krzymanska-Olejnik E, McDonagh T, Haigis M, Yaffe MB, Guarente L. SIRT1 shows no substrate specificity in vitro. J Biol Chem. 2005;280:9780–9785. doi: 10.1074/jbc.M414080200. [DOI] [PubMed] [Google Scholar]
- 24.Borra MT, Langer MR, Slama JT, Denu JM. Substrate specificity and kinetic mechanism of the Sir2 family of NAD+-dependent histone/protein deacetylases. Biochemistry. 2004;43:9877–9887. doi: 10.1021/bi049592e. [DOI] [PubMed] [Google Scholar]
- 25.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 26.Songyang Z, Blechner S, Hoagland N, Hoekstra MF, Piwnica-Worms H, Cantley LC. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr Biol. 1994;4:973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- 27.Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. A new type of synthetic peptide library for identifying ligand-binding activity. Nature. 1991;354:82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- 28.Furka A, Sebestyen F, Asgedom M, Dibo G. General method for rapid synthesis of multicomponent peptide mixtures. Int J Pept Protein Res. 1991;37:487–493. doi: 10.1111/j.1399-3011.1991.tb00765.x. [DOI] [PubMed] [Google Scholar]
- 29.Hu YJ, Wei Y, Zhou Y, Rajagopalan PT, Pei D. Determination of substrate specificity for peptide deformylase through the screening of a combinatorial peptide library. Biochemistry. 1999;38:643–650. doi: 10.1021/bi9820412. [DOI] [PubMed] [Google Scholar]
- 30.Nefzi A, Ostresh JM, Yu Y, Houghten RA. Combinatorial chemistry: libraries from libraries, the art of the diversity-oriented transformation of resin-bound peptides and chiral polyamides to low molecular weight acyclic and heterocyclic compounds. J Org Chem. 2004;69:3603–3609. doi: 10.1021/jo040114j. [DOI] [PubMed] [Google Scholar]
- 31.Falciani C, Lozzi L, Pini A, Bracci L. Bioactive peptides from libraries. Chem Biol. 2005;12:417–426. doi: 10.1016/j.chembiol.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 32.Olivos HJ, Bachhawat-Sikder K, Kodadek T. Quantum dots as a visual aid for screening bead-bound combinatorial libraries. Chembiochem. 2003;4:1242–1245. doi: 10.1002/cbic.200300712. [DOI] [PubMed] [Google Scholar]
- 33.Chan WC, Nie S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 34.Watson A, Wu X, Bruchez M. Lighting up cells with quantum dots. Biotechniques. 2003;34:296–300. 302–293. doi: 10.2144/03342bi01. [DOI] [PubMed] [Google Scholar]
- 35.Youngquist RS, Fuentes GR, Lacey MP, Keough T. Generation and Screening of Combinatorial Peptide Libraries Designed for Rapid Sequencing by Mass Spectrometry. J. Am. Chem. Soc. 1995;117:3900–3906. [Google Scholar]
- 36.Burgess K, Liaw AI, Wang N. Combinatorial technologies involving reiterative division/coupling/recombination: statistical considerations. J Med Chem. 1994;37:2985–2987. doi: 10.1021/jm00045a001. [DOI] [PubMed] [Google Scholar]
- 37.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson MD, Schmidt MT, Oppenheimer NJ, Denu JM. Mechanism of nicotinamide inhibition and transglycosidation by Sir2 histone/protein deacetylases. J Biol Chem. 2003;278:50985–50998. doi: 10.1074/jbc.M306552200. [DOI] [PubMed] [Google Scholar]
- 39.Wu KL, Khan S, Lakhe-Reddy S, Jarad G, Mukherjee A, Obejero-Paz CA, Konieczkowski M, Sedor JR, Schelling JR. The NHE1 Na+/H+ exchanger recruits ezrin/radixin/moesin proteins to regulate Akt-dependent cell survival. J Biol Chem. 2004;279:26280–26286. doi: 10.1074/jbc.M400814200. [DOI] [PubMed] [Google Scholar]
- 40.Nagashima M, Shiseki M, Miura K, Hagiwara K, Linke SP, Pedeux R, Wang XW, Yokota J, Riabowol K, Harris CC. DNA damage-inducible gene p33ING2 negatively regulates cell proliferation through acetylation of p53. Proc Natl Acad Sci U S A. 2001;98:9671–9676. doi: 10.1073/pnas.161151798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sawaya R, Shuman S. Mutational analysis of the guanylyltransferase component of Mammalian mRNA capping enzyme. Biochemistry. 2003;42:8240–8249. doi: 10.1021/bi034396d. [DOI] [PubMed] [Google Scholar]
- 42.Biskup C, Bohmer A, Pusch R, Kelbauskas L, Gorshokov A, Majoul I, Lindenau J, Benndorf K, Bohmer FD. Visualization of SHP-1-target interaction. J Cell Sci. 2004;117:5165–5178. doi: 10.1242/jcs.01397. [DOI] [PubMed] [Google Scholar]
- 43.Cao X, Sudhof TC. A transcriptionally [correction of transcriptively] active complex of APP with Fe65 and histone acetyltransferase Tip60. Science. 2001;293:115–120. doi: 10.1126/science.1058783. [DOI] [PubMed] [Google Scholar]
- 44.Burkle A, Diefenbach J, Brabeck C, Beneke S. Ageing and PARP. Pharmacol Res. 2005;52:93–99. doi: 10.1016/j.phrs.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 45.Blander G, Zalle N, Daniely Y, Taplick J, Gray MD, Oren M. DNA damage-induced translocation of the Werner helicase is regulated by acetylation. J Biol Chem. 2002;277:50934–50940. doi: 10.1074/jbc.M210479200. [DOI] [PubMed] [Google Scholar]
- 46.Comai L, Li B. The Werner syndrome protein at the crossroads of DNA repair and apoptosis. Mech Ageing Dev. 2004;125:521–528. doi: 10.1016/j.mad.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 47.Khan AN, Lewis PN. Unstructured conformations are a substrate requirement for the Sir2 family of NAD-dependent protein deacetylases. J Biol Chem. 2005 doi: 10.1074/jbc.M508247200. [DOI] [PubMed] [Google Scholar]
- 48.Langley E, Pearson M, Faretta M, Bauer UM, Frye RA, Minucci S, Pelicci PG, Kouzarides T. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. Embo J. 2002;21:2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Borra MT, Smith BC, Denu JM. Mechanism of human SIRT1 activation by resveratrol. J Biol Chem. 2005;280:17187–17195. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- 50.Bodanszky M. Principles of Peptide Synthesis. 2nd ed. Springer-Verlag; Germany: 1993. [Google Scholar]
- 51.King DS, Fields CG, Fields GB. A cleavage method which minimizes side reactions following Fmoc solid phase peptide synthesis. Int J Pept Protein Res. 1990;36:255–266. doi: 10.1111/j.1399-3011.1990.tb00976.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.