Abstract
Effort has been made to identify protective antigens in order to develop a recombinant vaccine against leptospirosis. Several attempts failed to conclusively demonstrate efficacy of vaccine candidates due to the lack of an appropriate model of lethal leptospirosis. The purposes of our study were: (i) to test the virulence of leptospiral isolates from Brazil, which are representative of important serogroups that cause disease in humans and animals; and (ii) to standardize the lethal dose 50% (LD50) for each of the virulent strains using a hamster (Mesocricetus auratus) model. Five of seven Brazilian isolates induced lethality in a hamster model, with inocula lower than 200 leptospires. Histopathological examination of infected animals showed typical lesions found in both natural and experimental leptospirosis. Results described here demonstrated the potential use of Brazilian isolates as highly virulent strains in challenge experiments using hamster as an appropriate animal model for leptospirosis. Furthermore these strains may be useful in heterologous challenge studies which aim to evaluate cross-protective responses induced by subunit vaccine candidates.
Keywords: Leptospira, leptospirosis, lethal dose, isolation, animal model, virulence
1. INTRODUCTION
Leptospirosis is an infectious disease caused by pathogenic leptospires that are transmitted directly or indirectly to humans and animals. Leptospirosis occurs worldwide but it is more commonly associated with tropical and subtropical areas [1]. The disease is found mainly wherever humans come into contact with the urine of infected animals or an urine-contaminated environment [2]. Exposure to leptospires may be associated to water sports, sporadic exposure in urban areas of developed countries, outbreaks associated with rain season and floods in urban areas with poor sanitary conditions, and it also occours as an endemic disease in rural areas in Latin America and Southeast Asia [2-4]. The clinical presentation of human infection ranges from olygosymptomatic or undifferentiated febrile illness to severe forms of Weil’s syndrome, with an overall 10-15% case fatality, and severe pulmonary hemorrhagic syndrome (SPHS), with ≥50% case fatality [5,6].
Whole genome sequences are now available for Leptospira interrogans serovars Lai and Copenhageni and L. borgpetersenii serovar Hardjo [7-9]. Both China [9] and Brazil [8] have made research in leptospirosis a priority in order to address this emerging public health problem. In the case of Brazil, a national multiinstitutional initiative led to the whole genome sequencing of L. interrogans serovar Copenhageni, the agent for epidemics of severe leptospirosis in Brazilian urban centers [10,11]. A major goal has been the use of genome information to identify targets for a subunit based vaccine [12]. Ideally an effective vaccine would induce induce cross-protection against the range of serovars of public health importance. Encorauging data using recombinant proteins and DNA vaccines in different animal models has already been published [13-19].
Brazilian strains of serovar Copenhageni isolated from humans have been used in recent studies on pathogenesis and proteomics [20-27]. However, there is a wide diversity of Leptospira serovars that constitute etiological agents of leptospirosis, specialy in rural settings. To date, there is little information on virulence of pathogenic isolates using a standard animal model. This is a critical issue for the evaluation of vaccine candidates that elicit potential cross immunoprotection response.
In this context, we tested the virulence of Leptospira isolates obtained from humans, dogs and from a mouse, representative of serogroups important for public health and veterinary areas. In addition, we performed LD50 experiments to demonstrate the suitability of these virulent strains for challenge assays using low inocula in the hamster (Mesocricetus auratus) model. This strategy aimed to develop a standard method for comparison of virulence that would make it easier to design and interpret immunoprotection experiments.
2. MATERIALS AND METHODS
2.1. Bacteria
All leptospires were cultivated in liquid Ellinghausen-McCullough-Johnson-Harris (EMJH) medium (Difco Labratories) at 29 °C, and leptospires were counted in a Petroff-Hauser counting chamber (Fisher Scientific) as previously described [28]. L. interrogans serovar Copenhageni strain Fiocruz L1-130 [11] and six isolates from Pelotas city, South of Brazil, previously serogrouped (Table 1), were passaged in hamster, and stored at -70 °C until use. In order to minimize variability on virulence due to re-adaption to culture medium, each isolate received a specific notation as function of the number of in vivo and in vitro passages. Thus, for standardization and reproducibility of future experiments, Fiocruz L1-130 was passaged in hamsters four times and then three times in EMJH medium. Aliquots were passaged four times in liquid medium prior to their use as a low-passage-number isolate for the infection experiments. Other strains had a distinct number of passages in hamsters and in vitro (Table 1).
TABLE 1.
Strains used in experiments
| Species | Serogroup | Strain | Source | Sample | Number of passages |
||
|---|---|---|---|---|---|---|---|
| In vitro pre-hamster | Hamster passages | In vitro post-hamster | |||||
| L. interrogans | Icterohaemorrhagiae | L1-130 | Human | Blood | 4 | 4 | 7 |
| L. interrogans | Djasiman | Isoton | Human | Blood | 31 | - | - |
| L. interrogans | Australis | Skoll | Mouse | Kidney | 7 | - | - |
| L. interrogans | Canicola | Kito | Dog | Urine | 8 | 2 | 5 |
| L. noguchii | Australis | Hook | Dog | Kidney | 27 | 4 | 5 |
| L. noguchii | Bataviae | Cascata | Human | Blood | 5 | 2 | 5 |
| L. noguchii | Autumnalis | Bonito | Human | Blood | 16 | 2 | 6 |
2.2. Animals and experimental challenge infections
Male and female Golden Syrian hamsters (Fiocruz/BA) were used in all experiments. For virulence evaluation of the clinical isolates, hamsters (n=2 in each group) weighing approximately 55 g, were infected intraperitoneally with 108 leptospires. All animals were monitored daily for the presence of clinical signs, including evidence of external hemorrhage, dehydration, ruffled hair coat, decreased activity and isolation. In these conditions, hamsters were euthanized and kidney tissue was cultured at 29 °C in liquid EMJH media without antibiotics. Leptospires were recovered from kidney tissue of moribund hamsters in order to obtain a challenge strain which would have reproducible virulence characteristics. After two or three sub-cultures in liquid media, each strain was distributed in aliquots of 1 ml and stored at -70 °C for future use. Only strains that proved to be virulent causing lethal disease in these screening experiments were further evaluated in LD50 experiments. Strains which failed to cause lethal disease in three experiments were regarded as avirulent. This project and all animal experiments were approved by the Committee for Animal Care and Use (CEUA/Fiocruz).
2.3. LD50 experimental design
For all experiments, nine-week old hamsters were infected intraperitoneally with 10 fold serial dilutions. Inocula of 105 to 100 organisms were tested. The number of animals for each inoculum was 4 or 8, depending on the availability of animal facilities (detailed information provided in table 2). Animals were monitored daily for clinical outcome until 28 days post-infection. After this, LD50 was calculated by the method of Reed and Muench [29]. Negative control animals were injected with the same volume of sterile EMJH media. All strains were evaluated in at least three independent experiments.
TABLE 2.
LD50 of virulent Leptospira strains
| Species | Strain | LD50 of individual experiments (number of animals used per inoculum) |
Mean LD50 (± SD) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Experiment 1 | Experiment 2 | Experiment 3 | |||||||
| F | M | F | M | F | M | F | M | ||
| L. interrogans | L1-130 | 56.2 (4) | 56.2 (4) | 165 (4) | 42.2 (4) | 100 (4) | 17 (4) | 105 (44.8) | 36.7 (16.6) |
| L. interrogans | Kito | 3.2 (2) | 3.2 (2) | 2.2 (4) | 1.0 (4) | 3.0 (4) | 3.3 (4) | 2.8 (0.54) | 2.5 (1.29) |
| L. noguchii | Cascata | 18.3 (4) | 21.5 (4) | 33.4 (4) | 50 (4) | 50 (2) | 100 (2) | 33.9 (15.9) | 57.2 (32.4) |
| L. noguchii | Hook | 100 (2) | 10 (2) | 68.1 (4) | 27.4 (4) | 178 (2) | 17.8 (2) | 115.4 (56.5) | 18.4 (8.71) |
| L. noguchii | Bonito | 1.78 (2) | 2.2 (2) | 3.16 (2) | 5.6 (2) | 3.2 (2) | 2.2 (2) | 2.7 (0.8) | 3.3 (2.0) |
SD = Standard deviation; F = Females; M = Males
2.4. Histopatologic analyses
In order to investigate whether inoculation with the strains could reproduce histopathological changes typical of experimental leptospirosis, we performed necropsies of the first two animals appearing moribund after infection by each strain and compared their histopathology with samples from two non infected hamsters of the same age euthanized nine days after inoculation of 1 ml sterile EMJH media. Neutral-buffered formalin fixed paraffin-embedded samples were sectioned and stained with Hematoxilin and Eosin (HE) and silver impregnation by Warthin-Starry technique [2].
3. RESULTS
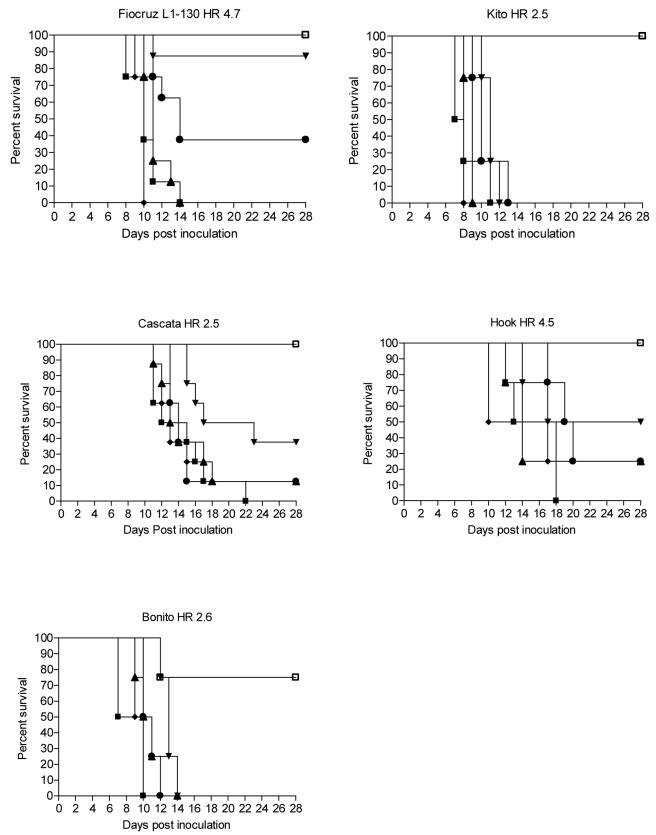
Strains Fiocruz L1-130, Kito, Cascata, Hook and Bonito (Table 1) produced lethal infection with inocula of 108 in all hamsters tested. Strains Isoton and Skoll did not cause disease or colonization in hamsters in three challenge experiments and therefore were regarded as avirulent strains. Kito and Bonito strains had the lowest LD50 (<10 leptospires). However, all virulent strains induced disease and led to death with an inoculum containing less than 200 leptospires. L. interrogans serovar Copenhageni strain Fiocruz L1-130 had a LD50 of 105 leptospires in females and 36.7 in males. L. interrogans serogroup Canicola strain Kito had a LD50 of 2.8 in females and 2.5 in males. L. noguchii serogroup Bataviae strain Cascata had a LD50 of 33.9 in females and 57.2 in males. L. noguchii serogroup Australis strain Hook had a LD50 of 115.4 in females and 18.4 in males. L. noguchii serogroup Autumnalis strain Bonito had a LD50 of 2.7 in females and 3.3 in males.
Clinical signs were observed from the 4th day post infection (d.p.i.) and the mean period of death of hamsters was 11 d.p.i. However, animals experimentally infected with strains belonging to L. interrogans died between 7 and 14 d.p.i. while strains belonging to L. noguchii caused death in hamsters 7 to 22 d.p.i. A survival curve from representative experiments is shown in figure 1.
Fig.1.
Kaplan-Meier analysis of the results from LD50 experiments with 5 virulent strains. In these five representative curves, challenge studies with Fiocruz L1-130 and Cascata strains were performed with eight hamsters per group, and challenge studies with Kito, Hook and Bonito strains were performed with four hamsters per group. Groups of hamsters were inoculated with 105 (■), 104 (◆), 103 (▲), 102 (●), 101 (▼) and 100 (□) leptospires. Survivors were followed for up to 28 days post infection.
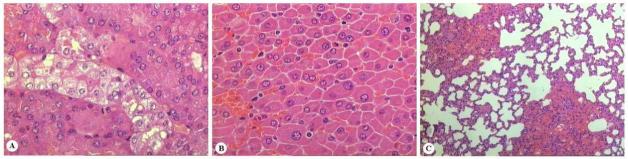
Hamsters infected with virulent strains developed acute lethal infection characterized by hepatic and renal complications. Acute damage of tubular epithelia with cell swelling in proximal segments was observed as a common feature in moribund hamsters before 10 d.p.i (Fig. 2A). In contrast, hamsters which died after 10 d.p.i. exhibited multifocal regenerative changes in tubular epithelia of renal cortex and moderate nephritis with infiltrates of leukocytes and histiocytes most often distributed around small arteries. All moribund animals exhibited marked dissociation of hepatic trabecula and had hepatocytes which undergone reactive changes such as cytoplasmic size variation, prominent nucleoli and binucleation (Fig. 2B). Macroscopic pulmonary and widespread bleeding was found in animals which received inocula of 103 or higher. Although gross hemorrhages were not detected during necropsy procedure of hamsters which received inocula of less than 103, microscopic foci of alveolar hemorrhage were observed in all animals inoculated with virulent strains (table 2). None of these features were seen in health control animals (Fig. 2C).
Fig. 2.
Typical lesions of leptospirosis in a 9 week old hamster dying eight days after the infection by strain FIOCRUZ L1-130. (A) Marked cell swelling of epithelial cells of proximal tubules (hematoxylin-eosin, 400x). (B) Diffuse loss of cohesion (liver-plate disarray) of liver cells (hematoxylin-eosin, 400x). (C) Microscopic foci of pulmonary hemorrhage (hematoxylin-eosin, 200x).
4. DISCUSSION
Increasing interest in the development of a recombinant leptospirosis vaccine has emerged in the past few years. The field was further stimulated by the whole genome sequencing of four strains from two pathogenic Leptospira species. Promising results have been reported by the use of recombinant proteins in hamster [13-15,19]. In addition, naked DNA and adenovirus used as a vaccine vector for a leptospiral antigen have also been evaluated in a gerbil model [18]. In these studies, the ability of sub-unit vaccine candidates to induce immunoprotection against heterologous serovars was not evaluated. However, a recent study reported heterologous protection in a gerbil model using plasmids encoding LipL32 (Hap1) from serovars Autumnalis and Grippotyphosa. Immunization with this construct conferred protection against L. interrogans serovar Canicola challenge [16]. A more complete evaluation of the capacity of vaccine candidates to induce cross-protective response requires a well-characterized panel of virulent strains representing serovars of public health and veterinary importance. We report here a suitable model for evaluation of vaccine candidates against a wider range of pathogenic serovars.
In our study, we performed challenge experiments in hamster model with five highly virulent Leptospira strains. Mice and hamster models have often required inocula as high as 106-108 leptospires to induce death [15,17]. Despite high doses, some strains are unable to induce mortality in all control animals. For example, challenge with a 108 inoculum of Pomona strain induced death in 25-57% of unvaccinated hamsters [15,30,31], which in turn complicates the conclusions on the effectivenes of a vaccine candidate. The need for high challenge doses of up to 106 organisms to produce lethal infection may be due to the non-susceptibility of the animal model (mouse) [17] or low virulence of the strains. Protective effects would be better evaluated using strains with an LD50 of <103 in a susceptible host, such as the hamster. In a previous evaluation of homologous protection using Fiocruz L1-130 strain, fragments of leptospiral immunoglobulin-like proteins prevented lethal disease in hamsters while unvaccinated controls showed a high rate of lethality (100% of 76 animals infected with 103 leptospiras) [14].
L. interrogans Copenhageni Fiocruz L1-130 strain was isolated from a patient identified during the flood-associated outbreak of leptospirosis in Salvador, Brazil, in 1996 [11]. This isolate was also the target of a multicenter initiative to sequence its genome [8]. Additionally, it has been used for studies on pathogenesis and leptospiral proteomics [20-21,26-27,32-34]. Serovar Copenhageni is the most common leptospiral isolate in Salvador and also in other urban areas of Brazil [10-11,35-36] and its low LD50 demonstrates that it is a highly virulent pathogen for hamsters, which has been used in previous immunoprotection experiments [14].
L. interrogans serovar Canicola is the second most important agent for urban leptospirosis in Brazil [10]. In our study, we could demonstrate it as a highly virulent strain for hamsters with a general LD50 of 2.57 leptospires. Serovar Canicola may cause severe disease in animals and has already been associated to a pulmonary hemorrhage human leptospirosis outbreak in rural areas of Nicaragua [37].
L. noguchii is a pathogenic species present in the American continent and has been recently reported in the South of Brazil [38]. It had been previously isolated from human, armadillo, toad, spiny rat, opossum, nutria, Mustela nivalis, cattle and sheep, showing a wide variety of domestic and wild hosts [2,38,39]. Strains Bonito and Cascata were isolated from human patients with clinical leptospirosis, while Hook strain was isolated from a stray dog with lethal leptospirosis. All strains were virulent for hamsters.
These human and animal virulent isolates were submitted to hamster passages before their use in LD50 experiments. Only strains Isoton and Skoll, which did not cause lethal disease and could not be recovered from hamster kidneys inoculated with 108 leptospiras, were not further evaluated. It is conceivable that the observed high virulence in LD50 experiments reflects a virulence artificially obtained by successive hamster passages (“hamster adaptation”), and not the natural virulence of each isolate. However, our goal in this study was to select distinct virulent strains to be used in hamster model regardless of the original virulence characteristics of the isolates.
The lack of virulence of strain Isoton, an isolate from the blood culture of a patient with severe leptospirosis might be explained by the successive in vitro propagation: 31 passages may have allowed mutations to occur rendering the strain avirulent. It has already been reported that high-passage strains can present impaired virulence [40]. On the other hand, although uncommon, some strains may be virulent in host animals but not in the experimental model as is the case for serovar Hardjo subtype A strains [7]. The Skoll strain was isolated from an asymptomatic mouse. In this case it is accepted that avirulent leptospires may colonize renal tissues of mammalian hosts [40,41]. Similarly, we have previously isolated the L. noguchii strain Caco from an asymptomatic sheep that failed to cause disease in the hamster model [38]. The lack of virulence in these strains precludes their use in immunoprotection experiments.
The hamster model reproduced pathologic findings observed in acute lethal forms of human and experimental leptospirosis, as previously described [42-46]. The presence of microscopic pulmonary hemorrhage in the absence of gross features has also been reported in humans and in guinea pigs [25,43]. The pattern of acute cell swelling in fulminant disease and a picture of multifocal regeneration tubular foci and interstitial nephritis in more prolonged illness have also been reported in humans [42]. Taken together, the reproducibility of acute lethal infection and target organ pathology makes the hamster model suitable for standardization of virulent strains and immunoprotection assays.
In conclusion, we characterized the virulence of five clinical isolates of Leptospira belonging to five different serogroups. These highly virulent strains are currently been used in experiments aiming at evaluating homologous and heterologous protection induced by killed whole-cell and recombinant vaccine candidates against acute lethal leptospirosis in the hamster model.
ACKNOWLEDGEMENTS
This work was supported by CAPES Foundation (Brazilian Government), Bio-Manguinhos and the Oswaldo Cruz Foundation (09224-7 and PDTIS RVR05), the Brazilian National Research Council (01.06.0298.00 3773/2005, 420067/2005), Research Support Foundation for the State of Bahia, and the National Institutes of Health (5 R01 AI052473, 2 D43 TW-00919).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
5. REFERENCES
- [1].World Health Organization . Human leptospirosis: guidance for diagnosis, surveillance and control. WHO Library Cataloguing-in-Publication Data 2003; [Google Scholar]
- [2].Faine SB, Adler B, Bolin C, Perolat P. Leptospira and leptospirosis. 2nd ed. MediSci; Melbourne, Australia: 1999. [Google Scholar]
- [3].Levett PN. Leptospirosis. Clin Microbiol Rev. 2001;14(2):296–326. doi: 10.1128/CMR.14.2.296-326.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Vinetz JM. Leptospirosis. Curr Opin Infect Dis. 2001;14(5):527–38. doi: 10.1097/00001432-200110000-00005. [DOI] [PubMed] [Google Scholar]
- [5].Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, Lovett MA, et al. Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis. 2003;3(12):757–71. doi: 10.1016/s1473-3099(03)00830-2. [DOI] [PubMed] [Google Scholar]
- [6].McBride AJ, Athanazio DA, Reis MG, Ko AI. Leptospirosis. Curr Opin Infect Dis. 2005;18(5):376–86. doi: 10.1097/01.qco.0000178824.05715.2c. [DOI] [PubMed] [Google Scholar]
- [7].Bulach DM, Zuerner RL, Wilson P, Seemann T, McGrath A, Cullen PA, et al. Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. PNAS USA. 2006;103(39):14560–65. doi: 10.1073/pnas.0603979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Nascimento AL, Ko AI, Martins EA, Monteiro-Vitorello CB, Ho PL, Haake DA, et al. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J Bacteriol. 2004;186(7):2164–72. doi: 10.1128/JB.186.7.2164-2172.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ren SX, Fu G, Jiang XG, Zeng R, Miao YG, Xu H, et al. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature. 2003;422(6934):888–93. doi: 10.1038/nature01597. [DOI] [PubMed] [Google Scholar]
- [10].Pereira MM, Matsuo MG, Bauab AR, Vasconcelos SA, Moraes ZM, Baranton G, et al. A clonal subpopulation of Leptospira interrogans sensu stricto is the major cause of leptospirosis outbreaks in Brazil. J Clin Microbiol. 2000;38:450–52. doi: 10.1128/jcm.38.1.450-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ko AI, Galvao Reis M, Ribeiro Dourado CM, Johnson WD, Jr., Riley LW, Salvador Leptospirosis Study Group Urban epidemic of severe leptospirosis in Brazil. Lancet. 1999;354(9181):820–5. doi: 10.1016/s0140-6736(99)80012-9. [DOI] [PubMed] [Google Scholar]
- [12].Thongboonkerd V. Proteomics in leptospirosis research: towards molecular diagnostics and vaccine development. Expert Rev Mol Diagn. 2008;8(1):53–61. doi: 10.1586/14737159.8.1.53. [DOI] [PubMed] [Google Scholar]
- [13].Seixas FK, da Silva EF, Hartwig DD, Cerqueira GM, Amaral M, Fagundes MQ, et al. Recombinant Mycobacterium bovis BCG expressing the LipL32 antigen of Leptospira interrogans protects hamsters from challenge. Vaccine. 2007;26(1):88–95. doi: 10.1016/j.vaccine.2007.10.052. [DOI] [PubMed] [Google Scholar]
- [14].Silva EF, Medeiros MA, McBride AJ, Matsunaga J, Esteves GS, Ramos JG, et al. The terminal portion of leptospiral immunoglobulin-like protein LigA confers protective immunity against lethal infection in the hamster model of leptospirosis. Vaccine. 2007;25(33):6277–86. doi: 10.1016/j.vaccine.2007.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Palaniappan RU, McDonough SP, Divers TJ, Chen CS, Pan MJ, Matsumoto M, et al. Immunoprotection of Recombinant Leptospiral Immunoglobulin-Like Protein A against Leptospira interrogans Serovar Pomona Infection. Infect Immun. 2006;74(3):1745–50. doi: 10.1128/IAI.74.3.1745-1750.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Branger C, Chatrenet B, Gauvrit A, Aviat F, Aubert A, Bach JM, et al. Protection against Leptospira interrogans sensu lato challenge by DNA immunization with the gene encoding hemolysin-associated protein 1. Infect Immun. 2005;73(7):4062–9. doi: 10.1128/IAI.73.7.4062-4069.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Koizumi N, Watanabe H. Leptospiral immunoglobulin-like proteins elicit protective immunity. Vaccine. 2004;22(1112):1545–52. doi: 10.1016/j.vaccine.2003.10.007. [DOI] [PubMed] [Google Scholar]
- [18].Branger C, Sonrier C, Chatrenet B, Klonjkowski B, Ruvoen-Clouet N, Aubert A, et al. Identification of the hemolysis-associated protein 1 as a cross-protective immunogen of Leptospira interrogans by adenovirus-mediated vaccination. Infect Immun. 2001;69(11):6831–38. doi: 10.1128/IAI.69.11.6831-6838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Haake DA, Mazel MK, McCoy AM, Milward F, Chao G, Matsunaga J, et al. Leptospiral outer membrane proteins OmpL1 and LipL41 exhibit synergistic immunoprotection. Infect Immun. 1999;67(12):6572–82. doi: 10.1128/iai.67.12.6572-6582.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Athanazio DA, Santos CS, Santos AC, McBride FW, Reis MG. Experimental infection in tumor necrosis factor alpha receptor, interferon gamma and interleukin 4 deficient mice by pathogenic Leptospira interrogans. Acta Trop. 2008;105(1):95–8. doi: 10.1016/j.actatropica.2007.09.004. [DOI] [PubMed] [Google Scholar]
- [21].Athanazio DA, Silva EF, Santos CS, Rocha GM, Vannier-Santos MA, McBride AJ, et al. Rattus norvegicus as a model for persistent renal colonization by pathogenic Leptospira interrogans. Acta Trop. 2008;105(2):176–80. doi: 10.1016/j.actatropica.2007.10.012. [DOI] [PubMed] [Google Scholar]
- [22].Matsunaga J, Sanchez Y, Xu X, Haake DA. Osmolarity, a Key Environmental Signal Controlling Expression of Leptospiral Proteins LigA and LigB and the Extracellular Release of LigA. Infect Immun. 2005;73(1):70–78. doi: 10.1128/IAI.73.1.70-78.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Nally JE, Chow E, Fishbein MC, Blanco DR, Lovett MA. Changes in lipopolysaccharide O antigen distinguish acute versus chronic Leptospira interrogans infections. Infect Immun. 2005;73(6):3251–60. doi: 10.1128/IAI.73.6.3251-3260.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Nally JE, Whitelegge JP, Aguilera R, Pereira MM, Blanco DR, Lovett MA. Purification and proteomic analysis of outer membrane vesicles from a clinical isolate of Leptospira interrogans serovar Copenhageni. Proteomics. 2005;5(1):144–52. doi: 10.1002/pmic.200400880. [DOI] [PubMed] [Google Scholar]
- [25].Nally JE, Chantranuwat C, Wu XY, Fishbein MC, Pereira MM, Da Silva JJ, et al. Alveolar septal deposition of immunoglobulin and complement parallels pulmonary hemorrhage in a guinea pig model of severe pulmonary leptospirosis. Am J Pathol. 2004;164(3):1115–27. doi: 10.1016/S0002-9440(10)63198-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Matsunaga J, Barocchi MA, Croda J, Young TA, Sanchez Y, Siqueira I, et al. Pathogenic Leptospira species express surface-exposed proteins belonging to the bacterial immunoglobulin superfamily. Mol Microbiol. 2003;49(4):929–45. doi: 10.1046/j.1365-2958.2003.03619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Barocchi MA, Ko AI, Reis MG, McDonald KL, Riley LW. Rapid translocation of polarized MDCK cell monolayers by Leptospira interrogans, an invasive but nonintracellular pathogen. Infect Immun. 2002;70(12):6926–32. doi: 10.1128/IAI.70.12.6926-6932.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Faine S. Guidelines for the control of leptospirosis. World Health Organization; Geneva: 1982. [Google Scholar]
- [29].Reed LJ, Muench HA. Simple method of determining fifty percent endpoints. Am J Hyg. 1938;27:494–97. [Google Scholar]
- [30].Faisal SM, Yan W, Chen CS, Palaniappan RU, McDonough SP, Chang YF. Evaluation of protective immunity of Leptospira immunoglobulin like protein A (LigA) DNA vaccine against challenge in hamsters. Vaccine. 2008;26(2):277–87. doi: 10.1016/j.vaccine.2007.10.029. [DOI] [PubMed] [Google Scholar]
- [31].Chang YF, Chen CS, Palaniappan RU, He H, McDonough SP, Barr SC, et al. Immunogenicity of the recombinant leptospiral putative outer membrane proteins as vaccine candidates. Vaccine. 2007;25(48):8190–7. doi: 10.1016/j.vaccine.2007.09.020. [DOI] [PubMed] [Google Scholar]
- [32].Spichler A, Ko AI, Silva EF, De Brito T, Silva AM, Athanazio D, et al. Reversal of renal tubule transporter downregulation during severe leptospirosis with antimicrobial therapy. Am J Trop Med Hyg. 2007;77(6):1111–9. [PubMed] [Google Scholar]
- [33].Cullen PA, Xu X, Matsunaga J, Sanchez Y, Ko AI, Haake DA, et al. Surfaceome of Leptospira spp. Infect Immun. 2005;73(8):4853–63. doi: 10.1128/IAI.73.8.4853-4863.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gamberini M, Gomez RM, Atzingen MV, Martins EA, Vasconcellos SA, Romero EC, et al. Whole-genome analysis of Leptospira interrogans to identify potential vaccine candidates against leptospirosis. FEMS Microbiol Lett. 2005;244(2):305–13. doi: 10.1016/j.femsle.2005.02.004. [DOI] [PubMed] [Google Scholar]
- [35].Barocchi MA, Ko AI, Ferrer SR, Faria MT, Reis MG, Riley LW. Identification of new repetitive element in Leptospira interrogans serovar copenhageni and its application to PCR-based differentiation of Leptospira serogroups. J Clin Microbiol. 2001;39(1):191–5. doi: 10.1128/JCM.39.1.191-195.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Romero EC, Billerbeck AE, Lando VS, Camargo ED, Souza CC, Yasuda PH. Detection of Leptospira DNA in patients with aseptic meningitis by PCR. J Clin Microbiol. 1998;36(5):1453–5. doi: 10.1128/jcm.36.5.1453-1455.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Trevejo RT, Rigau-Perez JG, Ashford DA, McClure EM, Jarquin-Gonzalez C, Amador JJ, et al. Epidemic leptospirosis associated with pulmonary hemorrhage-Nicaragua, 1995. J Infect Dis. 1998;178(5):1457–63. doi: 10.1086/314424. [DOI] [PubMed] [Google Scholar]
- [38].Silva ÉF, Brod CS, Cerqueira GM, Bourscheidt D, Seyffert N, Queiroz A, et al. Isolation of Leptospira Noguchii from sheep. Vet Microbiol. 2007;121:144–49. doi: 10.1016/j.vetmic.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Brenner DJ, Kaufmann AF, Sulzer KR, Steigerwalt AG, Rogers FC, Weyant RS. Further determination of DNA relatedness between serogroups and serovars in the family Leptospiraceae with a proposal for Leptospira alexanderi sp. nov. and four new Leptospira genomospecies. Int J Syst Bacteriol. 1999;49(Pt 2):839–58. doi: 10.1099/00207713-49-2-839. [DOI] [PubMed] [Google Scholar]
- [40].Haake DA, Walker EM, Blanco DR, Bolin CA, Miller MN, Lovett MA. Changes in the surface of Leptospira interrogans serovar Grippotyphosa during in vitro cultivation. Infect Immun. 1991;59:1131–1140. doi: 10.1128/iai.59.3.1131-1140.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ristow P, Bourhy P, McBride FW, Figueira CP, Huerre M, Ave P, et al. The OmpA-Like Protein Loa22 Is Essential for Leptospiral Virulence. PLoS Pathog. 2007;3(7):e97. doi: 10.1371/journal.ppat.0030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Arean VM. The pathologic anatomy and pathogenesis of fatal human leptospirosis (Weil’s disease) Amer J Path. 1962;40:393–423. [PMC free article] [PubMed] [Google Scholar]
- [43].Arean VM. Studies on the pathogenesis of leptospirosis. II. A clinicopathologic evaluation of hepatic and renal function in experimental leptospiral infections. Lab Invest. 1962;11:273–88. [PubMed] [Google Scholar]
- [44].De Brito T, Freymuller E, Hoshino S, Penna DO. Pathology of the kidney and liver in the experimental leptospirosis of the guinea-pig. A light and electron microscopy study. Virchows Arch Pathol Anat Physiol Klin Med. 1966;341(1):64–78. doi: 10.1007/BF00959245. [DOI] [PubMed] [Google Scholar]
- [45].De Brito T, Machado MM, Montans SD, Hoshino S, Freymuller E. Liver biopsy in human leptospirosis: a light and electron microscopy study. Virchows Arch Pathol Anat Physiol Klin Med. 1967;342(1):61–9. doi: 10.1007/BF00956849. [DOI] [PubMed] [Google Scholar]
- [46].Pereira MM, Da Silva JJ, Pinto MA, Da Silva MF, Machado MP, Lenzi HL, et al. Experimental leptospirosis in marmoset monkeys (Callithrix jacchus): a new model for studies of severe pulmonary leptospirosis. Am J Trop Med Hyg. 2005;72(1):13–20. [PubMed] [Google Scholar]