Abstract
In our previous work, we discovered potent HSV-1 inhibitory activity arising from sulfated form of lignin, a highly networked natural biopolymer composed of substituted phenylpropanoid monomers (Raghuraman et al. Biomacromolecules 2005, 6, 2822). We present here detailed characterization of the viral inhibitory properties of this interesting macromolecule. The inhibition was proportional to the average molecular weight of the lignin sulfate preparation with IC50 values in the range of 17 nM to 5 μM against HSV-1 and HSV-2, and 29 nM to 763 nM against HIV-1. Cytotoxicity studies displayed selectivity indices in the range of 14 to 31 suggesting reasonably good difference between activity and toxicity for polymeric preparations. Comparative molecular modeling studies suggest that lignin sulfate may contain certain structural features that mimic the three-dimensional organization of sulfate groups in heparan sulfate, thereby providing a plausible basis for its anti-viral activity. The combination of strongly hydrophobic (–Ar) and strongly hydrophilic (–OSO3−) groups in lignin sulfate makes this chemically modified biopolymer an interesting molecule for further work.
Keywords: Lignin sulfate, Heparan sulfate mimics, Anti-viral activity, Molecular modeling, HIV, HSV
Introduction
Growing evidence suggests that heparan sulfate, a proteoglycan present ubiquitously on nearly all cells, plays an important role in the initial attachment of enveloped viruses to target cells.1–6 For example, herpes simplex virus (HSV) interaction with target cells involves cell surface heparan sulfate interacting with several viral envelope proteins, especially glycoproteins gB, gC and gD.7–10 Likewise, gp120 and Tat of human immunodeficiency virus-1 (HIV-1) bind to heparan sulfate to mediate the internalization of the virus.11–13
Glycosaminoglycan heparan sulfate (HS) is a 1→4-linked linear polysaccharide of glucosamine (GlcNp) and glucuronic acid (GlcAp) residues, in which the majority of GlcNp residues are N-acetylated.14,15 In addition, epimerization of some GlcAp residues to iduronic acid (IdoAp) and sulfation at the 2-, 3- or 6-positions of GlcNp and 2-position of IdoAp introduce significant structural complexity and diversity in the HS biopolymer. Despite these large number of sequences, individual protein–HS interactions may involve specific binding sequences, as illustrated by HS–gD interaction of HSV-1, which appears to be mediated by a rare 3-O-sulfated GlcNp residue.8,10,16 Similar structural specificity has not yet been shown for HIV, however size selectivity is apparent.13,17
Several sulfated carbohydrate including heparin, dextran sulfate, fucoidans, and sulfated galactans have been found to inhibit HSV and HIV because of their structural similarity to HS.17–22 Recent studies on sulfated derivatives of E. coli K5 polysaccharides demonstrate good HIV-1 Tat protein antagonist activity.23 Thus, we reasoned that sulfated non-polysaccharide scaffolds should also exhibit viral antagonist activity, similar to that displayed by the sulfated polysaccharides. A potential advantage with sulfated non-polysaccharide molecules would be their relatively easy chemical or chemo-enzymatic synthesis.
Our initial attempt to derive small sulfated non-saccharide molecules as viral antagonists led to the serendipitous discovery of a biological macromolecule that was found to inhibit HSV-1 entry into cells.24 Elucidation of the structure of the active principle demonstrated it to be a sulfated derivative of lignin, a polymer made up of repeating phenylpropanoid units.24 Lignin is abundantly available in nature, especially from plants and vegetables, and is a complex, heterogeneous organic scaffold, which is radically different from the polysaccharide backbone of HS.25,26 In this note, we characterize the HSV-1, HSV-2 and HIV-1 inhibition and cytotoxic properties of lignin sulfate and demonstrate using comparative molecular modeling that certain structural features present in this interesting biopolymer may mimic HS structures, thus providing a basis for the observed viral inhibition property.
Experimental Methods
Chemicals, cells and viruses
Lignin sulfate and morin sulfate were prepared as described earlier.24 β-Galactosidase substrate, o-nitrophenyl β-D-galactopyranoside (ONPG), was from Pierce (Rockford, IL). High purity water, obtained from NERL Diagnostics (RI, USA), was used in all experiments. Dr. Patricia Spear (Northwestern University) provided HeLa cells and the HSV reporter viruses listed here. HSV-1 and HSV-2 virus strains carrying the lacZ gene of E. coli and capable of expressing β-galactosidase as a reporter of entry included HSV-1(KOS) gL86 and HSV-2(333).27,28 MT-2 cells, HIV-1IIIB- infected H9 cells (H9/HIV-1IIIB), and the HIV-1IIIB isolate were obtained from the NIH AIDS Research and Reference Reagent Program.
HSV-1 and HSV-2 virus infection assay
Assays for infection of cells were based on quantitation of β-galactosidase expressed by the mutant HSV viral genome containing the lacZ gene, as described earlier.8,10 HeLa cells were grown in 96-well tissue culture dishes (2–4×104 cells/well), washed after 16 h of growth, and exposed to 10 plaque forming units (PFU)/cell of the HSV virus in 50 μL of phosphate-buffered saline (PBS) containing glucose and 1% calf serum (PBS-G-CS) for 6 h at 37 °C. To test for inhibitory activity, the sulfated compounds were simultaneously added to this 50 μL medium in varying amounts ranging from 0.2 μg to 1.6 ng. Following incubation, the cells were solubilized in 100 μL of PBS containing 0.5% NP-40 and 10 mM ONPG. The initial rate of hydrolysis of the substrate was monitored spectrophotometrically at 410 nm, which corresponds to the concentration of the β-galactosidase within HeLa K-1 cell. The initial rate of hydrolysis of the substrate in the absence of any added sulfated molecule formed the control and assigned a value of 100% HSV infection. Assays were performed in duplicate and the mean value used for calculation of IC50, the concentration of inhibitor that reduces HSV infection by 50%.
HIV-1-mediated cell fusion and p24 assays
For detection of inhibition of HIV-1-mediated cell fusion, a dye transfer assay was used, as previously described.29 Briefly, H9/HIV-1IIIB cells were labeled with a fluorescent reagent, 2′,7′-bis(2-carboxyethyl)-5(and 6)-carboxyfluorescein acetoxyethyl ester (BCECF-AM, Molecular Probes, Inc., Eugene, OR), and then incubated with MT-2 cells (ratio = 1:10) in 96-well plates at 37 °C for 2 h in the presence or absence of the lignin sulfate samples. The fused and unfused BCECF-labeled HIV-1-infected cells were counted under an inverted fluorescence microscope (Zeiss, Germany) with an eyepiece micrometer disk. The percentage of inhibition of cell fusion and the IC50 values were calculated as previously described.29 For detection of inhibition of HIV-1 infection of MT-2 cells using the p24 antigen assay, 1×104 MT-2 cells were infected with HIV-1IIIB (100 TCID50) in the presence of lignin sulfate at graded concentrations, followed by incubation at 37 °C overnight. The culture media were changed and cells were cultured for 4 days before collection of supernatants for measuring the p24 antigen by ELISA, as described earlier.30,31
In vitro cytotoxicity of lignin sulfate samples
The in vitro cytotoxicity for MT-2 cells of lignin sulfates was determined in 96-well plates using the XTT dye to measure cell viability in the absence of virus. 5% Tritox X-100 (10 mL) was added to the wells corresponding to positive controls (P), and 10 mL of medium was added to wells corresponding to negative controls (N). The percent cytotoxicity was calculated using the formula [(E − N)/(P − N)]× 100, where E represents the experimental data in the presence of lignin sulfates. The concentration corresponding to 50% cytotoxicity (CC50) for MT-2 cells was calculated using the Calcusyn computer program from which the selectivity index (SI = CC50/IC50) was calculated.
Molecular modeling studies on heparan sulfate disaccharides and lignin sulfate dimers
Structures of LS dimers were built in Sybyl 6.9.2, assigned Gasteiger – Hûckel charges and minimized using the Tripos force field at dielectric constant of 80 subject to the gradient termination criterion of 0.01 kcal/mol-Å. The sulfur and oxygen atoms in the sulfate groups were assigned S.O2, O.CO2, and O.3 atom type. A total of 6 β-O-4- and β-5-linked lignin dimers containing three sulfate groups each were modeled. The modeling of HS disaccharides followed the protocol used in our earlier report.32 Briefly, 15 disaccharides, of the naturally occurring 23 disaccharides, containing at least three anionic groups were modeled. The structural differences between these 15 disaccharides are mainly determined by the pattern of sulfation and epimer (IdoAp) conformation. The ϕH/ψH torsion angles in these disaccharides were obtained from crystal structure studies, as extensively reviewed in our computational study of HS hexasaccharides.32 Thus, the values were constrained at the average value of 47.5° and 13°, respectively, using a force constant of 0.01 kcal mol−1 deg−2. Following minimization, each structure was analyzed for distance between its anionic groups (carboxylates and sulfates). The carbon and sulfur atoms in the carboxylate and sulfate groups, respectively, were used for inter-anionic distance analysis. Triangular arrays of inter-anionic distances in LS dimers were prepared and compared to that for HS disaccharides. A LS dimer was considered a mimic of a HS disaccharide if each distance in the triangular array matched within ± 1 Å.
Results
Structure of Lignin Sulfate
Lignin is one of the most abundant plant natural product, yet detailed structure of the molecule remains unclear.25,26 Lignin is a highly heterogeneous polydisperse polymer constituted of phenylpropanoid monomers, which are connected through β-O-4, β-5, β-β, or 5-5 linkages (Fig. 1). Of these inter-residue linkages, β-O-4 and β-5 linkages are reported to be most commonly present. Atmospheric pressure chemical ionization-mass spectrometry analysis suggested that our natural product also contained β-O-4-linked p-hydroxycinnamyl alcohol monomers with smaller proportions of β-β, β-5, and 5-5–linked structures.24 Chemical sulfation of the natural product using triethylamine–sulfur trioxide complex retains these inter-residue linkages, while introducing sulfate groups (–OSO3−) on the available alcoholic and phenolic –OH groups.24,33 Thus, chemically sulfated lignin is also a heterogeneous biomacromolecule consisting of a large number of diverse structures, which may be thought of as a lignin sulfate library. A characteristic feature of this library is the presence of both strongly hydrophilic (–OSO3−) and strongly hydrophobic (–Ar) groups. In this manner, the lignin sulfate (LS) library is dramatically different from the HS library, which has no aromatic features and minimal levels of hydrophobicity. However, as with all heterogeneous polymers, this library is a double-edged sword. While structural diversity present in the library enhances the probability of discovering biological activity, it also prohibits a definitive assignment of a specific structure(s) as the origin of this activity.
Figure 1.
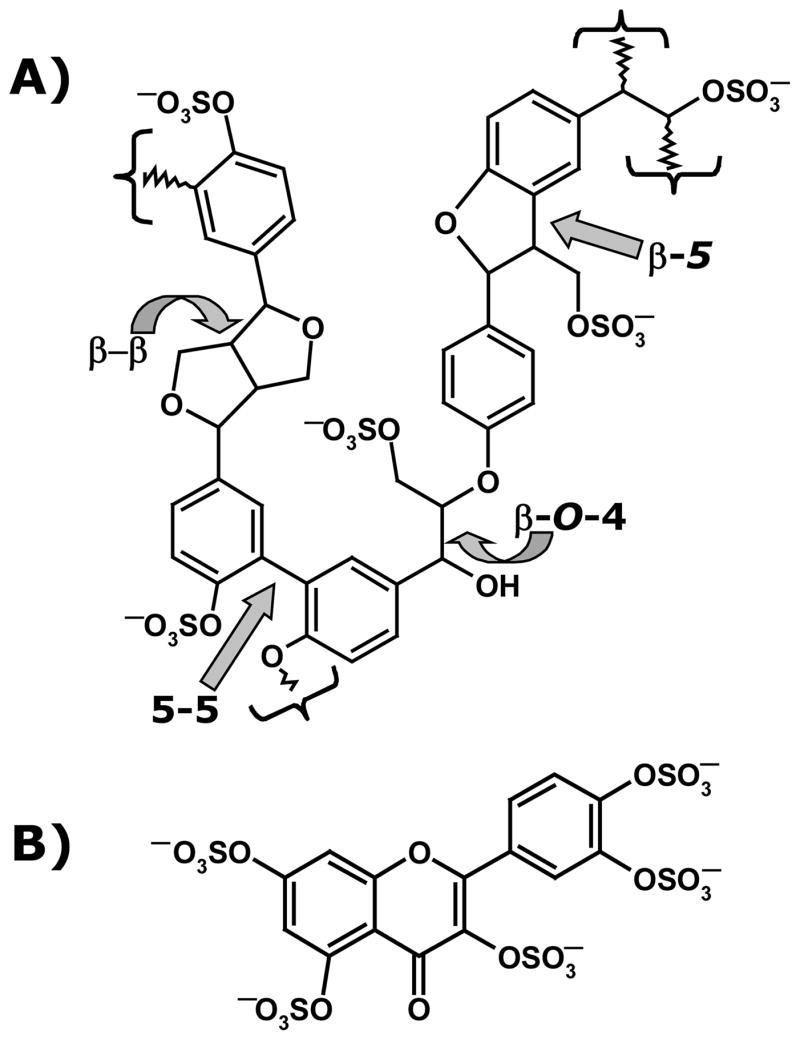
A representation of the structure of lignin sulfate (A) and morin sulfate MoS (B). β-O-4, β-β, β-5 and 5-5 in (A) indicate types of inter-monomer linkages, while curly brackets show the polymerization points. Positions α, β, 4 and 5, which constitute linkage positions, are marked.
Size Fractionation and Sulfation Level of Lignin Sulfate
Size exclusion chromatography (SEC) of LS indicated polymeric chains with a wide range of molecular weights −1.5 to 40 kDa. To gain insight into chain length dependence and elucidate whether smaller LS structures possess anti-viral activity, the polymer was fractionated into five fractions using a combination of centrifugal membrane filtration and SEC chromatography.24 Using a calibration curve prepared with polysulfonate standards, the average molecular weight (MR) of the five fractions were found to be 39.4, 14.9, 5.9, 2.5 and 1.9 kDa. The sulfation level of each fraction was calculated from its elemental composition, especially its sulfur content (~12% w/w), and found to be nearly identical (not shown). This indicates an average composition of approximately 1 sulfate group per p-hydroxycinnamyl monomer. This level of negative charge density is significantly lower than that of heparin, which typically contains ~1.8 anionic (–COO− and –OSO3−) groups per monomer,34 while it is equivalent to the average charge density of 1.0 to 1.4 found in HS.35,36
Lignin Sulfate Fractions Inhibit Cellular Entry of HSV
The ability of LS fractions to inhibit entry of HSV-1 and HSV-2 particles was studied in a well-established viral infection assay, in which the internalized viral particle is quantified indirectly through the β-galactosidase activity expressed by its genome.10,13,27,28 This assay involves the exposure of a constant dose of the virus, mutant strains of HSV-1 and HSV-2 containing the lacZ gene, to HeLa cells for 6 hours at 37 °C in the presence of sulfated inhibitors at several graded concentrations. Following incubation, the β-galactosidase activity of the internalized virus is measured spectrophotometrically, which is directly proportional to the level of viral particles that have entered the cells.
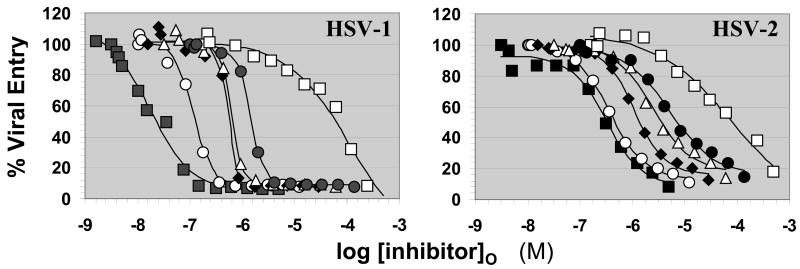
All five fractions of LS showed a concentration dependent inhibition of cellular entry of HSV type 1 and 2 (Fig. 2). We also tested morin sulfate (MoS), a small sulfated organic molecule, which we had synthesized earlier.33 MoS is a small homogeneous molecule containing 5 sulfate groups and thus represents a high charge density reference. The response to increasing LS concentration was found to be sigmoidal and when fit to the standard dose–response equation gave the concentration of the inhibitor required for 50% inhibition (IC50) (Table 1). The IC50 value for HSV-1 inhibition decreases from 1.45 μM for the smallest LS with MR of 1.9 kDa to 17 nM for 39.4 kDa LS. In contrast, the IC50 value for MoS was found to be 112 μM. Likewise, the IC50 values for HSV-2 inhibition were found to be between 0.32 and 5.0 μM for LS fractions, while that for MoS was 79 μM.
Figure 2.
Inhibition of HSV-1 and HSV-2 entry into HeLa cells by lignin sulfate. HeLa cells were exposed to the virus for 6 h at 37 °C in the presence and absence of LS fractions in varying amounts. Following incubation, the cells were solubilized and the initial rate of hydrolysis of the β-galactosidase substrate, ONPG, was monitored spectrophotometrically to determine % viral entry, as previously described.10,11,26,27 Labels ■, ○, ◆, △, and ●, correspond to lignin sulfate fractions with MR values of 39.4, 14.8, 5.9, 2.5, and 1.9 kDa, respectively, while □ corresponds to MoS.
Table 1.
Molecular weight dependence of IC50 values for LS-dependent inhibition of HSV entry into HeLa cells.
| IC50a | |||
|---|---|---|---|
| MR | HSV-1 | HSV-2 | |
| (kDa) | (μM) | (μM) | |
| LS1 | 39.4 | 0.017 | 0.32 |
| LS2 | 14.8 | 0.12 | 0.40 |
| LS3 | 5.9 | 0.53 | 1.0 |
| LS4 | 2.5 | 0.60 | 2.5 |
| LS5 | 1.9 | 1.45 | 5.0 |
| MoS | 0.8 | 112 | 79 |
Values are mean of two experiments.
Lignin Sulfate Fractions Inhibit HIV-1 Mediated Cell-Cell Fusion
Inhibition of cell fusion activity of HIV-1 was studied by incubating fluorophore-Am-labeled HIV-1IIIB-infected H9 cells with MT-2 cells in the presence and absence of varying concentrations of LS, as described earlier.30,31,37,38 In this well-established assay, the fluorescence of the infected cells, which fuse with the target cells, is used as the reporter of infectivity. Following incubation for 2 hours at 37 °C, the fused and unfused cells were counted in an inverted fluorescence microscope to determine percent inhibition. As the concentration of each LS fraction increased, the fusion of the two types of cells decreased in a sigmoidal manner (not shown), from which the IC50 value was calculated (Table 2). The IC50 values increase gradually from 60 nM for LS1 to 763 nM for LS5. These values are in the same range as those for the inhibition of cellular entry of HSV-1 (Table 1). As in the HSV study, MoS was found to be a poor inhibitor of HIV-1-mediated cell fusion.
Table 2.
IC50 values for inhibition of HIV-1 mediated cell fusion, entry into MT-2 cells, and cytotoxic effect (CC50a) of LS fractions.b
| MR(kDa) | IC50 (nM)
|
CC50a(nM) | Selectivity Indexd | ||
|---|---|---|---|---|---|
| Cell fusion | p24c | ||||
| LS1 | 39.4 | 60 | 29 | 603 | 20.8 |
| LS2 | 14.8 | 105 | 97 | 1340 | 13.8 |
| LS3 | 5.9 | 246 | 236 | 3834 | 16.2 |
| LS4 | 2.5 | 384 | 400 | 10280 | 25.7 |
| LS5 | 1.9 | 763 | 358 | 10945 | 30.6 |
| MoS | 0.8 | >250,000 | >125,000 | nde | - |
concentration of inhibitor that reduces number of viable cells by 50% in the absence of infection.
ratio of CC50 to IC50 determined in the p24 assay
not determined
Lignin Sulfate Fractions Inhibit HIV-1
To determine whether LS fractions inhibit HIV-1, we utilized the standard p24 antigen assay.30,38 Protein p24 is a major internal structural protein of HIV-1, which has been routinely used as a marker of infection. Briefly, MT-2 cells were exposed to a fixed dose of HIV-1IIIB in the presence of increasing concentrations of LS fractions. Following overnight incubation at 37 °C, the culture medium was changed and the cells left undisturbed for 4 days before collection of the supernatant medium for p24 detection by ELISA.30,37,38 As observed in cell fusion assay, HIV-1 was found to be inhibited by LS fractions (Table 2). The IC50 values range from 29 nM to 400 nM and generally correlate well (1–2-fold difference) with those measured in the cell fusion assay.
Cytotoxic Effect of Lignin Sulfate Fractions
To assess whether the anti-viral activities of LS arise from its cytotoxic activity, we measured cell viability on the sixth day without viral exposure in the presence of varying concentrations of LS, as previously described.30 The cells were exposed to XTT tetrazoline dye and the concentration of dye internalized by viable cells was measured spectrophotometrically at 450 nm. The concentration corresponding to 50% cytotoxicity (CC50) for MT-2 cells was found to be in the range of 0.6 to 11 μM, which is nearly 14 to 31-fold higher than the IC50 of LS fractions (Table 2). This suggests a reasonably good selectivity index for sulfated lignin. Interestingly, the cytotoxicity of LS is found to be proportional to the MR of the polymer and appears to level off for smaller species.
Comparative Molecular Modeling of Lignin Sulfate Dimers and Heparan Sulfate Disaccharides
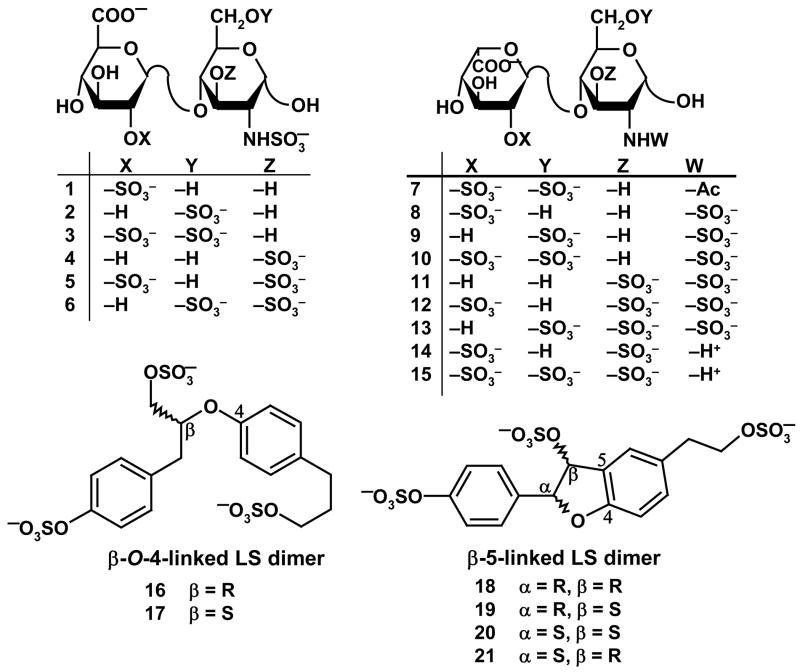
To determine whether certain LS structures mimic HS sequence(s), we resorted to a comparison of the three-dimensional orientation of sulfate groups by the two strikingly different scaffolds. More specifically, the ability of the LS scaffold to place three sulfate groups in an identical spatial orientation as three anionic groups (carboxylate or sulfate) present in the smallest HS repeat sequence, a disaccharide, was compared using molecular modeling. We hypothesized that LS dimers would mimic HS disaccharides based on their approximately equal size as well as similar negative charge density (~1–1.3 anions per monosaccharide).35,36
Of the 23 possible natural HS disaccharides, 15 contain at least 3 anionic groups (Fig. 3). Our previous molecular modeling study suggests that HS sequences adopt an average backbone conformation with similar ϕH/ψH torsion angles and the structural differences between the individual sequences are mainly a function of sulfation pattern and uronic acid epimer conformation.32 Thus, the ϕH/ψH angles and 1C4/2SO conformations of IdoAp residue were extracted from co-crystal structure of HS sequences and used to construct the 15 disaccharides targeted in this study. Likewise, 6 different tri-sulfated LS dimers based on β-O-4- and β-5-linkages are possible (Fig. 3). These LS dimers were constructed in silico using the small organic molecule builder in Sybyl and their structures optimized using standard energy minimization routines. Following the construction of both sets of dimers – HS and LS – inter-anion distances were calculated. The inter-anion distance set revealed that selected LS dimers positioned sulfate groups in a manner identical to that possible with HS disaccharides. For example, the αR,βS and αS,βR enantiomers of the β-5-linked LS dimer, i.e., molecules 21 and 19 (Fig. 3) were found to mimic HS disaccharides 2 and 1, respectively. Figure 4 shows the correspondence between the three sulfate groups present in each dimeric pair. Likewise, β-O-4-linked LS dimers 16 and 17 (Fig. 3) were found to mimic HS disaccharides 13 and 5, respectively (not shown). Yet, not all LS dimers studied were found to mimic HS disaccharides as well as not all HS structures could be mimicked by LS. For example, the αS,βS-linked β-5 LS dimer 20 did not appear to mimic any of the 15 HS disaccharides. Overall, comparative molecular modeling based on first principles demonstrated that selected LS structures are likely to mimic certain HS sequences.
Figure 3.
Structures of dimeric units of HS and LS studied for comparative molecular modeling. Six trisulfated β-O-4 and β-5-linked LS dimers were compared with 15 HS disaccharides that contain at least three anionic (–COO− and –OSO3−) groups.
Figure 4.
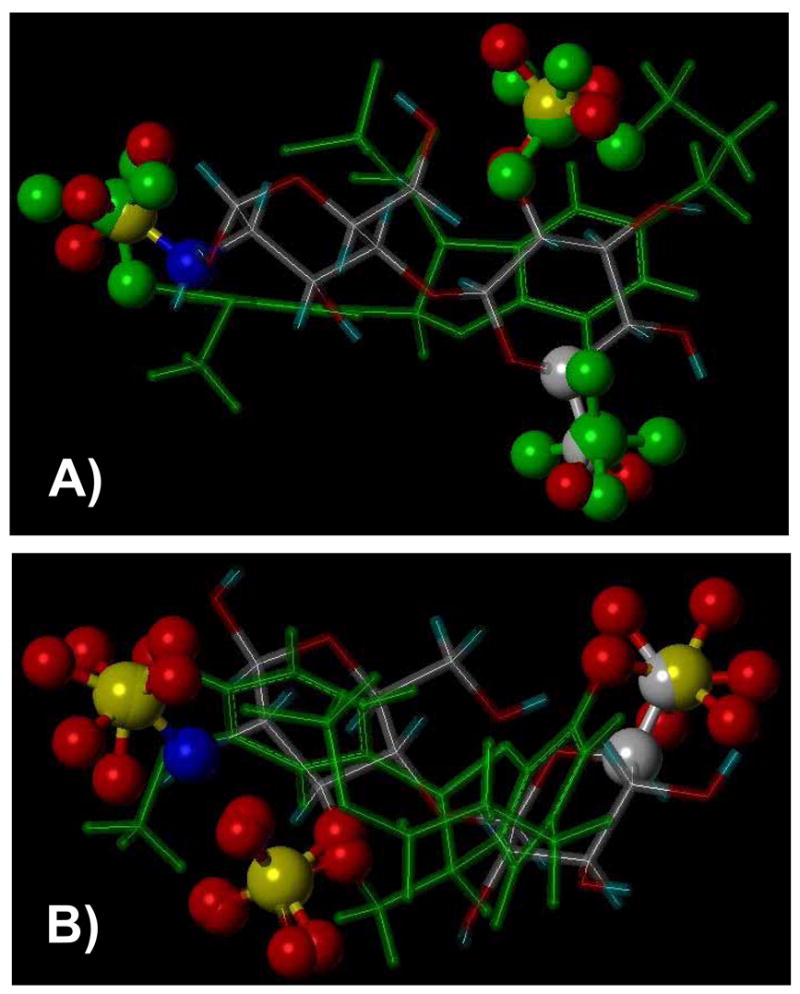
Comparison of three-dimensional orientation of anionic groups of LS dimers 19 and 21 in green sticks (see Fig. 3 for structures) with HS disaccharides 1 and 2 in atom type sticks, respectively. A) shows a comparison between 21 and 2, while B) shows comparison between 19 and 1. Note the identical orientation of three –OSO3− groups in each pair (shown as ball and stick).
Discussion
Our serendipitous discovery24 that lignin sulfate is an antagonist of HSV-1 led to the reasoning that the biomacromolecule is likely to inhibit other enveloped viruses, e.g., HSV-2 and HIV-1. This expectation was borne out of the possibility that LS, a heterogeneous, polydisperse, sulfated polymer, mimics heparan sulfate, a major cellular receptor which recognizes viral glycoproteins, including gB, gC, gD of HSV,2,3,5,7–10 and gp120 and Tat of HIV.1,4,6,11–13
This work demonstrates that polymeric LS is a reasonably good antagonist of HSV-1, HSV-2 and HIV-1. For the viruses studied, the inhibition was dependent on the molecular weight of LS with the heaviest chain possessing highest potency. This activity is comparable to the nM activity of soluble HS and heparin.18 Yet, an interesting observation is that the lighter chains (MR 1.9 kDa) display significant antagonism, e.g., an IC50 value of 1.5 and 5.0 μM against HSV and ~0.36 μM against HIV. Except for the size, the heavier and lighter chains are structurally equivalent.24 Thus, either the longer chain interacts with additional amino acid residues on the target protein in comparison to the shorter chain, or that it has a statistical advantage. Studies with more homogeneous species, perhaps synthetic oligomers, may clarify this feature.
The work reveals additional interesting aspects of viral antagonism. Firstly, MoS containing five sulfate groups in its small size (0.8 kDa, Fig. 1) is 16 to 78-fold weaker inhibitor of HSV entry and at least 300-fold weaker inhibitor of HIV entry than the smallest lignin sulfate (MR 1.9 kDa). The presence of five sulfate groups in MoS introduces massive anionic character in the molecule. In comparison, LS has much lower charge density. Thus, it is likely that the activity of LS arises from its structural features, rather than from the general recognition of its few sulfate groups. Secondly, the IC50 values decrease for both HSV-1 and HSV-2 as the MR of lignin sulfate samples increase (Table 1). However, the decrease is not parallel (not shown). For example, the potency for HSV-1 inhibition increases ~85-fold with the MR, while only 16-fold increase in potency was found for HSV-2. This indicates a difference in the activity of LS against the two viruses, an observation also found to be true with heparin.39
The heterogeneous and polydisperse nature of LS implies a large number of distinct structures in these preparations. The average molecular weight of lignin sulfate monomers is ~200 Da. Thus, an average 1.9 kDa chain is expected to consist of 4 to 5 dimers. The possibility of several inter-residue linkages – β-O-4, β-5, β-β and 5-5 – in these chains introduce significant complexity in identification of discrete structures responsible for anti-viral activity. It may be possible to identify potent sequences in this heterogeneous mixture using a combination of affinity and ion-exchange chromatographies, however the target HSV/HIV proteins remain unknown at present resulting in considerable ambiguity. Another approach that may yield useful sequence information is modeling. Comparative molecular modeling of a limited set of structures shows that certain LS sequences may mimic selected HS sequences by orienting appropriate sulfate groups in nearly identical manner (Fig. 4). These results indicate that LS may be a reasonably good mimic of HS, although high-affinity sequence information is still a matter of future work. A specific advantage with the LS structure, in comparison the HS scaffold, is that it is an aromatic, hydrophobic skeleton, which is readily amenable to chemo-enzymatic synthetic approach and structural modification.
Acknowledgments
This work was supported by the NIH (RO1 HL069975 and R41 HL081972) and AHA – National Center (EIA 0640053N).
Footnotes
Abbreviations: GlcAp, glucuronic acid residue; GlcNp, glucosamine residue; HIV, human immunodeficiency virus; HS, heparan sulfate; HSV, herpes simplex virus; IdoAp, iduronic acid residue; LS, lignin sulfate; MoS, morin sulfate;
References
- 1.Ugolini S, Mondor I, Sattentau QJ. Trends Microbiol. 1999;7:144–149. doi: 10.1016/s0966-842x(99)01474-2. [DOI] [PubMed] [Google Scholar]
- 2.Spear PG, Eisenberg RJ, Cohen GH. Virology. 2000;275:1–8. doi: 10.1006/viro.2000.0529. [DOI] [PubMed] [Google Scholar]
- 3.Shukla D, Spear PG. J Clin Invest. 2001;108:503–510. doi: 10.1172/JCI13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallay P. Microbes Infect. 2004;6:617–622. doi: 10.1016/j.micinf.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 5.Spear PG. Cell Microbiol. 2004;6:401–410. doi: 10.1111/j.1462-5822.2004.00389.x. [DOI] [PubMed] [Google Scholar]
- 6.Fittipaldi A, Giacca M. Adv Drug Deliv Rev. 2005;57:597–608. doi: 10.1016/j.addr.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 7.Herold BC, Visalli RJ, Susmarski N, Brandt CR, Spear PG. J Gen Virol. 1994;75:1211–1222. doi: 10.1099/0022-1317-75-6-1211. [DOI] [PubMed] [Google Scholar]
- 8.Shukla D, Liu J, Blaiklock P, Shworak NW, Bai X, Esko JD, Cohen GH, Eisenberg RJ, Rosenberg RD, Spear PG. Cell. 1999;99:13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 9.Cheshenko N, Herold BC. J Gen Virol. 2002;83:2247–2255. doi: 10.1099/0022-1317-83-9-2247. [DOI] [PubMed] [Google Scholar]
- 10.Tiwari V, Clement C, Duncan MB, Chen J, Liu J, Shukla D. J Gen Virol. 2004;85:805–809. doi: 10.1099/vir.0.19641-0. [DOI] [PubMed] [Google Scholar]
- 11.Zhang YJ, Hatziioannou T, Zang T, Braaten D, Luban J, Goff SP, Bieniasz PD. J Virol. 2002;76:6332–6342. doi: 10.1128/JVI.76.12.6332-6343.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Argyris EG, Kulkosky J, Meyer ME, Xu Y, Mukhtar M, Pomerantz RJ, Williams KJ. Virology. 2004;330:481–486. doi: 10.1016/j.virol.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 13.Vives RR, Imberty A, Sattentau QJ, Lortat-Jacob H. J Biol Chem. 2005;280:21353–21357. doi: 10.1074/jbc.M500911200. [DOI] [PubMed] [Google Scholar]
- 14.Esko JD, Lindahl U. J Clin Invest. 2001;108:169–173. doi: 10.1172/JCI13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rabenstein DL. Nat Prod Rep. 2002;19:312–331. doi: 10.1039/b100916h. [DOI] [PubMed] [Google Scholar]
- 16.Liu J, Shriver Z, Pope RM, Thorp SC, Duncan MB, Copeland RJ, Raska CS, Yoshida K, Eisenberg RJ, Cohen G, Linhardt RJ, Sasisekharan R. J Biol Chem. 2002;277:33456–33467. doi: 10.1074/jbc.M202034200. [DOI] [PubMed] [Google Scholar]
- 17.Nishimura SI, Kai H, Shinada K, Yoshida T, Tokura S, Kurita K, Nakashima H, Yamamoto N, Uryu T. Carbohydr Res. 1998;306:427–433. doi: 10.1016/s0008-6215(97)10081-7. [DOI] [PubMed] [Google Scholar]
- 18.Witvrouw M, De Clercq E. Gen Pharmacol. 1997;29:497–511. doi: 10.1016/s0306-3623(96)00563-0. [DOI] [PubMed] [Google Scholar]
- 19.Ponce NM, Pujol CA, Damonte EB, Flores ML, Stortz CA. Carbohydr Res. 2003;338:153–165. doi: 10.1016/s0008-6215(02)00403-2. [DOI] [PubMed] [Google Scholar]
- 20.Viveros-Rogel M, Soto-Ramirez L, Chaturvedi P, Newburg DS, Ruiz-Palacios GM. Adv Exp Med Biol. 2004;554:481–487. doi: 10.1007/978-1-4757-4242-8_69. [DOI] [PubMed] [Google Scholar]
- 21.Nyberg K, Ekblad M, Bergstrom T, Freeman C, Parish CR, Ferro V, Trybala E. Antiviral Res. 2004;63:15–24. doi: 10.1016/j.antiviral.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Damonte EB, Matulewicz MC, Cerezo AS. Curr Med Chem. 2004;11:2399–2419. doi: 10.2174/0929867043364504. [DOI] [PubMed] [Google Scholar]
- 23.Urbinati C, Bugatti A, Oreste P, Zoppetti G, Waltenberger J, Mitola S, Ribatti D, Presta M, Rusnati M. FEBS Lett. 2004;568:171–177. doi: 10.1016/j.febslet.2004.05.033. [DOI] [PubMed] [Google Scholar]
- 24.Raghuraman A, Tiwari V, Thakkar JN, Gunnarsson GT, Shukla D, Hindle M, Desai UR. Biomacromolecules. 2005;6:2822–2832. doi: 10.1021/bm0503064. [DOI] [PubMed] [Google Scholar]
- 25.Reale S, Di Tullio A, Spreti N, De Angelis F. Mass spectrometry in the biosynthetic and structural investigation of lignins. Mass Spectrom Rev. 2004;23:87–126. doi: 10.1002/mas.10072. [DOI] [PubMed] [Google Scholar]
- 26.Davin LB, Lewis NG. Curr Opin Biotechnol. 2005;16:407–415. doi: 10.1016/j.copbio.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 27.Montgomery RI, Warner MS, Lum BJ, Spear PG. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 28.Warner MS, Geraghty RJ, Martinez WM, Montgomery RI, Whitbeck JC, Xu R, Eisenberg RJ, Cohen GH, Spear PG. Virology. 1998;246:179–189. doi: 10.1006/viro.1998.9218. [DOI] [PubMed] [Google Scholar]
- 29.Jiang S, Lin K, Strick N, Neurath AR. Biochem Biophys Res Commun. 1993;195:533–538. doi: 10.1006/bbrc.1993.2078. [DOI] [PubMed] [Google Scholar]
- 30.Debnath AK, Radigan L, Jiang S. J Med Chem. 1999;42:3203–3209. doi: 10.1021/jm990154t. [DOI] [PubMed] [Google Scholar]
- 31.Jiang S, Lin K, Lu M. J Virol. 1998;72:10213–10217. doi: 10.1128/jvi.72.12.10213-10217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raghuraman A, Mosier PD, Desai UR. J Med Chem. 2006;49:3553–3562. doi: 10.1021/jm060092o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gunnarsson GT, Desai UR. Bioorg Med Chem Lett. 2003;13:579–583. doi: 10.1016/s0960-894x(02)01055-7. [DOI] [PubMed] [Google Scholar]
- 34.Vongchan P, Warda M, Toyoda H, Toida T, Marks M, Linhardt RJ. Biochim Biophys Acta. 2005;1721:1 –8. doi: 10.1016/j.bbagen.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 35.Warda M, Gouda EM, Toida T, Chi L, Linhardt RJ. Comp Biochem Physiol B. 2003;136:357–365. doi: 10.1016/j.cca.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 36.Warda M, Linhardt RJ. Comp Biochem Physiol B Biochem Mol Biol. 2006;143:37–43. doi: 10.1016/j.cbpb.2005.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang S, Lin K, Zhang L, Debnath AK. J Virol Meth. 1999;80:85–96. doi: 10.1016/s0166-0934(99)00041-5. [DOI] [PubMed] [Google Scholar]
- 38.Naicker KP, Jiang S, Lu H, Ni J, Boyer-Chatenet L, Wang LX, Debnath AK. Bioorg Med Chem. 2004;12:1215–1220. doi: 10.1016/j.bmc.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 39.Trybala E, Liljeqvist JA, Svennerholm B, Bergstrom T. J Virol. 2000;74:9106–9114. doi: 10.1128/jvi.74.19.9106-9114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]