Abstract
Motivation: Gene clustering and gene set-based functional analysis are widely used for the analysis of expression profiles. The development of a comprehensive method jointly combining the two methods would allow for greater biological insights.
Results: We developed a software package, PathCluster for gene set-based clustering via an agglomerative hierarchical clustering algorithm. The distances between predefined gene sets are illustrated in a dendrogram in which the relationships between gene sets can be visually assessed. Valuable biological insights can be obtained according to the type of gene sets, e.g. coordinated action of molecular functions (functional gene sets) and putative motif synergy (promoter gene set) in a biological process. The combined use of gene sets further enables the interrogation of different biological themes and their putative relationships, such as function-versus-regulatory motif or drug-versus-function. PathCluster can also be used for knowledge-based sample partitioning or class categorization for clinical purposes. With extended applicability, PathCluster will facilitate the gleaning of meaningful biological insights and testable hypotheses in the contexts of given expression profiles.
Availability: PathCluster executable files can be freely downloaded at http://www.systemsbiology.co.kr/PathCluster/.
Contact: yejun@catholic.ac.kr
1 BACKGROUND
The objective of gene clustering is to group genes with similar expression patterns or that are expressed in a coordinated manner (Eisen et al., 1998). Subsequent functional enrichment analysis can provide clues as to which molecular functions or annotation categories are associated with individual gene clusters using biological knowledge. Despite its potential utility, the treatment of gene clusters as exclusive units may raise a number of practical concerns in subsequent functional analysis. For example, a large list of candidate functionalities is obtained as the number of clusters increases, thus making it difficult to compare the results between clusters or to establish appropriate significance thresholds considering multiple testing adjustments. Also, the performance of enrichment analysis is profoundly dependent on prior clustering result, which varies considerably according to the cluster methods and parameter settings. More importantly, the potential relationships between gene sets or clusters are difficult to identify in conventional settings. The integration of a priori knowledge of gene set information in clustering may be an appropriate solution to these problems (Rapaport et al., 2007); however, there are currently no available user-friendly tools that implement this alternate algorithm.
Thus, we developed a software package, PathCluster, which utilize an agglomerative hierarchical clustering algorithm for gene set-based clustering. In a given expression profile, the distance matrix is constructed between gene sets and illustrated as a dendrogram. The relationship between gene sets can be visually assessed in the results, thereby facilitating the construction of an association map between diverse annotation categories. The related algorithms are implemented in a freely available software package. Major functionalities of PathCluster are summarized as follows:
Gene set-based hierarchical clustering and visualization of the results with user-friendly graphic interface,
Identification of potential relationship between gene sets; putative interaction between molecular functions or synergism between regulatory motif sequences,
Revealing previously unknown links between different annotation categories in terms of gene sets; function-versus-regulatory motif or drug-versus-function,
Function-based class categorization of disease samples.
2 HIERARCHICAL CLUSTERING OF GENE SETS
Two strategies can be employed to determine the expression similarities or distances between gene sets. First, individual gene sets can be scored for the mean expression of belonging genes or enrichment scores derived from non-parametric (GSEA) or parametric version (PAGE) of gene set enrichment algorithms (Cheadle et al., 2007). The matrix of gene set scores with respect to the samples can be used to calculate the gene set distance and hierarchical clustering. Alternatively, the distance between two gene sets can be calculated directly as a mean correlation level of all possible gene pairs, each of which represents one possible gene-to-gene match between corresponding gene sets. When dealing with large gene sets and when the overlapping genes between gene sets have peculiar interests (especially the case of promoter gene sets), the mean correlation can also be calculated only for the gene pairs within overlapping genes between gene sets. Detailed descriptions of the metrics utilized and examples are available in the online manual at the PathCluster homepage (http://www.systemsbiology.co.kr/PathCluster/Manual.pdf).
PathCluster provides default gene sets covering four kinds of gene annotation categories; molecular functions, the association with regulatory motifs corresponding to transcription factors or miRNA, as well as drug treatment-related expression changes. In addition, gene sets from public databases such as MSigDB or user-defined custom query sets can be readily included in the gene set reference, in order to ensure the versatility of the method.
3 BIOLOGICAL APPLICATION
3.1 Associated molecular functions or regulatory motif sequences in a biological process
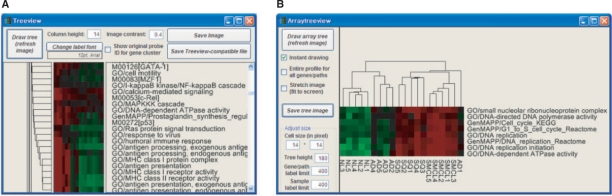
Using functional gene sets, PathCluster can identify the putative associations between molecular functions, thereby providing clues on coordinated action of specific functions in a given expression profile. Similarly, in the case of promoter gene sets, PathCluster can identify the putative motif synergy between cis-regulatory motifs or corresponding transcription factors delineating the regulatory crosstalks in a transcriptional regulatory network. Moreover, using combined gene sets with different annotation categories, previously unknown, novel links can be revealed. In erythropoiesis-related expression profiles, a number of functionalities related with immunity and the major histocompatibility complex are observed in a cluster (Fig. 1A). Within the cluster, signal-related functionalities (Ras protein signal transduction and MAPKKK cascade) as well as sequence motifs corresponding transcription factors of GATA-1 and c-Rel (a component of NK-κB) were also observed indicative of their potential interactions during erythropoiesis. This strategy can be also applied to other combinations of gene sets to reveal novel links between different biological themes such as function versus drug and function versus miRNA.
Fig. 1.
Screenshots of PathCluster. (A) An example of analysis using publicly available expression profiles representing human erythroid differentiation (Keller et al., 2006). The dendrogram shows a clustering of immune-related functional annotations as well as signal-related functionalities and relevant sequence. (B) The function-based classification of human lung cancer samples (Bhattacharjee et al., 2001). Four histological subtypes of lung cancer samples (normal, NL; adenocarcinoma, AD; squamouse cell carcinoma, SQ; small cell carcinoma, SMCL) are distinguished at the gene set-based expression level.
3.2 Function-based sample classification
Knowledge-driven or function-based class categorization has recently emerged as a highly challenging subject. This strategy has already been employed to identify the functional relationships in a large cancer-derived expression compendium or to elucidate drug-signature relationships for clinical benefits (Wong et al., 2008). Adopting a user-friendly platform and extended reference of gene sets, PathCluster provides a platform for the classification or molecular diagnosis of clinical samples, also allowing for the interrogation of diverse biological knowledge in terms of gene sets.Figure 1B shows that function-based classification can successfully distinguish between the three lung cancer subtypes, including normal tissues. In this cluster, eight cancer-related functions are specifically up-regulated in small cell lung cancer and squamous cell carcinoma of the lung.
ACKNOWLEDGEMENTS
This work is supported by the grant of the Korea Health 21 R&D Project, Ministry of Health & Welfare, Republic of Korea (0405-BC02-0604-0004) and (01-PJ3-PG6-01GN07-0004).
Conflict of Interest: none declared.
REFERENCES
- Bhattacharjee A, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc. Natl Acad. Sci. USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheadle C, et al. GSMA: gene set matrix analysis, an automated method for rapid hypothesis testing of gene expression data. Bioinform. Biol. Insights. 2007;1:49–62. [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, et al. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MA, et al. Transcriptional regulatory network analysis of developing human erythroid progenitors reveals patterns of coregulation and potential transcriptional regulators. Physiol. Genomics. 2006;28:114–128. doi: 10.1152/physiolgenomics.00055.2006. [DOI] [PubMed] [Google Scholar]
- Rapaport F, et al. Classification of microarray data using gene networks. BMC Bioinformatics. 2007;8:35. doi: 10.1186/1471-2105-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DJ, et al. Revealing targeted therapy for human cancer by gene module maps. Cancer Res. 2008;68:369–378. doi: 10.1158/0008-5472.CAN-07-0382. [DOI] [PubMed] [Google Scholar]