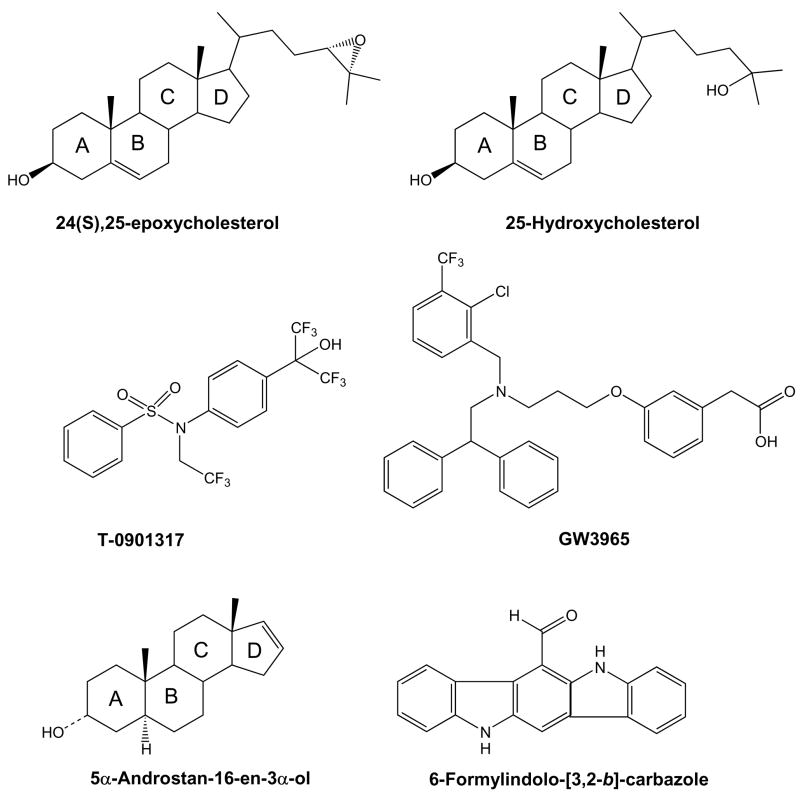
Abstract
Liver X receptors (LXRs) are key regulators of lipid and cholesterol metabolism in mammals. Little is known, however, about the function and evolution of LXRs in non-mammalian species. The present study reports the cloning of LXRs from African clawed frog (Xenopus laevis), Western clawed frog (Xenopus tropicalis), and zebrafish (Danio rerio), and their functional characterization and comparison with human and mouse LXRs. Additionally, an ortholog of LXR in the chordate invertebrate Ciona intestinalis was cloned and functionally characterized. Ligand specificities of the frog and zebrafish LXRs were very similar to LXRα and LXRβ from human and mouse. All vertebrate LXRs studied were activated robustly by the synthetic ligands T-0901317 and GW3965 and by a variety of oxysterols. In contrast, Ciona LXR was not activated by T-0901317 or GW3965 but was activated by a limited number of oxysterols, as well as some androstane and pregnane steroids. Pharmacophore analysis, homology modeling, and docking studies of Ciona LXR predict a receptor with a more restricted ligand-binding pocket and less intrinsic disorder in the ligand-binding domain compared to vertebrate LXRs. The results suggest that LXRs have a long evolutionary history, with vertebrate LXRs diverging from invertebrate LXRs in ligand specificity.
Keywords: oxysterols, nuclear hormone receptors, Urochordata, structure-activity relationship, molecular models, androstanes, pregnanes
1. Introduction
Liver X receptor (LXR) α (NR1H3) and β (LXRβ; NR1H2) are members of the nuclear hormone receptor (NR) superfamily of ligand-activated transcription factors. NRs work in concert with co-activators and co-repressors to regulate gene expression [1]. NRs share a modular domain structure, which includes, from N-terminus to C-terminus, a modulatory A/B domain, the DNA-binding domain (DBD; C domain), the hinge D domain, the ligand-binding domain (LBD; E domain) and a variable C-terminal F domain [1]. LXRs, like other members of the NR1 family, function as permissive heterodimers with the retinoid X receptors (NR2B1, 2B2, and 2B3). LXRs were originally classified as ‘orphan’ NRs but are perhaps best referred to now as ‘adopted orphans’ [2], following the identification of endogenous ligands, namely oxysterols such as 24(S)-hydroxycholesterol and 24(S),25-epoxycholesterol [3,4], oxysterol metabolites [5], and some bile acids [6].
LXRs are key regulators of lipid and cholesterol metabolism [2]. More recently, LXRs have been shown to regulate uterine contractility [7]. In all mammals whose genomes have been sequenced so far, two distinct LXR genes are found. LXRα is typically detected at high levels in macrophages, adipose tissues, kidney, lung, and spleen; in contrast, LXRβ is expressed at similar levels in a wide variety of tissues, the basis for an alternative name for this receptor as ‘ubiquitous receptor’ [8]. Based on the sequenced genomes of chicken, pufferfish (fugu; Takifugu rubripes) [9], freshwater pufferfish (Tetraodon nigroviridis), zebrafish (Danio rerio), and Western clawed frog (Silurana or Xenopus tropicalis), non-mammalian species appear to generally have only a single LXR gene.
For non-mammalian species, the pufferfish LXR has been the subject of the most detailed study [9]. Pufferfish LXR is more closely related to mammalian LXRα genes by sequence similarity, although the pattern of tissue expression more closely resembles mammalian LXRβ genes in the ubiquity of expression, including expression in brain, gill, gut, heart, ovary, and liver [9]. The current sequence data suggests that a single LXR gene duplicated before mammalian evolution or early in mammalian evolution [9]. If this hypothesis is correct, then one of the duplicated genes maintained ubiquitous tissue expression (LXRβ) while the other (LXRα) carried out specific roles in cholesterol and lipid metabolism with more restricted expression in adipose tissue, liver, and macrophages.
So far, the ligand specificities of non-mammalian LXRs have not been reported. To this end, we cloned and functionally expressed the LXR genes from three model non-mammalian species, African clawed frog (Xenopus laevis), Western clawed frog (Xenopus tropicalis), and zebrafish (Danio rerio). We compared the specificities for ligand activation of these LXRs to human and mouse LXRα and LXRβ. We also sought to probe the evolution of LXR in invertebrates by studying the chordate invertebrate sea squirt (Ciona intestinalis), a member of Urochordata, a subphylum that may contain the closest extant invertebrate relatives to modern vertebrates [10]. The completed genome of Ciona intestinalis revealed a gene that is an apparent ortholog to vertebrate LXRs [11]. From the Ghost database of Ciona intestinalis Genomic and cDNA Resources (http://ghost.zool.kyoto-u.ac.jp/indexr1.html), cDNA clone IDs cigd011h11 and cieg096k22 correspond to this putative ‘Ciona LXR’ (ciLXR). This expression profile of these cDNAs based on expressed sequence tag counts shows high expression in gonadal tissue and neural complex, and lower expression in blood cells, eggs, cleaving embryos, gastrulae/neurulae, tailbud embryos, young adult animals, and mature adult animals. We cloned and expressed this ciLXR to determine how similar this receptor is to its vertebrate orthologs with respect to activation by ligands. We also used molecular modeling studies to compare and contrast the ciLXR to human LXRs.
2. Materials and methods
2.1. Chemicals
The sources of the chemicals were as follows: fexaramine, GW3965, GW4064, glycocholic acid, taurocholic acid (Sigma-Aldrich, St. Louis, MO, USA); T-0901317 (Axxora, San Diego, CA, USA); 5α-petromyzonol (5α-cholan-3α,7α,12α,24-tetrol), petromyzonol sulfate, 3-ketopetromyzonol sulfate (Toronto Research Chemical, Inc., North York, ON, Canada); Nuclear Receptor Ligand Library (76 compounds known as ligands of various nuclear hormone receptors; BIOMOL International, Plymouth Meeting, PA, USA). Other than those described above, steroids and bile salts were obtained from Steraloids (Newport, RI, USA). As previously described [12,13], 5α-cyprinol and 5α-cyprinol 27-sulfate were isolated from Asiatic carp (Cyprinus carpio) bile, 5β-scymnol and 5β-scymnol 27-sulfate were isolated from Spotted eagle ray (Aetobatus narinari) bile, and taurine-amidated 5β-cholestan-3α,7α,12α-triol-27-oic acid was isolated from the bile of the American alligator (Alligator mississippiensis). 5β-Cyprinol was generously provided by the laboratory of Dr. Alan F. Hofmann (University of California – San Diego, CA, United States).
2.2. Animals
Xenopus tropicalis frogs were obtained from NASCO (Fort Atkinson, WI, USA). Adult Ciona intestinalis were purchased from Marine Biological Laboratory (Woods Hole, MA, USA). All animal studies were carried out in accordance with the Declaration of Helsinki and the Guide for the Care and Use of Laboratory Animals as adopted and promulgated by the U.S. National Institutes of Health. All vertebrate animal studies were approved by the University of Pittsburgh Institutional Animal Care and Use Committee.
2.3. Analysis of bile
Biliary contents were dissolved and diluted in methanol and analyzed using ESI-MS on a PE Sciex API III (Perkin Elmer, Alberta, Canada) triple-quadrupole tandem mass spectrometer modified with a nanoESI source from Protana A/S (Odense, Denmark) When operating in the negative mode, the following voltages were used: ISV 600, IN 110, ORI 90. A curtain gas of ultrapure nitrogen was pumped into the interface at a rate of 0.6 L/min to aid evaporation of solvent droplets and to prevent particulate matter from entering the analyzer. Medium-sized palladium-coated borosilicate glass capillaries from Protana A/S were used for sample delivery. Collision gas-induced fragmentation used for structural identification was performed with ultrapure argon as a collision gas. Precursor ion spectra were acquired by scanning the first quadrupole, while collisions with argon in the second quadrupole produced dissociated ions. The third quadrupole was used to mass select the fragment ion. Spectra were the result of averaging from 10 to 100 scans, depending on the scan time and the number of scans necessary to obtain a sufficient signal-to-noise ratio.
Conjugated bile acids were analyzed by high-performance liquid chromatography (HPLC) using a modification of a previously reported technique [14]. An octadecylsilane column (RP C-18, Beckman Instruments, Fullerton, CA) was used with isocratic elution at 0.75 mL/min. The eluting solution was composed of a mixture of methanol and 0.01 M KH2PO4 (67% v/v), adjusted to an apparent pH of 5.3 with H3PO4. Bile acids were quantified by measuring their absorbance at 204 nm. Bile acid amidates (taurine and glycine) have similar extinction coefficients. Bile acids were tentatively identified by matching their relative retention times with those of known standards.
2.4. Cell lines and cell culture
The creation of a HepG2 (human liver) cell line stably expressing the human Na+-taurocholate cotransporter (NTCP) has been previously reported [12]. HepG2-NTCP cells were grown in modified Eagle’s medium-α containing 10% fetal bovine serum and 1% penicillin/streptomycin. The cells were grown at 37ºC in 5% CO2. The Xenopus laevis A6 kidney cell line (ATCC, Manassus, VA, USA) was grown in 75% NCTC 109 medium, 15% distilled water, and 10% fetal bovine serum at 26ºC in 2% CO2. The zebrafish ZFL liver cell line (ATCC) was grown in 50% Leibovitz’s L-15 medium with 2 mM L-glutamine, 35% Dulbecco’s modified Eagle’s medium with 4.5 g/L glucose and 4 mM L-glutamine, 15% Ham’s F-12 with 1 mM L-glutamine supplemented with 0.15 g/L sodium bicarbonate, 15 mM HEPES, 0.01 mg/mL insulin (Sigma-Aldrich), 50 ng/mL recombinant human epidermal growth factor (Sigma-Aldrich), and 5% fetal bovine serum. ZFL cells were grown at 28ºC in room air. Except as noted above, all media and media supplements for the HepG2, A6, and ZFL cell lines were obtained from Invitrogen (Carlsbad, CA, USA).
2.5. Cloning and molecular biology
The LBDs of human LXRα (hLXRα), human LXRβ (hLXRβ), mouse LXRα (mLXRα), and mouse LXRβ (mLXRβ) were cloned by PCR from the following IMAGE clones: hLXRα– IMAGE clone 5744574 (ATCC # 10436197); hLXRβ– IMAGE clone 6469687 (ATCC # 10436559); mLXRα– IMAGE clone 4163109 (ATCC # MGC-13756); and mLXRβ– IMAGE clone 6829527 (ATCC # 10699107). The LBDs of the Xenopus laevis LXR (xlLXR), Xenopus tropicalis (xtLXR), zebrafish LXR (zfLXR), and Ciona intestinalis LXR (ciLXR) were cloned by reverse-transcriptase PCR from RNA extracted from the following sources: xlLXR – A6 cell line; xtLXR – liver from adult X. tropicalis; zfLXR – ZFL cell line; and ciLXR – total RNA extracted from stomach, intestine, and pharynx of adult C. intestinalis. The LBDs of the hLXRα (amino acid residues 163-447), hLXRβ (residues 154-461), mLXRα (residues 161-445), mLXRβ (residues 145-446), xlLXR (residues 154-441), xtLXR (residues 154-441), zfLXR (residues 130-412), and ciLXR (residues 152-435) were inserted into the pM2-GAL4 vector to create GAL4/LBD chimeras suitable for study of ligand activation [12,15].
2.6. Co-transfections and transactivation assays
Ligand activation of LXRs was determined by a luciferase-based functional assay using the HepG2-NTCP cells as previously described [12,15]. The basic methodology for the luciferase reporter assays in 96-well format was as follows. On day 1, cells were seeded onto 96-well white opaque plates at 30,000 cells/well. On day 2, the medium was exchanged, and cells were transfected using calcium phosphate precipitation. The reporter plasmid was tk-UAS-Luc, which contains GAL4 DNA binding elements driving luciferase expression. For hLXRα, hLXRβ, mLXRα, mLXRβ, xlLXR, and xtLXR, 75 ng/well of LXR plasmid was co-transfected with 50 ng/well of the reporter tk-UAS-Luc and 20 ng/well of pSV-β-galactosidase (Promega, Madison, WI, USA). For zfLXR and ciLXR, 100 ng/well of LXR plasmid was co-transfected with 67 ng/well of the reporter tk-UAS-Luc and 20 ng/well of pSV-β-galactosidase. For experiments involving sulfated bile salts or steroids, human organic anion transporting protein (SLC21) was co-transfected at 10 ng/well to facilitate bile salt uptake. On day 3, the cells were washed with Hanks’ buffered salt solution (Invitrogen) and then exposed to medium containing the ligands or vehicle to be tested. The medium utilized charcoal-dextran-treated fetal bovine serum (Hyclone, Logan, UT, USA). Each drug concentration was performed at least in quadruplicate and repeated in separate experiments for a total of at least three times. On day 4, the cells were washed with Hanks’ buffered salt solution and then exposed to 150 μL lysis buffer (Reporter Lysis Buffer, Promega). Separate aliquots were taken for measurement of β-galactosidase activity (Promega) and luciferase activity (Steady-Glo, Promega).
To facilitate more reliable cross-species comparisons, complete concentration-response curves for ligands were determined in the same microplate as determination of response to a maximal activator. This allows for determination of relative efficacy, ε, defined as the maximal response to test ligand divided by maximal response to a reference maximal activator (note than ε can exceed 1). For mLXRα, mLXRβ, and zfLXR, 5 μM T-0901317 was used as the maximal activator. For hLXRα, hLXRβ, xlLXR, and xtLXR, 10 μM T-0901317 was used as the maximal activator. For ciLXR, 50 μM 5α-androst-16-en-3α-ol (androstenol) was used as the maximal activator. All comparisons to maximal activators were done within the same microplate. Luciferase data were normalized to the internal β-galactosidase control and represent means ± SD of the assays.
2.7. Pharmacophore models and molecular modeling studies
Common feature pharmacophore modeling was performed as described previously [16,17]. Briefly, using the X-ray crystal structure 1P8D [18] from the RCSB Protein Data Bank (PDB), the ligand epoxycholesterol was extracted, and bond orders were corrected. This molecule conformation was used as a template. Similarly T-0901317 was extracted from the X-ray crystal structure 1PQC [19] and used as an alternate template. All other ligands were imported as sdf files and up to 255 conformations were generated with the Best conformer generation method, allowing a maximum energy difference of 20 kcal/mol with Discovery Studio Catalyst 1.7 (Accelrys, San Diego, CA). Initially 6-formylindolo-[3,2-b]-carbazole (6-FC) was aligned with epoxycholesterol and T-0901317 separately with the HIPHOP algorithm. In addition, in order to try to understand the human versus Ciona LXR LBD differences we generated HIPHOP models with the same 7 molecules including actives and inactives (Supplementary Table 1). During model construction we requested hydrogen bond acceptor, hydrogen bond donor, hydrophobic, and excluded volume features.
A structural model of the LBD of the Ciona LXR (from residues 217 to 454) was constructed using the Insight II Homology Module (Accelrys, Inc., San Diego, CA) from the published crystal structure of human LXR in complex with epoxycholesterol (PDB ID = 1P8D) [18] as the modeling template. Several energy minimization-based refinement procedures were implemented on the initial model and the quality of the final model was confirmed by the WHATIF-Check program. The volume of the LBP for hLXRβ and ciLXR were determined using CASTp (http://sts.bioengr.uic.edu/castp/calculation.php) [20].
Molecular docking studies were performed on the two synthetic LXR agonist (T-0901317 and GW3965) and two cholesterol compounds (epoxycholesterol and 25-hydroxycholesterol) using the GOLD docking program [21]. During the docking process, the protein was held fixed while full conformational flexibility was allowed for ligands. For each ligand 30 independent docking runs were performed to achieve the consensus orientation in the ligand binding pocket.
2.8. Calculation of predicted protein structural disorder
Disorder prediction of protein sequences were performed using the PONDR VL3H algorithm [22] available at http://www.ist.temple.edu/disprot/predictor/php. The disorder calculations for each amino acid residue are available as Supplementary Table 2 and summarized in Supplementary Table 3.
2.9. Phylogenetic analysis
The following sequences were used for sequence comparisons and phylogenetic analysis (Ensembl accession nos. are from available at http://www.ensembl.org): human LXRα (GenBank accession no. NM_005693), chimpanzee LXRα (GenBank accession no. XM_521906), rhesus monkey LXRa (Ensembl accession no. ENSMMUT00000021881), cow LXRα (Ensembl accession no. ENSBTAT00000014131), dog LXRα (Ensembl accession no. ENSCAFT00000014022), elephant LXRα (Ensembl accession no. ENSLAFT00000009267), mouse LXRα (GenBank accession no. NM_013839), lesser hedgehog tenrec LXRα (Ensembl accession no. ENSETET00000000414), pig LXRα (GenBank accession no. DQ059138), duck-billed platypus LXRα (Ensembl accession no. ENOANT00000010003), chicken LXR (GenBank accession no. AF492498), Xenopus laevis LXR (GenBank accession no. BC074169), Xenopus tropicalis LXR (Ensembl accession no. ENSXETT00000000613), pufferfish LXR (Ensembl accession no. NEWSINFRUT00000136830), medaka LXR (Ensembl accession no. ENSORLT00000001582), stickleback fish LXR (Ensembl accession no. ENSGACT00000022713), Tetraodon nigriviridis LXR (Ensembl accession no. GSTENT00018165001), zebrafish LXR (GenBank accession no. NM_001017545), Ciona intestinalis LXR (Ensembl accession no. ENSCINT00000014778), purple sea urchin LXR (GenBank accession no. XM_774904), human LXRβ (GenBank accession no. NM_007121), rhesus monkey LXRβ (Ensembl accession no. ENSMMUT00000032951), cow LXRβ (Ensembl accession no. ENSCAFT00000004229), dog LXRβ (Ensembl accession no. ENSCAFT00000005340), mouse LXRβ (GenBank accession no. NM_009473), human FXR (GenBank accession no. NM_005123), dog FXR (Ensembl accession no. ENSCAFT00000010998), mouse FXR (GenBank accession no. NM_009108), opossum FXR (Ensembl accession no. ENSMODT00000006279), platypus FXR (Ensembl accession no. ENOANT00000008880), chicken FXR (GenBank accession no. AF492497), Xenopus laevis FXR (GenBank accession no. AF456451), Xenopus tropicalis FXR (Ensembl accession no. ENSXETT00000046366), pufferfish FXR (Ensembl accession no. NEWSINFRUT0000143035), medaka FXR (Ensembl accession no. ENSORLT00000014125), Tetraodon nigroviridis FXR (Ensembl accession no. GSTENT00022799001), zebrafish FXR (Ensembl accession no. ENSDART00000061794), Ciona intestinalis FXR (GenBank accession no. BR000116), Ciona savignyi FXR (Ensembl accession no. ENSCAVT00000012600), ixotid tick (Amblyomma americanum) ecdysone receptor (GenBank accession no. AF020187). Sequences were aligned using ClustalW software (http://www.ebi.ac.uk/clustalw) and manually adjusted as needed. An alignment of LXR sequences is provided in Supplementary Fig. 1. Phylogeny was inferred using parsimony analysis by PAUP*4.0-beta for UNIX/LINUX (Sinauer Associates, Sunderland, MA, USA) with the ixotid tick ecdysone receptor used as the outgroup. A heuristic search of 100 replicates of random addition plus tree-bisection-reconnection branch swapping was used; to estimate support, 10,000 bootstrap replicates were analyzed.
3. Results
3.1. Ligand specificity of vertebrate LXRs
To compare ligand activation of mammalian and non-mammalian LXRs, the LBDs of hLXRα, hLXRβ, mLXRα, mLXRβ, xlLXR, xtLXR, and zfLXR were cloned and inserted into the pM2-GAL4 vector to create LBD/GAL4 ‘chimeric’ receptors. The chimeric receptors were then used together with the reporter plasmid tk-UAS-Luc in a luciferase-based reporter assay described previously [15]. An initial screening of possible ligands for these receptors revealed that T-0901317 and GW3965 (see Fig. 1 for chemical structures), both previously reported as agonists of mammalian LXRs [23,24], also robustly activated the two Xenopus LXRs, as well as the zfLXR. T-0901317 was selected as the reference ‘maximal’ activator for the vertebrate LXRs in this study; the maximal activation of all other tested compounds at these receptors was then compared to T-0901317.
Fig. 1.
Chemical structures of key compounds in this study. The number of the steroid rings is indicated for the steroidal compounds.
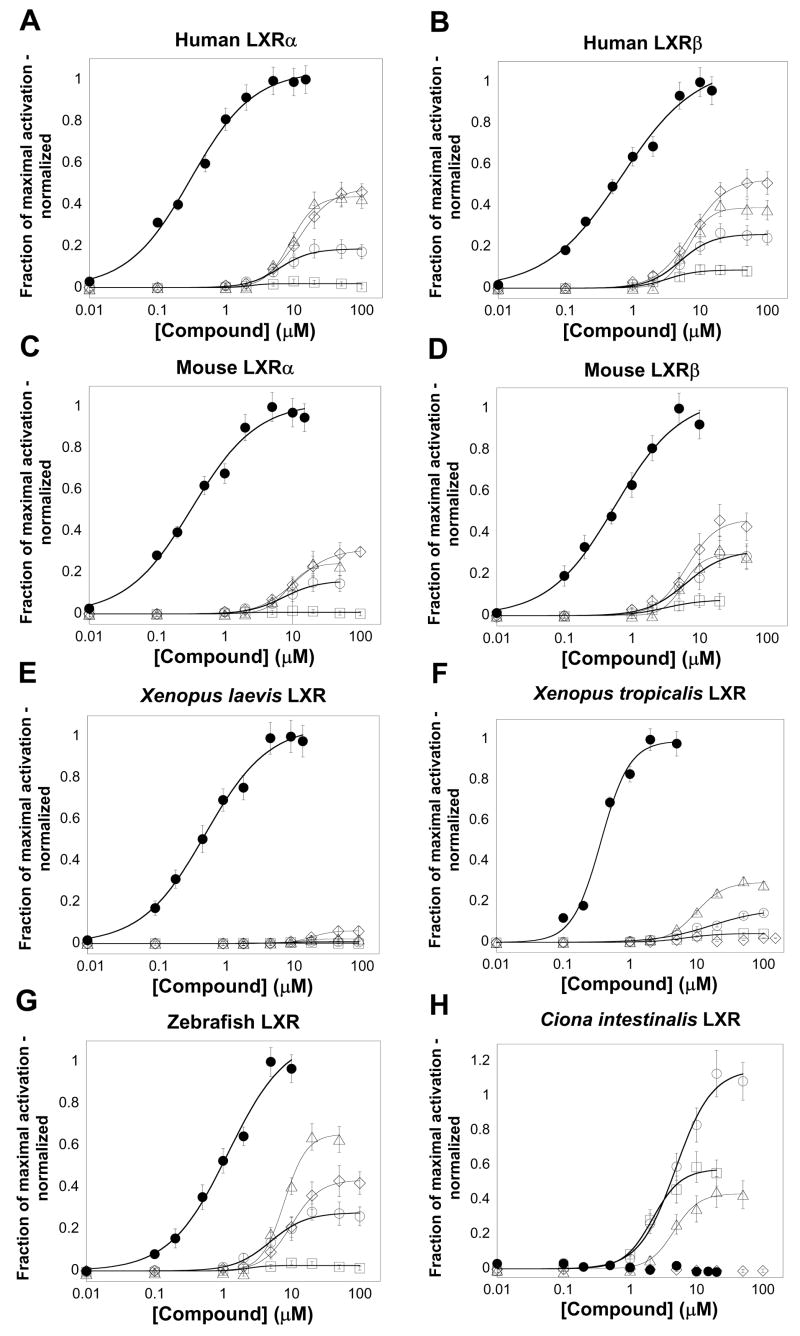
Similar to previous reports [3,25], the ligand specificities of human and mouse LXRα and LXRβ are very similar (Fig. 2A–D; Table 1). These receptors were activated by a number of oxysterols including 20(S)-hydroxycholesterol, 22(R)-hydroxycholesterol, 24(S)-hydroxycholesterol, 25-hydroxycholesterol, 24(S),25-epoxycholesterol, and 24-ketocholesterol with EC50 values in the low micromolar range. The maximal effects of the oxysterols was in general lower than that of T-0901317 in the assay used (Fig. 2A–D; Table 1). xlLXR, xtLXR, and zfLXR were also activated by the same oxysterols as the mammalian LXRα and LXRβ with the exception that 20(S)-hydroxycholesterol did not activate the non-mammalian LXRs (Fig. 2E–G; Table 2). None of the vertebrate LXRs were activated by the farnesoid X receptor (FXR; NR1H4) agonists farnesol, fexaramine, or GW4046 (Tables 1 and 2).
Fig. 2.
Concentration-response curves for activation of LXRs by T-0901317 and oxysterols. The ordinate represents activation of LXR, relative to vehicle control, and normalized to the maximal activator (5 μM T-0901317 for mLXRα, mLXRβ, and zfLXR; 10 μM T-0901317 for hLXRα, hLXRβ, xlLXR, and xtLXR; 50 μM androstenol for ciLXR). The compounds tested were T-0901317 (●), 24(S)-hydroxycholesterol (○), 25-hydroxycholesterol (□), 24(S),25-epoxycholesterol (⋄), and 24-ketocholesterol (Δ). (A-D,F,G) hLXRα, hLXRβ, mLXRα, mLXRβ, xtLXR, and zfLXR show very similar patterns of activation by the five compounds. (E) For xlLXR, the efficacy of the oxysterols was much lower than that of T-0901317. (H) ciLXR was not activated by T-0901317 but was activated by three of the oxysterols.
Table 1.
Effects of compounds on human and mouse LXRs. Comparison of EC50 and relative efficacy (ε) values for human and mouse LXRs. Efficacy is compared to T-0901317 as the maximal activator.
| Common name | Significance | Human LXRα EC50 (μM) | Human LXRα ε | Human LXRβ EC50 (μM) | Human LXRβ ε | Mouse LXRα EC50 (μM) | Mouse LXRα ε | Mouse LXRβ EC50 (μM) | Mouse LXRβ ε |
|---|---|---|---|---|---|---|---|---|---|
| T-0901317 | Synthetic LXR ligands | 0.31 ± 0.24 | 1.0a | 0.67 ± 0.054 | 1.0a | 0.33 ± 0.028 | 1.0a | 0.56 ± 0.03 | 1.0a |
| GW3965 | 0.99 ± 0.07 | 0.66 | 0.71 ± 0.09 | 0.86 | 1.1 ± 0.10 | 0.63 | 0.61 ± 0.04 | 0.85 | |
| Paxilline | 4.7 ± 0.28 | 0.08 | 2.7 ± 0.18 | 0.01 | 5.4 ± 0.27 | 0.08 | 4.5 ± 0.33 | 0.01 | |
| Cholesterol | Oxysterols | No effect | No effect | No effect | No effect | ||||
| 7α-Hydroxycholesterol | No effect | No effect | No effect | No effect | |||||
| 20(S)-Hydroxycholesterol | 10.0 ± 0.60 | 0.03 | 9.3 ± 0.79 | 0.08 | 10.5 ± 0.74 | 0.02 | 10.3 ± 0.77 | 0.09 | |
| 22(S)-Hydroxycholesterol | No effect | No effect | No effect | No effect | |||||
| 22(R)-Hydroxycholesterol | 5.4 ± 0.32 | 0.03 | 5.2 ± 0.31 | 0.08 | 5.2 ± 0.44 | 0.02 | 4.9 ± 0.25 | 0.09 | |
| 24(S)-Hydroxycholesterol | 6.1 ± 0.52 | 0.19 | 5.1 ± 0.26 | 0.26 | 7.3 ± 0.66 | 0.16 | 5.7 ± 0.29 | 0.30 | |
| 25-Hydroxycholesterol | 4.3 ± 0.34 | 0.03 | 3.2 ± 0.27 | 0.09 | 3.5 ± 0.21 | 0.02 | 3.1 ± 0.22 | 0.08 | |
| 24(S),25-Epoxycholesterol | 11.9 ± 0.59 | 0.48 | 7.4 ± 0.52 | 0.53 | 10.9 ± 0.88 | 0.31 | 6.4 ± 0.49 | 0.07 | |
| 24-Ketocholesterol | 9.0 ± 0.63 | 0.44 | 6.5 ± 0.55 | 0.39 | 8.5 ± 0.25 | 0.25 | 6.4 ± 0.58 | 0.31 | |
| 5β-Cholestan-3α,7α,12α-triol | Bile salt intermediate | No effect | No effect | No effect | No effect | ||||
| 5α-Petromyzonolb | Bile salts from cartilaginous fishes | No effect | No effect | No effect | No effect | ||||
| 5α-Petromyzonol 24-sulfate | No effect | No effect | No effect | No effect | |||||
| 3-Ketopetromyzonol sulfate | No effect | No effect | No effect | No effect | |||||
| 5β-Scymnolc | No effect | No effect | No effect | No effect | |||||
| 5β-Scymnol 27-sulfate | No effect | No effect | No effect | No effect | |||||
| 5α-Cyprinold | Major bile salts for zebrafish | No effect | No effect | No effect | No effect | ||||
| 5α-Cyprinol 27-sulfate | No effect | No effect | No effect | No effect | |||||
| 5β-Cyprinol | Major bile salt of Xenopus frogs | No effect | No effect | No effect | No effect | ||||
| Farnesol | FXR ligands | No effect | No effect | No effect | No effect | ||||
| Fexaramine | No effect | No effect | No effect | No effect | |||||
| GW4064 | No effect | No effect | No effect | No effect | |||||
| TTNPBe | No effect | No effect | No effect | No effect | |||||
| 5α-Androstan-3α-ol | Miscellaneous | No effect | No effect | No effect | No effect | ||||
| 5α-Androsten-16-en-3α-ol | No effect | No effect | No effect | No effect | |||||
| Diindoylmethane | No effect | No effect | No effect | No effect | |||||
| Ecdysone | No effect | No effect | No effect | No effect | |||||
| 6-Formylindolo-[3,2,b]-carbazole | 3.1 ± 0.2 | 0.11 | 0.99 ± 0.1 | 0.17 | 5.0 ± 0.4 | 0.16 | 2.1 ± 0.1 | 0.22 |
Maximal reference compound (efficacy = 1.0); efficacy of other compounds are relative to the maximal reference compound.
5α-Cholan-3α,7α,12α,24-tetrol
5β-Cholestan-3α,7α,12α,24,26,27-hexol
5α-Cholestan-3α,7α,12α,26,27-pentol
4-[(E)-2-(5,6,7,8-Tetrahydro-5,5,8,8-tetramethyl-2-naphthalenyl)-1-propenyl]benzoic acid
Table 2.
Effects of compounds on non-mammalian LXRs. Comparison of EC50 and relative efficacy (ε) values for non-mammalian LXRs. Efficacy is compared to 5α-androst-16-en-3α-ol for Ciona intestinalis LXR and T-0901317 for the other LXRs.
| Common name | Significance | Xenopus laevis LXRα EC50 (μM) | Xenopus laevis LXRα ε | Xenopus tropicalis LXRα EC50 (μM) | Xenopus tropicalis LXRβ ε | Zebrafish LXRα EC50 (μM) | Zebrafish LXRα ε | Ciona intestinalis LXRβ EC50 (μM) | Ciona intestinalis LXRβ ε |
|---|---|---|---|---|---|---|---|---|---|
| T-0901317 | Synthetic LXR ligands | 0.55 ± 0.033 | 1.0a | 0.54 ± 0.03 | 1.0a | 1.7 ± 0.09 | 1.0a | No effect | |
| GW3965 | 1.8 ± 0.11 | 0.85 | 0.85 ± 0.06 | 0.70 | 1.7 ± 0.06 | 0.73 | No effect | ||
| Paxilline | 5.2 ± 0.42 | 0.07 | ~5 | < 0.05 | 5.3 ± 0.37 | 0.01 | No effect | ||
| Cholesterol | Cholesterol and oxysterols | No effect | No effect | No effect | No effect | ||||
| 7α-Hydroxycholesterol | No effect | No effect | No effect | No effect | |||||
| 20(S)-Hydroxycholesterol | 11.6 ± 0.58 | 0.01 | No effect | 9.1 ± 0.64 | 0.06 | No effect | |||
| 22(S)-Hydroxycholesterol | No effect | No effect | No effect | No effect | |||||
| 22(R)-Hydroxycholesterol | 3.3 ± 0.23 | 0.01 | 9.8 ± 0.77 | 0.08 | 4.9 ± 0.25 | 0.06 | No effect | ||
| 24(S)-Hydroxycholesterol | 8.3 ± 0.66 | 0.01 | 11.9 ± 0.97 | 0.14 | 4.9 ± 0.39 | 0.28 | 4.8 ± 0.47 | 1.16 | |
| 25-Hydroxycholesterol | 3.0 ± 0.24 | 0.01 | 3.2 ± 0.24 | 0.04 | 2.3 ± 0.18 | 0.03 | 2.2 ± 0.34 | 0.58 | |
| 24(S),25-Epoxycholesterol | 20.2 ± 1.7 | 0.06 | 5.4 ± 0.35 | 0.27 | 9.9. ± 0.84 | 0.43 | > 10 | ~0.5 | |
| 24-Ketocholesterol | 13.4 ± 1.2 | 0.02 | 10.3 ± 0.84 | 0.30 | 7.8 ± 0.63 | 0.65 | 4.6 ± 0.37 | 0.44 | |
| 5β-Cholestan-3α,7α,12α-triol | Bile salt intermediate | No effect | No effect | No effect | No effect | ||||
| 5α-Petromyzonolb | Bile salts from cartilaginous fishes | No effect | No effect | No effect | No effect | ||||
| 5α-Petromyzonol 24-sulfate | No effect | No effect | No effect | No effect | |||||
| 3-Ketopetromyzonol sulfate | No effect | No effect | No effect | No effect | |||||
| 5β-Scymnolc | No effect | No effect | No effect | No effect | |||||
| 5β-Scymnol 27-sulfate | No effect | No effect | No effect | No effect | |||||
| 5β-Cyprinold | Major bile salt | No effect | No effect | No effect | No effect | ||||
| 5α-Cyprinol 27-sulfate | for zebrafish | No effect | No effect | No effect | No effect | ||||
| 5β-Cyprinol | Major bile salt of Xenopus frogs | No effect | No effect | No effect | No effect | ||||
| Farnesol | FXR ligands | No effect | No effect | No effect | No effect | ||||
| Fexaramine | No effect | No effect | No effect | No effect | |||||
| GW4064 | No effect | No effect | No effect | No effect | |||||
| TTNPBe | No effect | No effect | No effect | No effect | |||||
| 5α-Androstan-3α-ol | Miscellaneous | No effect | No effect | No effect | ~100 | ~1.6 | |||
| 5α-Androsten-16-en-3α-ol | No effect | No effect | No effect | 58.3 ± 4.7 | 1.0a | ||||
| Diindoylmethane | No effect | No effect | No effect | No effect | |||||
| Ecdysone | No effect | No effect | No effect | No effect | |||||
| 6-Formylindolo-[3,2,b]-carbazole | 12.4 ± 1.1 | 0.18 | 13.1 ± 1.3 | 0.13 | 13.9 ± 1.2 | 0.13 | 38.1 ± 2.9 | 16.2 |
Maximal reference compound (efficacy = 1.0); efficacy of other compounds are relative to the maximal reference compound.
5α-Cholan-3α,7α,12α,24-tetrol
5β-Cholestan-3α,7α,12α,24,26,27-hexol
5α-Cholestan-3α,7α,12α,26,27-pentol
4-[(E)-2-(5,6,7,8-Tetrahydro-5,5,8,8-tetramethyl-2-naphthalenyl)-1-propenyl]benzoic acid
In mammals, LXRs are activated by a limited number of bile acids (particularly 6α-hydroxylated bile acids) [6], whereas the related FXRs are activated by a broader diversity of bile acids such as cholic acid (CA) or chenodeoxycholic acid (CDCA) [2]. The bile salts of non-mammalian vertebrates can differ from that of the 24-carbon (C24) bile acids found in the majority of mammals [26,27]. For example, the 27-carbon (C27) bile alcohol 5α-cyprinol (5α-cholestan-3α,7α,12α,26,27-pentol) 27-sulfate is known to be the dominant constituent of zebrafish bile [28]. The bile of Xenopus laevis frogs has also been examined previously and been shown to contain predominantly 5β-cyprinol sulfate [29]. To complement these earlier studies, we also analyzed bile aspirated from gallbladders of adult X. tropicalis frogs by electrospray ionization-mass spectrometry (ESI-MS) and found the presence of ions with m/z ratios consistent with the bile being a mixture of a C27 pentahydroxylated bile alcohol sulfate (likely 5β-cyprinol sulfate or a closely related bile alcohol sulfate) and taurine-conjugated C24 and C27 bile acids (Fig. 3A). HPLC analysis showed the presence of compounds that eluted with the retention times characteristic of taurocholic acid (13.9 min), taurochenodeoxycholic acid (24.8 min), and taurodeoxycholic acid (28.3 min) (Fig. 3B). None of the vertebrate LXRs were activated by a variety of other bile salts tested, including C24 bile acids typically found in mammals (e.g., CDCA, CA, lithocholic acid, and their glycine, taurine, or sulfated conjugates) or evolutionarily ‘early’ bile alcohols and their sulfated conjugates, such as 5α-cyprinol, 5α-cyprinol 27-sulfate, 5β-cyprinol, 5β-scymnol (5β-cholestan-3α,7α,12α,24,26,27-hexol), or 5β-scymnol 27-sulfate (Tables 1 and 2). In contrast, we have found that X. laevis and zebrafish FXRs were activated by some bile alcohol sulfates (E.J. Reschly, M.D. Krasowski, unpublished data). Thus, in humans and mice as well as zebrafish and Xenopus frogs, LXRs are primarily activated by oxysterols while FXRs are primarily activated by bile salts.
Fig. 3.
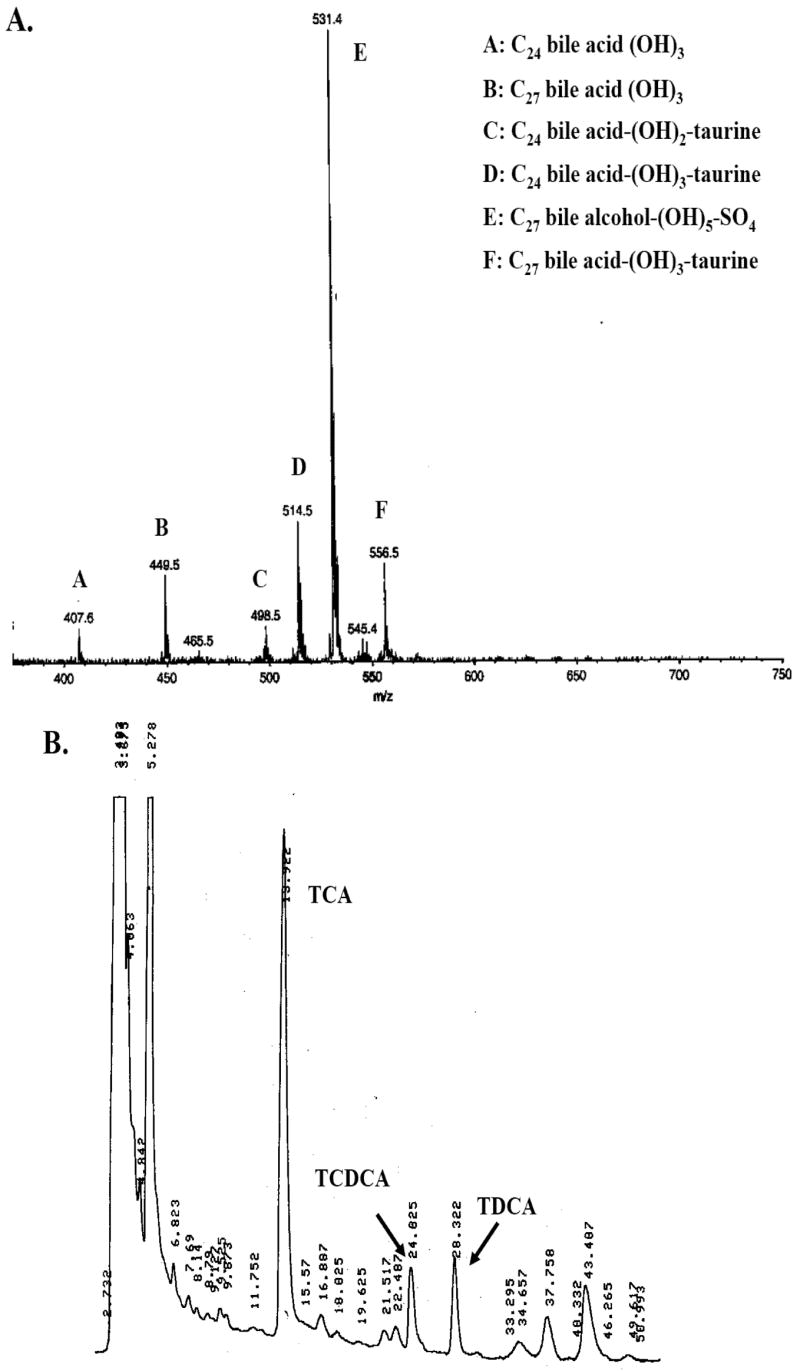
Analysis of Xenopus tropicalis bile. (A) ESI/MS analysis of Xenopus tropicalis bile. From an extensive library generated from analysis of many vertebrate bile specimens, the major ions are annotated with probable matches indicating the number of carbon atoms (24 or 27), whether the compound is a bile acid or bile alcohol, number of hydroxyl groups, as well as absence or presence of conjugation (taurine for acids, sulfate for alcohols). The dominant ion corresponds to the m/z ratio expected for a penta-hydroxylated C27 bile alcohol sulfate (e.g., cyprinol sulfate); in addition, m/z ratios consistent with trihydroxylated unconjugated and taurine-conjugated C24 and C27 bile acids are also present. (B) HPLC analysis of X. tropicalis bile shows the likely presence of taurocholic acid (13.9 min), taurochenodeoxycholic acid (24.0 min), and taurodeoxycholic acid (28.3 min).
In mammals, GW3965 has been reported as a selective ligand for LXRs relative to other NRs (although it does not distinguish between LXRα and LXRβ) [23]. We also tested GW3965 on Xenopus laevis and zebrafish FXRs, vitamin D receptors (VDRs, NR1I1), and pregnane X receptors (PXRs, NR1I2) and found no effect of this compound on these receptors; in contrast, T-0901317 activated the zebrafish FXR in the low micromolar range (E.J. Reschly, M.D. Krasowski, unpublished data). We also found that paxilline, a fungal metabolite reported to activate LXRα and LXRβ [30], also activated zebrafish, Xenopus laevis, and Xenopus tropicalis LXRs, although with much lower efficacy than T-0901317 (Tables 1 and 2).
3.2. Phylogenetic analysis of the Ciona intestinalis LXR ortholog
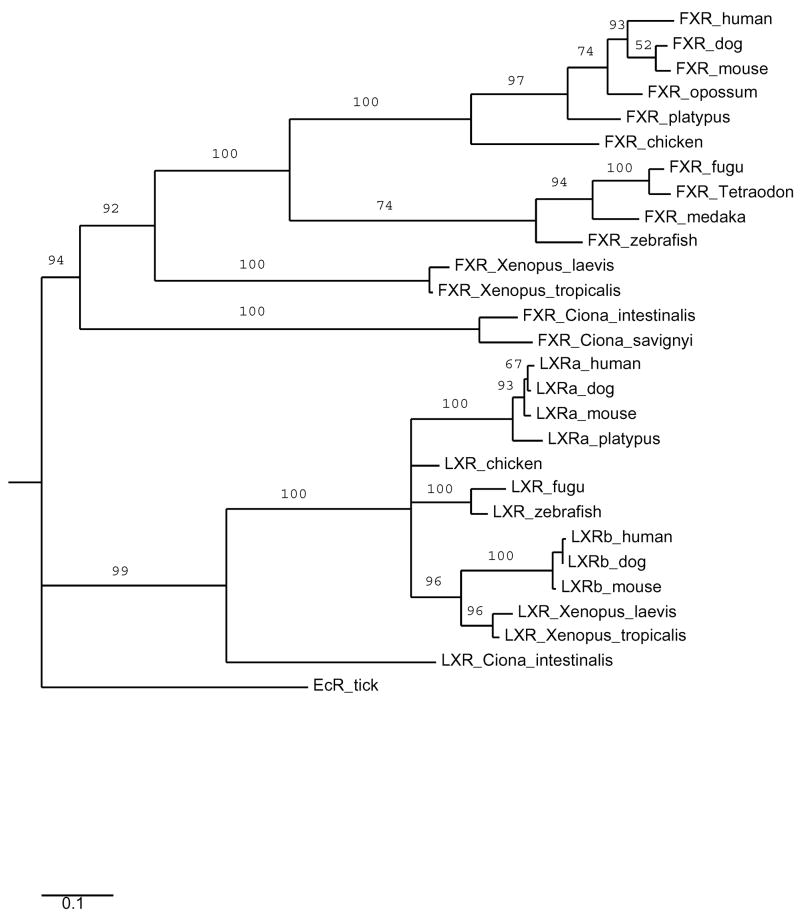
Sequencing of the Ciona intestinalis genome revealed two genes that have close sequence similarity to vertebrate NR1H genes [11]. By reciprocal BLAST analysis, one gene (ciLXR; Ensembl database accession no. ENSCINT00000014778) appears orthologous to vertebrate LXRs while the other (ciFXR; GenBank accession no. BR000116) appears orthologous to vertebrate FXRs (NR1H4 and H5). Recently, the draft genome of the purple sea urchin (Strongylocentrotus purpuratus) genome revealed four NR1H-like genes, one of which is most closely related by sequence comparison to LXRs (GenBank accession no. XM_774904; hereafter referred to as ‘sea urchin LXR’) [31]. These reports indicate that LXR or LXR-like genes are present in invertebrates.
The ciLXR had the following percent sequence identities to the DBDs and LBDs of LXRs and FXRs: hLXRα (70.6%, DBD; 54.2%, LBD), zfLXR (73.5%, DBD; 53.4%, LBD), human FXR (67.6%, DBD; 34.1%, LBD), zebrafish FXR (67.6%, DBD; 36.7%, LBD), Ciona FXR (77.6%, DBD; 29.2%, LBD), and sea urchin LXR (66.2%, DBD; 49.1%, LBD). The ciFXR had the following percent sequence identities to the DBDs and LBDs of LXRs and FXRs: human FXR (77.6%, DBD; 39.7%, LBD), chicken FXR (76.1%, DBD; 38.0%, LBD), zebrafish FXR (77.6%, DBD; 36.7%, LBD), human LXRα (70.1%, DBD; 26.2%, LBD), zebrafish LXR (70.1%, DBD; 27.5%, LBD), Ciona LXR (77.6%, DBD; 29.2%, LBD), and sea urchin LXR (64.2%, DBD; 24.3%, LBD). A sequence alignment of the DBD and LBD of seven vertebrate LXRαs, two vertebrate LXRβs, ciLXR, and purple sea urchin LXRs is found in Supplementary Fig. 1. The phylogeny of ciLXR and ciFXR in relation to vertebrate LXRs and FXRs was inferred using parsimony analysis (Fig. 4). The results are also consistent with the assignment of Ciona LXR and FXR as orthologs to vertebrates LXRs and FXRs, respectively.
Fig. 4.
Complete maximum parsimony phylogeny. Branch labels indicate bootstrap percentages. The putative Ciona intestinalis LXR ortholog clearly clusters with the vertebrate LXRs.
3.3. Ligand specificity of the Ciona intestinalis LXR ortholog
The LBD of ciLXR was cloned by PCR from RNA pooled from pharynx, stomach, and intestine of adult animals and inserted into the pM2-GAL4 vector. Unlike the vertebrate LXRs, ciLXR was not activated by T-0901317, GW3965, or paxilline. Several oxysterols, including 24(S)-hydroxycholesterol and 24(S),25-epoxycholesterol, as well as 5α-androst-16-en-3α-ol (androstenol) and 5α-androstan-3α-ol (androstanol) activated ciLXR (Fig. 2H), with androstenol chosen as the reference maximal activator. The pattern of oxysterol activation of ciLXR is different than that of the vertebrate LXRs. Even more distinctive was the activation of ciLXR by androstenol and androstanol, compounds that were inactive at all of the vertebrate LXRs tested (Tables 1 and 2). Androstenol was chosen as the reference maximal activator. Additional screening of a 76 compound library of known NR ligands revealed that 6-formylindolo-[3,2-b]-carbazole (6-FC; see Fig. 1 for chemical structure), a tryptophan photoproduct and high-affinity aryl hydrocarbon receptor agonist [32], robustly activated the ciLXR at micromolar concentrations with maximal effect much greater than that of androstenol (EC50 = 38.1 μM, ε = 16.2 relative to androstenol). 6-FC also activated all the vertebrate LXRs (Tables 1 and 2).
3.4. Conservation of ligand-binding residues in the LXRs
As detailed in Materials and Methods, partial or complete sequence data is available for 10 mammalian LXRαs, 8 non-mammalian vertebrate LXRs, 5 mammalian LXRβs, and two invertebrate LXRs (Ciona intestinalis and sea urchin). High-resolution crystal structures of LXRs bound to various agonists have been published for human LXRα [33], mouse LXRα [34], and human LXRβ [18,19,35]. From these structures, a total of 31 amino acid residue positions have been shown to interact with ligands in LXRα and/or LXRβ. The amino acid residues at these 31 positions are absolutely conserved among all 23 vertebrate genes analyzed; that is, no amino acid differences were found across species at any of the ‘ligand-binding residues’, even between LXRαs and LXRβs. This suggests that the endogenous ligands for LXRs vary little across vertebrate species, at least among teleost fish, amphibians, birds, and mammals.
In contrast, some of these 31 ligand-binding residues of mammalian LXRs are not conserved between vertebrate LXRs and the invertebrate LXRs or closely related invertebrate ecdysone receptors (NR1H1). Of these 31 residues, 21 are conserved in ciLXR, 13 in sea urchin LXR, and 12 in either Drosphila melanogaster ecdysone receptor (GenBank accession no. NM_165465) or ixodid tick ecdysone receptor (GenBank accession no. AF020187). Only one study has determined the structure of an LXR bound to an endogenous ligand [18]. The published structure of human LXRβ bound to 24(S),25-epoxycholesterol identified six amino acid residues as particularly critical in interactions with the A and D rings of the epoxycholesterol. Only two of these six amino acid residues are conserved between ciLXR and vertebrate LXRs; three of six are conserved between purple sea urchin LXR and vertebrate LXRs (Supplementary Table 4). None of the three amino residues of hLXRβ that interact with the D ring of epoxycholesterol are conserved in ciLXR; in fact, each difference markedly changes the amino acid properties (Ser-278 to Ile; Glu-281 to Leu; Arg-319 to Gln). For Glu-281 of hLXRβ, the corresponding residue in ciLXR, sea urchin LXR, and six ecdysone receptors is leucine, indicating a change in the vertebrate sequence compared to the likely ancestral residue. For Arg-319 of hLXRβ, the ecdysone receptors and sea urchin LXR also have arginine while ciLXR has a glutamine; in this case, the ciLXR diverges from the probable ancestral sequence (Supplementary Table 4). These changes may underlie some of the differences in pharmacology determined between ciLXR and vertebrate LXRs and suggest that if the true endogenous ligands to ciLXR are steroidal compound(s), they may be different from LXR ligands found in vertebrates and possibly other invertebrates such as insects and crustaceans that express ecdysone receptors.
3.5. Pharmacophore analysis of human and Ciona LXRs
Initially we were interested in the alignment of 6-FC with 24(S),25-epoxycholesterol in hLXRβ using the common feature alignment HIPHOP method (Fig. 5A). This yielded a model with a hydrogen bond acceptor (HBA) that corresponds to hLXRβ residue number His-435 based on the crystal structure of hLXRβ [18]. Note that the planar 6-FC also manages to interact with the HBA. Alignment of 6-FC with T-0901317 in the orientation from a different crystal structure of hLXRβ [19] suggests a different pharmacophore (Fig. 5B) in which His-435 serves as a hydrogen bond donor (HBD; this residue is known from structural studies as having the capability of serving as either a HBD or HBA) [18]. 6-FC does not map well to this HBD feature in the pharmacophore. In addition, no HBA was shown to map to the sulfonamide group of T-0901317 in the X-ray structure [18]. Additional HIPHOP models for hLXRβ and ciLXR based on multiple active and inactive molecules revealed that epoxycholesterol appears to fit in a 180° flipped orientation in ciLXR (Fig. 5C) compared with hLXRβ (Fig. 5D). This suggests that the His-435 position may be preferentially a HBD in ciLXR. Also, the addition of two excluded volumes which appear in the ciLXR pharmacophore (Fig. 5C) suggests that the ciLXR LBD may have a more restrictive ligand-binding pocket (LBP) than hLXRβ.
Fig. 5.
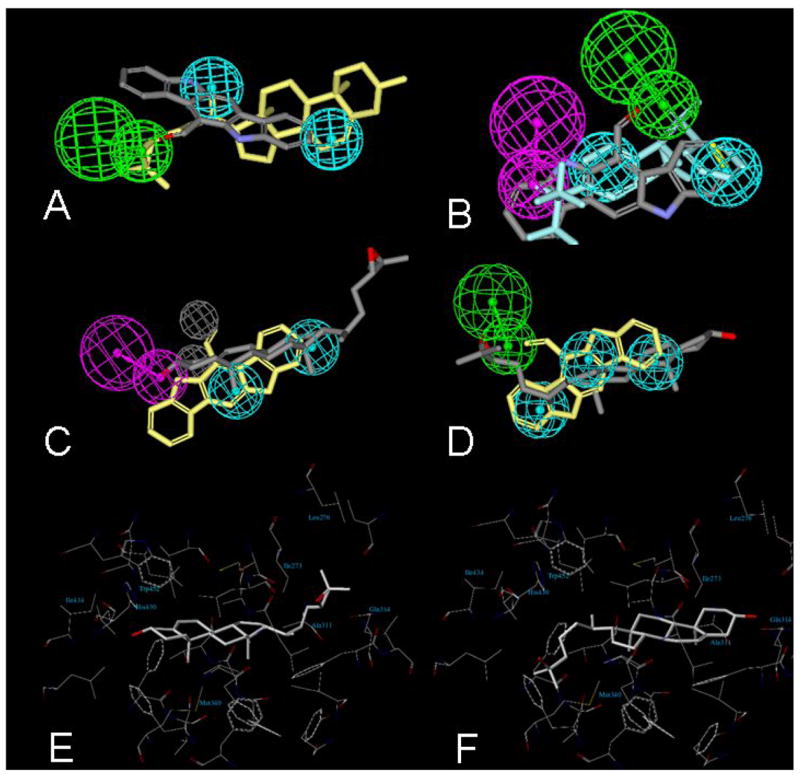
Molecular modeling studies of human LXRβ and Ciona LXR. HIPHOP alignments of 6-formylindolo-[3,2-b]-carbazole with (A) 24(S),25-epoxycholesterol and (B) T-0901317. Green indicates hydrogen bond acceptor while cyan denotes hydrophobic features. (C) HIPHOP model for ciLXR based on multiple active and inactive molecules. (D) HIPHOP model for hLXRα based on multiple active and inactive molecules. The color schemes for both (C) and (D) are as follows: epoxycholesterol – grey; 6-formylindolo-[3,2,b]-carbazole – yellow; hydrogen bond acceptor – green; hydrophobic features – cyan; hydrogen bond donor – purple; excluded volumes – grey. (E) Docking of epoxycholesterol into the predicted binding pocket of ciLXR generated by a homology model. The orientation of epoxycholesterol is flipped 180° relative to that seen in a crystal structure of hLXRβ [18]. Epoxycholesterol has good complementarity to the pocket, but the hydrogen bond interaction of the 3-hydroxyl group of epoxycholesterol seen in the ηυμα ΛΞPβ crystal structure is not present. F, Docking of 25-hydroxycholesterol into the predicted binding pocket of ciLXR generated by a homology model. The 3-hydroxyl group of 25-hydroxycholesterol forms a hydrogen bond with Gln-314 and the tail part of the 25-hydroxycholesterol has extensive hydrophobic contacts with the binding pocket.
3.6. Homology modeling and docking studies of Ciona LXR
To gain additional insight into the ligand selectivity of ciLXR, a homology model of this receptor was created using hLXRβ as a template (Supplementary Fig. 2). Compared with hLXRβ, the ciLXR LBD is very similar; however, the latter is predicted to have a smaller (ciLXR is 908 Å3 versus hLXRβ is 1198 Å3) and more hydrophobic ligand binding pocket (LBP). The alteration in the size and properties of the LBP clearly would affect the relative binding affinity to various ligands. Our docking studies confirmed that two synthetic LXR agonists, T-0901317 and GW3965, cannot fit into the LBP of the ciLXR. For these two ligands, the substitution Leu-345 (hLXRβ) to Met-340 (ciLXR) seems to be very critical. In hLXRβ, Leu-345 engages in van der Waals interactions with the trifluoromethyl group in T-0901317 and the chloro-trifluoromethyl benzyl group in GW3965. However, the longer side chain of Met-340 in the ciLXR occupies significantly more volume at this position; therefore, the LBP of ciLXR cannot accommodate these synthetic ligands, consistent with the experimental results. In contrast, epoxycholesterol can also activate ciLXR similar to hLXRβ. Interestingly, docking studies predict that epoxycholesterol adopts a flipped orientation in the ciLXR compared to hLXRβ (Fig. 5E). This observation is consistent with the pharmacophore modeling described above. In this flipped orientation, epoxycholesterol has better hydrophobic complementarity to the pocket; however, relative to hLXRβ, a critical hydrogen bonding interaction with epoxycholesterol at the 3 position is lost. Molecular docking results therefore suggest that epoxycholesterol would show lower binding affinity to ciLXR than to hLXRβ, consistent with the experimental results (Tables 1 and 2).
As shown in Fig. 5F, the binding orientation of 25-hydroxycholesterol in the ciLXR LBP is similar to epoxycholesterol in the hLXRβ crystal structure [18]. The 3-hydroxy group of 25-hydroxycholesterol forms a hydrogen bond with Gln-314, and the tail part of 25-hydroxycholesterol has extensive hydrophobic contacts with the binding pocket of ciLXR. The independent molecular modeling approaches (pharmacophore analysis, homology modeling, and ligand-receptor docking) used here to compare the hLXRβ and ciLXR binding pockets thus appear to be quite convergent in their findings, suggesting a more restrictive and hydrophobic LBP for ciLXR.
3.7. Intrinsic disorder analysis
An important factor in protein interactions with ligands or other proteins is presence of intrinsic structural disorder [22,36]. To assess whether disorder may account for pharmacological differences between the LXRs from different species, intrinsic disorder of the amino acid residues were predicted using the PONDR VL3H algorithm [22] and summarized by the percentage of residues with probability of disorder greater than 50% (Supplementary Table 2). Disorder probabilities were generally very similar across all LXRα and LXRβ sequences analyzed, whether analyzed by domain (DBD or LBD) or full-length protein sequence (Supplementary Table 3). One major difference was that ciLXR had no prediction of disorder in the first part of the LBD that includes the long helix-1 and the interloop region between helix-1 and helix-3, a region that is difficult to resolve crystallographically due to unclear electron density [19,33]. The low prediction of disorder in the ciLXR LBD contrasts markedly with all other LXRs analyzed, including that from the purple sea urchin (Supplementary Table 3). Interestingly, predicted disorder in the LBD was a better predictor for overall protein disorder in LXRβ (r2 = 0.90) than LXRα (r2 = 0.63). The results are consistent with differences in structural disorder possibly contributing to differences in pharmacologic specificity.
4. Discussion
LXRs are part of the NR1H subfamily that also includes FXRs (NR1H4 and H5) and the ecdysone receptors (NR1H1; found exclusively in invertebrates). The receptors in the NR1H subfamily all respond to steroidal compounds: FXRs to bile salts, LXRs to oxysterols (and limited numbers of bile acids), and ecdysone receptors to ecdysteroids such as α-ecdysone [7,(5α)-cholesten-2β,3β,14α,22(R),25-pentol-6-one]. The major endogenous ligands identified so far for mammalian LXRs are oxysterols such as 24(S)-hydroxycholesterol [3,4]. In this report, we determine in detail the ligand specificity of LXRs from three model non-mammalian species, X. laevis, X. tropicalis, and zebrafish. Compared to human and mouse LXRαs and LXRβs, the zebrafish LXR and the two Xenopus LXRs have very similar ligand specificity, consistent with the high degree of sequence conservation of the LBD of the LXR across species. The high degree of conservation of the vertebrate LXRs contrasts with divergence of FXRs across species. For example, the Xenopus laevis FXR has markedly different pharmacology from mammalian FXRs and also has a novel insert in the LBD not found in human or rodent FXRs [37].
A related group of receptors is the NR1I subfamily which includes PXR (NR1I2), vitamin D receptor (VDR; NR1I1), and the constitutive androstane receptor (CAR; NR1I3). These NR1I receptors can also bind steroidal compounds including pregnane and androstane steroids (PXR, CAR), bile salts (PXR, VDR), and vitamin D derivatives (VDR). The evolutionary origins of the NR1H and 1I subfamilies are unknown. Even though some of the ligands for the 1H and 1I receptors, such as bile salts and vitamin D, are thought to be vertebrate innovations, orthologs to these receptors are found in C. intestinalis and other invertebrates. In addition to the Ciona LXR analyzed in this report, comparative genomics reveals an FXR ortholog and a single ortholog to VDR/PXR/CAR [11]. The preliminary draft of the purple sea urchin (Strongylocentrotus purpuratus) genome shows four NR1H-like genes, some of which have high expression in early embryogenesis, suggesting developmental and/or protective functions. No putative orthologs to vertebrate NR1I genes have been identified so far in the sea urchin genome [31].
One of the major innovations in vertebrate evolution was changes in the use of cholesterol and other lipids [38]. The steroidal composition of invertebrates often differs significantly from vertebrates. Recent research has begun to uncover the use of steroidal compounds as ligands for NRs in invertebrates. For example, bile acid-like ligands have recently been elegantly characterized for the nematode Caenorhabditis elegans to a nuclear hormone receptor (DAF-12) that has closest sequence similarity to vertebrate NR1H and 1I receptors [39]. There have been few detailed studies of steroidal compounds in C. intestinalis. One study reported that the major sterols of adult C. intestinalis were cholest-5-en-3β-ol (68.7%), 24-methyl-cholest-5-en-3β-ol (7.3%), 24-ethyl-cholesta-5,22-dien-3β-ol (6.3%), and 24-methyl-cholesta-5,22-dien-3β-ol (5.3%); thirty carbon sterols found in some other invertebrates were not detected [40]. Another study found dehydroepiandrosterone, cortisone, and cortisol in testes, and cortisone in C. intestinalis ovary. Incubation of C. intestinalis testicular tissue with radiolabeled progesterone and pregnenolone resulted in formation of dehydroepiandrosterone, testosterone, and deoxycorticosterone, demonstrating the presence of enzymes that can interconvert steroid hormones. Similar incubations of ovarian tissue led to formation of 17α-hydroxypregnenolone, dehydroepiandrosterone, androstenedione, testosterone, and deoxycorticosterone [41]. A more recent report identified 3α,4β,7α,26-tetrahydroxycholestane-3,26-disulfate, a compound with some resemblance to vertebrate bile salts, as a chemoattractant for Ciona sperm [42].
In summary, the results of our studies show that the ligand specificities of LXRs from human, mouse, Xenopus frogs, and zebrafish are very similar. This is consistent with the high degree of sequence conservation of the LXR ligand-binding domain across vertebrates. In contrast, an LXR ortholog from the chordate invertebrate Ciona intestinalis has different ligand specificity from the vertebrate LXRs, and this is exemplified by our computational analyses, suggesting that the endogenous ligands for the invertebrate LXRs may be a different, although possibly structurally related, group of molecules. Our novel use of calculated intrinsic disorder [22,36] suggests that following analysis of 21 sequences of LXRs across species, only Ciona LXR demonstrated a dramatic difference in LBD predicted disorder, a factor that could impact protein-protein and protein-ligand interactions. Our results also suggest that, like the estrogen receptors [43], LXRs may have an extensive evolutionary history in invertebrates. One hypothesis would be that a single ancestral LXR gene present in invertebrates diverged early in vertebrate evolution in response to differences in lipid metabolism between invertebrates and vertebrates. Our findings suggest that the natural ligands for Ciona LXR may be steroidal compounds or closely related structures. Future research will focus on identification of ligands for Ciona NRs.
Supplementary Material
Supplemenary Table 1 (suppl_table1.pdf): Activity of compounds for HIPHOP analysis.
Supplementary Table 2 (suppl_table2.xls): Predicted disorder data.
Supplementary Table 3 (suppl_table3.pdf): Summary of predicted disorder data.
Supplementary Table 4 (suppl_table4.pdf): Conservation of ligand-binding residues across species.
Supplementary Fig. 1 (suppl_fig1.pdf): Sequence of alignment of LXRs
Supplementary Fig. 2 (suppl_fig2.pdb): Structure pdb file of homology model of Ciona intestinalis LXR.
Acknowledgments
The authors thank the laboratory of Dr. Alan F. Hofmann (University of California – San Diego, CA, United States) for providing 5β-cyprinol. SE acknowledges Dr. David Lawson for initial help providing information on the disorder analysis approach.
This study was supported by National Institutes of Health grant K08-GM074238 (to MDK), the Competitive Medical Research Fund from the University of Pittsburgh Medical Center (to MDK), and USEPA-funded Environmental Bioinformatics and Computational Toxicology Center (ebCTC), under STAR Grant number GAD R 832721-010 (to NA and WJW).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This PDF receipt will only be used as the basis for generating PubMed Central (PMC) documents. PMC documents will be made available for review after conversion (approx. 2–3 weeks time). Any corrections that need to be made will be done at that time. No materials will be released to PMC without the approval of an author. Only the PMC documents will appear on PubMed Central -- this PDF Receipt will not appear on PubMed Central.
References
- 1.Steinmetz ACU, Renaud JP, Moras D. Binding of ligands and activation of transcription by nuclear receptors. Annu Rev Biophys Biomol Struct. 2001;30:329–359. doi: 10.1146/annurev.biophys.30.1.329. [DOI] [PubMed] [Google Scholar]
- 2.Kalaany NY, Mangelsdorf DJ. LXRs and FXR: the Yin and Yang of cholesterol and fat metabolism. Annu Rev Physiol. 2006;68:159–191. doi: 10.1146/annurev.physiol.68.033104.152158. [DOI] [PubMed] [Google Scholar]
- 3.Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. An oxysterol signalling pathway mediated by the nuclear receptor LXRα α. Nature. 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- 4.Lehmann JM, Kliewer SA, Moore LB, Smith-Oliver TA, Oliver BB, San JL, Sundseth SS, Winegar DA, Blanchard DE, Spencer TA, Willson TM. Activation of the nuclear receptor LXR by oxysterols defines a new hormone response pathway. J Biol Chem. 1997;272:3137–3140. doi: 10.1074/jbc.272.6.3137. [DOI] [PubMed] [Google Scholar]
- 5.Song C, Liao S. Cholestenoic acid is a naturally occurring ligand for liver X receptor alpha. Endocrinology. 2000;141:4180–4184. doi: 10.1210/endo.141.11.7772. [DOI] [PubMed] [Google Scholar]
- 6.Song C, Hiipakka RA, Liao S. Selective activation of liver X receptor alpha by 6α-hydroxy bile acids and analogs. Steroids. 2000;65:423–427. doi: 10.1016/s0039-128x(00)00127-6. [DOI] [PubMed] [Google Scholar]
- 7.Mouzat K, Prod’homme M, Volle DH, Sion B, Dechelotte P, Gauthier K, Vanacker JM, Lobaccaro JM. Oxysterol nuclear receptor LXRβ regulates cholesterol homeostasis and contractile functions in mouse uterus. J Biol Chem. 2007;282:4693–4701. doi: 10.1074/jbc.M606718200. [DOI] [PubMed] [Google Scholar]
- 8.Song C, Kokontis JM, Hiipakka RA, Liao S. Ubiquitous receptor: a receptor that modulates gene activation by retinoic acid and thyroid hormone receptors. Proc Natl Acad Sci USA. 1994;91:10809–10813. doi: 10.1073/pnas.91.23.10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maglich JM, Caravella JA, Lambert MH, Willson TM, Moore JT, Ramamurthy L. The first completed genome sequence from a teleost fish (Fugu rubripes) adds significant diversity to the nuclear receptor superfamily. Nucleic Acids Res. 2003;31:4051–4058. doi: 10.1093/nar/gkg444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- 11.Yagi K, Satou Y, Mazet F, Shimeld SM, Degnan B, Rokhsar D, Levine M, Kohara Y, Satoh N. A genomewide survey of developmentally relevant genes in Ciona intestinalis. III. Genes for Fox, ETS, nuclear receptors and NFκB. Devel Genes Evol. 2003;213:235–244. doi: 10.1007/s00427-003-0322-z. [DOI] [PubMed] [Google Scholar]
- 12.Krasowski MD, Yasuda K, Hagey LR, Schuetz EG. Evolution of the pregnane X receptor: adaptation to cross-species differences in biliary bile salts. Mol Endocrinol. 2005;19:1720–1739. doi: 10.1210/me.2004-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goto T, Holzinger F, Hagey LR, Cerrè C, Ton-Nu HT, Schteingart CD, Steinbach JH, Shneider BL, Hofmann AF. Physicochemical and physiological properties of 5α-cyprinol sulfate, the toxic bile salt of cyprinid fish. J Lipid Res. 2003;44:1643–1651. doi: 10.1194/jlr.M300155-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Rossi SS, Converse JL, Hofmann AF. High pressure liquid chromatography analysis of conjugated bile acids in human bile: simultaneous resolution of sulfated and unsulfated lithocholyl amidates and the common conjugated bile acids. J Lipid Res. 1987;28:589–595. [PubMed] [Google Scholar]
- 15.Krasowski MD, Yasuda K, Hagey LR, Schuetz EG. Evolutionary selection across the nuclear hormone receptor superfamily with a focus on the NR1I subfamily (vitamin D, pregnane X, and constitutive androstane receptors) Nucl Recept. 2005;3:2. doi: 10.1186/1478-1336-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ekins S, Chang C, Mani S, Krasowski MD, Reschly EJ, Iyer M, Kholodovych V, Ai N, Welsh WJ, Sinz M, Swaan PW, Patel R, Bachmann K. Human pregnane X receptor antagonists and agonists define molecular requirements for different binding sites. Mol Pharmacol. 2007;72:592–603. doi: 10.1124/mol.107.038398. [DOI] [PubMed] [Google Scholar]
- 17.Ekins S, Mirny L, Schuetz EG. A ligand-based approach to understanding selectivity of nuclear hormone receptors PXR, CAR, FXR, LXRα, and LXRβ. Pharm Res. 2002;19:1788–1800. doi: 10.1023/a:1021429105173. [DOI] [PubMed] [Google Scholar]
- 18.Williams S, Bledsoe RK, Collins JL, Boggs S, Lambert MH, Miller AB, Moore J, McKee DD, Moore L, Nichols J, Parks D, Watson M, Wisely B, Willson TM. X-ray crystal structure of the liver X β ligand binding domain: regulation by a histidine-tryptophan switch. J Biol Chem. 2003;278:27138–27143. doi: 10.1074/jbc.M302260200. [DOI] [PubMed] [Google Scholar]
- 19.Färnegårdh M, Bonn T, Sun S, Ljunggren J, Ahola H, Wilhelmsson A, Gustafsson JA, Carlquist M. The three-dimensional structure of the liver X receptor β reveals a flexible ligand-binding pocket that can accommodate fundamentally different ligands. J Biol Chem. 2003;278:38821–38828. doi: 10.1074/jbc.M304842200. [DOI] [PubMed] [Google Scholar]
- 20.Liang J, Edelsbrunner H, Woodward C. Anatomy of protein pockets and cavities: measurement of binding site geometry and implications for ligand design. Protein Sci. 1998;7:1884–1897. doi: 10.1002/pro.5560070905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J Mol Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 22.Peng K, Vucetic S, Radivojac P, Brown CJ, Dunker AK, Obradovic Z. Optimizing long intrinsic disorder predictors with protein evolutionary information. J Bioinform Comput Biol. 2005;3:35–60. doi: 10.1142/s0219720005000886. [DOI] [PubMed] [Google Scholar]
- 23.Collins JL, Fivush AM, Watson MA, Galardi CM, Lewis MC, Moore LB, Parks DJ, Wilson JG, Tippin TK, Binz JG, Plunket KD, Morgan DG, Beaudet EJ, Whitney KD, Kliewer SA, Willson TM. Identification of a nonsteroidal liver X receptor agonist through parallel array synthesis of tertiary amines. J Med Chem. 2002;45:1963–1966. doi: 10.1021/jm0255116. [DOI] [PubMed] [Google Scholar]
- 24.Schultz JR, Tu H, Luk A, Repa JJ, Medina JC, Li L, Schwendner S, Wang S, Thoolen M, Mangelsdorf DJ, Lustig KD, Shan B. Role of LXRs in control of lipogenesis. Genes Devel. 2000;14:2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janowski BA, Grogan MA, Jones SA, Wisely GB, Kliewer SA, Corey EJ, Mangelsdorf DJ. Structural requirements of ligands for the oxysterol liver X receptors LXRα and LXRβ. Proc Natl Acad Sci USA. 1999;96:266–271. doi: 10.1073/pnas.96.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moschetta A, Xu F, Hagey LR, van Berge Henegouwen GP, van Erpecum KJ, Brouwers JF, Cohen JC, Bierman M, Hobbs HH, Steinbach JH, Hofmann AF. A phylogenetic survey of biliary lipids in vertebrates. J Lipid Res. 2005;46:2221–2232. doi: 10.1194/jlr.M500178-JLR200. [DOI] [PubMed] [Google Scholar]
- 27.Une M, Hoshita T. Natural occurrence and chemical synthesis of bile alcohols, higher bile acids, and short side chain bile acids. Hiroshima J Med Sci. 1994;43:37–67. [PubMed] [Google Scholar]
- 28.Farber SA, Pack M, Ho SY, Johnson DL, Wagner DS, Dosch R, Mullins MC, Hendrickson HS, Hendrickson EK, Halpern ME. Genetic analysis of digestive physiology using fluorescent phospholipid reporters. Science. 2001;292:1385–1388. doi: 10.1126/science.1060418. [DOI] [PubMed] [Google Scholar]
- 29.Anderson IG, Haslewood GAD, Oldham RS, Amos B, Tökés L. A more detailed study of bile salt evolution, including techniques for small-scale identification and their application to amphibian biles. Biochem J. 1974;141:485–494. doi: 10.1042/bj1410485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bramlett KS, Houck KA, Borchert KM, Dowless MS, Kulanthaivel P, Zhang Y, Beyer TP, Schmidt R, Thomas JS, Michael LF, Barr R, Montrose C, Eacho PI, Cao G, Burris TP. A natural product ligand of the oxysterol receptor, liver X receptor. J Pharmacol Exp Ther. 2003;307:291–296. doi: 10.1124/jpet.103.052852. [DOI] [PubMed] [Google Scholar]
- 31.Goldstone JV, Hamdoun A, Cole BJ, Howard-Ashby M, Nebert DW, Scally M, Dean M, Epel D, Hahn ME, Stegeman JJ. The chemical defensome: environmental sensing and response genes in the Strongylocentrotus purpuratus genome. Dev Biol. 2006;300:366–384. doi: 10.1016/j.ydbio.2006.08.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rannug A, Rannug U, Rosenkranz HS, Winqvist L, Westerholm R, Agurell E, Grafström AK. Certain photooxidized derivatives of tryptophan bind with very high affinity to the Ah receptor and are likely to be endogenous signal substances. J Biol Chem. 1987;262:15422–15427. [PubMed] [Google Scholar]
- 33.Svensson S, Östberg T, Jacobsson M, Norström C, Stefansson K, Hallén D, Johansson IC, Zachrisson K, Ogg D, Jendeberg L. Crystal structure of the heterodimeric complex of LXRα and RXRβ ligand-binding domains in a fully agonistic conformation. EMBO J. 2003;22:4625–4633. doi: 10.1093/emboj/cdg456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaye MC, Krawiec JA, Campobasso N, Smallwood A, Qiu C, Lu Q, et al. Discovery of substituted maleimides as liver X receptor agonists and determination of a ligand-bound cyrstal structure. J Med Chem. 2005;48:5419–5422. doi: 10.1021/jm050532w. [DOI] [PubMed] [Google Scholar]
- 35.Hoerer S, Schmid A, Heckel A, Budzinski RM, Nar H. Crystal structure of the human liver X receptor β ligand-binding domain in complex with a synthetic agoinist. J Mol Biol. 2003;334:853–861. doi: 10.1016/j.jmb.2003.10.033. [DOI] [PubMed] [Google Scholar]
- 36.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets: the roles of intrinsic disorder in protein interaction networks. FEBS Lett. 2005;270:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 37.Seo YW, Sanyal S, Kim HJ, Won DH, An JY, Amano T, Zavacki AM, Kwon HB, Shi YB, Kim WS, Kang H, Moore DD, Choi HS. FOR, a novel orphan nuclear receptor related to farnesoid X receptor. J Biol Chem. 2002;277:17836–17844. doi: 10.1074/jbc.M111795200. [DOI] [PubMed] [Google Scholar]
- 38.Nes WR, Nes WD. Lipids in evolution. New York: Plenum Press; 1980. [Google Scholar]
- 39.Motola DL, Cummins CL, Rottiers V, Sharma KK, Li T, Li Y, Suino-Powell K, Xu HE, Auchus RJ, Antebi A, Mangelsdorf DJ. Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell. 2006;124:1209–1223. doi: 10.1016/j.cell.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 40.Voogt PA, van Rheenan JWA. On the sterols of some ascidians. Arch Intl Physiol Biochim. 1975;83:563–572. doi: 10.3109/13813457509071400. [DOI] [PubMed] [Google Scholar]
- 41.Delrio G, D’Istria M, Milone M, Chieffi G. Identification and biosynthesis of steroid hormones in the gonads of Ciona intestinalis. Experientia. 1971;27:1348–1350. doi: 10.1007/BF02136733. [DOI] [PubMed] [Google Scholar]
- 42.Yoshida M, Murata M, Inaba K, Morisawa M. A chemoattractant for ascidian spermatozoa is a sulfate steroid. Proc Natl Acad Sci USA. 2002;99:14831–14836. doi: 10.1073/pnas.242470599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thornton JW, Need E, Crews D. Resurrecting the ancestral steroid receptor: ancient origin of estrogen signaling. Science. 2003;301:1714–1717. doi: 10.1126/science.1086185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemenary Table 1 (suppl_table1.pdf): Activity of compounds for HIPHOP analysis.
Supplementary Table 2 (suppl_table2.xls): Predicted disorder data.
Supplementary Table 3 (suppl_table3.pdf): Summary of predicted disorder data.
Supplementary Table 4 (suppl_table4.pdf): Conservation of ligand-binding residues across species.
Supplementary Fig. 1 (suppl_fig1.pdf): Sequence of alignment of LXRs
Supplementary Fig. 2 (suppl_fig2.pdb): Structure pdb file of homology model of Ciona intestinalis LXR.